Abstract
Recent work revealed a new class of molecular machines called molecular sleds, which are small basic molecules that bind and slide along DNA with the ability to carry cargo along DNA. Here, we performed biochemical and single-molecule flow stretching assays to investigate the basis of sliding activity in molecular sleds. In particular, we identified the functional core of pVIc, the first molecular sled characterized; peptide functional groups that control sliding activity; and propose a model for the sliding activity of molecular sleds. We also observed widespread DNA binding and sliding activity among basic polypeptide sequences that implicate mammalian nuclear localization sequences and many cell penetrating peptides as molecular sleds. These basic protein motifs exhibit weak but physiologically relevant sequence-nonspecific DNA affinity. Our findings indicate that many mammalian proteins contain molecular sled sequences and suggest the possibility that substantial undiscovered sliding activity exists among nuclear mammalian proteins.
INTRODUCTION
Cells encode genes and the information needed to regulate their expression in the molecular structure of their DNA genomes. Factors that read, write and regulate access to this information must engage the physical and chemical properties of DNA in vivo. In mammalian cells, gene regulation is complex and requires the coordinated action of many protein factors. Protein molecules that function in the nucleus are tagged with nuclear localization signal (NLS) peptides that enable their transport from the cytosol. About 5000 human proteins have been observed or predicted to be localized in the nucleus of cells (1). Many of these proteins function on DNA or chromatin, and some have been shown to be capable of sliding along the genome using specialized DNA-binding interfaces (2–8).
A new class of molecular machines called molecular sleds was recently described (9,10). Molecular sleds are small basic molecules such as peptides that bind and slide along DNA and can translocate cargo, for example a protein molecule, along DNA. Sliding is driven by thermal diffusion and is insensitive to DNA sequence (11–13). Previously, sliding activity was only known in DNA-binding proteins with well-structured DNA binding interfaces that were believed to be essential for the fast sliding activity that can support facilitated diffusion (2,14–17). Some of these proteins contain stable α-helical modules of 20–60 amino acids that can bind in the major groove of B-form DNA (18). von Hippel et al. showed that synthetic 15 amino acid peptides can be induced to form α-helicies upon sequence nonspecific binding to B-form DNA, and that this binding required specific i → i + 3 or + 4 spacing of basic amino acids to enable electrostatic interactions between the α-helical peptide and the DNA major groove (19). This work collectively reinforces the dogma that binding and sliding activity requires a precision-tuned DNA-binding interface entailing precise registration or phasing of charged residues (16,17,19,20).
In contrast, pVIc, the first molecular sled discovered, is only 11 amino acids long, forms no stable structure on its own, and displays consecutive charged residues that are unfavorable for DNA binding as an α-helix (9). pVIc functions as an NLS of the adenoviral protein pVI and plays a role in the viral life cycle by regulating one-dimensional (1D) biochemical interactions among DNA-bound viral proteins via its robust sliding activity (9,10). Tiny peptidyl molecular sleds like pVIc with NLS-like basic residue sequences were found to slide along DNA as fast as 26.0 ± 1.8 M(bp2/s), many times faster than the fastest-sliding proteins, with the exception of the pVIc–AVP complex in which the pVIc molecular sled constitutes the DNA-binding interface (8–10).
The discovery of the pVIc molecular sled and sliding activity in two homologous human peptides (9) raised important questions: (i) how widespread could DNA sliding activity be among the many nuclear proteins in mammalian cells? (ii) how do molecular sleds function? Here, we take the first steps in answering these questions by identifying the peptide functional groups that play roles in sliding in pVIc, defining a minimal model sled and characterizing its function, determining which sorts of primary amino acid sequences support DNA sliding activity in molecular sleds, and proposing a model for molecular sled sliding activity.
MATERIALS AND METHODS
Chemicals
All unlabeled and N-terminal dye labeled peptides were purchased from the Biopolymer & Proteomics Laboratory at MIT (with >85% purity). Tetramethylrhodamine(TMR)-maleimide was purchased from Anaspec. Cy3B-maleimide was purchased from GE Healthcare Life Sciences. Sulfhydryl-labeled peptides were prepared by reacting TMR- (or Cy3B)-maleimide with unlabeled peptides, and the final products were purified by HPLC and confirmed by MALDI analysis. pVIc-Cy3B was a gift from Dr Walter Mangel at Brookhaven National lab. DAPI (with ≥98% purity) was purchased from Sigma. Hoechst 33258 (with ≥98% purity) was purchased from Santa Cruz Biotechnology. λ-DNA was purchased from New England Biolabs and a DNA oligo with a biotin attached, 5′-GGGCGGCGACCTAAAAAAAAAAA-biotin-3′ was ordered from Integrated DNA Technologies, Inc. A 30-mer oligo (5′-GACGACTAGGACGACGACGAGGATGACGAC-3′) and its complementary strand were purchased from IDT and annealed together to form a double-stranded 30-mer for binding studies.
Flow cell construction
Flow cells were constructed by sandwiching a double-sided tape with pre-cut channels between poly(ethylene glycol) (PEG) functionalized coverslip and a PDMS slab containing inlet and outlet holes (see peg coverslip preparation protocol in the supplementary text). The biotin-λ-DNA was made by ligating a DNA oligo with a biotin attached to the 5′ overhang of λ-DNA (see protocol by Schroeder et al. (21)), and then immobilized onto the flow channel surface by utilizing biotin and streptavidin chemistry.
Single molecule flow stretching experiments
Peptide samples were infused at 10–1000 pM concentrations at rates of 25 ml/h (at such a high flow rate, the contribution of DNA fluctuations to the apparent motion of DNA-bound molecules along the DNA is negligible under our assay conditions; see Supplementary Figure S9). The assay buffer consisted of 10 mM phosphate buffer, 2–20 mM NaCl, 50 μM EDTA, 20 mM ethanol, 5% (v/v) glycerol, 0.01% Tween-20 and 1% (v/v) β-mercaptoethanol and the pH was adjusted by adding aliquots of concentrated NaOH solution. This titration caused the sodium ion concentration of buffer solutions at different pHs to be different, resulting in buffers with ionic strengths ranging from 15 mM at pH 6.5 to 31 mM at pH 8.63. Individual dye-labeled molecules were imaged by using a home-built total internal reflection fluorescence microscope based on a Nikon Ti-E body with a Hamamatsu ORCA-Flash 4.0 V1 camera. We used customized single particle tracking software to process the raw image data (see Single Particle Tracking Software Details in the Supplementary Text).
Fluorescence polarization (FP) experiments
Steady-state fluorescence polarization (FP) measurements were performed using a SpectraMax M5 plate reader from Molecular Devices. Samples were prepared in a 96-well plate. To determine Kd values, the fluorophore-labeled peptide concentrations were held at 10 nM and 30 bp dsDNA titrated from 3000 to 0 nM. The excitation/emission/cut-off wavelengths were set as 544/ 590/ 570 nm (or 590 nm; both generate identical equilibrium dissociation constants, Kds from fits). The Kd values were estimated by fitting the FP data to a one-to-one stoichiometry binding model (see Supplementary Figure S5A for details).
RESULTS
KRRR is the functional core of pVIc
pVIc (GVQSLKRRRCF) contains four consecutive basic amino acids (KRRR) that we hypothesized were sufficient for sliding activity despite the fact that tetrapeptides are too small to form stable secondary structures. To test this idea, we performed standard single molecule flow stretching experiments for 1D diffusion (2,22) of the tetrapeptide (labeled on its N terminus). We observed that the tetrapeptide is indeed capable of fast diffusion along DNA with an estimated 1D diffusion constant, D1, of 10.5 ± 0.7 (standard error) M(bp2/s) (see Figure 1).
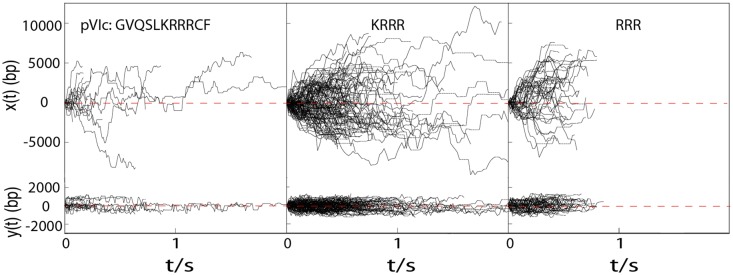
Figure 1.
Diffusion of TMR-pVIc, TMR-KRRR and TMR-RRR along flow stretched λ-DNA molecules in 2 mM NaCl buffer. x(t) and y(t) are the displacements along and transverse to λ-DNA, respectively. For N-terminal labeled peptides, no significant change in D1 was observed between pH 6.5 and pH 7.4, and thus, diffusion trajectories observed at pH 6.5 and pH 7.4 were combined to give more accurate estimation of D1. Dotted horizontal lines indicate missing points resulting from dye blinking.
In principle, 1D diffusion can take place by two mechanisms, sliding and hopping. Sliding consists of 1D translocation along the DNA in continuous contact with the DNA, while hopping occurs when molecules move by repeated dissociation from the DNA and rebinding to DNA at nearby locations in fast local equilibrium. The salt concentration dependence of the 1D diffusion constant is a useful method to differentiate sliding from hopping for molecules that bind with an electrostatic contribution to their DNA binding interaction energy (2). For sliding, D1 is expected to be independent of salt concentration, while D1 is expected to increase sharply with salt concentration if a hopping mechanism contributes significantly to 1D translocation. This is because increasing the salt concentration will lower the sequence-nonspecific DNA affinity of protein, decrease its residence time on DNA during 1D translocation events, and thus give rise to an apparent increase in D1. The D1 of KRRR in 20 mM NaCl buffer (see Supplementary Figure S4) is not significantly higher than in 2 mM NaCl buffer, indicating that KRRR translocates on DNA by sliding in our assay. The D1 of KRRR is somewhat lower than that of equivalently prepared pVIc (N-terminal TMR labeled pVIc slides with D1 = 14.7 ± 1.5 M(bp2/s)). We also performed FP experiments to determine the affinities of these peptides for DNA (see Figure 2B), discovering that the equilibrium dissociation constant (Kd) of KRRR (197 ± 40 nM) is comparable to that of pVIc (352 ± 50 nM) (Figure 2B).
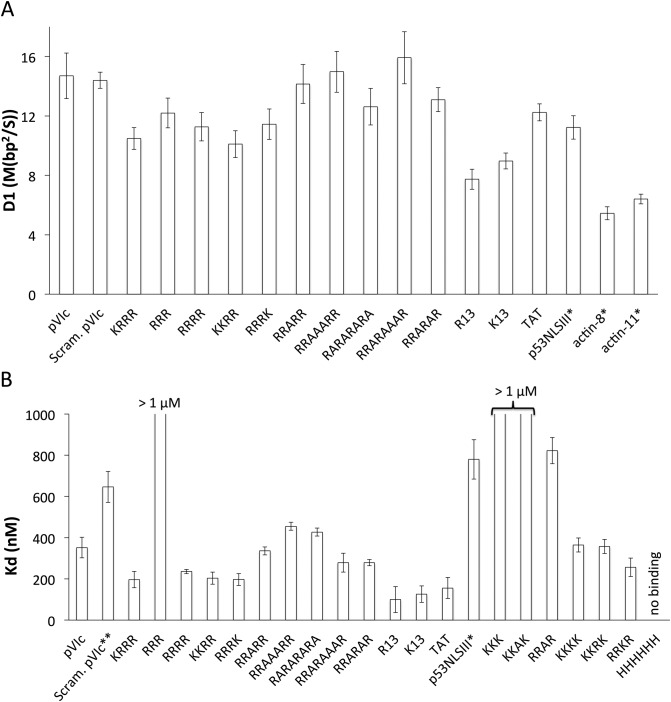
Figure 2.
(A) The estimated D1 values for a panel of different peptides sliding on DNA in 2 mM NaCl buffer. TMR or Cy3 were conjugated to the N-terminus of all peptides except for actin-8 and actin-11 in which Cy3B was conjugated to the cysteine residues present in these peptides. D1 values measured at pH 7.4 and 6.5 were combined for all peptides except for actin-8 and actin-11 whose D1 values were measured only at pH 6.5. All the error bars are standard errors of the mean. On average, over 50 trajectories were analyzed to estimate each D1 value. (B) Equilibrium dissociation constants, Kd values of different peptides binding to a 30 bp dsDNA in 2 mM NaCl buffer at pH 7.4, determined by fluorescence polarization measurements. TMR was conjugated to the N-terminus. * Result from Mangel et al. (9). ** Result from personal communication with Dr. Walter Mangel. All the error bars are standard errors of the mean. The Kd values were averaged from at least three independent measurements. See Supplementary Table S1 for D1 and Kd values.
Tripeptides can slide along DNA
We wondered whether even smaller peptides could bind DNA and slide along it. To determine whether tripeptides can slide, we further truncated pVIc to RRR. The DNA affinity of RRR is substantially decreased (Kd > 1 μM) in comparison to KRRR (see Figure 2B), which reduces the observable event frequency in the single-molecule assay. Nonetheless, we observed RRR sliding events and estimated D1 for RRR at 12.2 ± 1.0 M(bp2/s), in-line with the D1 values for pVIc and KRRR. Altogether, it is clear that KRRR forms the functional core of the pVIc molecular sled and that tiny peptide molecular sleds like RRR can recapitulate the sliding activity of pVIc.
The sliding activity of molecular sleds is robust to primary amino acid sequence variation
Is there a primary amino acid sequence ‘code’ for molecular sleds? To answer this question, we tested peptides with varying amino acid sequences in a series of experiments.
First, we tested a peptide with the same amino acids as pVIc but in a scrambled sequence, SFRRCGLRQVK (N-terminally labeled), observing that this peptide is capable of sliding on DNA with an estimated D1 of 14.4 ± 0.5 M(bp2/s), essentially identical to that of similarly labeled pVIc (Figure 2A). This result indicates that the sequence of residues in pVIc is not critical for sliding activity.
To systematically probe for effects of primary amino acid sequence on sliding, we examined the effect of variable spacing of the basic amino acids by separating arginine residues with intervening alanine residues. To our surprise, RRRR, RRARR, RRAAARR, RARARARA, RRARAAAR, RRARAR not only exhibited similar Kd values (Figure 2B) but also similar D1 values (D1 = 11.3 ± 1.0, 14.2 ± 1.3, 15.0 ± 1.4, 12.6 ± 1.2, 15.9 ± 1.8 and 13.1 ± 0.8 M(bp2/s), respectively) that approximate the values measured for KRRR and RRR. The spacing-independence of binding affinity and diffusion constant values show that these tiny NLS-like molecular sleds interact with duplex DNA by a mechanism distinct from natural and synthetic peptides that bind the DNA major groove as α-helices and are highly sensitive to residue spacing (19). Robust binding and sliding activity in peptides with interleaved alanine residues indicate that no particular configuration of side chains or the peptide backbone specific to consecutive basic amino acids is required for 1D diffusion activity.
Next, we compared the 1D diffusion activity of lysine- and arginine-containing peptides to determine if one type of basic side chain supported sliding to a greater degree than the other. Again using peptides with four basic amino acids, we varied the number and order of lysine and arginine residues and measured D1 values. Figure 2 shows that the measured D1 values of RRRR, KRRR, RRRK and KKRR are essentially identical (D1 = 11.3 ± 1.0, 10.5 ± 0.7, 11.4 ± 1.0 and 10.1 ± 0.9 M(bp2/s), respectively). FP measurements show that the DNA binding affinities of the lysine-containing peptides are very similar to those measured for the peptides with four arginine residues in the alanine-spacer experiments (see Figure 2B). These results (and those below for longer peptides) indicate that the interactions between positively charged side chains and DNA needed for sliding can be made with cationic moieties having either electronically localized (K) or delocalized (R) charges. We also examined hexahistidine, finding that it did not bind to DNA at pH 7.4, most likely due to deprotonation of its side chains at this pH.
So far, all the peptides tested that contained four basic (R or K) side chains bound to DNA with similar Kd values (200–500 nM) and diffused along DNA with similar D1 values (10–15 M(bp2/s)). Shortening the peptide to three amino acids had a predictable effect on Kd due to the reduced number of basic functional groups but little effect on D1. To determine the effects of increasing the length of the peptides, we measured Kd and D1 values for poly(K) and poly(R) peptides of 13 residues. These longer basic peptides bind DNA somewhat more strongly, with Kd values of ∼100 nM (see Figure 2B). Figure 2 shows that the measured D1 values of (K)13 and (R)13 are 7.7 ± 0.7 and 8.9 ± 0.5 M(bp2/s), respectively, 10–20% lower than the D1 values determined for corresponding tetra-basic peptides. Unlike the shorter peptides, these D1 values rise by ∼ 50% in 20 mM NaCl buffer (see Supplementary Figure S6). These results suggest that (K)13 and (R)13 translocate on DNA by a different mechanism than the peptides with short basic cores, for example by a mechanism wherein the most mobile configurations for sliding entail partial engagement between the peptide basic side chains and the DNA. Our salt-dependence results do not formally distinguish a hopping model from an inchworm model (where portions of the peptide undergo hopping) or models wherein which a fixed subset of basic side chains engage with the DNA, but are consistent with the hypothesis that the fully-engaged tridecapeptides slide more slowly than peptides with four or fewer DNA-engaged basic residues.
Cell-penetrating peptides can slide along DNA
Based on our results, it seems likely that many natural basic protein motifs can bind and slide along DNA. Here we tested a canonical cell-penetrating peptide (CPP) from TAT, a trans-activator protein of HIV-1. TAT also functions in activating viral gene expression inside host cells (including cross-reactivity with viruses other than HIV-1), suppressing the immune system, modulating apoptosis, inducing AIDS-related cancer, causing neurotoxicity, promoting RNA annealing and interacting with membranes (23). TAT contains a CPP and a NLS sequence in its basic domain (amino acids 48–60, GRKKRRQRRRPPQ) (24). This arginine rich sequence is also required for transactivating the HIV-1 long terminal repeat promoter by binding to the transactivation response region RNA (25).
We observed that the TAT basic domain peptide is capable of translocating along DNA with an estimated D1 of 12.2 ± 0.6 M(bp2/s). Previous work showed that the nuclear localization sequence III of p53 (STSRHKKLMFKTE, p53NLSIII) can slide on DNA (9). The TAT and p53 peptides exhibit essentially identical D1 values (Figure 2A) in spite of their diverging DNA affinities (see Figure 2B). This disparity, like the pVIc pH dependence presented below, suggests that dynamics of the bound peptide affect the diffusion constant for sliding.
There is a substantial likelihood that many nuclear proteins slide along DNA
Natural protein sequences homologous to pVIc including p53NLSIII and the NLS-like C-terminal peptides from β-actin (SIVHRKCF and SGPSIVHRKCF) have been shown to function as molecular sleds (see D1 values in Figure 2A) (9). Here we found that molecular sleds are extremely robust to variation in peptide amino acid content and primary amino acid sequence. Knowing that molecular sleds can carry protein cargo (9), the amino acid sequence insensitivity of molecular sleds is important because many nuclear proteins have surface-accessible basic peptide sequences that are variable, but likely to support sliding activity on DNA in light of the amino acid sequence insensitivity shown here. We expect that any classical monopartite (K-K/R-X-K/R) or bipartite NLS motif (26) can support DNA binding and sliding activity if the signal peptide is surface-exposed.
The N-terminal amino group modulates rapid sliding activity
To examine the impact of dye labeling on sliding, we tested the effect of different label positions and dye types on the D1 of pVIc. Figure 3 shows that N-terminal labeling decreases the D1 value of pVIc to one-half the value for cysteine side chain-labeled pVIc that retains a free N-terminal amine. Similar functionalization effects were observed with K13 and R13 (see Supplementary Figure S6). With N-terminal labeling, the dye identity (Rhodamine versus Cyanine) has no effect on D1 (Figure 3), while with cysteine side chain labeling, pVIc-Cy3B exhibits a slightly higher D1 than does pVIc–TMR (27). To determine whether D1 of the N-terminal conjugates was affected by blockage of the N-terminal free amine, or alternatively, the presence of N-terminal dye molecules, we acetylated the pVIc N-terminus to alter the properties of the N-terminal amine without introducing a dye molecule at that position. Figure 3B (pH 7.4) shows that the D1 values of cysteine-labeled acetyl-pVIc decrease by a factor of two relative to those of pVIc, in-line with the N-terminal labeling results above and strongly implicating the presence of a free N-terminal primary amine in the large D1 value of the fast-sliding side-chain-labeled pVIc. The free N-terminal amine shows little impact on the DNA affinity of pVIc (the measured Kd values of pVIc–TMR and TMR–pVIc are 262 ± 88 nM and 352 ± 50 nM, respectively).
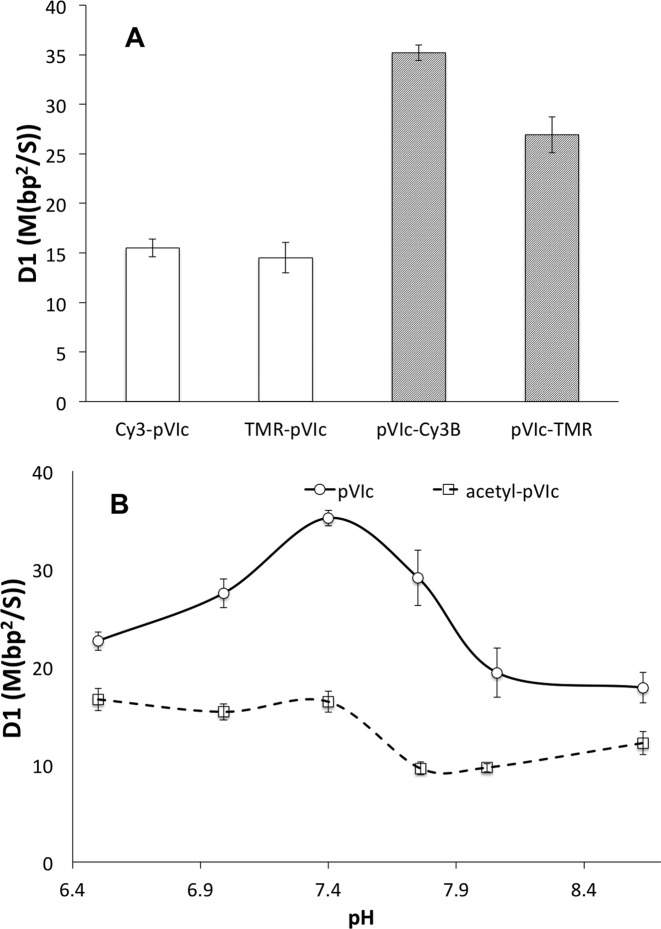
Figure 3.
(A) The estimated D1 values of Cy3-pVIc, TMR-pVIc, pVIc-Cy3B and pVIc-TMR in 2 mM NaCl buffer at pH 7.4. TMR or Cy3 was conjugated to the N-terminus or Cys residues. (B) The pH dependence of D1 of pVIc (solid line) and acetyl-pVIc (dashed line) in 2 mM NaCl buffer. Cy3B was conjugated to Cys residues. See Supplementary Table S2 for D1 values. All the error bars are standard errors of the mean. On average, over 50 trajectories were analyzed to estimate each D1 value.
pVIc sliding is pH-dependent
We noticed an extremely high D1 value for pVIc–Cy3B, 35.1 ± 0.8 M(bp2/s), when collecting the data in Figure 3A, where we standardized the pH at 7.4. This value is more than 30% larger than that previously reported for pVIc–Cy3B at pH 6.5 (26.0 ± 1.8 M(bp2/s) (9)), which had been the highest diffusion constant measured for sliding along DNA by a polypeptide. Observing the 2-fold effects of N-terminal labeling and pH shift on the diffusion constant for sliding, we decided to systematically characterize the pH dependence of D1 values for pVIc and N-terminally acetylated pVIc (both labeled with Cy3B on the cysteine side chain). Regarding pH, the D1 values of both peptides are modulated significantly between pH 7 and 8, with pVIc showing a strong peak between 7.0 and 7.75 (Figure 3B). It is tempting to speculate that the N-terminal amine drives the pH dependence of pVIc sliding activity because of all the chemical functional groups in pVIc it has the pKa (∼8) predicted to be closest to the range where D1 is modulated. However, we would expect juxtaposition of the N-terminus near the DNA to raise the pKa of this amine somewhat higher, and although the 2-fold reduction in the baseline diffusion constant is observed in acetylated pVIc, significant modulation of D1 is found in the range pH 7–8 for the acetylated peptide despite the dramatic reduction in pKa (to −0.5) expected when the N-terminal amine is converted to an amide by acetylation (Figure 3B).
The reduction in D1 upon acetylation of the N-terminal amine and the mismatch between D1 modulation as a function of pH and the pKa of the N-terminus of pVIc argue against explanations of these effects based on equilibrium ionization states. It is particularly surprising that eliminating a potentially cationic group from the peptide by acetylation would strongly decrease D1 for two reasons: (i) the cationic group has a potential to interact with DNA and increase friction for sliding, and (ii) we observed smaller changes in D1 when comparing results among tri-, tetra- and trideca-basic peptide molecular sleds. Thus, changes in proton exchange kinetics or hydrogen bonding dynamics are the most likely explanation of the reduction in D1 upon N-terminal functionalization and the observed pH-dependence of D1. We note that in a protein context, C-terminal and internal basic polypeptide sequences would have an N-terminal amide rather than an amine and be better modeled by the acetylated peptide molecular sled than the peptide molecular sled with a free N-terminal amine group.
DNA minor groove binding molecules strongly affect peptide sliding
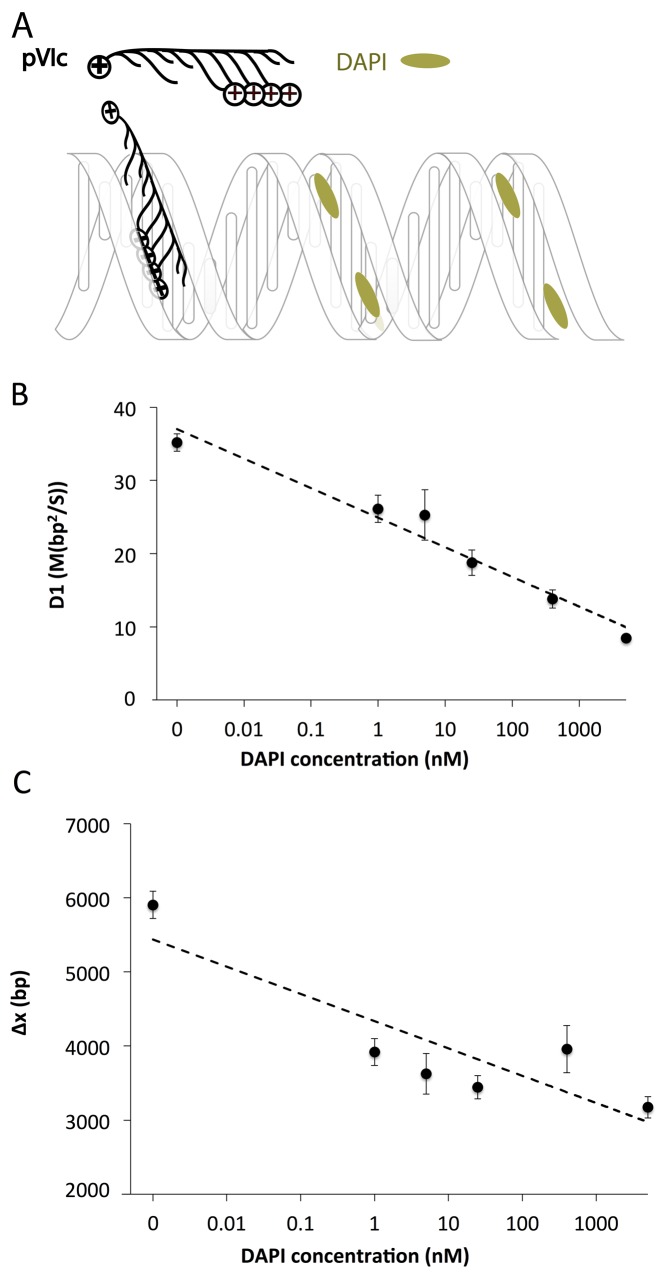
In structural studies, lysine and arginine are often found making favorable interactions with the focused electric field found in the minor groove of DNA (28). Other DNA-binding small molecules such as polyamides are also known to bind in the minor groove of DNA (29). To determine whether molecular sleds slide in the DNA minor groove, we studied the effects of the DNA minor groove binder DAPI (30) on pVIc sliding. We observed the D1 of pVIc to decrease by 2-fold as the DAPI concentration increases from 0 to 5 μM (Figure 4B). The average displacement of pVIc along DNA also exhibits a decreasing trend with increasing DAPI concentration (Figure 4C). These results indicate that DAPI molecules interfere with sliding of pVIc. FP measurements indicate that DAPI inhibits pVIc binding to DNA (Supplementary Figure S8C). Similar ‘roadblock’ effects were observed by the addition of Hoechst 33 258, another DNA minor groove binder (31) (Supplementary Figures S7 and S8C). Although we were unable to identify a suitable major-groove blocker to serve as a negative control, the DAPI and Hoechst results suggest that molecular sleds slide in the DNA minor groove.
Figure 4.
(A) A schematic of pVIc sliding on DNA in the presence of DAPI; DAPI is in dynamic equilibrium between DNA bound and unbound states. (B) The DAPI effect on D1 of pVIc in 2 mM NaCl buffer at pH 7.4. (C) The DAPI effect on total diffusion distance along λ-DNA, Δx. Dashed lines are linear trend lines. Cy3B was conjugated to Cys residues. The estimated Kd of DAPI to a 30mer ds-DNA is ∼13 nM (see Supplementary Figure S8A). See Supplementary Table S3 for D1 and Δx values. All the error bars are standard errors of the mean. At least 20 trajectories were analyzed to estimate each D1 value.
DISCUSSION
Previous work regarding facilitated diffusion and sliding along DNA focused exclusively on proteins and fragments of proteins involved in DNA metabolism and gene regulation with the exceptions of recent work on adenovirus proteins and poly-amidoamine dendrimers (10,32,33). Not until very recently were the first molecular sleds reported (9). The discovery of small, non α-helical peptides with sliding activity proved that large, well-structured DNA-binding interfaces with specific phasing of interfacial residues are not required for robust sliding activity. Here, we carried out a systematic investigation of molecular sled structure-function relationships, gaining new insight into the mechanism by which these molecules slide along DNA and the potential for facilitated diffusion activity by mammalian proteins that contain basic motifs.
We identified the functional core of the first molecular sled, pVIc, to be the tetrapeptide KRRR and showed that even the tripeptide RRR has sliding activity. We did not attempt sliding experiments with RR as the very low predicted DNA affinity (Kd estimated at >5000 nM by extrapolation along the curve in Supplementary Figure S5B) would challenge data collection using our assay. The observation of sliding activity in tiny peptides that cannot form stable secondary structures rules out specific phasing of charged residues as a requirement for rapid sliding activity and allows us to recognize that a variety of basic small molecules may have sliding activity on DNA. We examined the peptide primary sequence dependence of 1D diffusion and discovered that 1D diffusion is highly robust to primary sequence variation, with observed diffusion constants falling in a relatively narrow range between 8 and 16 M(bp2)/s (N-terminally labeled peptides). These results indicate that sliding activity is likely conserved across adenovirus serotypes where there is primary sequence variation in pVIc, including the AD8 serotypes where only three basic residues are present (9).
Our finding that essentially all arginine- and lysine-containing peptides, including natural NLS and CPP sequences, slide at physiological pH has strong implications. Protein motifs such as those we demonstrated here to have 1D diffusion activity are common in the mammalian proteome, and enriched in the nuclear proteome. For example, there are 12,313 and 10,362 (K/R)8 ≥n≥4X8-n basic molecular sled motifs among 5065 and 4235 predicted nuclear proteins in the human and mouse proteomes, respectively (see Supplementary Table S4). If even a small fraction of basic motif-containing proteins are sliding-active, there would be many proteins capable of sliding along nuclear DNA in cells. Furthermore, it seems probable that a number of nuclear proteins with solvent-accessible basic sequences but no known role in binding the genome actually bind DNA and can slide along it to carry out functions that we have not yet recognized. Future studies will reveal the protein context-dependence of molecular sled activity and the DNA-binding and sliding activities of NLS-containing mammalian proteins.
In contrast with the lack of primary sequence dependence of D1, we observed that the N-terminal amine exhibits strong effects on D1. Converting the N-terminal amine to an amide either by coupling to an organic dye or by reaction with acetic anhydride dramatically reduces D1. In addition, the D1 of pVIc exhibits a peak near pH 7.4, where we observed an extraordinarily high diffusion constant for sliding along DNA, 35.1 ± 0.8 M(bp2/s) (pVIc with a free amino terminus and dye molecule cargo). These observations are not easily explained by equilibrium electrostatic models, but constitute a basis on which the 1D sliding activity of pVIc can be regulated.
Incongruity between equilibrium electrostatics and sliding dynamics was observed previously in pH-dependent sliding studies of the DNA repair protein human oxoguanine DNA glycosylase (hOgg1)(2). hOgg1 has a single basic residue in its DNA-binding interface (Histidine 270) and exhibits a reduction in D1 above pH 7.5 when this residue is mutated to alanine. This observation was mysterious, because titrating the interfacial histidine to a neutral and less bulky form would be expected to reduce the interaction with DNA and ‘increase’ D1. As here with the pVIc peptide, interpretation of the hOgg1 data appealed to dynamics to explain these unexpected observations, although a specific mechanism was not proven. The parallel pH-dependence between the short peptides studied here and the hOgg1 DNA binding protein underscores the utility of molecular sleds as simplified model systems for mechanistic studies that are relevant to the functionality of much larger DNA-binding protein molecules.
It is unknown whether sliding by molecular sleds occurs in the DNA minor or major groove. Recent studies suggested that nonspecific protein–DNA interaction is predominantly electrostatic (34). Honig et al. have shown that the electrostatic potential in the DNA minor groove is more negative and more continuous (28,35) than that in the major groove due to electrostatic focusing (36), and thus we expect will be more favorable for binding and transport of the basic side chains borne by molecular sleds. The disruption of molecular sled sliding activity we observed by blockade of the minor groove is consistent with models for sliding that involve substantial minor groove binding and sliding by molecular sleds.
Though molecular sleds were only investigated under in vitro conditions here, we expect molecular sleds to function in vivo as well (10). Under physiological conditions with higher salt concentration, the DNA affinities of molecular sleds will be decreased, but the effective DNA concentration in the nucleus of cells is so high that DNA binding will still occur (see Supplementary Text calculations). The mean in vivo sliding length of molecular sleds will be shorter than observed here in vitro at low salt concentration due to shorter DNA binding lifetimes and obstruction by other DNA-bound proteins (14,37) (see quantitative estimation in the supplementary text). At the same time, the probability of molecular sleds quickly rebinding to DNA after dissociation is high, enabling efficient search and obstacle bypass (14).
Altogether, we have shown that DNA binding and sliding activity by peptide molecular sleds is a robust phenomenon across a wide range of basic polypeptide sequences, including essentially all known NLS sequences as well as other natural basic polypeptides such as CPPs. In addition to the direct relevance for sliding by natural basic peptides and proteins containing such polypeptide sequences, peptide molecular sleds offer a convenient model system that recapitulates key features of the structured-interface-based protein sliding activity that has been the focus of studies to date on facilitated diffusion. We expect that molecular sleds will find biotechnological application in molecular bioengineering that takes advantage of the properties characterized here.
Supplementary Material
Acknowledgments
We thank Tony Kulesa, Robin Kirkpatrick and Evangelos Gatzogiannis for assistance with instrumentation testing and setup. We also thank Tony Kulesa for his help with writing and evaluating the customized single particle tracking software. We thank Dr Walter Mangel at Brookhaven National lab for important discussions and anonymous peer reviewers for thoughtful and detailed feedback that improved this manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Broad/MIT startup funding and a Career Award at the Scientific Interface (to P.C.B.) from the Burroughs Wellcome Foundation supported this work. Funding for open access charge: Startup funding and MIT.
Conflict of interest statement. The Broad Institute may elect to file patent applications that include aspects of the work presented here.
REFERENCES
- 1.Rastogi S., Rost B. LocDB: experimental annotations of localization for Homo sapiens and Arabidopsis thaliana. Nucleic Acids Res. 2011;39:D230–D234. doi: 10.1093/nar/gkq927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blainey P.C., van Oijen A.M., Banerjee A., Verdine G.L., Xie X.S. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc. Natl. Acad. Sci. U.S.A. 2006;103:5752–5757. doi: 10.1073/pnas.0509723103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Etson C.M., Hamdan S.M., Richardson C.C., van Oijen A.M. Thioredoxin suppresses microscopic hopping of T7 DNA polymerase on duplex DNA. Proc. Natl. Acad. Sci. U.S.A. 2010;107:1900–1905. doi: 10.1073/pnas.0912664107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ponferrada-Marin M.I., Roldan-Arjona T., Ariza R.R. Demethylation initiated by ROS1 glycosylase involves random sliding along DNA. Nucleic Acids Res. 2012;40:11554–11562. doi: 10.1093/nar/gks894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J.J., Zhang Z.J., Li L., Chen B.C., Revyakin A., Hajj B., Legant W., Dahan M., Lionnet T., Betzig E., et al. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell. 2014;156:1274–1285. doi: 10.1016/j.cell.2014.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dikic J., Menges C., Clarke S., Kokkinidis M., Pingoud A., Wende W., Desbiolles P. The rotation-coupled sliding of EcoRV. Nucleic Acids Res. 2012;40:4064–4070. doi: 10.1093/nar/gkr1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leith J.S., Tafvizi A., Huang F., Uspal W.E., Doyle P.S., Fersht A.R., Mirny L.A., van Oijen A.M. Sequence-dependent sliding kinetics of p53. Proc. Natl. Acad. Sci. U.S.A. 2012;109:16552–16557. doi: 10.1073/pnas.1120452109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blainey P.C., Luo G.B., Kou S.C., Mangel W.F., Verdine G.L., Bagchi B., Xie X.S. Nonspecifically bound proteins spin while diffusing along DNA. Nat. Struct. Mol. Biol. 2009;16:U1224–U1234. doi: 10.1038/nsmb.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mangel W.F., McGrath W.J., Xiong K., Graziano V., Blainey P.C. ‘Molecular sled’- vehicle of 11-amino acids that facilitates biochemical interactions via sliding components along DNA. Nat. Commun. 2015;7:10202. doi: 10.1038/ncomms10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blainey P.C., Graziano V., Perez-Berna A.J., McGrath W.J., Flint S.J., Martin C.S., Xie X.S., Mangel W.F. Regulation of a viral proteinase by a peptide and DNA in one-dimensional space IV. Viral proteinase slides along DNA to locate and process its substrates. J. Biol. Chem. 2013;288:2092–2102. doi: 10.1074/jbc.M112.407460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berg O.G., Winter R.B., von Hippel P.H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981;20:6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 12.Winter R.B., Berg O.G., von Hippel P.H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 3. The Escherichia coli lac repressor–operator interaction: kinetic measurements and conclusions. Biochemistry. 1981;20:6961–6977. doi: 10.1021/bi00527a030. [DOI] [PubMed] [Google Scholar]
- 13.Winter R.B., von Hippel P.H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 2. The Escherichia coli repressor–operator interaction: equilibrium measurements. Biochemistry. 1981;20:6948–6960. doi: 10.1021/bi00527a029. [DOI] [PubMed] [Google Scholar]
- 14.Hammar P., Leroy P., Mahmutovic A., Marklund E.G., Berg O.G., Elf J. The lac repressor displays facilitated diffusion in living cells. Science. 2012;336:1595–1598. doi: 10.1126/science.1221648. [DOI] [PubMed] [Google Scholar]
- 15.Sidorova N.Y., Muradymov S., Rau D.C. Differences in hydration coupled to specific and nonspecific competitive binding and to specific DNA binding of the restriction endonuclease BamHI. J. Biol. Chem. 2006;281:35656–35666. doi: 10.1074/jbc.M608018200. [DOI] [PubMed] [Google Scholar]
- 16.Breyer W.A., Matthews B.W. A structural basis for processivity. Protein Sci. 2001;10:1699–1711. doi: 10.1110/ps.10301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeltsch A., Wenz C., Stahl F., Pingoud A. Linear diffusion of the restriction endonuclease EcoRV on DNA is essential for the in vivo function of the enzyme. EMBO J. 1996;15:5104–5111. [PMC free article] [PubMed] [Google Scholar]
- 18.Talanian R.V., McKnight C.J., Kim P.S. Sequence-specific DNA binding by a short peptide dimer. Science. 1990;249:769–771. doi: 10.1126/science.2389142. [DOI] [PubMed] [Google Scholar]
- 19.Johnson N.P., Lindstrom J., Baase W.A., von Hippel P.H. Double-stranded DNA templates can induce alpha-helical conformation in peptides containing lysine and alanine: functional implications for leucine zipper and helix-loop-helix transcription factors. Proc. Natl. Acad. Sci. U.S.A. 1994;91:4840–4844. doi: 10.1073/pnas.91.11.4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agre P., Johnson P.F., Mcknight S.L. Cognate DNA binding specificity retained after leucine zipper exchange between GCN4 and C/EBP. Science. 1989;17:922–926. doi: 10.1126/science.2530632. [DOI] [PubMed] [Google Scholar]
- 21.Schroeder C.M., Blainey P.C., Kim S., Xie X.S. Hydrodynamic Flow-stretching Assay for Single-Molecule Studies of Nucleic Acid–Protein Interactions. NY: Cold Spring Harbor Laboratory Press; 2008. [Google Scholar]
- 22.van Oijen A.M., Blainey P.C., Crampton D.J., Richardson C.C., Ellenberger T., Xie X.S. Single-molecule kinetics of lambda exonuclease reveal base dependence and dynamic disorder. Science. 2003;301:1235–1238. doi: 10.1126/science.1084387. [DOI] [PubMed] [Google Scholar]
- 23.Romani B., Engelbrecht S., Glashoff R.H. Functions of Tat: the versatile protein of human immunodeficiency virus type 1. J. Gen. Virol. 2010;91:1–12. doi: 10.1099/vir.0.016303-0. [DOI] [PubMed] [Google Scholar]
- 24.Futaki S., Suzuki T., Ohashi W., Yagami T., Tanaka S., Ueda K., Sugiura Y. Arginine-rich peptides—an abundant source of membrane-permeable peptides having potential as carriers for intracellular protein delivery. J. Biol. Chem. 2001;276:5836–5840. doi: 10.1074/jbc.M007540200. [DOI] [PubMed] [Google Scholar]
- 25.Dandekar D.H., Ganesh K.N., Mitra D. HIV-1 Tat directly binds to NFkappaB enhancer sequence: role in viral and cellular gene expression. Nucleic Acids Res. 2004;32:1270–1278. doi: 10.1093/nar/gkh289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lange A., Mills R.E., Lange C.J., Stewart M., Devine S.E., Corbett A.H. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J. Biol. Chem. 2007;282:5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ranjit S., Levitus M. Probing the interaction between fluorophores and DNA nucleotides by fluorescence correlation spectroscopy and fluorescence quenching. Photochem. Photobiol. 2012;88:782–791. doi: 10.1111/j.1751-1097.2012.01121.x. [DOI] [PubMed] [Google Scholar]
- 28.Rohs R., West S.M., Sosinsky A., Liu P., Mann R.S., Honig B. The role of DNA shape in protein-DNA recognition. Nature. 2009;461:1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dervan P.B., Edelson B.S. Recognition of the DNA minor groove by pyrrole-imidazole polyamides. Curr. Opin. Struct. Biol. 2003;13:284–299. doi: 10.1016/s0959-440x(03)00081-2. [DOI] [PubMed] [Google Scholar]
- 30.Wilson W.D., Tanious F.A., Barton H.J., Jones R.L., Strekowski L., Boykin D.W. Binding of 4′,6-diamidino-2-phenylindole (DAPI) to GC and mixed sequences in DNA: intercalation of a classical groove-binding molecule. J. Am. Chem. Soc. 1989;111:5008–5010. [Google Scholar]
- 31.Baily C., Colson P., Henichart J.-P., Houssier C. The diferent binding modes of Hoechst 33258 to DNA studied by electric linear dichroism. Nucleic Acids Res. 1993;21:3705–3709. doi: 10.1093/nar/21.16.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ficici E., Andricioaei I. On the possibility of facilitated diffusion of dendrimers along DNA. J. Phys. Chem. B. 2015;119:6894–6904. doi: 10.1021/acs.jpcb.5b02090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Graziano V., Luo G.B., Blainey P.C., Perez-Berna A.J., Mcgrath W.J., Flint S.J., Martin C.S., Xie X.S., Mangel W.F. Regulation of a viral proteinase by a peptide and DNA in one-dimensional space II. adenovirus proteinase is activated in an unusual one-dimensional biochemical reaction. J. Biol. Chem. 2013;288:2068–2080. doi: 10.1074/jbc.M112.407312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalodimos C.G., Biris N., Bonvin A.M.J.J., Levandoski M.M., Guennuegues M., Boelens R., Kaptein R. Structure and flexibility adaptation in nonspecific and specific protein-DNA complexes. Science. 2004;305:386–389. doi: 10.1126/science.1097064. [DOI] [PubMed] [Google Scholar]
- 35.Jayaram B., Sharp K.A., Honig B. The electrostatic potential of B-DNA. Biopolymers. 1989;28:975–993. doi: 10.1002/bip.360280506. [DOI] [PubMed] [Google Scholar]
- 36.Honig B., Nicholls A. Classical electrostatics in biology and chemistry. Science. 1995;268:1144–1149. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- 37.Li G.W., Berg O.G., Elf J. Effects of macromolecular crowding and DNA looping on gene regulation kinetics. Nat. Phys. 2009;5:294–297. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.