Abstract
Bacterial genomes are transcribed by DNA-dependent RNA polymerase (RNAP), which achieves gene selectivity through interaction with sigma factors that recognize promoters, and transcription factors (TFs) that control the activity and specificity of RNAP holoenzyme. To understand the molecular mechanisms of transcriptional regulation, the identification of regulatory targets is needed for all these factors. We then performed genomic SELEX screenings of targets under the control of each sigma factor and each TF. Here we describe the assembly of 156 SELEX patterns of a total of 116 TFs performed in the presence and absence of effector ligands. The results reveal several novel concepts: (i) each TF regulates more targets than hitherto recognized; (ii) each promoter is regulated by more TFs than hitherto recognized; and (iii) the binding sites of some TFs are located within operons and even inside open reading frames. The binding sites of a set of global regulators, including cAMP receptor protein, LeuO and Lrp, overlap with those of the silencer H-NS, suggesting that certain global regulators play an anti-silencing role. To facilitate sharing of these accumulated SELEX datasets with the research community, we compiled a database, ‘Transcription Profile of Escherichia coli’ (www.shigen.nig.ac.jp/ecoli/tec/).
INTRODUCTION
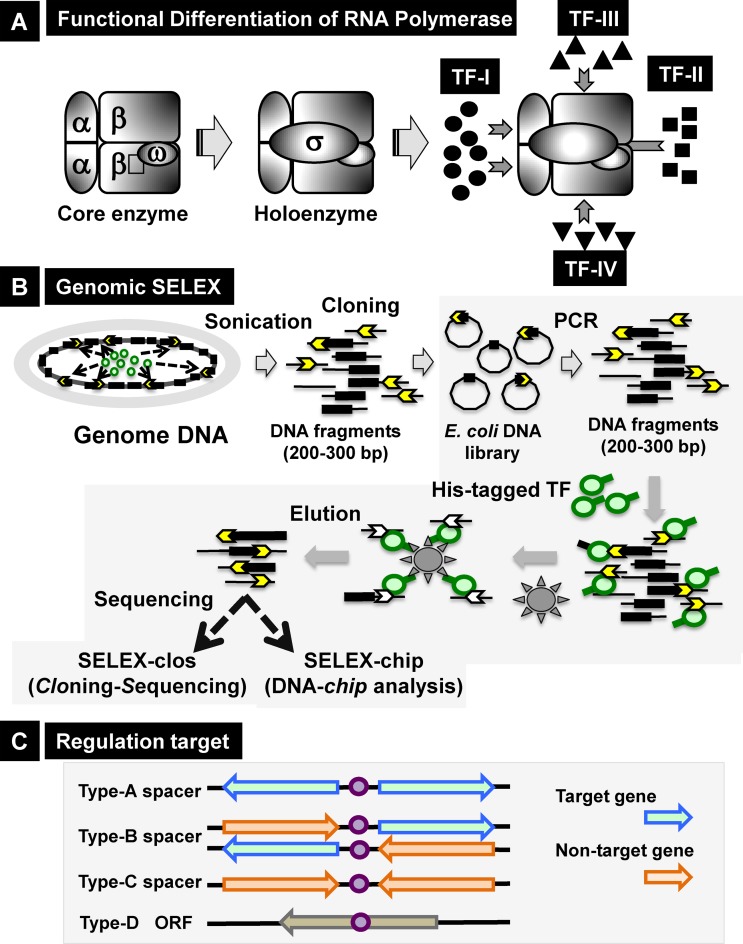
Bacteria constantly monitor the physical, chemical and biological conditions in the environment and adapt to these conditions by modifying the expression pattern of their genomes. Transcription, the primary step gene expression, is carried out by a single species of RNA polymerase (RNAP). Transcription pattern is modulated by controlling the utilization of a limited number of RNAPs among the whole set of genes in the genome (1,2). The RNAP core enzyme with the subunit composition α2ββ′ω possesses RNA polymerization activity but lacks the ability to initiate transcription. For promoter recognition and transcription initiation, another subunit, the sigma (σ) factor, is required. Transcriptionally active RNAP holoenzyme (or transcriptase) is formed after association of the core enzyme with the σ subunit (3–5). The model prokaryote Escherichia coli harbors seven species of the σ subunit, each recognizing a specific set of promoters (6–8). Each resultant RNAP holoenzyme is able to recognize a specific set of promoters, referred to the constitutive promoters, in the absence of additional transcription factors (TFs) and initiate transcription from the constitutive promoters (9). The promoter selectivity of RNAP holoenzyme is, however, modulated through interplay with another set of regulatory proteins, referred to as TFs (1,2). Most TFs associate with DNA targets (usually located near promoters), interact with promoter-bound RNAP, and modulate transcription from their target promoters (Figure 1A).
Figure 1.
Outline of genomic SELEX screening system. (A) Functional differentiation of RNAP. The core enzyme, with subunit structure α2ββ′ω, has RNA polymerization activity but cannot recognize promoters. After binding one of the sigma (σ) subunits, the holoenzyme acquires the ability to initiate transcription from constitutive promoters. The promoter selectivity of RNAP is further modulated through interactions with DNA-binding TFs that bind to their target DNA near promoters. Each DNA-bound TF interacts with a subunit of promoter-bound RNAP. Based on the contact subunit, TFs are classified into four groups (1,2). (B) Genomic SELEX system for identification of regulatory targets of TFs. A plasmid library of Escherichia coli genomic DNA was constructed using collections of genomic DNA fragments 200–300 bp in length. The mixture of genome DNA segments can be regenerated by PCR. All 285 E. coli TFs were expressed in His-tagged form by addition of a His6 sequence to either the N- or C-terminus, and then affinity purified. Mapping of TF-bound DNA segments on the E. coli genome was carried out by either SELEX-clos (cloning-sequencing) or SELEX-chip (DNA tiling array analysis) method. (C) The binding sites of TFs on the genome were classified into four types: type-A, spacer of bidirectional transcriptional units; type-B, spacer upstream of one transcription unit but downstream of another transcription unit; type-C, spacer downstream of both transcription units; and type-D, inside open reading frames. Regulatory targets of TFs were predicted based on the locations of their binding sites.
Complete genome sequences have allowed prediction of the full repertoire of TFs (1–2,10–11). In E. coli, a total of 285 TFs have been identified and classified into 54 families based on their DNA-binding motifs (2,12) (Table 1; for details see Supplementary Table S1). When bound to target DNA sites, these proteins interact directly with one of the RNAP subunits (13–15). There is a tight correlation between the mode of transcriptional action and the contact subunit (class I, II, III or IV) (Figure 1A) (1,2). Binding of TFs at specific target sites near promoters plays an essential role in ensuring effective protein–protein interactions by increasing the local concentration of pairing proteins at promoter regions. To date, 70–80% of the estimated 285 TFs in E. coli have been linked to at least one regulatory target gene; however, the whole set of target promoters, genes or operons has been identified for only a small number of TFs; moreover, the regulatory targets and regulatory functions remain unknown for about a quarter of E. coli TFs, referred to as Y-gene TFs. Consequently, we currently have only fragmentary knowledge of the genome-wide transcriptional regulation even for E. coli, the best-characterized model prokaryote. In order to obtain detailed insight into the molecular basis of genome regulation, it is absolutely necessary to determine the targets and regulatory modes of all predicted TFs.
Table 1. DNA-binding transcription factors (TF) in Escherichia coli.
| Family | No. | Characterized TF | Uncharacterized TF |
|---|---|---|---|
| AraC | 32 | Ada, AdiY, AppY, AraC, ChbR, EnvY, EutR, FeaR, FliZ, FrvR, GadW, GadX, LsrR, MarA, MelR, NimR, RclR, RhaR, RhaS, Rob, SoxS, XylR, YdeO, YkgA, YqhC | YbcM, YdiP, YidL, YiiF, YijO, YneL, YpdC |
| ArgR | 1 | ArgR | |
| ArsR | 3 | ArsR, FeoC, YgaV | |
| AsnC | 3 | AsnC, Lrp | YbaO |
| BirA | 1 | BirA | |
| BolA | 1 | BolA | |
| CadC | 2 | CadC | YqeI |
| CaiF | 1 | CaiF | |
| CbpA | 1 | CbpA | |
| CdaR | 1 | CdaR | |
| CitB | 3 | CitB, DctR, DcuR | |
| Crp | 4 | Crp, Fnr, YeiL | YaiV |
| DeoR | 13 | AgaR, DeoR, DeoT, FucR, GatR, GlpR, SgcR, SrlR, UlaR | YdjF, YfjR, YgbI, YihW |
| DicC | 1 | DicC | |
| DnaA | 1 | DnaA | |
| Dps | 1 | Dps | |
| DtxR* | 1 | MntR | |
| Fis | 1 | Fis | |
| FlhC | 1 | FlhC | |
| FlhD | 1 | FlhD | |
| FrmR | 2 | FrmR, RcnR | |
| Fur | 2 | Fur, Zur | |
| GntR | 25 | CsiR, DgoR, ExuR, FadR, FarR, FrlR, GlcC, LctR, LgoR, McbR, MtlR, NanR, PaaX, PdhR, PhnF, UxuR, YdfH, YggD, YqjI | YdcR, YegW, YidP, YieP, YihL, YjiR |
| GutM | 1 | GutM | |
| H-NS | 2 | Hns, StpA | |
| HU | 4 | HupA, HupB, IhfA, IhfB | |
| IclR | 8 | AllR, IclR, KdgR, MhpR, RhmR, YiaJ, | YagI, YjhI |
| LacI | 15 | AscG, Cra, CscR, CytR, EbgR, GalR, GalS, GntR, IdnR, LacI, MalI, PurR, RbsR, TreR | YcjW |
| LexA | 1 | LexA | |
| LuxR* | 20 | BglJ, CsgD, EvgA, FimZ, GadE, MalT, MatA, NarL, NarP, RcsA, RcsB, SdiA, UhpA, UvrY, YgeH, YjjQ | YahA, YgeK, YhjB, YqeH |
| LysR | 47 | AbgR, AllS, ArgP, Cbl, CynR, CysB, Dan, DmlR, DsdC, GcvA, HcaR, HdfR, IlvY, LeuO, LrhA, LysR, MetR, Nac, NhaR, OxyR, PerR, PgrR, QseA, QseD, TdcA, XapR, YeiE, YfeR, YidZ | YafC, YahB, YbdO, YbeF, YbhD, YcaN, YdcI, YdhB, YeeY, YfeC, YfeD, YfiE, YgfI, YhaJ, YhjC, YiaU, YneJ, YnfL |
| LytR* | 2 | YehT, YpdB | |
| MarR | 5 | EmrR, MarR, SlyA | YdaW, YtfH |
| MerR* | 5 | BluR, CueR, MlrA, SoxR, ZntR | |
| MetJ | 1 | MetJ | |
| ModE | 1 | ModE | |
| MraZ | 1 | MraZ | |
| NagC | 3 | Mlc, NagC | YphH |
| NikR | 1 | NikR | |
| NrdR | 1 | NrdR | |
| NsrR | 1 | NsrR | |
| NtrC | 8 | AtoC, NorR, NtrC, PrpR, PspF, QseF, RtcR, ZraR | |
| OgrK | 1 | OgrK | |
| OmpR | 14 | ArcA, BaeR, BasR, CpxR, CreB, CusR, KdpE, OmpR, PhoB, PhoP, QseB, RstA, TorR, YedW | |
| PepA | 1 | PepA | |
| PutA | 1 | PutA | |
| RpiR | 4 | HexR, MurR, RpiR | YfhH |
| SfsA | 2 | SfsA, SfsB | |
| TdcR | 1 | TdcR | |
| TetR* | 15 | AcrR, BdcR, BetI, ComR, EnvR, FabR, GusR, NemR, RcdA, RutR, SlmA | YagA, YbiH, YcfQ, YjdC |
| TrpR | 1 | TrpR | |
| TyrR | 4 | DhaR, FhlA, HyfR, TyrR | |
| Xre* | 13 | DicA, DinJ, HigA, MqsA, NadR, PuuR, RacR, SutR, YgiV | YbaQ, YddM, YheO, YiaG |
| 54 | 285 | 227 | 58 |
*Thse six families are named using TF symbols from bacteria other than E. coli. DtxR, diphtheria toxin repressor of Corynebacterium diphtheriae; LuxR, luminescence activator of Vibrio fisheri; LytR, autolysin regulator of Streptococcus mutans; MerR, metal regulatory protein of R100 plasmid; TetR, tetracycline repressor of Tn10; and Xre, xenobiotic-response-element recognition protein.
Most of the 285 E. coli TFs recognize and bind to specific DNA sequences. However, it is difficult to identify the whole set of in vivo TF-binding sites in the genome, mainly because of competition between DNA-binding proteins, including the abundant nucleoid proteins that cover the entire E. coli genome. In addition, the target selectivity of TFs is modulated in vivo by molecular interaction with specific effector ligands and/or by covalent modification of TFs by phosphorylation and acetylation. For accurate and quick search for DNA sequences that are recognized by a defined form of TF under a fixed condition, therefore, we developed the in vitro ‘Genomic SELEX’ system (16). In the original SELEX (systematic evolution of ligands by exponential enrichment) system, DNA–protein complexes are isolated from mixtures of a test DNA-binding protein and synthetic oligonucleotides with all possible sequences, followed by sequencing of protein-bound DNA fragments (17,18). As the size of the probes increases, the number of probe species increases; consequently, it becomes difficult to solubilize all probes at the effective concentrations required for protein binding. To overcome this solubility problem, a mixture of genomic DNA fragments is used in place of synthetic oligonucleotide mixtures, because the binding sites of the TFs of interest are located in the E. coli genome (16,19–20). Using the improved genomic SELEX system (Figure 1B), we initiated a systematic search for DNA sequences recognized by each of the seven sigma subunits and 285 DNA-binding TFs from E. coli. The sequences of protein-bound SELEX DNA fragments are determined using two procedures, SELEX-clos (cloning and sequencing) and SELEX-chip (mapping by tiling array consisting of 43 450 species of 60-base oligonucleotides probe aligned at ∼45-bp intervals along the E. coli genome). Genomic SELEX screening is particularly useful for identifying regulatory targets of uncharacterized TFs because it is difficult to predict targets from phenotypic screening of TF mutants. To date, this method has been used to identify the regulatory functions of more than 10 hitherto uncharacterized Y-gene TFs, including Dan (renamed from YgiP) (21), NemR (renamed from YdhM) (22), NimR (renamed from YeaM) (23), PgrR (renamed from YcjZ) (24), RcdA (renamed from YbjK) (25), RutR (renamed from YcdC) (26,27) and SutR (renamed from YdcN) (28).
EcoCyc (29,30) and RegulonDB (31,32) are currently the most comprehensive databases of the E. coli regulatory network. Most of our published data obtained by SELEX screening of regulatory targets have been deposited into these databases. However, the regulatory targets of E. coli TFs listed in these databases were assigned with varying levels of accuracy: some were predicted in silico simply based on the presence of sequences similar to the recognition sequence of a given TF, but are not supported by experimental confirmation. One serious problem with current TF databases is that the data were obtained using various E. coli strains grown in different types of media, under different conditions, using various techniques, by a number of different laboratories. Since we found five different types of the rpoS gene encoding the stationary phase-specific σ subunit of RNAP in a single and the same E. coli K-12 W3110 strain from eleven different laboratories (33), the heterogeneity of RpoS, a key regulator of transcription, has been widely recognized in a number of laboratory stocks of E. coli from all over the world (34). At present, massive diversification is recognized in the genome of wild-type E. coli lab strains, leading to difference in the regulatory mode of genome transcription (35). To overcome this problem, we have undertaken a long-term project aimed at identifying the regulatory targets of all 285 TFs from a single E. coli strain, K-12 W3110, using the same experimental systems in a single research group. This is the first summary report of a total of 156 SELEX patterns of a set of 116 TFs. Using the collection of data sets, we elucidated a number of novel concepts regarding transcriptional regulation in E. coli, some of which are described in this report. In order to make the useful information open to the public, the SELEX screening data are assembled into a database, entitled ‘Transcription Profile of E.coli’ (TEC).
MATERIALS AND METHODS
Genomic SELEX screening
Genomic SELEX screenings of 116 TFs and SELEX-chip analyses were conducted as previously described (16). In brief, a collection of 200–300-bp DNA fragments of the E. coli W3110 genome was prepared by polymerase chain reaction (PCR) using an E. coli DNA plasmid library (16) as the template and a set of primers, EcoRV-F and EcoRV-R, that hybridize to vector pBR322 at EcoRV junctions. This collection includes ∼105 different random DNA fragments, covering about 6.5-fold the length of E. coli genome. The size of DNA fragments was determined so as to allow binding of all the hitherto characterized E. coli TFs. The standard protocol for genomic SELEX was as follows: (i) a mixture of 5 pmol of genome DNA fragments and 10 pmol of His-tagged TF in binding buffer [10 mM Tris–HCl (pH 7.8 at 4°C), 3 mM Mg acetate, 150 mM NaCl and 1.25 mg/ml bovine serum albumin] was incubated for 30 min at 37°C [note that in the initial screening, TFs were added in excess to find the binding sites even with weak affinity]; (ii) The DNA–TF mixture was applied to a Ni-NTA column; (iii) To remove unbound DNA, the column was washed three times with binding buffer containing 10 mM imidazole; and (iv) DNA–TF complexes were eluted with elution buffer containing 200 mM imidazole. DNA was isolated from the DNA–TF complexes and PCR-amplified using EcoRV-F and EcoRV-R primers. This SELEX cycle was repeated at least twice and up to eight times for some TFs, to enrich TF-binding DNA. At each SELEX step, a gel-shift assay was carried out to examine enrichment of specific DNA segments with high affinity to the test TF.
The activity and specificity of most E. coli TFs are modulated by covalent modifications, such as phosphorylation, or interaction with low-molecular weight effectors (2). Therefore, the genomic SELEX screening of TF-binding sequences was also performed in the presence of effector ligands. Among the 116 TFs examined in this study, some were analyzed in the presence and absence of effector ligands; as a result, the total number of SELEX patterns increased to 156.
Identification of TF-bound DNA
To identify DNA segments recovered by genomic SELEX screening, we employed the SELEX-chip (tilling array) system. For this purpose, we used a DNA tiling array consisting of 43 450 species of 60-base probes spaced at an average interval of 45 bp along the E. coli genome. Using this array, the 200–300-bp SELEX fragments should bind to three or more consecutive probes. The TF-bound SELEX DNA fragment and the original DNA library are labeled with Cy5 and Cy3, respectively. The height of each peak is calculated relative to the combined fluorescence intensities of all probes within the highest peak. The height of TF-bound peaks correlates with the affinity of test TF to the corresponding probe sequence. The fluctuation level of background DNA was <2-fold as estimated after SELEX experiment in the absence of TF addition. Under the resolution of this genomic SELEX system herein employed, however, the exact binding sites and their orientations within promoter/operator regions remain unknown.
The TF-binding sites within the E. coli genome were classified into four types depending on their positions relative to transcriptional units (Figure 1C): type-A, upstream of bidirectional transcription units; type-B, upstream of one transcription unit but downstream of another; type-C, downstream of two transcription units; and type-D, inside open reading frames. TFs bound to type-A spacers could regulate one or both bidirectional transcription units, whereas TFs bound to type-B spacers should regulate transcription in only one direction.
Construction of TEC database
The collection of datasets from SELEX screening was assembled into a single database, entitled ‘TEC’. The data in the TEC database are managed by a PostgreSQL relational database system. Web applications were developed on an Apache Tomcat web server. Software for data processing and drawing maps were written in gcc, Perl or Java. BioProspector and WebLogo, furnished under the MIT license, can be used to analyze the consensus sequence of each TF.
RESULTS AND DISCUSSION
Classification of TFs
When we started this project in the mid-1990s, nothing was known of the structure and function for more than 100 DNA-binding TFs in E. coli K-12. Since we published the first classification of E. coli TFs (12), the regulatory functions have been identified for some of the uncharacterized TFs, and in addition, several additional proteins with DNA-binding activity have been shown to participate in transcriptional regulation. Accordingly the classification list of DNA-binding TFs has been revised twice (1,2). Based on recent progress in this field, including the systematic screening using the improved genomic SELEX system described herein, we propose a revised version of the TF classification including 285 DNA-binding TFs in E. coli K-12 W3110 (Table 1; for details see Supplementary Table S1). Fifty-eight out of the 285 TFs still remain uncharacterized. Genomic SELEX screening system is, in particular, useful for the identification of regulatory targets for hitherto uncharacterized TFs, referred to Y-gene TFs.
Besides these DNA-binding TFs, about 10 species of TFs, including transcription termination and anti-termination factors such as Rho, NusA and GreA can be isolated as complexes with RNAP in the absence of DNA. These RNAP-binding TFs without DNA-binding affinity are not included in the revised list of TFs.
Genomic SELEX search for TF regulatory targets
Genomic SELEX screening aimed at identification of the binding sites of all seven RNAP sigma factors and all 285 TFs from E. coli K-12 W3110 is currently underway. For instance, the full set of 669 constitutive promoters recognized by RpoD holoenzyme was identified by SELEX screening and already published (9,36). For SELEX screening of DNA sequences recognized by TFs, DNA–TF complexes were recovered by affinity purification using His-tagged TFs. Most TFs retain their binding activity on specific target sequences after addition of the His-tag at the N- or C-terminus. However, in some cases, such as OmpR, the addition of a His-tag at the N-terminus interferes with the recognition specificity of known target sequences, and in this case, His-tag was added to the C-terminus (37). The cycle of SELEX screening was repeated at least twice, but in some cases, up to eight cycles were necessary for enrichment of specific DNA segments as detected by gel electrophoresis.
The distribution of TF-bound DNA sequences along the E. coli genome was analyzed using a tiling array (16). In the case of E. coli TFs, target promoters, genes and operons are located in close proximity to TF-binding sites. Thus, the regulatory targets of each TF were estimated based on the binding site of the test TF (Figure 1C). In some cases, the identification of TF-bound DNA segments was identified using the cloning and sequencing procedure (the SELEX-clos system) (see the reference list in Table 2). The number of clones identified by the SELEX-clos procedure correlates with the affinity for the test TF. The combination of SELEX-clos and SELEX-chip patterns provides a more reliable set of regulatory targets for a given TF. In this report, we describe results of the SELEX screening for a total of 116 TFs (84 characterized and 32 uncharacterized) (Supplementary Table S2), of which the SELEX screening results have already been reported for 19 TFs (note that SELEX screening of SdiA was performed in the absence and present of four different effectors) after experimental confirmation of the predicted targets (Table 2).
Table 2. Regulation targets of transcription factor (TF).
| TF | Effector | Family | No. TF binding sites | No.regulation targets [A] | No.known targets [B] | Increase [A]/[B] | Ref |
|---|---|---|---|---|---|---|---|
| AscG | LacI | 13 | 13–19 | 4 | 3.3∼4.8 | (91) | |
| BasR | Acetyl-P | OmpR | 38 | 33–47 | 11 | 3.0∼4.3 | (92) |
| BetI | Choline | RutR | 1 | 2 | 1 | 2.0 | Figure 2A |
| CitB | CitB | 11 | 11–15 | 5 | 2.2–3.0 | (93) | |
| Cra | LacI | 194 | 193–296 | 40 | 4.8–7.4 | (16,58) | |
| CRP | cAMP | CRP | 277 | 192–290 | 190 | 1.0–1.5 | (57) |
| LeuO | LysR | 140 | 131–173 | 15 | 8.7–11.5 | (60) | |
| Lrp | AsnC | 314 | 218–296 | 43 | 5.1–6.9 | (63) | |
| NanR | GntR | 2 | 3 | 3 | ∼1.0 | Figure 2C | |
| NemR | RutR | 2 | 4 | 2 | ∼2.0 | (22) | |
| NimR | AraC | 1 | 2 | 2 | ∼1.0 | (23) | |
| OmpR | Acetyl-P | OmpR | 26 | 21–32 | 12 | 1.8–2.1 | (37) |
| PgrR | LacI | 11 | 8–11 | 4 | 2.0–2.75 | (24) | |
| RbsR | LacI | 5 | 3–4 | 2 | 1.5–2.0 | (94) | |
| RcdA | RutR | 20 | 11–17 | 8 | 1.4–2.1 | (25) | |
| RutR | RutR | 40 | 35–60 | 7 | 5.0–8.6 | (26) | |
| SdiA | SdiA | 31 | 20–27 | 3 | 6.7–9.0 | (41) | |
| SdiA | HSL-A | SdiA | 43 | 32–54 | 3 | 10.7–18.0 | (41) |
| SdiA | HSL-F | SdiA | 26 | 17–29 | 3 | 5.7–9.7 | (41) |
| SdiA | HSL-K | SdiA | 37 | 26–41 | 3 | 8.7–13.7 | (41) |
| SdiA | HSL-OC | SdiA | 34 | 25–44 | 3 | 8.3–14.7 | (41) |
| SutR | DinJ | 17 | 13–15 | 4 | 3.3–3.8 | (28) | |
| UlaR | Acetyl-P | DeoR | 2 | 3 | 2 | 1.0–1.5 | Figure 2B |
| The regulation targets of these TFs were predicted by Genomic SELEX screening and afterward experimentally confirmed. Number of the known targets are from EcoCyc. | |||||||
| Nucleoid protein | |||||||
| TF | Effector | Family | No. TF binding sites | No.regulation target operons | No. targets (EcoCyc) | Ref | |
| Dan | LysR | 688 | (21) | ||||
| Fis | Fis | 1269 | Figure 3A | ||||
| HNS | HNS | 987 | (60); Figure 3C | ||||
| IHF | HU | 813 | Figure 3B | ||||
The binding sites of these nucleoid proteins were identified by using the in vitro SELEX screening system.
The affinity and specificity of DNA-binding of E. coli TFs are mostly modulated by either protein phosphorylation or interactions with low-molecular-weight effectors. E. coli contains 285 species of DNA-binding TF (Table 1), of which 30–33 are organized into two-component systems (TCSs) (38,39), each consisting of a signal sensor that can auto-phosphorylate its specific Asp residue (HK) and a response regulator (RR) that can phosphorylate its own His residue using the HK-bound phosphate. In most cases, RR phosphorylation can take place in vivo using acetyl phosphate (AcP) as a substrate. Therefore, for TFs belonging to TCS RRs, SELEX screening was performed in the presence or absence of high concentrations of acetyl phosphate. In this report, we provide SELEX screening data for 11 TCS RRs, including the hitherto uncharacterized YpdB (Table 2 and Supplementary Table S2).
On the other hand, the activity and specificity of about 90% of other E. coli TFs are regulated through molecular interactions with low-molecular weight effector ligands. For 106 TFs under the control of known effectors, SELEX screening was performed in the presence or absence of the appropriate effector ligands. For some TFs, the involvement of multiple effectors in activity control has been established and for TFs in this group, SELEX was performed in the presence of individual effectors such as Arg and Lys for ArgP (40) or HSL-A, HSL-F, HSL-K and HSL-OC for SdiA (41). For some TFs, the activity and specificity of DNA recognition are modulated through protein–protein interactions. For instance, SELEX screening of the nucleoid protein H-NS was performed in the presence or absence of its small partner protein Hha (42). As a result, the total number of SELEX patterns for effector-regulated TFs increased to 156 (Supplementary Table S2).
Prediction of regulatory targets of TFs
The whole set of regulatory target promoters, genes and operons for each TF was predicted based on the distribution of TF-binding sites along the entire E. coli genome, as determined by SELEX patterns (see Figure 1C for the classification of TF-binding sites). SELEX patterns, however, always include background noise, for instance, arising from TF binding to non-specific DNA sequences and incomplete removal of unbound DNA segments during SELEX screening. Therefore, the initial screening of the targets of each TF was carried out by setting a fixed but low cut-off for all TFs (Supplementary Table S2). The binding sites of 116 TFs (156 SELEX patterns) were located within 641 type-A spacers between bidirectional transcription units, which are thus predicted to regulate a minimum of 641 (in cases of one direction transcription) and a maximum of 1282 units (in cases of bidirectional transcription) (Supplementary Table S3, type-A spacer). Based on the known operon organization, the total number of genes under the control of 641 type A-spacer-associated TFs is 2176 (average, 3.39 genes per TF). By contrast, a total of 1212 TFs bind to promoter-regions of type-B spacers, which should regulate unidirectional transcription units including 1858 genes (average, 1.53 genes per TF) (Supplementary Table S3, type-B spacer). One surprising finding of this research is that a total of as many as 1223 TF-binding sites was identified within type-B spacers inside operons (Supplementary Table S3, type-B spacer). A marked difference in the number of TF-binding peaks was observed among the 143 SELEX patterns, ranging from a single peak to more than 1000 peaks. Likewise, a marked difference in TF-binding intensity was observed, with a ∼10 000-fold difference (after back-ground subtraction) in the peak height between the highest and the lowest values. Both the number and the height of TF-binding peak vary after resetting the cut-off level.
The background level differed among the 156 SELEX patterns. To achieve a more accurate estimation of regulatory targets, care was taken to set the background level for each TF. One important criterion employed for background setting was the inclusion of known targets, allowing a more accurate estimation of specific TF-binding sites and regulatory targets. Depending on the research purpose, the background can be freely controlled by using the TEC database (see below). The set of regulatory targets described in Supplementary Table S2 represents the list of estimation that is still useful for detailed analysis of the regulatory function of each TF. For confirmation of the exact regulatory targets, we employ several experimental approaches such as in vitro assays of DNA-TF interaction (by gel retardation assay and DNase foot-printing) and in vivo assays of promoter regulation by the test TFs (using reporter assays, northern blot and RT-PCR of the target RNAs). The regulatory targets have been experimentally confirmed for a total of 19 TFs (Table 2). For these TFs, the number of regulatory target promoters (and operons) increased from 2- to 20-folds than those listed in the databases such as RegulonDB and EcoCyc (Table 2). The list of promoters identified by the SELEX screening system herein employed represents the potential promoters that are recognized in the presence of excess TF alone in the absence of other DNA-binding proteins. The number of target promoters should decrease in vivo with decrease in the level of functional TF and in the presence of competing proteins.
Classification of TFs
Early molecular genetic studies suggested that E. coli promoters are regulated by a single specific regulatory protein, either a repressor or an activator; a classic example is regulation of the lac operon by LacI repressor (43). Accordingly, most TFs in E. coli are believed to be ‘local’ TFs that control transcription of a specific gene or a small number of transcription units (44,45). Until recently, only a few were classified as global regulators that influence the expression of a large number of transcription units belonging to different metabolic pathways, thereby exhibiting pleiotropic phenotypes (10,46–47). The set of global TFs includes CRP (cAMP receptor protein), FNR (fumarate and nitrate reduction), ArcA (anoxic redox control), Fur (ferric uptake regulation), Lrp (leucine-responsive regulatory protein) and NarL (nitrate/nitrite response regulator) (48). Based on the whole sets of regulatory targets for the 116 TFs (and 156 SELEX patterns) listed in this report, we next reexamined the classification of TFs with respect to the number of regulatory targets.
Single-target transcription factors (local regulators)
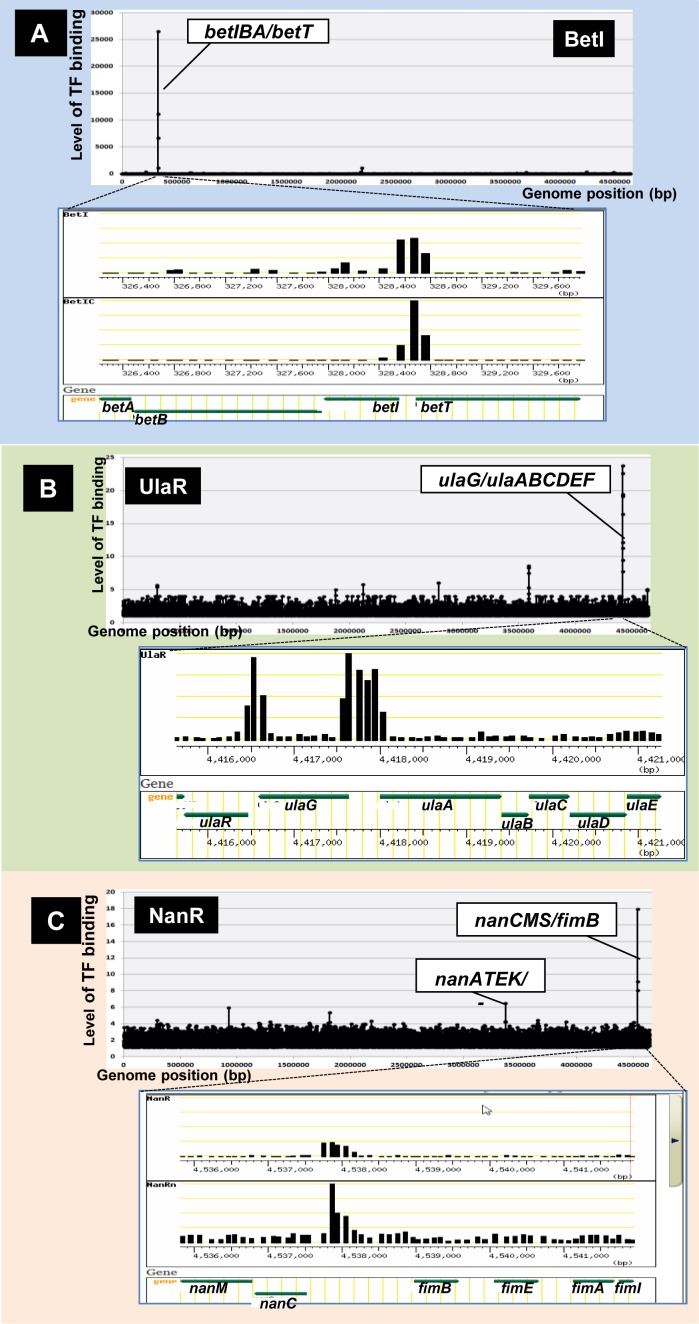
Among the 116 TFs (156 SELEX patterns) examined, 24 (20%) regulate fewer than 10 genes. This group of local regulators includes BetI (betaine inhibitor), NorR (NO reduction and detoxification regulator), NanR (N-acetyl-neuraminic acid regulator), NimR (2-imidazole regulator) and UlaR (utilization of L-ascorbate operon regulator). For instance, BetI (betaine inhibitor) binds to a single site within the spacer of bidirectional transcription units betIBA (encoding BetI itself and enzymes for glycine betaine biosynthesis from choline) and betT (encoding choline:H+ symporter) (Figure 2A: genome-wide pattern in upper panel, expanded local pattern in lower panel). Expression of the bet operon is regulated in vivo by the effector choline (49), but BetI is a unique regulator that remains bound to DNA even in the presence of choline (50). NanR (N-acetyl-neuraminic acid regulator) was identified in the spacer between nanCM and fimB, and upstream of nanATEK (Figure 2C). NanR activates transcription of both nanCM and nanATEK operons, together encoding proteins involved in transport and metabolism of N-acetyl-neuraminic acid (51,52), but NanR also represses transcription of fimB, which encodes a Fim switch protein (51,52). NimR, a typical single-target regulator, regulates divergent transcription of nimT, which encodes a transporter of the anti-bacterial agent 2-imidazole, (23). UlaR is the repressor of the divergent ula operon involved in transport and catabolism of L-ascorbate (53). The high-affinity binding site of UlaR is located within the spacer of bidirectional transcription units ulaG and ulaABCDEF (Figure 2B), both of which are involved in transport and utilization of L-ascorbic acid. In addition, a low-level peak was identified upstream of the ulaR gene itself, indicating that it regulates its own expression.
Figure 2.
Single-target TFs. After in vitro SELEX screening of DNA-binding sequences of purified TFs, their regulatory targets were predicted based on their binding sites, as noted in Figure 1C. Among 156 SELEX patterns examined for 116 TFs, a small number of TFs regulated only one specific target operon. Some representative SELEX patterns are shown: (A) BetI: (B) UlaR; and (C) NanR. For each SELEX pattern, the upper panel shows the genome-wide distribution of TF-binding sites, whereas the lower panel shows the expanded local region of a TF-binding site [note that the expanded patterns were retrieved from TEC (Transcription Profile of Escherichia coli) database]. The Y-axis indicates the fluorescent intensity of SELEX fragment binding to each probe relative to that of library DNA. The Y-axis in each expanded image indicates the level of each peak relative to that of the highest peak.
Multi-target transcription factors (global regulators)
On the other hand, Genomic SELEX screening revealed that most TFs herein examined regulate multiple targets, ranging from 10 to 100 operons. Among the TFs controlling more than 100 targets, two sets of TFs were shown to act as global regulators of metabolism: CRP (cAMP receptor protein) and Cra (catabolite repressor and activator) for carbon metabolism, LeuO (leucine regulator) and Lrp (leucine response regulatory protein) for nitrogen metabolism (Table 3).
Table 3. Overlapping of binding sites between silencer H-NS and global regulators.
| [A] Overlapping of H-NS binding sites with global regulators | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Type-A spacer (A) | Type-B spacer (B) | ||||||||||||
| TF | Total no. H-NS sites | No. H-NS sites | Overlap TF | No. TF sites | Overlapping (%) | No. H-NS sites | Overlap TF | No. TF sites | Overlapping (%) | Total no. H-NS (A + B) | Total no. overlap TF sites | Overlapping (%) | Ref |
| H-NS | 987 | 137 | CRP | 37 | 27.0 | 294 | CRP | 36 | 12.2 | 431 | 73 | 16.9 | (60) |
| H-NS | 987 | 137 | LeuO | 31 | 22.6 | 294 | LeuO | 56 | 19.1 | 431 | 87 | 20.2 | (60) |
| H-NS | 987 | 137 | Lrp | 36 | 26.3 | 294 | Lrp | 61 | 20.7 | 431 | 97 | 22.5 | (60) |
| [B] Overlapping of global TF-binding sites with silencer H-NS | |||||||||||||
| Type-A spacer (A) | Type-B spacer (B) | ||||||||||||
| TF | Total no. TF sites | No. TF sites | Overlap H-NS sites | No. H-NS sites | Overlapping (%) | No. TF sites | Overlap H-NS sites | No. H-NS sites | Overlapping (%) | Total no. TF sites (A + B) | Total no. overlap H-NS sites | Overlapping (%) | Ref |
| CRP | 374 | 142 | H-NS | 37 | 26.0 | 141 | H-NS | 36 | 25.5 | 283 | 73 | 25.8 | (57) |
| Leu | 140 | 42 | H-NS | 31 | 73.8 | 76 | H-NS | 56 | 73.7 | 118 | 87 | 73.7 | (60) |
| Lrp | 314 | 78 | H-NS | 36 | 46.2 | 140 | H-NS | 61 | 43.6 | 218 | 97 | 44.5 | (63) |
Focused three TFs were set bold in this table.
The availability of carbon in the environment influences the expression pattern of multiple genes in E. coli in various ways. The cAMP receptor protein CRP, also called catabolite gene activator protein (CAP), is the best-characterized global regulator of genes involved in transport and utilization of carbon sources (54–56). Lack of glucose activates synthesis of cAMP, which associates with CRP to convert it into an active regulator of transcription. Based on the SELEX pattern of CRP in the presence of cAMP, CRP was predicted to regulate minimum 119 and maximum 219 target operons (57). Taken together with the hitherto identified targets, a total of 283–425 operons were predicted to be under the control of cAMP–CRP complex (Table 3). The primary role of cAMP-CRP is control of genes involved in both uptake of carbon sources and metabolism downstream of glycolysis, including the TCA cycle and aerobic respiration. On the other hand, Cra (renamed from FruR, another global regulator of carbon metabolism) was determined to bind a total of 173 sites, allowing prediction of regulatory targets between minimum 70 and maximum 104 (16,58). Cra was shown to act as an activator of most genes encoding enzymes involved in gluconeogenesis, the TCA cycle, and the glyoxylate shunt pathway, as well as a repressor of the genes encoding the Entner–Doudoroff pathway and glycolysis. Derepression of glycolysis genes takes place when the repressor Cra is inactivated after interaction with inducers such as D-fructose-1-phosphate and D-fructose-1,6-bisphosphate. In the presence of glucose, the intracellular concentrations of these inducers increase, and they interact with Cra to prevent it from binding to the target operons.
Genomic SELEX screening revealed that two global regulators, LeuO and Lrp, regulate a large set of genes including those involved in N metabolism. Leucine, a metabolic signal that controls the level of amino acids, affects expression of multiple E. coli genes. LeuO, one of the Leu sensors, was originally identified as a regulator of genes involved in Leu biosynthesis (59). Genomic SELEX screening identified at least 140 LeuO-binding sites in the E. coli genome (60), of which 118 are in promoter regions involved in gene regulation. Again the total number of regulatory targets increased, from 15 known targets to 118 (60), of which 74% overlap with the binding sites of H-NS, the universal silencer of stress-response genes (Table 3). This finding indicates that one important function of LeuO is anti-silencing of H-NS-mediated repression of a set of unnecessary genes, including some toxic genes transferred from other organisms. Another Leu sensor, Lrp, regulates genes involved in nutrient transport, biosynthesis and catabolism of amino acids, as well as utilization of amino acids for varieties of cellular functions (61,62). Using Genomic SELEX screening, we identified 314 Lrp-binding sites on the E. coli genome, and predicted 218–296 regulatory targets (Table 3) (63). Consistent with the role of Lrp in sensing leucine availability, multiple genes involved in nitrogen metabolism and genes encoding components of the translation system appear to be under the direct control of Lrp. In addition, a variety of stress-response genes that respond to nutrient availability were also included in the Lrp regulon. With respect to the wide range of genome regulation, the role of Lrp as a key regulator of N-source utilization is similar to that of CRP in carbon utilization.
Nucleoid transcription factors (bifunctional regulators)
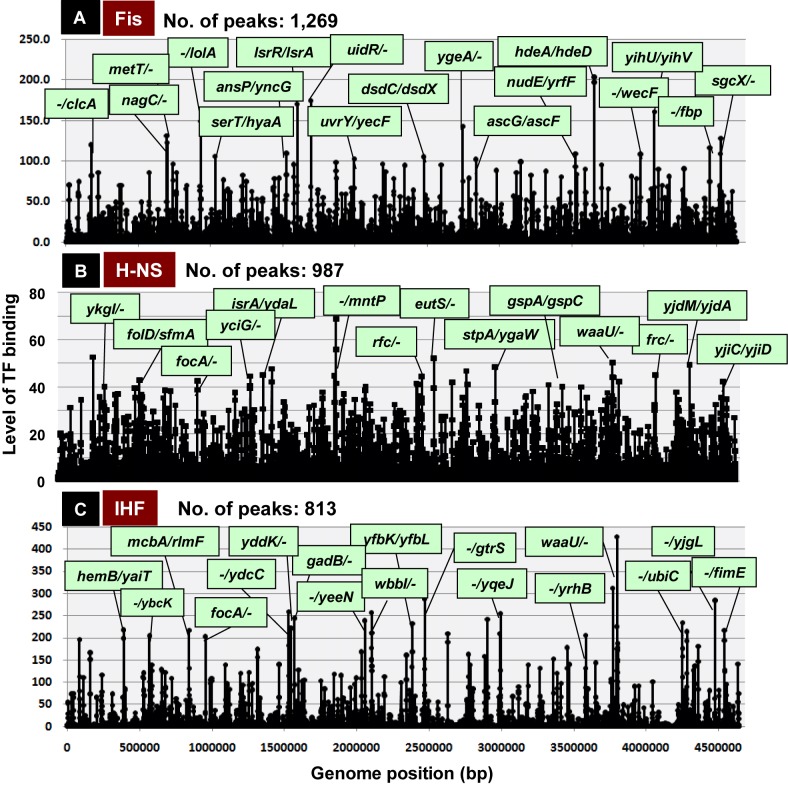
In the E. coli nucleoid, two groups of nucleoid protein are present: universal nucleoid proteins (UNPs) that always remain in the nucleoid, and growth phase- and growth condition-specific nucleoid proteins (GNPs) that appear only at specific phases of cell growth (12,64). All of these nucleoid proteins are bifunctional and play both architectural roles in folding and packaging of genomic DNA and regulatory roles in genome functions (12,64–66). We subjected all of the nucleoid proteins to SELEX screening. IHF is abundant during all growth phases, at concentrations ranging from 6000 dimers per cell in log phase to 3000 dimers per cell in stationary phase (67,68). Genomic SELEX screening revealed 813 IHF-binding sites in the E. coli genome (Figure 3C). The list of IHF-binding targets supports its dual role, i.e. an architectural role in DNA supercoiling and DNA duplex destabilization, and a regulatory role in genome functions controlling processes such as DNA replication, recombination, and the expression of multiple genes (12,69–70). H-NS is present in many copies per cell in exponential growth phase, but is much less abundant in stationary phase (12,65,71). H-NS is a general silencer of a large number of unused genes, with a strong preference for horizontally acquired genes (72–74). Genetic SELEX screening revealed that the E. coli genome contains 987 H-NS binding sites (Figure 3B). Almost all the genes under silencing control by H-NS are included in the list of H-NS binding targets.
Figure 3.
Multi-target TFs. SELEX screening of TF-binding sequences revealed that a set of the bifunctional nucleoid proteins with both architectural roles in genome folding and regulatory role in transcription bind up to ∼1000 sites along the entire Escherichia coli genome. The number of binding peaks was: 1269 for Fis (A); 987 for H-NS (B); and 813 for IHF (C). High-level binding sites of these nucleoid proteins within type-A and type-B spacers are shown under green background. In the case of bidirectional transcription units within type-A spacers, the first genes on both sides are shown, whereas in the case of type-B spacers, the first genes in one transcriptional direction are shown. The Y-axis indicates the fluorescent intensity of SELEX fragment binding to each probe relative to that of library DNA.
The GNP group of nucleoid proteins includes Fis (preferentially expressed in exponentially growing phase), Dps (expressed only in the stationary phase) and Dan (expressed under anaerobic conditions), all of which participate in the organization and maintenance of nucleoid structure through direct contact with specific sequences in the genome. Marked growth-coupled changes are observed in the intracellular levels of these nucleoid proteins (67,68). Fis is a typical GNP involved in activation of the growth-related genes (75). SELEX screening identified 1269 Fis-binding sites (Figure 3A), including multiple genes necessary for high-rate growth such as components of the translation system. The stationary phase-specific nucleoid protein Dps binds in vitro to a total of 624 sites, but as in the case of other nucleoid proteins, it forms clusters on the genome through cooperative protein–protein interactions when its concentration increases. The uncharacterized nucleoid protein YgiP, renamed Dan, is virtually absent during aerobic growth, but its levels increase under anaerobic conditions, when it binds 688 sites (21). Overall each nucleoid protein has a large number (∼1000) primary binding sites on the E. coli genome.
Novel concepts of the genome regulation using SELEX data
Multi-factor promoters
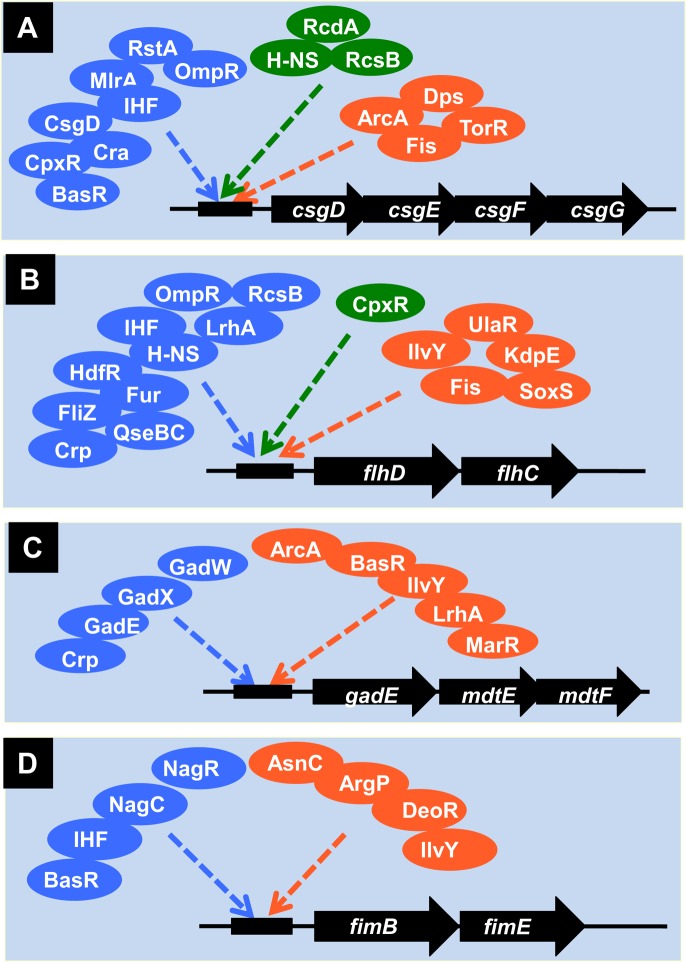
Current promoter databases indicate that ∼50% of E. coli promoters are under the control of a specific regulator, whereas the remaining genes are regulated by two or more TFs. Genomic SELEX search, however, revealed that most E. coli promoters carry binding sites for multiple TFs, each of which monitors a different environmental condition or a different metabolic state. The involvement of multiple TFs may be employed in fine tuning of genomic transcription. One well-characterized example is the regulation of promoter of the csgD gene encoding the master regulator of biofilm formation. The expression of csgD is controlled by more than 10 TFs, each monitoring a specific environmental factor or condition that is effective in induction of biofilm formation (2,32,76–77) (Figure 4A). In parallel with the SELEX screening, a systematic search for multi-TF promoters is being carried out using the PS-TF (promoter-specific TF) screening system (41).
Figure 4.
Multi-TF promoters. After SELEX screening of regulatory targets for a number of TFs, multiple TFs were shown to bind the same promoters. (A) Promoter of the csgDEFG operon, which encodes CsgD, the master regulator of biofilm formation and the biofilm components CsgEFG. (B) Promoter of the flhDC operon, which encodes the master regulator of flagella formation. (C) Promoter of the gadE-mdtEF operon, which encodes the acid-stress response regulator and multi-drug efflux system proteins. (D) Promoter of the fimBE operon, which encodes the recombinase for fimbriae switching. TFs shown in blue indicate those hitherto listed in RegulonDB (29,30) and EcoCyc (31,32) whereas TFs shown in green were identified by this research team and published, but not listed in these databases: H-NS, RcdA and RcdB for the csgG promoter (2,32,76–77); and CpxR for the flhD promoter (2,32,78–79). TFs shown in orange are those identified by the SELEX screening (Ishihama, A., unpublished).
A set of the genes that are involved in construction of the flagella, the key apparatus of bacterial motility for planktonic growth, is controlled by the master regulator FlhDC (78). The flhDC operon promoter is also under the direct control of multiple TFs (Figure 4B), each monitoring a different environmental condition and factor (32,78–79). The gadE operon encodes the master regulator (GadE) of pH homeostasis and multidrug efflux system (MdtEF) (80,81). In concert with the wide-range of regulation, the gadE promoter is under the control of more than nine TFs (32,80–81) (Figure 4C). As in the case of csgD, flhD and gadE operons, the fimBE operon encoding the fim switch recombinase for type-I fimbriae expression is controlled by various environmental signals through at least eight TFs (Figure 4D) (32,52).
Multi-factor operons
During the search for TF-binding sites by SELEX screening, we identified a set of TFs that bind inside specific operons (type-D binding site; see Figure 1) (2,27). For instance, Cra (renamed from FruR), one of the global regulators controlling the whole set of genes encoding enzymes for central carbon metabolism, binds to a total of 170 sites on the E. coli genome, of which 100 sites are located on open reading frames (type-D site, see Figure 1) (57). This unique pattern of Cra binding implies that Cra is involved in regulation, either activation or repression, of flanking genes organized downstream of the same operons.
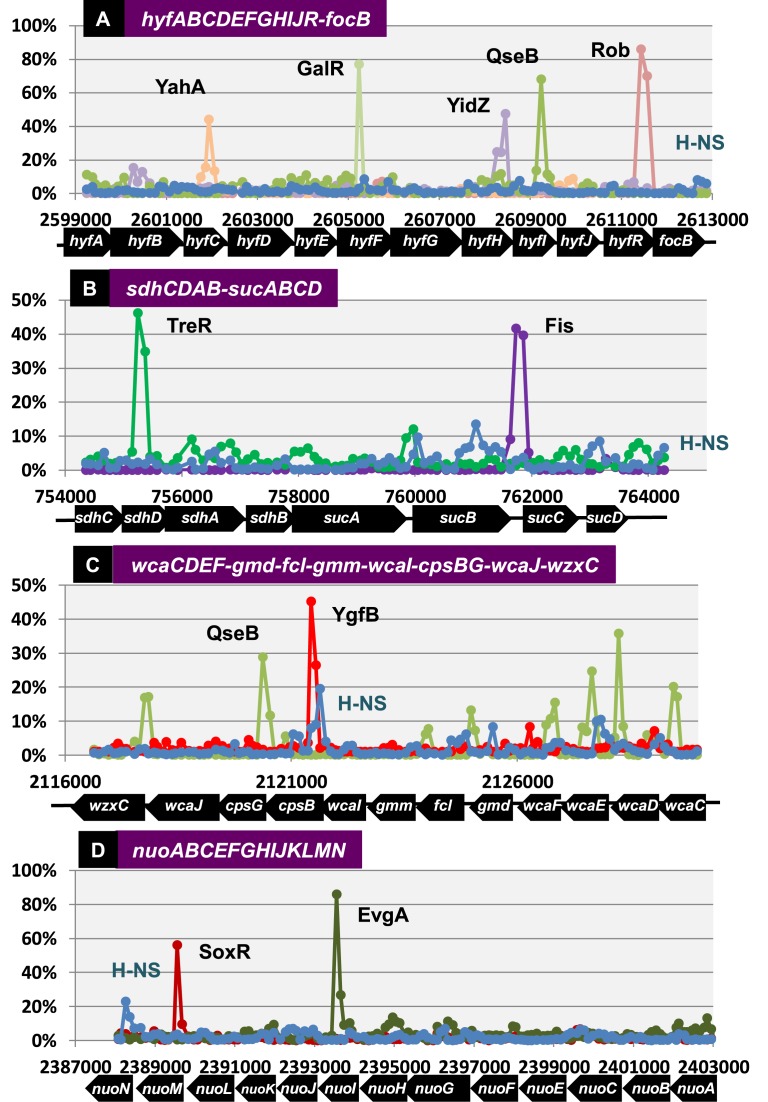
The assembly of SELEX patterns allows identification of operons that contain TF-binding sites inside a single and the same operon. For instance, the hyfA operon is composed of 12 genes: 5′-proximal 10 genes encoding all 10 subunits of the hydrogenase that interacts with formate dehydrogenase (FdhF) to produce the active formate hydrogenlyase complex; the hyfR gene encoding a regulator of hydrogenase 4; and the 3′-proximal focB gene coding for a formate transporter (82). At least five different TFs bind along this 13 581-bp operon, including YahA (TF with c-di-GMP phosphodiesterase activity), GalR (galactose repressor), YidZ (anaerobiosis- and NO-response TF), QseB (quorum-sensing TCS response regulator) and Rob (Mar-Sox-Rob group dual regulator) (Figure 5A). Inside the 3228-bp fusion operon, including the 5′-proximal sdhCDAB genes encoding quinone oxidoreductase and the 3′-proximal sucABCD genes encoding 2-oxoglutaratet decarboxylase (83), two regulators, TreR (trehalose repressor) and Fis (nucleoid-architectural regulator of growth-related genes), bind within the sdhCDAB operon and Fis within the sucABCD operon, respectively (Figure 5B). Inside the 13 390-bp wca operon, which includes 12 genes involved in synthesis of the capsular polysaccharide export apparatus, the binding sites of QseB (QS regulator) and YgfB (uncharacterized TF) were identified (Figure 5C). Likewise, the binding sites of at least two regulators, SoxR (superoxide response regulator) and EvgA (acid response TCS regulator), were identified within the 15 024-bp nuoA operon, which encodes the NADH dehydrogenase I complex (53) (Figure 5D). We hypothesize that TFs located inside operons play as-yet-unidentified roles in transcription initiation from internal promoters and/or transcription continuation, including transcriptional pausing, elongation, termination, anti-termination.
Figure 5.
Biding of TFs inside operons. After SELEX screening of DNA-binding sequences for a total of116 TFs, some operons were found to carry the binding sites for specific TFs. Some representative operons are shown: (A) hyfA operon (13 581 bp); (B) sdhC operon (9872 bp); (C) wcaC operon (13 390 bp); and (D) nuoA operon (15 024 bp). Some TFs with clear binding peaks are shown, but there were several small peaks within all four operons. The binding level of the general silencer H-NS is also shown (blue line) even though the peak height is low. The fluorescent intensity of SELEX fragment binding to each probe relative to that of library DNA was determined, and is shown as the relative value to that of the highest peak.
Novel TF network between silencers and anti-silencers
Nucleoid structuring protein (H-NS) plays a key role in silencing of a set of genes acquired through horizontal gene transfer, including pathogenicity islands (84,85). Gene silencing is now recognized to expand to the set of E. coli genes that are not essential for growth under laboratory culture conditions. After extensive comparison of binding sites between LeuO and H-NS, we realized that among 140 LeuO-binding sites, as many as 133 (95%) overlapped with the H-NS-binding sites (60). Among a total of 118 LeuO-binding promoters (type-A and type-B spacers; see Figure 1), 87 (74%) overlapped with H-NS-binding sites (Table 3). Our findings support the role of LeuO in antagonizing H-NS-mediated silencing. In fact, the order-of-addition experiment indicated that LeuO did not bind to its targets when LeuO was added after addition of silencer H-NS (60). Extending along these lines, we examined the possible overlap of other global regulators with H-NS binding. To our surprise, among a total of 431 high-level H-NS-binding sites that were identified in promoter regions, 97 (22.5%) and 73 (16.9%) promoters harbor the binding sites of Lrp and CRP, respectively (Figure 6). Including LeuO (20.2%), overall more than 50% of H-NS-binding sites were found to overlap with the binding sites of these three global regulators (LeuO, Lrp and CRP). Based on these findings, we propose that one key role of global regulators is anti-silencing, e.g. antagonism of the general silencer H-NS. The search for global regulators involved in anti-silencing of other H-NS binding sites is in progress.
Figure 6.
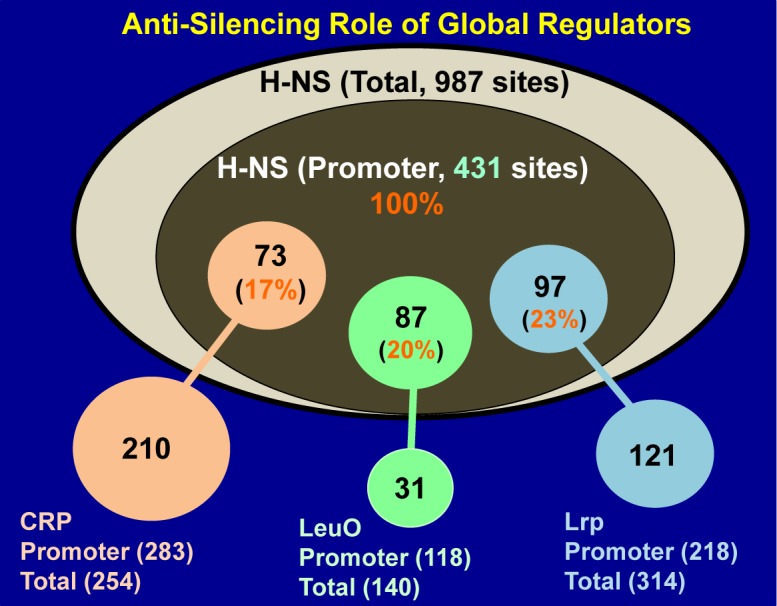
Network involving the silencer H-NS and the anti-silencers Crp, LeuO and Lrp. Among the 987 binding sites of the nucleoid protein H-NS (see Figure 3B), 431 are located near promoter regions within type-A and type-B spacers, where H-NS acts as a general silencer of gene expression. These H-NS binding sites overlap with binding sites of global regulators: Crp (73 sites), LeuO (87 sites) and Lrp (97 sites). High-level overlap of binding sites between the silencer H-NS and the global regulators Crp, LeuO and Lrp indicates that these global regulators act as anti-silencers, and that each is involved in derepression of a large set of target operons (for details see Table 3).
Transcription profile of Escherichia coli: TEC database
The SELEX screening data were assembled into a database entitled ‘Transcription Profile of E.coli’ (TEC), which is open to the public. The first version of this TEC database compiles SELEX-chip analysis data of 123 TFs herein described. TEC is complementary to the established databases, EcoCyc (29,30), RegulonDB (31,32) and CollectTF (86). The most important and unique property of this database is that all data were obtained from a single strain of E. coli K-12 W3110, using the same experimental systems, and by a single research group. Therefore, the data are quite reliable and useful, especially for comparisons among different TFs. The TEC database is the first to provide both locations and intensity of TF binding across the E. coli genome. Therefore, it has enormous potential to provide new insight and facilitate discoveries, especially for TFs whose targets have not yet been identified. TEC provides a number of functions, some of which are discussed below.
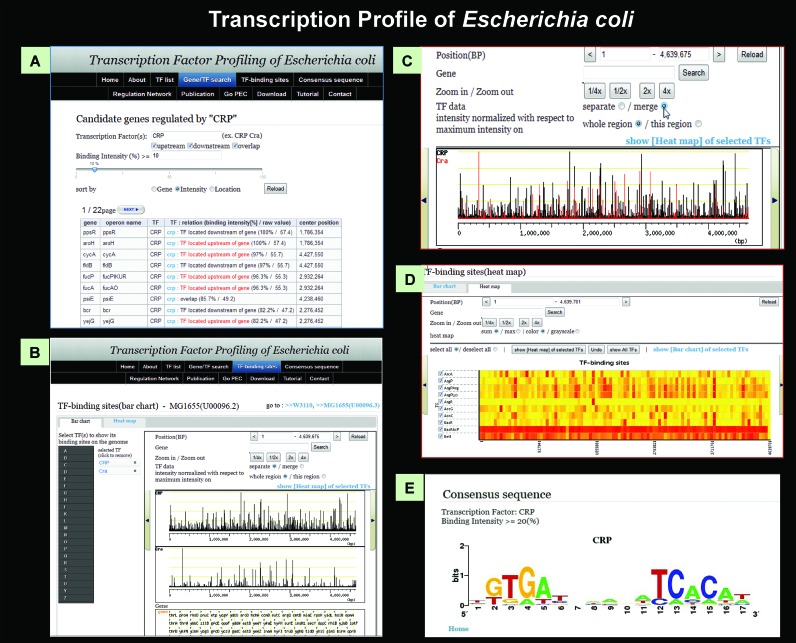
Search for TF-binding sites and regulatory targets
The genome-wide distribution of TF-binding sites can be displayed on a single pattern (Figure 7A), and candidate genes and operons under the control of a specified TF can be retrieved from this pattern. The list of regulated genes and operons can also be narrowed down by changing the cut-off value of TF-binding intensity. By selecting a set of genes under the control of a specific TF, a bar chart can be generated in which the vertical axis corresponds to the binding strength between the TF and probes and the horizontal axis represents genome location (Figure 7B). For identification of binding sites of multiple TFs, the binding patterns of specified TFs can be assembled in a single chart (Figure 7C). From two to ten TFs can also be selected on the same screen, and multiple bar charts will be displayed. For comparison, users can merge these into one chart with different colors by clicking the ‘merge’ button (Figure 7C). In addition to the bar charts, TEC provides a heat map view, allowing visualization of binding intensities among test TFs (Figure 7D).
Figure 7.
TEC database. All SELEX patterns described herein are compiled in the TEC database. Using this database, various types of analysis are possible: (A) search for the list of regulatory targets under direct control of each TF. (B) The location of binding peaks of a test TF along the entire E. coli genome can be visualized at various scales, from the whole genome to restricted areas. (C) The relative location of binding sites of different TFs can be visualized on various scales. (D) Heat map of binding intensity of test TFs along the entire E. coli genome. (E) Search for the consensus sequence of TF binding, using the whole set of TF-binding sequences.
Search for TF consensus sequence
Using the sequence collection of binding regions of TF of interest, the consensus sequence of TF binding can be analyzed with the open-source tool ‘BioProspector’, and shown in WebLogo style (Figure 7E). For this purpose, all 500 bp sequences centered on a probe are prepared in advance. Following selection of a test TF and a cut-off value for TF-binding level, only continuous probes exceeding the set values are automatically extracted. Then the probe with the highest intensity from each continuous probe set is collected and sequences of the final collection are imported into the BioProspector software.
Search for regulatory networks
Using the Regulatory Network page of TEC, the list of regulated genes can be retrieved as compressed zip files(s), including .eda, .sif and .txr files, and the TF network can then be created using ‘Cytoscape’. The levels of TF peaks can be modulated by controlling the cut-off value. This function was successfully used to identify the regulatory network involving the H-NS silencer and global regulators with anti-silencing activity (see Figure 6).
Future of TEC database
Based on advances in strategies for whole-genome studies, several attempts have been made to identify the full set of regulatory targets by comparing transcriptome patterns between wild-type and mutants lacking or over-producing specific TFs (87,88). However, the majority of genes detected in this manner represent indirectly affected genes (1,2). A search for genomic sites associated with TFs in vivo has been carried out using chromatin immunoprecipitation (ChIP)-chip assays (89,90). As noted above, the in vivo binding of TFs within nucleoids interferes with the binding of competing proteins, but it is worthwhile to compare the list of potential TF-binding sites identified in vitro by SELEX-chip and the actual binding sites within nucleoids in vivo, detected by ChIP-chip. For instance, among of a total of 70–80 binding sites in vitro of RutR (the key regulator of pyrimidine synthesis and degradation), only 10∼20 sites were found to be bound in vivo by RutR under normal growth conditions (1).
SELEX screening of all 285 TFs from E. coli is in progress, and we plan to add more SELEX data to the TEC database. In parallel with SELEX screening for target promoters, genes and operons of each sigma and TF, we initiated a systematic search of TFs involved in regulation of one specific promoter using a newly developed PS-TF (promoter-specific screening) system (41). The results of PS-TF screening will be added to TEC in near future. Using both SELEX screening and PS-TF screening data together, TEC will provide a comprehensive view of the genome-wide regulation of transcription in E. coli.
The interaction of TFs with target promoters, genes and operons depends on the intracellular concentration of each TF, which varies depending on environmental physical and chemical conditions, including the levels of nutrients, toxic compounds and cell–cell communication signals. The intracellular concentrations of TFs in E. coli K-12 W3110 under various growth conditions and at various growth phases will also be entered into the TEC database (67,68).
Supplementary Material
Acknowledgments
We thank all the members of the SELEX screening project team for all sigmas and TFs from E. coli K-12 W3110. In particular, Ayako Kori and Kayoko Yamada contributed to the expression and purification of all TF proteins.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of Genetics (to Y.Y.); MEXT Grants-in-Aid for Scientific Research [21241047, 18310133, 25430173 to A.I.]; MEXT Grant-in-Aid for Young Scientists [24710214 to T.S.]; Research Fund from IFO (Institute for Fermentation, Osaka) (to T.S.); MEXT Cooperative Research Program of Network Joint Research Center for Materials and Devices (to T.S., A.I.); MEXT-Supported Program for the Strategic Research Foundation at Private Universities 208-2012 [S0801037 to Hosei University]. Funding for open access charge: National Institute of Genetics.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ishihama A. Prokaryotic genome regulation: multi-factor promoters, multi-target regulators and hierarchic networks. FEMS Microbiol. Rev. 2010;34:628–645. doi: 10.1111/j.1574-6976.2010.00227.x. [DOI] [PubMed] [Google Scholar]
- 2.Ishihama A. Prokaryotic genome regulation: a revolutionary paradigm. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2012;88:485–508. doi: 10.2183/pjab.88.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Helmann J., Chamberlin M. Structure and function of bacterial sigma factors. Annu. Rev. Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- 4.Gross C.A., Lonetto M., Losick R. Bacterial sigma factors. In: McKnight SL, Yamamoto KR, editors. Transcriptional Regulation. NY: Cold Spring Harbour Press; 1992. pp. 129–176. [Google Scholar]
- 5.Ishihama A. Functional modulation of Escherichia coli RNA polymerase. Annu. Rev. Microbiol. 2000;54:499–518. doi: 10.1146/annurev.micro.54.1.499. [DOI] [PubMed] [Google Scholar]
- 6.Gruber T.M., Gross C.A. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 2003;57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- 7.Paget M.S., Helmann J.D. The sigma 70 family of sigma factors. Genome Biol. 2003;4:203. doi: 10.1186/gb-2003-4-1-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gourse R.L., Ross W., Rutherford S.T. General pathway for turning on promoters transcribed by RNA polymerase containing alternative sigma factors. J. Bacteriol. 2006;188:4589–4591. doi: 10.1128/JB.00499-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimada T., Yamazaki Y., Tanaka K., Ishihama A. The whole set of constitutive promoters recognized by RNA polymerase RpoD holoenzyme in Escherichia coli. PLoS One. 2014;9:e90447. doi: 10.1371/journal.pone.0090447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perez-Rueda E., Collado-Vides J. The repertoire of DNA-binding transcription regulators in Escherichia coli K-12. Nucleic Acids Res. 2000;28:1838–1847. doi: 10.1093/nar/28.8.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Babu M.M., Teichmann S.A. Evolution of transcription factors and the gene regulatory network in Escherichia coli. Nucleic Acids Res. 2003;31:1234–1244. doi: 10.1093/nar/gkg210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishihama A. The nucleoid: an overview. In: Böck A, Curtiss R III, Kaper JB, Karp PD, Neidhardt FC, Nystrom T, Slauch JM, Squires CL, Ussery D, editors. EcoSal—Escherichia coli and Salmonella: Cellular and Molecular Biology. Washington: ASM Press; 2009. [Google Scholar]
- 13.Ishihama A. Role of the RNA polymerase alpha subunit in tanscription activation. Mol. Microbiol. 1992;6:3283–3288. doi: 10.1111/j.1365-2958.1992.tb02196.x. [DOI] [PubMed] [Google Scholar]
- 14.Ishihama A. Protein-protein communication within the transcription apparatus. J. Bacteriol. 1993;175:2483–2489. doi: 10.1128/jb.175.9.2483-2489.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Busby S., Ebright R.H. Transcription activation by catabolite activator protein (CAP) J. Mol. Biol. 1999;293:199–213. doi: 10.1006/jmbi.1999.3161. [DOI] [PubMed] [Google Scholar]
- 16.Shimada T., Fujita N., Maeda M., Ishihama A. Systematic search for the Cra-binding promoters using genomic SELEX systems. Genes Cells. 2005;10:907–918. doi: 10.1111/j.1365-2443.2005.00888.x. [DOI] [PubMed] [Google Scholar]
- 17.Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 18.Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 19.Gold L., Brown D., He Y., Shtatland T., Singer B.S., Wu Y. From oligonucleotides shapes to genomic SELEX: novel biological regulatory loops. Proc. Natl. Acad. Sci. U.S.A. 1997;94:59–64. doi: 10.1073/pnas.94.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singer B.S., Shtatland T., Brown D., Gold L. Libraries for genomic SELEX. Nucleic Acids Res. 1997;25:781–786. doi: 10.1093/nar/25.4.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teramoto J., Yoshimura S. H., Takeyasu K., Ishihama A. A novel nucleoid protein of Escherichia coli induced under anaerobiotic growth conditions. Nucleic Acids Res. 2010;38:3605–3618. doi: 10.1093/nar/gkq077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Umezawa Y., Ogasawara H., Shimada T., Kori A., Ishihama A. The uncharacterized YdhM is the regulator of the nemA gene, coding N-ethylmaleimide reductase. J. Bacteriol. 2008;190:5890–5897. doi: 10.1128/JB.00459-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogasawara H., Ohe S., Ishihama A. Role of transcription factor NimR (YeaM) in sensitivity control of Escherichia coli to 2-nitroimidazole. FEMS Microbiol. Lett. 2015;362:1–8. doi: 10.1093/femsle/fnu013. [DOI] [PubMed] [Google Scholar]
- 24.Shimada T., Yamazaki K., Ishihama A. Novel regulator PgrR for switch control of peptidoglycan recycling in Escherichia coli. Genes Cells. 2013;18:123–134. doi: 10.1111/gtc.12026. [DOI] [PubMed] [Google Scholar]
- 25.Shimada T., Katayama Y., Kawakita S., Ogasawara H., Nakano M., Yamamoto K., Ishihama A. A novel regulator RcdA of the csgD gene encoding the master regulator of biofilm formation in Escherichia coli. Microbiologyopen. 2012;1:381–394. doi: 10.1002/mbo3.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimada T., Hirao K., Kori A., Yamamoto K., Ishihama A. RutR is the uracil/thymine-sensing master regulator of a set of genes for synthesis and degradation of pyrimdines. Mol. Microbiol. 2007;66:744–757. doi: 10.1111/j.1365-2958.2007.05954.x. [DOI] [PubMed] [Google Scholar]
- 27.Shimada T., Ishihama A., Busby S.J., Grainger D.C. The Escherichia coli RutR transcription factor binds at targets within genes as well as intergenic regions. Nucleic Acids Res. 2008;36:3950–3955. doi: 10.1093/nar/gkn339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamamoto K., Nakano M., Ishihama A. Regulatory role of transcription factor SutR (YdcN) in sulfur utilization in Escherichia coli. Microbiology. 2015;161:99–111. doi: 10.1099/mic.0.083550-0. [DOI] [PubMed] [Google Scholar]
- 29.Keseler I.M., Collado-Vides J., Gama-Castro S., Ingraham S., Paley S., Paulsen I.T., Peralta-Gil M., Karp P.D. EcoCyc: a compehensive database resource for Escherichia coli. Nucleic Acids Res. 2005;44:D334–D337. doi: 10.1093/nar/gki108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karp P.D., Weaver D., Paley S., Fulcher C., Kubo A., Kothari A., Krummenacker M., Subharaveti P., Weerasinghe D., Cama-Castro S., et al. The EcoCyc database. Ecosal Plus. 2014 doi: 10.1128/ecosalplus.ESP-0009-2013. doi:10.1128/ecosalplus.ESP-0009–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salgado H., Gama-Castro S., Martinez-Flores I., Diaz-Peredo E., Sanchez-Solano F., Peralta-Gil M., Garcita-Alonso D., Jimenez-Jacinto V., Santos- Zavaleta A., Bonavides-Martinez C., et al. RegulonDB (version 4.0): transcriptional regulation, operon organization and growth conditions. Nucleic Acids Res. 2006;32:D303–D306. doi: 10.1093/nar/gkh140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salgado H., Peralta-Gil M., Gama-Castro S., Santos-Zavaleta A., Muniz-Raccado L., Garcia-Sotelo J.S., Weiss V., Solano-Lira H., Martinez-Flores I., Medina-Revira A., et al. RegulonDB v8.0: omics data sets, evolutionary conservation, regulatory phrases, cross-validated gold standards an dmore. Nucleic Acids Res. 2013;41:D203–D213. doi: 10.1093/nar/gks1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jishage M., Ishihama A. Variation in RNA polymerase sigma subunit composition within different stocks of Escherichia coli W3110. J. Bacteriol. 1997;179:959–963. doi: 10.1128/jb.179.3.959-963.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bleibtreu A., Clermont O., Darlu P., Glodt J., Branger C., Picard B., Denamur E. The rpoS gene is predominantly inactivated during laboratory storage and undergoes source-sink evolution in Escherichia coli species. J. Bacteriol. 2014;196:4276–4284. doi: 10.1128/JB.01972-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cook H., Ussery D.W. Sigma factors in a thousand E. coli genomes. Environ. Microbiol. 2013;15:3121–3129. doi: 10.1111/1462-2920.12236. [DOI] [PubMed] [Google Scholar]
- 36.Conway T., Creecy J.P., Maddox S.M., Grissom J.E., Conkle T.L., Shadid T.M., Teramoto J., Miguet P.S., Shimada T., Ishihama A., et al. Unprecedented high-resolution view of bacterial operon architecture revealed by RNA sequencing. MBio. 2014;5 doi: 10.1128/mBio.01442-14. doi:10.1128/mBio.01442-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimada T., Takada H., Yamamoto K., Ishihama A. Expanded roles of the two-component response regulator OmpR in Escherichia coli: Genomic SELEX search for novel regulation targets. Genes Cells. 2015;20:915–931. doi: 10.1111/gtc.12282. [DOI] [PubMed] [Google Scholar]
- 38.Mizuno T. Compilation of all genes encoding two-component phosphotransfer signal transducers in the genome of Escherichia coli. DNA Res. 1997;4:161–168. doi: 10.1093/dnares/4.2.161. [DOI] [PubMed] [Google Scholar]
- 39.Yamamoto K., Hirao K., Oshima T., Aiba H., Utsumi R., Ishihama A. Functional characterization in vitro of all two-component signal transduction systems from Escherichia coli. J. Biol. Chem. 2005;280:1448–1456. doi: 10.1074/jbc.M410104200. [DOI] [PubMed] [Google Scholar]
- 40.Laishram R.S., Gowrishankar J. Envirornmental regulation operating at the promoter clearance step of bacterial transcription. Genes Dev. 2007;21:1258–1272. doi: 10.1101/gad.1520507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimada T., Shimada K., Matsui M., Kitai Y., Igarashi J., Suga H., Ishihama A. Roles of cell division control factor SdiA: recognition of quorum sensing signals and modulation of transcription regulation targets. Genes Cells. 2014;19:405–418. doi: 10.1111/gtc.12139. [DOI] [PubMed] [Google Scholar]
- 42.Nieto J.M., Madrid C., Miquelay E., Parra J.L., Rodriguez S., Juaraz A. Evidence for direct protein-protein interaction between members of the enterobacterial Hha/YmoA and H-NS families of proteins. J. Bacteriol. 2002;184:629–635. doi: 10.1128/JB.184.3.629-635.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reznikoff W.S. The lactose operon controlling elements: a complex paradigm. Mol. Microbiol. 1992;6:2419–2422. doi: 10.1111/j.1365-2958.1992.tb01416.x. [DOI] [PubMed] [Google Scholar]
- 44.Martinez-Antonio A., Collado-Vides J. Identifying global regulators in transcriptional regulatory networks in bacteria. Curr. Opin. Microbiol. 2003;6:482–489. doi: 10.1016/j.mib.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 45.Wei G.H., Liu D.P., Lian C.C. Charting gene regulatory networks: strategies, challenges and perspectives. Biochem. J. 2004;381:1–12. doi: 10.1042/BJ20040311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gottesman S. Bacterial regulation: global regulatory nework. Ann. Rev. Genet. 1984;18:415–441. doi: 10.1146/annurev.ge.18.120184.002215. [DOI] [PubMed] [Google Scholar]
- 47.Gutierrez-Rois R.M., Rosenblueth D.A., Loza J.A., Huerta A.M., Glasner J.D., Blattner F.R., Collado-Vides J. Regulatory network of Escherichia coli: consistency between literature and knowledge and microarray profiles. Genome Res. 2003;13:2435–2443. doi: 10.1101/gr.1387003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Balderas-Martinez Y.I., Savageau M., Saldago H., Perez-Rueda E., Morett E., Collado-Vides J. Transcription factors in Escherichia coli prefer the holo conformation. PLoS One. 2013;8:e65723. doi: 10.1371/journal.pone.0065723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lamark T., Rokenes T.P., McDougall J., Strom A.R. The complex bet promoters of Escherichia coli: regulation by oxygen (ArcA), choline (BetI), and osmotic stress. J. Bacteriol. 1996;178:1655–1662. doi: 10.1128/jb.178.6.1655-1662.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rkenes T.P., Lamark T., Strom A.R. DNA-binding properties of the BetI repressor protein of Escherichia coli: the inducer choline stimulates BetI-DNA complex formation. J. Bacterol. 1996;178:1663–1670. doi: 10.1128/jb.178.6.1663-1670.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kalivoda K.A., Steenbergen S.M., Vimr E.R., Plumbridge J. Regulation of sialic acid catabolism by the DNA binding protein NanR in Escherichia coli. J. Bacteriol. 2003;185:4806–4815. doi: 10.1128/JB.185.16.4806-4815.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sohanpal B.K., El-Labany S., Lahooti M., Plumbridge J.A., Blomfield I.C. Integrated regulatory responses of fimB to N-acetylneuraminic (sialic) acid and GlcNAc in Escherichia coli K-12. Proc. Natl. Acad. Sci. U.S.A. 2004;101:16322–16327. doi: 10.1073/pnas.0405821101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Campos E., Baldoma L., Aguilar J., Badia J. Regulation of expression of the divergent ulaG and ulaABCDEF operons involved in L-ascorbate dissimilation in Escherichia coli. J. Bacteriol. 2004;186:1720–1728. doi: 10.1128/JB.186.6.1720-1728.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kolb A., Busby S., Buc H., Garges S., Adhya S. Transcriptional regulation by cAMP and its receptor protein. Annu. Rev. Biochem. 1993;62:749–795. doi: 10.1146/annurev.bi.62.070193.003533. [DOI] [PubMed] [Google Scholar]
- 55.Harman J.G. Allosteric regulation of the cAMP receptor protein. Biochim. Biophys. Acta. 2001;1547:1–17. doi: 10.1016/s0167-4838(01)00187-x. [DOI] [PubMed] [Google Scholar]
- 56.Krueger S., Gregurick S., Shi Y., Wang S., Wladkowski B.D., Schwarz F.P. Entropic nature of the interaction between promoter and bound CRP mutants and RNA polymerase. Biochemistry. 2003;42:1958–1968. doi: 10.1021/bi026755v. [DOI] [PubMed] [Google Scholar]
- 57.Shimada T., Fujita N., Yamamoto K., Ishihama A. Novel roles of cAMP receptor protein (CRP) in regulation of transport and metabolism of carbon sources. PLoS One. 2011;6:e20081. doi: 10.1371/journal.pone.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shimada T., Yamamoto K., Ishihama A. Novel members of the Cra regulon involved in carbon metabolism in Escherichia coli. J. Bacteriol. 2011;193:649–659. doi: 10.1128/JB.01214-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hertzberg K.M., Gemmill R., Jones J., Calvo J.M. Cloning of an EcoR1-generated fragment of the leucine operon of Salmonella typhimurium. Gene. 1980;8:135–152. doi: 10.1016/0378-1119(80)90033-5. [DOI] [PubMed] [Google Scholar]
- 60.Shimada T., Bridier A., Briandet R., Ishihama A. Novel roles of LeuO in transcription regulation in E. coli: Antagonistic interplay with the universal silencer H-NS. Mol. Microbiol. 2011;82:378–397. doi: 10.1111/j.1365-2958.2011.07818.x. [DOI] [PubMed] [Google Scholar]
- 61.Ernsting B.R., Atkinson M.R., Ninfa A.J., Matthews R.G. Characterization of the regulon controlled by the leucine-responsive regulatory prootein in Escherichia coli. J. Bacteriol. 1992;174:1109–1118. doi: 10.1128/jb.174.4.1109-1118.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Calvo J.M., Matthews R.G. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol. Rev. 1994;58:466–490. doi: 10.1128/mr.58.3.466-490.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shimada T., Saito N., Maeda M., Tanaka K., Ishihama A. Expanded roles of Lrp in transcription regulation of the Eschericia coli genome: Genomic SELEX screening of the regulation targets. Microbial. Genomics. 2015 doi: 10.1099/mgen.0.000001. doi:10.1099/mgen.0.000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ishihama A. Modulation of the nucleoid, the transcription apparatus, and the translation machinery in bacteria for stationary phase survival. Genes Cells. 1999;4:135–143. doi: 10.1046/j.1365-2443.1999.00247.x. [DOI] [PubMed] [Google Scholar]
- 65.Dorman C.J. H-NS: a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2004;2:391–400. doi: 10.1038/nrmicro883. [DOI] [PubMed] [Google Scholar]
- 66.Macvanin M., Adhya S. Architectural organization in E. coli nucleoid. Biochim. Biophys. Acta. 2012;1819:830–835. doi: 10.1016/j.bbagrm.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Azam T.A., Iwata A., Nishimura A., Ueda S., Ishihama A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 1999;181:6361–6370. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ishihama A., Kori A., Koshio E., Yamada K., Maeda H., Shimada T., Makinoshima H., Iwata A., Fujita N. Intracellular concentrations of 65 species of transcription factors with known regulatory functions in Escherichia coli. J. Bacteriol. 2014;196:2718–2727. doi: 10.1128/JB.01579-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Freundlich M., Ramani N., Mathew E., Sirko A., Tsui P. The role of integration host factor in gene expression in Escherichia coli. Mol. Microbiol. 1992;6:2557–2563. doi: 10.1111/j.1365-2958.1992.tb01432.x. [DOI] [PubMed] [Google Scholar]
- 70.Swinger K.K., Rice P.A. Structure based analysis of HU-DNA binding. J. Mol. Biol. 2007;365:1005–1016. doi: 10.1016/j.jmb.2006.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tending C., Bertin P.N. H-NS in Gram-negative bacteria: a family of mutitfaceted proteins. Trends Microbiol. 2003;11:511–518. doi: 10.1016/j.tim.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 72.Atlung T., Ingmer H. H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 1997;24:7–17. doi: 10.1046/j.1365-2958.1997.3151679.x. [DOI] [PubMed] [Google Scholar]
- 73.Hengge-Aronis R. Interplay of global regulators and cell physiology in the general stress response of Escherichia coli. Curr. Opin. Microbiol. 1999;2:148–152. doi: 10.1016/S1369-5274(99)80026-5. [DOI] [PubMed] [Google Scholar]
- 74.Oshima T., Biville F. Functional identification of ygiP as a positive regulator of the ttdA-ttdB-ygjE operon. Microbiology. 2006;152:2129–2135. doi: 10.1099/mic.0.28753-0. [DOI] [PubMed] [Google Scholar]
- 75.Bradley M.D., Beach M.B., de Koning P.J., Pratt T.S., Osuna R. Effects of Fis on Escherichia coli gene expression during different growth stages. Microbiology. 2007;153:2922–2940. doi: 10.1099/mic.0.2007/008565-0. [DOI] [PubMed] [Google Scholar]
- 76.Ogasawara H., Yamada K., Kori A., Yamamoto K., Ishihama A. The E. coli csgD promoter: interplay between eight transcription factors. Microbiology. 2010;156:2470–2483. doi: 10.1099/mic.0.039131-0. [DOI] [PubMed] [Google Scholar]
- 77.Ogasawara H., Yamamoto K., Ishihama A. Regulatory role of MlrA in transcription activation of csgD, the master regulator of biofilm formation in Escherichia coli. FEMS Microbiol. Lett. 2010;312:160–168. doi: 10.1111/j.1574-6968.2010.02112.x. [DOI] [PubMed] [Google Scholar]
- 78.Dudlin O., Gelselmann J., Ogasawara H., Ishihama A., Lacour S. Repression of flagella genes in exponential phase by CsgD and CpxR, two crucial modulators of Escherichia coli biofilm formation. J. Bacteriol. 2013;196:707–715. doi: 10.1128/JB.00938-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ma Z., Zong S., Richard H., Tucker D.L., Conway T., Foster J.W. GadE activates glutamate decarboxylase-dependent acid resistance in Escherichia coli. Mol. Microbiol. 2003;49:130–1320. doi: 10.1046/j.1365-2958.2003.03633.x. [DOI] [PubMed] [Google Scholar]
- 80.Seo S.W., Kim D., O'Brien E.J., Szubin R., Palsson B.O. Decoding genome-wide GadEWX-transcriptional regulatory networks reveals multifaceted cellular responses to acid stress in Escherichia coli. Nat. Commun. 2015;6:7970. doi: 10.1038/ncomms8970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Soutourina O.A., Bertin P.N. Regulation of flagella expression in Gram-negative bacteria. FEMS Microbiol. Rev. 2003;27:505–523. doi: 10.1016/S0168-6445(03)00064-0. [DOI] [PubMed] [Google Scholar]
- 82.Park S.J., Chao G., Gunsalus R.P. Aerobic regulation of the sucABCD genes of Escherichia coli, which encode alpha-ketogkutarate dehydrogenase and succinyl coenzyme A synthetase: roles of ArcA, Fnr, and the upstream sdhCDAB promoter. J. Bacteriol. 1997;179:4138–4142. doi: 10.1128/jb.179.13.4138-4142.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bongaerts J., Zoske S., Weldner U., Unden G. Transcriptional regulation of the proton translocating NADH dehydrogenase genes (nuoA-N) of Escherichia coli by electron acceptors, electron donors and gene regulators. Mol. Microbiol. 1995;16:521–534. doi: 10.1111/j.1365-2958.1995.tb02416.x. [DOI] [PubMed] [Google Scholar]
- 84.Stoebel D.M., Free A., Dorman C.J. Anti-silencing: overcoming H-NS-mediated repression of transcription in Gram-negative enteric bacteria. Microbiology. 2008;154:2533–2545. doi: 10.1099/mic.0.2008/020693-0. [DOI] [PubMed] [Google Scholar]
- 85.Lucchini S., Rowley G., Goldberg M.D., Hurd D., Harrison M., Hinton J.C. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2006;2:e81. doi: 10.1371/journal.ppat.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Erill I. Every site counts: submitting transcription factor-biding site information through the CollectTF portal. J. Bacteriol. 2015;197:2454–2457. doi: 10.1128/JB.00031-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Richmond C.S., Glasner J.D., Mau R., Blattner F.R. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 1999;27:3821–3835. doi: 10.1093/nar/27.19.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tao H., Bausch C., Richmond C., Blattner F.R., Conway T. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J. Bacteriol. 1999;181:6425–6440. doi: 10.1128/jb.181.20.6425-6440.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bulyk M.L. DNA microarray technologies for measuring protein-DNA interactions. Curr. Opin. Biotechnol. 2006;17:422–430. doi: 10.1016/j.copbio.2006.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wade J.T., Struhl K., Busby S.J.W., Grainger D.C. Genomic analysis of protein-DNA interactions in bacteria: insights into transcription and chromosome organization. Mol. Microbiol. 2007;65:21–26. doi: 10.1111/j.1365-2958.2007.05781.x. [DOI] [PubMed] [Google Scholar]
- 91.Ishida Y., Kori A., Ishihama A. Participation of regulator AscG of the β–glucoside utilization operon in regulation of the propionate catabolism operon. J. Bacteriol. 2009;191:6136–6144. doi: 10.1128/JB.00663-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ogasawara H., Shinohara S., Ishihama A. Regulation targets of metal-response BasS-BasR two-component system in Escherichia coli: Modification of cell membrane. Microbiology. 2012;158:1482–1492. doi: 10.1099/mic.0.057745-0. [DOI] [PubMed] [Google Scholar]
- 93.Yamamoto K., Matsumoto F., Minagawa S., Oshima T., Fujita N., Ogasawara N., Ishihama A. Characterization of CitA-CitB signal transduction activating genes involved in anaerobic citrate catabolism in Escherichia coli. Biosci. Biotechnonl. Biochem. 2009;73:346–350. doi: 10.1271/bbb.80586. [DOI] [PubMed] [Google Scholar]
- 94.Shimada T., Kori A., Ishihama A. Involvement of the ribose operon repressor RbsR in regulation of purine nucleotide synthesis in Escherichia coli. FEMS Microbiol. Lett. 2013;344:159–165. doi: 10.1111/1574-6968.12172. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.