Abstract
Guanine-rich DNA strands can fold in vitro into non-canonical DNA structures called G-quadruplexes. These structures may be very stable under physiological conditions. Evidence suggests that G-quadruplex structures may act as ‘knots’ within genomic DNA, and it has been hypothesized that proteins may have evolved to remove these structures. The first indication of how G-quadruplex structures could be unfolded enzymatically came in the late 1990s with reports that some well-known duplex DNA helicases resolved these structures in vitro. Since then, the number of studies reporting G-quadruplex DNA unfolding by helicase enzymes has rapidly increased. The present review aims to present a general overview of the helicase/G-quadruplex field.
INTRODUCTION
G-quadruplex structures
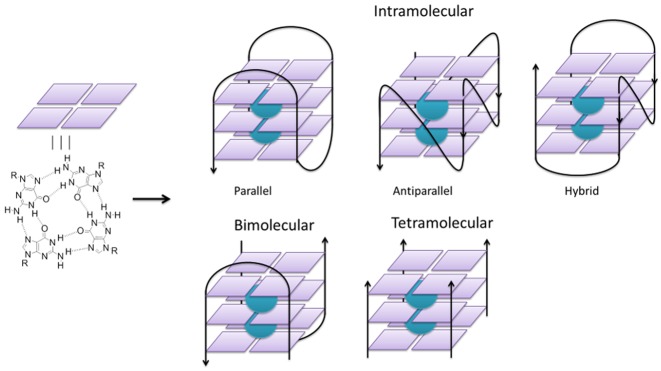
It has been long known that guanine-rich DNA strands can fold in vitro into non-canonical DNA structures called G-quadruplexes (G4). G4 structures are based on the stacking of several G-quartets, which consist each of four guanine bases held together by Hoogsteen-type hydrogen bonding, further stabilized by the presence of cations (generally monovalent) in the central channel of the G4 helix (Figure 1). A G4 motif can be formed by one or several DNA strands with the strands in parallel or antiparallel orientations. The structural diversity, folding properties and stabilities of G-quadruplex DNA have been extensively studied, both as a model for a secondary structure deviant from the canonical Watson–Crick DNA structure and as a pharmacological target for small molecules that have potential to impact gene expression.
Figure 1.
Schematic representations of a G-quadruplexes containing one (intramolecular), two (bimolecular) and four (tetramolecular) DNA strands. The two cations (green spheres) in the central channel stabilize the structure.
For many years after the first characterization of G4, there was general scepticism that G4 formed in vivo. During the past decade, however, mounting evidence indicates that intramolecular G4 motifs are biologically relevant structures. Recently, D. Rhodes and H.J. Lipps have published a comprehensive review on the experimental data supporting role of G4 in replication, transcription or telomere biology (1). Under certain circumstances (e.g. recombination between sister chromatids, telomeric DNA transactions), inter-strand G4 may also arise and be biologically important, in addition to intramolecular G4 (2,3). Two bioinformatics studies suggested that there are ∼370 000 regions in the human genome that can potentially form G4 structures (4,5). This figure was recently re-evaluated using a novel algorithm (6) and use of high-resolution sequencing–based methods, indicating that the number of regions in the human genome with potential to form G4 is significantly greater, with 716 310 distinct potential G4 structures identified (7). Interestingly, these putative G4 are not distributed randomly in the genome. Sequences with G4-forming potential are often found near the replication origins of higher eukaryotes. The majority of the 250 000 human replication origins are close to G4 motifs (8–10), suggesting that the formation of stable G4 structures participates in initiation of replication. Additionally, the promoter regions of numerous genes are characterized by the presence of putative G-quadruplex forming sequences in humans, arguing for a potential role in transcription regulation (11). In humans, potential G-quadruplex forming regions are not exclusively found in nuclear DNA sequences but are also found in mitochondrial DNA sequences (12,13).
From a structural viewpoint, intramolecular G-quadruplex DNA motifs differ depending on the number of G-quartets, the nature and lengths of the interconnecting loops, and the nature of the cation present in the central channel (Figure 1). The polarity of the conformation (parallel, antiparallel or hybrid) enlarges the diversity of DNA G-quadruplex structures that may originate from a single sequence. In fact, many G-rich DNA oligonucleotides can fold into multiple G4 structures, which can be present simultaneously and in thermodynamic equilibrium, and there is a close relation between the number of species coexisting simultaneously and the polarity of the DNA strands involved in the G4 motif (14).
Although G4 DNA and potential involvement of DNA helicases will be emphasized in this review, RNA and hybrid DNA–RNA sequences are also able to fold into highly stable G-quadruplexes (15–17), arguing that these structures could affect all functions of nucleic acids in the cell. The importance of RNA G4 and RNA helicases is beginning to emerge. RNA G-quadruplexes are less structurally diverse than their DNA counterparts, as mostly parallel polarity has been reported for these structures. Another distinguishing characteristic of RNA G4 is their remarkable stability when compared with DNA. The same oligonucleotide sequence usually leads to more stable G4 structures when formed by pure ribonucleotide strands than when formed by deoxyribonucleotide strands. Several recent excellent reviews cover various G4-RNA aspects such as structure, pharmacology or recognition (16,18–19).
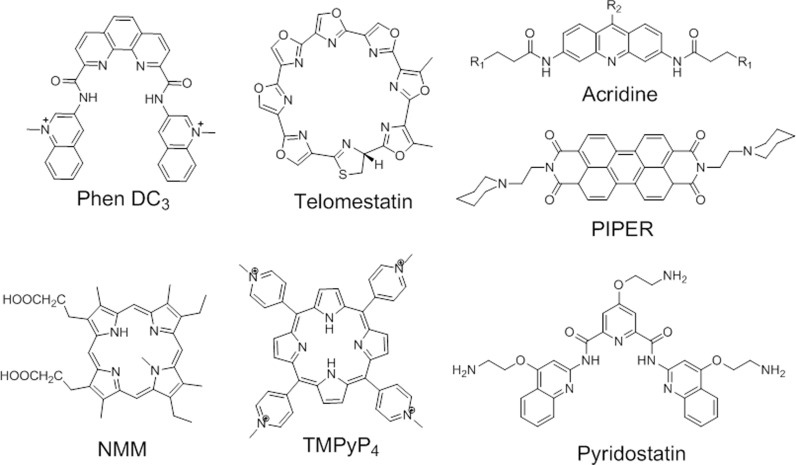
In addition to the extensive investigation of G-quadruplex motifs and their biological roles, chemists have synthesized an ever-growing library of small molecules capable of binding selectively to G4 structures (20). These G4-ligands are generally small aromatic molecules that bind G4 structure with high affinity, showing little or no interaction with duplex DNA (Figure 3 shows a few examples of commonly used ligands). Many of the reports discussed in the present review employed some of these G4-ligands in order to demonstrate G4-dependent biological activities.
Figure 3.
Examples of G4 ligands.
G4 helicase proteins
Helicases are molecular motors that use the energy of nucleoside triphosphate hydrolysis in order to impact nucleic acid structure (21–24). The unwinding of DNA and RNA double helices by helicases is often required before cellular machineries can act on the nucleic acid substrate. Helicases are involved in practically all aspects of cell metabolism: replication, repair, recombination, transcription, chromosome segregation and telomere maintenance (25–28). Although the main task attributed to helicase enzymes is the catalytic formation of single-stranded nucleic acids, some helicases also have the ability to catalyse the opposite reaction in vitro, namely to reanneal complementary DNA strands (29). In addition, increasing evidence suggests that some helicases are involved in the active resolution of other non-canonical DNA structures such as triplexes (30,31), four-stranded Holliday junctions (HJs) (32) and G4s. The mechanism employed by helicase enzymes to separate the two strands from a duplex DNA (or to unwind a G4 motif) involves a complex reaction pathway that has been widely discussed in the literature (see ref (33) for a comprehensible review).
It has become increasingly evident that helicases are an important class of proteins in human disease, such as cancer, and in ageing (34–37). Helicases play critical roles in DNA damage response or DNA repair pathways to help cells cope with or correct endogenous or exogenously induced DNA damage and replication errors. When such helicase-dependent pathways are deficient, the coding information and genomic integrity is compromised resulting in not only defective replication and transcription but also mutagenesis, cellular senescence and death, carcinogenesis, ageing at the tissue and organismal levels, and neurological problems. Helicases preserve genome homeostasis by not only their catalytic action on structured DNA but also via their protein interactions with other factors that govern nucleic acid processing or signal transduction events that help to mediate the appropriate cellular response. Therefore, understanding the nucleic acid substrates acted upon by helicases and how the transactions involving helicases are important for genome homeostasis has become a priority in the field. In the current review, we will highlight the molecular and cellular roles of helicases known to resolve the G-quadruplex structures that are now widely believed to arise during nucleic acid transactions in vivo.
Why helicases are needed for G4 unwinding?
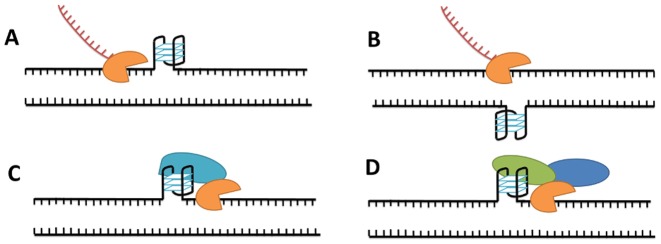
Accumulating evidence suggests that G-quadruplex structures are present in living cells (38). In certain contexts G4 structures may have beneficial roles (e.g. telomere capping, regulation of replication initiation or transcription), but it is commonly agreed that G4 formation can be detrimental to certain processes such as the progression of the replication fork. During DNA replication and transcription, helicases separate individual strands of the DNA double helix, at least transiently. At this point, guanine-rich strands can potentially fold into stable G-quadruplexes. The role of these non-canonical structures could be either stimulatory (acting as DNA binding sites for regulatory factors) or inhibitory (acting as barriers or disrupting a double-stranded binding site) (Figure 2) (39). Whatever their role, G4s must be unfolded during the replication or transcription of the G-rich strand and to correctly refold the DNA double helix. Therefore, highly stable G-quadruplex structures could act as kinetic traps that alter efficiency of replication, transcription or duplex reannealing. In fact, the presence of the complementary C-rich sequence is not always sufficient to unfold a stable G-quadruplex structure within a reasonable timeframe, and some extra element may be required for the correct refolding of the duplex DNA. Even under conditions in which duplexes are the more thermodynamically favoured species, G4 can be long-lived enough to act as kinetic traps.
Figure 2.
Possible roles of DNA G-quadruplex in transcription by RNA polymerase (orange) (39). (A) Blocking transcription: transcription is inhibited due to the formation of a stable G-quadruplex. (B) Facilitating transcription: formation of a stable G4 in the complementary strand enhances transcription. (C) Stimulating transcription: G-quadruplex binds proteins (blue) that stimulate the transcription process. (D) Repressing transcription: G-quadruplex binds protein that repress transcription directly (green) or via other proteins (blue).
The first indication of how stable G4 structures could be unfolded enzymatically came in the late 1990s, when reports showed that some well-known duplex DNA helicases resolved G4s in vitro (40–43). Since then, the number of studies reporting G-quadruplex DNA unfolding by helicase enzymes has rapidly increased. The present review aims to present a general overview of the helicase/G4 field.
Reported G4 helicases
Based on their amino acid sequences, DNA helicases are classified into six superfamilies (SF) (23), and this classification will be used in the present review. The SF1 Pif1 helicase and SF2 helicases RecQ, FANCJ, Bloom syndrome protein (BLM) and Werner syndrome protein (WRN) are the best characterized G4 helicases; thus this review will pay particular attention to these two families. Less studied G4 helicases, such as RTEL1 and DNA2, are also discussed in this review. Finally, we will briefly discuss RNA helicases known to act on G4 RNA. Table 1 shows the families of G4-helicase enzymes discussed in this review.
Table 1. Helicase enzymes reported in this review.
| Superfamily | Subfamily | Helicase name | Substratea | Directionality |
|---|---|---|---|---|
| SF1 | Pif1 | DNA | 5′ → 3′ | |
| DNA2 | DNA | 5′ → 3′ | ||
| SF2 | Fe-S helicases | FANCJ | DNA | 5′ → 3′ |
| DDX11 | DNA | 5′ → 3′ | ||
| RTEL1 | DNA | 5′ → 3′ | ||
| RecQ | BLM | DNA | 3′ → 5′ | |
| WRN | DNA | 3′ → 5′ | ||
| Yeast Sgs1 | DNA | 3′ → 5′ | ||
| Bacterial RecQ | DNA | 3′ → 5′ | ||
| DHX9 | RNA and DNA | 3′ → 5′ | ||
| SF3 | SV40 T-ag | DNA | 3′ → 5′ | |
| SF4 | Twinkle | mtDNA | 5′ → 3′ | |
| SF5 | RHAU | RNA and DNA | 3′ → 5′ |
aDNA and RNA stands for the oligonucleotide substrates employed in the investigations cited in the present review, thus selectivity towards DNA versus RNA is not demonstrated.
SUPERFAMILY 1
Helicase Pif1
Pif1 is a founding member of the SF1 family of DNA helicases. Genetic studies in yeast showed that the two yeast Pif1 family helicases, Pif1 and RRM3, are region-specific helicases, active in the nucleus at telomeres and at the rDNA locus (44), respectively. Pif1 is also present in mitochondria, where it was originally discovered (45).
Pif1 is an inhibitor of telomerase. At the mechanistic level, it is believed that Pif1 removes telomerase by directly displacing telomerase from DNA ends (both from telomeres and from double-stranded breaks), due to its robust ability to unwind RNA–DNA hybrids (46,47). Pif1 was first recognized to be a G4 processing enzyme through a serendipitous observation in yeast that was then confirmed in vitro. Ribeyre et >al. showed that in absence of Pif1, a repeated array of the G-rich human minisatellite CEB1 was genetically unstable in yeast growing exponentially, leading to a 100-fold increase in gross chromosomal rearrangements at the repeated locus (48,49). A mutated version of this array, unable to form G4 structures, was much less sensitive to the absence of the helicase in the nucleus. By melting experiments, it was shown that the CEB1 sequence in solution is able to form G4 structures (49), which were recently shown to be a ‘snapback’ parallel structure. This parallel conformation involves a dimeric G-quadruplex, where each subunit adopts a 3-tetrads parallel scaffold formed by three guanine columns and one snapback fragment (with two conservative G's and one G randomly occupied by several guanines) (50). In bulk enzymatic experiments, Pif1 was shown to unwind a tetra-molecular G4 formed by the CEB1 sequence at a similar apparent rate that it unwinds RNA–DNA hybrids (49). In addition, the potent G4 binder PhenDC3 (51), a bisquolinium compound, inhibits G4 unwinding by Pif1 in vitro and growing wild-type (WT) cells in its presence mimics Pif1 deficiency (52). These studies in an artificial genetic system were confirmed by analysis of Pif1 residency sites on the natural yeast genome. Pif1 acts in regions with the potential to form G-quadruplex in the Saccharomyces cerevisiae genome at the end of S-phase, pointing towards a post-replicative role in processing remaining G4 structures (53). In the absence of Pif1, replication fork progression is slowed down at the vicinity of potential G4 sites, and Pif1 deficiency increases the rate of gross chromosomal rearrangement at several G4 forming sequences (48,54). Pif1 was originally thought to be a general guardian of double-stranded breaks through its inhibition of telomerase binding, but these recent experiments indicate that Pif1 also prevents deleterious events that lead to DNA breakage through its processing of G-rich secondary structures during or after replication (53,55). Linking these two activities (telomerase and G4) will certainly be an interesting development of future molecular genetic studies of Pif1.
The first in vitro studies demonstrating Pif1 activity on G4 DNA were done in bulk assays using tetramolecular G-quadruplexes (56). With these types of substrates, the G4 resolvase activity of helicases is easily monitored by electrophoresis of reaction products on native polyacrylamide gels, given the difference in mobility between tetramolecular substrates and single-stranded DNA products. Analyses of this type of multi-molecular substrate are unlikely to recapitulate Pif1 activities in vivo, as Pif1 is believed to act mainly on intramolecular G4 species. Several studies have therefore been conducted to assay Pif1 helicase activity on intramolecular substrates by single molecule Förster resonance energy transfer (smFRET) assays (57), bulk FRET assays (58) and fluorescence-based assays (59,60) using well defined G-quadruplex forming sequences. These studies are facilitated given the relatively wide range of buffer conditions in which Pif1 is active in vitro; the enzyme works as well in sodium- or potassium-containing buffers (58,60). From these studies, some mechanistic characteristics of Pif1 G4-processing activities are emerging.
Pif1 appears to bind tightly to G4 structures, but the energy required to open G-quadruplexes is higher that the energy necessary for translocase activity (58,59). Several studies have proposed that Pif1 dimerizes upon DNA binding and that this activity potentiates Pif1 translocase activity through double-stranded DNA substrates (57–58,61). It will be interesting in the future to address the mechanistic implications of Pif1 dimerization and to determine whether monomer cooperativity is an important factor of Pif1 regulation in the cell. Slow binding of Pif1 to G4 substrates would kinetically facilitate the formation of Pif1 dimers, which would in turn explain the observation that Pif1 unwinding of double-stranded DNA in vitro is enhanced by the presence of an upstream G4 structure (59).
Another pending question is whether Pif1 has any preference for parallel or antiparallel G4 fold. This question was briefly addressed in two recent studies. Mendoza et al. analysed Pif1 unwinding of parallel (e.g. cMyc promoter) and antiparallel (e.g. human telomere) G4 structures (60). It was found that both types of structures are processed with similar rates, arguing against the recognition of a particular fold. A similar conclusion was reached in the single-molecule study performed by Zhou et >al. (57). Since DNA helicases are not expected to work in isolation within the cell, but as multi-protein complexes, there remains the possibility that various G4 folds are recognized by different G4 binders in the cell as a first step towards their processing by a helicase. Future work on proteins that preferentially bind to particular G4 conformations (e.g. Sub1 (62) and STM1 (63)) will likely be necessary to address this question.
DNA2 helicase-nuclease
The Dna2 gene was originally identified in a screening study of genes involved in DNA replication (64). The Dna2 protein exhibits several biochemical activities: it possesses a single strand DNA-specific endonuclease activity (both on 5′ and 3′ single-stranded overhangs) (65) and a helicase enzymatic activity with a 5′ to 3′ translocation direction (66). Dna2 plays an essential role in processing flaps during maturation of Okazaki fragments (67), and increasing evidence suggests that Dna2 has also important roles in DNA replication stress response and DNA repair (68). In vitro assays proved that Dna2 can recognize and resolve telomeric G-quadruplex structures through cleavage at the G4 site (69). The study was complemented by in vivo studies employing mouse cells. Reduction of levels of Dna2 results in telomere replication defects and high levels of fragile telomeres and sister telomere associations. The frequency of these telomere defects was found to increase moderately when cells were treated with G4-ligands such as TMPyP4 and TMS (Figure 3) (69). Thus, the role of Dna2 in the processing of DNA structures such as G4 in 5′ flaps, the purported activity of Dna2 related to G4 during general genome replication, may also come into play in the metabolism of G4 structures that arise at chromosome ends.
SUPERFAMILY 2 DNA HELICASES
Fe-S helicases
Fe-S helicases contain an iron–sulphur cluster that has an essential capacity to accept and donate electrons (70). This cluster can strongly bind to organic substrates or proteins containing electron-rich oxygen and nitrogen atoms. The iron–sulphur moiety also plays an important role during protein folding, stabilizing an important domain of the enzyme (71). Eukaryotic Fe-S family members include helicases XPD, FANCJ, RTEL1 and DDX11.
Fanconi anemia group J helicase (FANCJ)
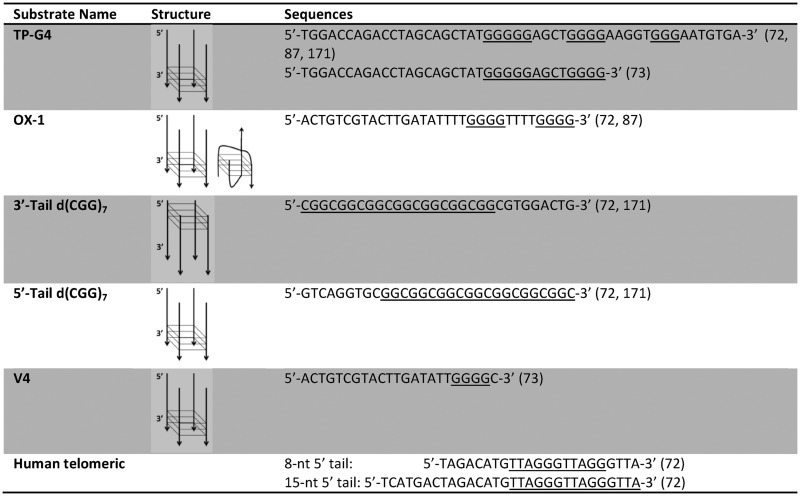
Initial reports on the FANCJ helicase subfamily were focused on the unwinding reactions of tetra-stranded G4 structures formed by the murine immunoglobulin switch sequence (TP-G4), the Oxytrichia nova telomere (OX1), and an oligonucleotide V4 containing a four-guanine run (Table 2) (72,73). Radiolabelled oligonucleotides were used to identify and quantify the helicase reaction carried out in both K+ and Na+ buffers. The FANCJ-catalysed unwinding of G4 structures is adenosine triphosphate (ATP)-dependent, and the reaction yield is proportional to protein concentration and incubation time (72,73). When the helicase assay was performed in sodium-containing buffer FANCJ efficiently unwound OX1 and V4 substrates, but TP-G4 structure was a clear obstacle for the protein (73). This difference may result from differences in stabilities of the various G-quadruplexes. OX1 folds into a bimolecular G4 in potassium, whereas in Na+ the predominant form is tetra-molecular, thus—and counter-intuitively—the potassium form of G4-DNA substrate presents a less stable obstacle for the protein and therefore showed a greater FANCJ helicase activity.
Table 2. Commonly used G4 substrates for helicase studies in vitro.
|
The directionality of the FANCJ enzyme is 5′ to 3′ for both G4 and duplex DNA unwinding. This polarity was analysed by comparing the activity of this helicase against a four-stranded parallel G4 system formed by d(CGG)7 flanked by an 8-nt tail located either at 5′end (5′-Tail d(CGG)7 in Table 2) or 3′end (3′-Tail d(CGG)7 in Table 2) (72): the activity of FANCJ is negligible on the 3′-Tail d(CGG)7 substrate, but the G4 formed by 5′-Tail d(CGG)7 is efficiently unwound. Additionally, the length of the 5′ tail affects FANCJ helicase activity (72). The substrate with a 15-nt 5′ tail (Table 2) is more efficiently unfolded by FANCJ than the substrate containing a shorter 8-nt 5′ tail. The helicase-promoted G4 opening activity was also compared to Watson–Crick duplex unwinding activity: FANCJ is more active on G4s, suggesting that FANCJ's preference to unwind G4 may be important for its role during replication of G-rich DNA sequences.
The effect of selective G4-ligands on the helicase-promoted reaction was also studied in an early report. The activity of FANCJ is significantly reduced by the presence of the G4-ligand telomestatin (TMS, Figure 3) (72). Other G4 binders, including proteins, have the same effect. The presence of the MutSα (MSH2/MSH6) heterodimer, which binds strongly to G-quadruplex structures (74), reduces the catalytic activity of FANCJ. In contrast, proteins such as Replication Protein A (RPA), which binds to single-stranded DNA, stimulate the G4 unwinding reaction. This protein is not considered a helicase enzyme and in vitro studies showed that RPA unfolds G4 structures in the absence of ATP (75–77). During the replication process, RPA has a key role in ensuring that the two strands do not refold into duplex. In vitro assays showed that RPA prevents the formation of G-quadruplex in the lagging strand during telomeric replication (76,78). RPA appears therefore as a major partner of helicases in promoting the processing of G4 structures arising in single strand intermediates during DNA replication.
In eukaryotic cells, mounting evidence suggests that FANCJ unwinding of G4 DNA is necessary for maintenance of genomic stability. In nematodes with mutations in the FANCJ orthologue dog-1, deletions in the genome are proposed to be a consequence of guanine-rich secondary structures that perturb normal lagging strand synthesis. It was hypothesized that the transient single-strands that arise on the lagging strand templates form G4 structures that impede the normal replication machinery. Based on these data, the Lansdorp lab conjectured that DOG-1 has a specialized duty to resolve G4 DNA (79). Recent experimental data suggest that an unresolved replication fork barrier blocking G4 DNA structure in the dog-1 mutant persists through mitotic divisions, causing gaps that are converted to double strand breaks and promote genomic instability at each generation, with consequences during organogenesis (80).
Extrapolation of the Caenorhabditis elegans dog-1 mutant phenotype to human cells deficient in FANCJ was made independently by two laboratories. First, FANCJ-deficient cells (either cells of patients with Fanconi anemia (FA), which results from a mutation in the gene encoding FANCJ, or human cells depleted of FANCJ by RNA interference) are hypersensitive to the G4 ligand TMS, which induces apoptosis and DNA damage (72). An important observation made in this work was that FA patient cells mutated in both copies of the genes encoding FANCA (a member of the FA core complex) or FANCD2 (a key player in the activation step of the FA interstrand crosslink repair pathway) did not display hypersensitivity to TMS, consistent with a model in which FANCJ helicase has a unique role outside the classic FA pathway to resolve G-quadruplexes that arise in the human genome. In another study that same year, FANCJ-deficient cells were shown to accumulate deletions at predicted G4 motifs (73). Further evidence for the specificity of FANCJ among the Fe-S cluster helicases for a role in G4 DNA metabolism was provided by the observations that only FANCJ efficiently resolves intramolecular G4 DNA substrates and that deficiencies in the related XPD and DDX11 Fe-S helicases did not render human cells sensitive to the anti-proliferative and DNA damage-inducing effects of TMS (81). Consistent with the cell-based assays, purified human DDX11 or archaeal XPD helicases very poorly resolved intramolecular G4 DNA substrates under conditions that the enzymes efficiently unwound more conventional forked duplex DNA substrates (81).
Turning to a model vertebrate cell-based system amenable to genetic analysis, chicken DT40 cells provided fresh mechanistic insight into the functional roles of the conserved FANCJ orthologue BRIP1. It was originally reported that expression of human FANCJ in DT40 cells deficient in BRIP1 rescue the mutant cells from sensitivity to DNA damaging agents (82), including the G4 ligand TMS (83). Later, transcription profiling of the BRIP1-deficient DT40 cells demonstrated a genetic interaction between brip1 and the genes encoding the trans-lesion polymerase REV1 and the RecQ DNA helicases WRN and BLM. All are necessary to maintain epigenetic stability at G4 DNA motifs (84). Another study showed that BRIP1 facilitates replication through TMS-stabilized G4 DNA sequences, thereby suppressing heterochromatin spreading (85). Consistent with these observations supporting an important role of FANCJ in G4 DNA metabolism, FANCJ-deficient DT40 cells exposed to TMS accumulate G4 structures as demonstrated by elevated nuclear immunofluorescent staining using a monoclonal antibody specific for G4 DNA (86).
DDX11 helicase
DDX11 is also called DEAH box protein 11, ChlR1, Ctf1 and Mcm12. DDX11 resolves G4 and other non-canonical DNA structures such as triplex DNA (31). Although DDX11 belongs to the same protein family as FANCJ, it has a clearly different behaviour against G-quadruplex DNA substrates. This difference in G4-resolving activity is evidenced by activities toward a tetra-stranded parallel G-quadruplex DNA substrate formed by TP-G4 sequence (Table 2). Less than 5% of the TP-G4 substrate was unwound by DDX11 (87) compared to nearly complete unwinding by FANCJ (72). This difference is remarkable considering that both helicases belong to the same family and contain the conserved Fe-S domain. DDX11 does efficiently unwind two-stranded anti-parallel G4 DNA substrates (referred to as G2’). The preferential ability of DDX11 to resolve the two-stranded G4 substrate likely reflects the greater affinity of the helicase for the G2’ DNA molecule compared to a four-stranded G4 substrate. Although the biological significance of DDX11 resolution of G2’ DNA is unclear, it is conceivable that the G4-resolving capacity may be important for resolution of replicated DNA molecules or during recombination between sister chromatids (88). In support of this idea, the replication fork complex factor Timeless/Tim operates in the same fork recovery pathway as DDX11, physically binds to DDX11 and stimulates its unwinding activity on several DNA substrates, including a bimolecular anti-parallel G4 DNA structure (89).
Regulator of telomere elongation helicase 1 (RTEL1)
Evidence for the involvement of RTEL1 (also known as NHL, DKCA4, DKCB5 and C20orf41) in G4 processing comes mainly from cell-based experiments. In vitro experiments showed that purified RTEL1 is able to unwind an intermolecular G4 substrate made from the human telomeric sequence, and that the ATPase activity of the helicase is required for unwinding to occur (90).
Boulton's lab showed that in the absence of RTEL1, T-loops are not correctly resolved during DNA replication. This results in a complete loss of the telomere circle (90). Further analysis was then completed by the group of De Lange, who employed mouse cells to study the specific role of RTEL1 in replication (91). They found that RTEL1 binds to the replisome through interaction with a DNA clamp protein known as proliferating cell nuclear antigen (PCNA). If this binding is disrupted, the telomere replication is adversely affected, resulting in replication fork instability, reduced replication fork extension rates and increased origin usage. This study therefore proved a link between RTEL1 and telomere replication. Indeed, individuals with inherited mutations in the gene encoding helicase RTEL1 exhibit Hoyeraal-Hreidarsson syndrome, a severe form of Dyskeratosis congenita characterized by telomere dysfunction (92,93).
RTEL1 has been suggested originally to be involved in G-rich DNA metabolism based on its sequence similarity with dog-1 and the presence of the RTEL1 gene in a genomic location linked to telomere length in mice. It was shown by the Lansdorp group that Rtel1 deficiency is embryonically lethal in mice, and that telomere elongation indeed compromised in Rtel−/− ES cells (94). The mechanistic link between G-rich regions and RTEL1 activity was further confirmed in a study by the de Lange lab, where it was shown that shutting down RTEL1 expression increases telomere fragility in a similar way as the BLM helicase (95). In agreement with a link between RTEL1 and telomere replication, individuals with inherited mutations in RTEL1 exhibit Hoyeraal-Hreidarsson syndrome, a severe form of Dyskeratosis congenital characterized by telomere dysfunction (92,93). It was proposed that a RTEL1 telomere-associated role was the removal of a barrier to the replication fork machinery, most likely a G4 structure (95). This hypothesis is strengthened by the observation that treating RTEL1 deficient cells with the G4 ligand TMPyP4 increases the telomere fragility (91).
RecQ helicases
RecQ helicases are a large family of proteins found in bacteria and eukaryotes. RecQ enzymes have a conserved domain of about 450 amino acids that includes seven motifs conserved in many different classes of helicases. These include the DExH box domain, a characteristic feature of this family which comprises the DEAD, DEAH and DExH subgroups. RecQ helicases are involved in several important functions in the cell such as preserving DNA integrity during replication (96). In human cells there are five RecQ family members; three (BLM, WRN and RECQ4), are of special interest because of their involvement in inherited disorders associated with a predisposition to cancer and premature ageing (97).
The G4 opening catalysed by the RecQ group of enzymes is well characterized. As in RecQ-promoted duplex unwinding, ATP is required and opening proceeds in the 3′→5′ direction for denaturing G4 structures. The quadruplex unfolding process first involves the binding between the DNA structure and the RecQ C-terminal protein domain (RQC). The RQC domain is present in RecQ helicases such as BLM and WRN and it is responsible for the recognition of various DNA structures (98,99). Initial reports on this helicase family were performed using simple tetra-stranded G-quadruplex motifs (42,72,73); however, reactions with more complex substrates such as biologically relevant intramolecular G-quadruplexes have also been studied (100,101).
BLM helicase
The gene encoding the BLM helicase is mutated in patients with Bloom syndrome (102). Patients with this syndrome suffer from genomic instability and have a predisposition to cancer development. The BLM enzyme was one of the first human helicases ever reported to resolve G4 DNA. In 1998, the laboratories of I. D. Hickson and N. Maizels demonstrated that this protein unwinds intermolecular G4 structures in a 3′ to 5′ direction as previously observed for duplex DNA (42). As found for FANCJ-mediated helicase reactions, BLM preferentially binds to G4 structures over duplex DNA. The preference of BLM for G4 systems was also proved by comparing the helicase reaction with more ‘branched’ nucleic acids such as HJs (103). When the enzyme is incubated in the presence of a HJ system and G4, the preferred DNA substrate is always the G-quadruplex. The remarkable difference in activity against G4 and HJ was correlated with a 15-fold higher binding affinity for G4 over HJ (103).
BLM activity is inhibited by G4 ligands. G4 organic ligands such as acridines and porphyrins (Figure 3) decrease the activity of BLM significantly (104). This inhibitory effect was first proposed to be the result of G4 stabilization; however, these ligands also inhibit duplex unwinding, suggesting that it results from an uncharacterized effect of the ligand on BLM activity.
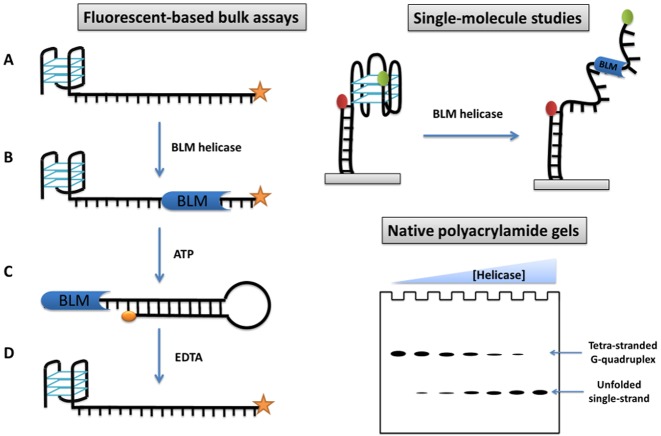
These previous studies were using tetramolecular G4 DNA as substrates. As for other helicases, the studies have moved more recently towards more physiologically relevant intramolecular substrates and single molecule studies, in order to get insights into the structural and kinetic mechanism of G4 unfolding. The unfolding of intramolecular G4 structures by BLM has been studied in real-time employing fluorophore-labelled DNA substrates (Figure 4). Tan et al. compared the unfolding of a G4-forming oligonucleotide and a duplex DNA in the presence of a 10-fold excess of protein (100). This analysis showed that BLM unfolds G-quadruplexes less efficiently than it does duplexes, proving that G-quadruplexes are indeed blocks for enzyme progression. These results contrast with previous reports based on bulk studies and intermolecular quadruplex, which showed that intermolecular G4 structures are better substrates for BLM and the yeast helicase Sgs1, with at least 10× higher unwinding efficiency than dsDNA. This apparent discrepancy is likely due to the differences in the conformations of intra- and intermolecular G-quadruplexes. This highlighted that intermolecular G4 DNA and intramolecular G-quadruplexes are very different substrates, which may be unwound with different efficiencies by various G4 helicases.
Figure 4.
Schematic representation of most common helicase assays reported, based on: fluorophore-labelled DNA substrate (100); single-molecular FRET (57,105–106,110) and electrophoresis gel (see ref (40,42,72–73,103–104,118) for some examples).
BLM requires a 3′ single-stranded overhang in order to load onto a DNA substrate (42). The minimum length of this tail was determined employing fluorophore-labelled DNA substrates (Figure 4). The telomeric d(TTAGGG) repetitive sequence was used as a substrate; a 6-nt tail allowed the helicase to bind to the G4 substrate (101). As expected, the absence of a 3′ overhang led to the total suppression of the helicase activity.
The unwinding of telomeric G4 by BLM has also been studied using single-molecule FRET (smFRET), providing important mechanistic insights into how the BLM helicase is able to process intramolecular G-quadruplexes. For this purpose a truncated protein containing the RecQ-core of BLM was constructed and incubated with the human telomeric sequence (105). In these experiments 3′ overhang lengths from 2- to 15-nt were evaluated: in agreement with bulk studies, a 6-nt minimum overhang was found. The activity of the helicase enzyme was also evaluated as a function of ionic strength of the solution. Increasing the presence of monovalent cations increased the stability of the G-quadruplex and thus reduced the efficiency of the helicase-mediated G4 unfolding reaction, confirming that a G-quadruplex provides a roadblock for the progression of the helicase. Interestingly, Budhatokhi et al. (105) also showed that if the intramolecular G4 is followed by a single-strand region as small as 5–10 nt, BLM is able to process the G-quadruplex without requiring ATP hydrolysis. This is unique to BLM since in the same assay, the Pif1 helicase is not able to process the quadruplex without ATP. The mechanistic reason behind this property was elucidated with a smFRET study from the Rothenberg lab (106), in which they showed how the two RecQ-family protein domains, the RQC and the Helicase-RNase D C-terminal (HRDC) domains (107), are important for the G4 unwinding activity, and also provided a rationale to explain ATP-independent G4 unwinding. Analysis of smFRET traces of intramolecular G4 substrates revealed that in presence of a single stranded DNA region downstream of the G4 block, the interaction between the HRDC domain and this single-stranded region favours G4 unfolding, due to the cooperative binding of the HRDC domain with the G4-binding RQC domain. In absence of a single-stranded region downstream of the G4 block, unfolding is very inefficient, even in the presence of ATP. If a single-stranded region is provided by a complementary strand to the G4 sequence, as would be encountered in a DNA bubble for example, the G4 processing activity of BLM is restored, owing to the interaction of the HRDC domain with the complementary strand. As a supplementary argument, the same study provided evidence that WRN, another RecQ helicase containing HRDC and RQC domains (see below), exhibits both ATP dependent and independent G4 processing, whereas RecQL5, which is devoid of HRDC and RQC domains, cannot process G4 substrates (either inter or intramolecular) (108,109). These results suggest that BLM (and other helicases possessing RQC and HRDC domains) process G4 substrates through cooperative interaction between the helicase, a G4 structure and proximal ssDNA downstream or opposite the G4 structure.
Recent work has suggested that BLM helicase has the ability to unfold G4 DNA by distinct mechanisms dependent on the structure of G4 encountered by the enzyme (110). Using a FRET-based assay, researchers of the Xi laboratory showed that BLM may either actively translocate along single-stranded DNA to resolve the G4 or may anchor at the junction of single-stranded and structured DNA and acts in a repetitive manner to unwind the adjacent G4 in a 3′ to 5′ direction, reeling in the single stranded DNA 5′ overhang by its 3′-5′ translocation activity. This is reminiscent (albeit in the other orientation), of the activity described for Pif1 by the Ha group (57) in a similar smFRET approach.
Werner syndrome helicase WRN
Like BLM, WRN belongs to the RecQ family. Patients with dysfunctional WRN suffer premature ageing (111). The observation of a physical interaction between BLM and WRN suggests a possible cooperation between these two enzymes in vivo (112). The first report showing the G4-unwinding activity of this enzyme was performed on the G-quadruplexes formed by repetitions of the d(CGG) trinucleotide (113). The formation of either a hairpin or G4 by this trinucleotide repeat is thought to be the cause for its genomic instability, making this sequence clearly biologically relevant. The ability of WRN to resolve G4 structures was demonstrated on intermolecular substrates formed by d(CGG)7. Formation of G4 from d(CGG)7 requires very long incubation times and further gel purification to remove single-stranded forms present in the solution. Expectedly, the enzyme requires the presence of Mg2+, and the unfolding reaction is ATP-dependent. In addition, as for the majority of helicase enzymes, WRN requires a single-stranded tail, and blunt-ended structures are found to be inert to the enzyme (113). The presence of a short 7-nt tail is sufficient to allow WRN to denature G4 more efficiently than it does duplexes (113). Subsequent work verified the requirement of a 3′ ssDNA tail for WRN to unwind a variety of G4 DNA substrates (114). WRN helicase activity has been also studied in presence of G4 ligands such as acridines (104). As found for BLM, acridine derivatives also have an inhibitory effect on duplex DNA substrates, arguing against a G4-based specific inhibition. At the mechanistic level, WRN has not be studied as thoroughly as BLM, probably owing to the difficulty to purify the enzyme.
In vivo studies have been also carried out to unveil the specific role of WRN in resolving G4 structures. The level of mRNAs in human fibroblasts deficient in several RecQ family helicases was compared to WT cells. This study found a specific correlation between the location of G4 sequences and the upregulated loci in wrn cells (115).
The group of Opresko used a forward mutagenesis assay based on the supF shuttle vector assay to study the fidelity of replication of episome-borne telomeric repeats in WT versus WRN knockdown cells. This study showed that the sequence (TTAGGG)6 is properly replicated in this vector in WT cells, but mutagenic in WRN-deficient cells. In the case of WRN knockdown, the frequency of large deletions and rearrangements within the shuttle vector increase significantly, pointing to a specific role of WRN in processing structures formed by these repeats (116). The inhibitory effect exhibited by G4 ligands in living cells has been also investigated by exposing human cells to the organic molecule 1-(propoxymethyl)-maleimide. The incubation of living cells with this ligand significantly decreased the growth and proliferation of the human cells inducing apoptosis in a WRN-dependent manner. In addition, the exposure resulted in elevated γ-H2AX and PCNA foci (117).
Yeast Sgs1
The S.cerevisiae Sgs1 helicase is a RecQ homologue. It is easily purified from yeast and has been used as a model for helicase studies in vitro. In the first report on Sgs1 activity on G4, Sun et al. showed how this helicase unfolds G-quadruplexes in an ATP-dependent manner and requires the presence of cofactors such as Mg2+ (43). The processivity of this protein is 3′ to 5′, and it requires the presence of a 3′ tail in the DNA substrate. The sequence studied in this first report contained a G4 motif composed of the S. cerevisiae telomere sequence with a 7-nt long 3′ overhang. The reaction time was found to be similar to that of G4 DNA unwinding catalysed by recombinant BLM helicase (42). Sgs1 catalyses more efficiently G4 unfolding than the unwinding of duplexes (43) or other DNA structures such as HJs (103).
The activity of this enzyme is also modulated by G4 ligands. The well-known G-quadruplex binder N-methyl mesoporphyrin IX (NMM, Figure 3) has a significant inhibitory effect on this enzyme. This reduction in activity is probably due to the increase in G4 stability (103). Other ligands reported to be G4-chaperones such as N,N′-Bis[2-(1-piperidino)ethyl]-3,4,9,10-perylenetetra-carboxylic diimide (PIPER, Figure 3) also block Sgs1 activity on tetra- and bi-molecular systems (118). Again, this inhibitory effect probably results from the stabilization of the G4 motif, which then becomes an obstacle for the enzyme (118). A direct or non-specific effect of the helicase was excluded as the helicase activity on duplex DNA substrates was not affected by the presence of these G4 ligands.
In vivo, however, evidence supporting a role of Sgs1 in processing G4 DNA has been scarce. For example, deletion of SGS1 had no effect on the genomic stability of G4-forming minisatellites in the yeast genetic assay developed by Nicolas et al. (49) (see discussion on Helicase Pif1 above). To our knowledge, G4 related effects of Sgs1 in vivo have been evidenced by the Johnson lab only. They showed that variation in the steady state mRNA levels in a sgs1Δ strains compared to a WT strain affect in a statistically significant manner genes possessing G4 forming motifs in their open reading frames (119). In another study, they showed that growth defects originating from compromised telomere capping are suppressed by deletion of Sgs1 or by the G4-stabilizing agent NMM (120). However, the mechanism involved remains to be characterized, and we are still lacking strong evidences for Sgs1 importance in G4-DNA processing, as compared to evidences accumulated for Pif1 in the same organism.
Bacterial RecQ
The Escherichia coli RecQ enzyme also resolves G4 structures in a ATP-dependent manner and in presence of divalent cations as cofactors (121). This helicase exhibits a similar behaviour against duplex and G-quadruplex DNA. As found for other RecQ helicases, NMM (Figure 3) is a highly specific inhibitor of RecQ activity on G-quadruplex substrates; NMM causes no reduction in helicase-mediated duplex unwinding. This supports the conclusion that the inhibition of G4 unwinding is the result of specific G4 stabilization (121).
SUPERFAMILY 3: SIMIAN VACUOLATING VIRUS 40 T-ANTIGEN (SV40 T-ag) HELICASE
The simian virus 40 (SV40) genome is a simple model system commonly employed for investigating eukaryotic replication. Indeed, the SV40 T-ag was one of the first helicase enzymes to show an unwinding activity against G4 (40). The unwinding activity of this tumour antigen enzyme was demonstrated using 32P-labelled tetra- and bi-molecular DNA systems. A complete study of the effect of G4 ligands on this helicase was performed by Kerwin et al. (122). They screened a small library of organic ligands for those that reduced helicase activity. The porphyrin NMM inhibited T-ag enzyme less effectively than it did other common helicases such as Sgs1, BLM and WRN. The IC50 value of NMM for SV40 T-ag helicase was about 95 μM, whereas the IC50 values for Sgs1, BLM and WRN helicases were in the low μM range (103,121). In contrast, a well-established—but poorly selective for G4 relative to duplex DNA—cationic porphyrin TMPyP4 did not reduce the G-quadruplex unwinding activity of T-ag, even at concentrations as high as 100 μM. This is a remarkable selectivity as TMPyP4 significantly affected the activity of several RecQ-like helicases such as BLM (103,104), WRN (104) and Sgs1 (103).
HELICASES THAT UNWIND BOTH RNA AND DNA G4 STRUCTURES
Superfamily 2 helicase DEAH box protein 9 (DHX9)
DHX9, which is also called RNA helicase A (RHA), leukophysin (LKP) and nuclear DNA helicase II (NDHII), unwinds DNA–DNA, RNA–RNA and DNA–RNA duplexes with a 3′ to 5′ polarity (123). It acts preferentially on RNA substrates and is capable of unwinding R-loops and G4 structures. DHX9 is a substrate-specific helicase with properties comparable to the WRN helicase. As WRN, DHX9 helicase resolves G-quadruplex-containing substrate (124). However, unlike WRN, DHX9 failed to unwind HJs. To confirm this remarkable difference, a 17-nt single-stranded extension was added to the HJ and DHX9 was still unable to unwind this substrate (124).
Superfamily 5 helicase RNA helicase associated with AU-rich element (RHAU)
RNA helicase associated with AU-rich element (RHAU) is also known as MLE-like protein 1 (MLEL1), G4 resolvase (G4R1) and DEAH box protein 36 (DHX36). This RNA helicase unwinds duplex RNA, RNA–DNA hybrids as well as other non-canonical DNA structures such as triplexes (125). In a 1997 report, the group of Steven A. Akman showed that the G4 unwinding properties of RHAU depend on Mg2+ and ATP (41). Subsequent studies have focused mostly on tetramolecular DNA substrates.
The activity of RHAU has been investigated on the G4 DNA systems listed in Table 3. These include the well-characterized murine immunoglobulin switch sequence TP-G4, the oligonucleotide sequence from the human 28S ribosomal RNA gene (rD4), and a sequence from the H2O2/Fe-mediated mutation in the supF gene of the reporter plasmid pZ189 (Z33 (126)) (127,128). The tetra-stranded G4-DNA substrates formed by these sequences were readily denatured by RHAU. A simple series of oligonucleotides with a single G-run of variable length (substrates containing 5, 6, 7 or 8 consecutive guanines) followed by a 15-nt poly(dA) 3′-flanking overhang (Table 3) was employed to study the influence of the number of G-quartets on the unwinding reaction catalysed by RHAU (128). As expected, increasing the number of G-quartets gradually reduced the RHAU-resolvase activity as a consequence of the increase in G4 stability. G4-ligands such as pyridostatin, Phen-DC3 and 12459 also significantly inhibited RHAU helicase unwinding of G4 substrates (128).
Table 3. Substrates used for studying RHAU G4-resolvase activity.
| Substrate name | Sequence |
|---|---|
| Zic1 | 5′-AAA AAA AAA AGG GT GGG GGG GCG GGG GAG GCC GGG GAA AAA AAA AA-3′ |
| c-Myc | 5′-GGC CGC TTA TGG GGA GGG TGG GGA GGG TGG GGA AGG TGG GGA GGA GAC TCA-3′ |
| Z33 | 5′-AAAGTGATGGTGGTGGGGGAAGGATTTCGAACCT-3′ |
| TP-G4 | 5′-TGGACCAGACCTAGCAGCTATGGGGGAGCTGGGGAAGGTGGGAATGTGA-3′ |
| rD4 | 5′-TTGAAAATCCGGGGGAGAGGGTGTAAATCTCG-3′ |
| 8G | 5′-TTAGGGGGGGGA15–3′ |
| 7G | 5′-TTAGGGGGGGA15–3′ |
| 6G | 5′-TTAGGGGGGA15–3′ |
| 5G | 5′-TTAGGGGGA15–3′ |
Akman and Vaughn showed that RHAU has an extraordinarily high affinity for DNA and RNA G-quadruplexes. RHAU showed a strong affinity for tetra-stranded RNA G-quadruplexes (Kd ∼ 39 pM) (129) as well as intramolecular DNA structures such as Zic1 and a c-Myc (Table 3, Kds of 6.3 and 4 pM, respectively) (130). These remarkable values suggest that RHAU might function to denature G-quadruplexes in cells (129). RIP-ChIP methods have been used to screen for RNA G4 motifs recognized by RHAU (131). RHAU acts as a chaperone to promote the formation of the P1 helix in the human telomerase RNA (132), which is in competition with quadruplex formation (133). The binding mode of this protein has also been characterized. The structure of RHAU contains a helicase core, which is responsible for the ATP hydrolysis and helicase activity, flanked on either side by N- and C-terminal extensions. The N-terminal extension is essential for G-quadruplex recognition (134–136).
G4 HELICASES AND TRANSCRIPTION OR TRANSLATION
RHAU appears to be involved in translational regulation. Interestingly, RHAU inhibits the expression of PITX1, a transcription factor, apparently by binding to a non-G4 region (137). In contrast, RHAU enhances the expression of the oncoprotein YY1. YY1 is a multifunctional protein with regulatory potential in tumorigenesis, which contains G4 sequences in both the promoter and the 5′-UTR of its gene. RHAU acts at the DNA level by unwinding the G4 formed in the promoter (138).
The activity of RHAU helicase can be also stimulated and assisted by the presence of other proteins. Vaughn et al. demonstrated a cooperative association between RHAU and the telomerase holoenzyme (139). This association leads to an increase of the telomerase activity. The authors speculate that RHAU unwinds the telomeric G4-structures allowing the telomerase to act on the single-strand sequence (139). The cytoplasmic protein Aven RGG/RG also showed a cooperative effect with RHAU helicase. Translation of MLL (the mRNA encoding the oncogene mixed lineage leukemia) is stimulated by Aven. The Aven RGG/RG motif selectively binds to G4 structures present in the coding regions of the MLL mRNAs, and the RHAU helicase denatures these G-quadruplexes (140).
Unexpectedly, Werner syndrome and Bloom syndrome mutant cells defective in RecQ helicases WRN and BLM, respectively, displayed preferential upregulation of genes characterized by predicted G-quadruplex forming sequences in non-coding regions, primarily promoters and first introns (115). It was surmised that G4 DNA may preclude normal nucleosome formation or the interaction of duplex DNA binding proteins that might repress transcription; however, this remains to be determined. In another work, the individual genes regulated by BLM were investigated by measuring the expression of mRNA and microRNA (miRNA) in fibroblasts (141). The results obtained in BLM-depleted cells and cells derived from Bloom syndrome patients (a syndrome caused by mutations in the gene encoding BLM protein) were found to be significantly different, and the difference in mRNA expression was correlated with the presence of G4 motifs. Although a precise understanding of how human helicases regulate gene expression in a G4-dependent manner is still lacking, molecular evidence is pointing toward a direct interaction of DNA helicases (e.g. BLM (141)) with G4 motifs residing near transcriptional start sites of differentially expressed genes. In the case of BLM, these differences between BLM-deficient and BLM-proficient cells would affect the abundance of messenger RNA molecules that regulate cell proliferation, survival and cancer as well as microRNA molecules influencing cancer and immune function.
However, a more recent study suggested a more complex relationship between expression of genes and their G4-forming potential that is dependent on the mutational status of the WRN and BLM genes (142). A model was presented that intramolecular G4 structures occurring just downstream of the transcriptional start site (within 140–270 bp) activate transcription, and that their resolution by the WRN and BLM helicases may attenuate G4-activation of transcription. However, G4 formation that occurs further downstream of the transcriptional start site may hinder RNA polymerase II transcription; in this case, G4 resolution by WRN and BLM helicases may alleviate transcription inhibition by G4.
The human helicases XPB and XPD of the general transcription factor complex were also found to have a role in transcription related to G4 structure. ChIP-Seq analysis of human cells showed that XPB and XPD binding sites overlap with G4 motifs, suggesting that these proteins are recruited to G4 structures that form in the genome (143). XPB and XPD were found to be enriched in the vicinity of transcriptional start sites of highly expressed genes; however, further mechanistic studies are required to advance our understanding of the functional importance of XPB/XPD targeting to genes implicated in signalling pathways or cancer. The potential involvement of the human RecQ DNA helicase RECQ1 in regulation of genes playing a role in tumorigenesis was investigated by the group of Sharma. Depletion of RECQ1 in HeLa or MDA-MB-231 cells reduced the expression of several genes that play key roles in cell migration, invasion and metastasis. Remarkably, the promoters of genes regulated by RECQ1 helicase showed a rich concentration of putative G4 sequences. In addition, chromatin immunoprecipitation assays showed the binding of RECQ1 to G4 motifs in the promoters regions of the genes regulated by RECQ1 (144). Although RECQ1 was reported to display very low to no activity on a variety of G4 DNA substrates in vitro (72), it remains to be understood if RECQ1's involvement in regulating cancer genes involves a direct effect of the helicase on G4-associated promoter elements.
G4 HELICASES AND REPLICATION
Recent studies have shown how G-quadruplex structures are unwound during DNA replication. In an in vivo approach, Sarkies et al. showed how FANCJ is able to maintain epigenetic stability near G4 DNA motifs through two independent mechanisms (145). First, FANCJ helicase and REV1 polymerase appear to cooperate: in this model, REV1 destabilizes G-quadruplex structures so that FANCJ can unwind the G4 from the opposite side of the G4 structure. More recently, the group of Eoff unveiled the mechanism of the G4-disrupting activity of REV1 (146). In a second mechanism, WRN or BLM helicases assist FANCJ unwinding activity. These enzymes have opposite polarities to FANCJ. FANCJ and BLM could act upon G-quadruplexes found in specific chromosomal regions under particular environmental or genetic conditions (147).
Evidence that FANCJ smooths replication fork progression through G4 DNA was provided by studies using Xenopus egg extracts and single-stranded plasmid DNA templates (148). Replication stalling within a few nucleotides from the G-quadruplex was greatly enhanced in the absence of FANCJ; moreover, G4 stabilization by the inclusion of a G-quadruplex binding ligand in the reaction mixture significantly increased the need for FANCJ to be present in order to alleviate replication block. Notably, stalling of the replisome proteins at sites of G4 DNA under conditions of FANCJ depletion did not occur when FANCD2 was depleted, indicating that FANCJ has a unique role in facilitating replication progression through G4 templates that is independent of its role in the classical FA pathway used for repair of interstrand crosslinks.
Role of yeast S. cerevisiae Pif1 and its essential Schizosaccharomyces pombe orthologue Pfh1 during replication was also assessed in several studies (53–55,149). Although this family of helicase has not been involved in general fork progression, in both these models, absence of these helicases leads to replication fork slowing and chromosomal fragility around G4 sites.
G4-HELICASES AND TELOMERES
The human telomeric DNA G-rich strand is able to fold into G-quadruplex structures. This (GGGTTA)n repeat is probably the most studied and characterized of G4 motifs. The presence of a single-stranded 3′ telomeric overhang allows telomeric G-quadruplex formation without opening a corresponding DNA duplex. Multiple topologies have been observed in biophysical studies of oligonucleotides with this sequence, and the human telomeric motif can be considered as a highly polymorphic G-quadruplex-prone repeat. G-quadruplexes have been proposed to play role in capping telomeres to preserve chromosome integrity (150).
The correct replication of telomeric DNA has been found to be significantly dependent on WRN and BLM helicases, which likely unwind the G-quadruplex motif during the replication process. Cells lacking WRN helicase lose telomeres from the lagging strand in single sister chromatids (151). More recent reports suggested that WRN may possess a greater unwinding activity for telomeric G-rich sequences than BLM (152). A single-molecule analysis, SMARD, was performed to study the effect of BLM helicase on telomeric replication. Telomeric synthesis in the leading strand replication forks was slower in BLM-deficient cells. In addition, fork progression was inhibited in the presence of the G4 ligand PhenDC3 (153).
These complementary studies suggest that these two RecQ helicases have specialized roles in resolving G4 motifs in the telomeric region, in the leading strand (BLM) and lagging strand (WRN) templates. Thus, a combination of BLM and WRN may be necessary for the efficient replication of the chromosome terminal regions, preventing telomeric loss and genomic instability. As noted in previous sections, in addition to BLM and WRN, RTEL1 and DNA2 helicases have been also reported to have key roles in resolving telomeric G4 structures. As discussed before, the group of Boulton has significantly investigated the key role of RTEL1 in maintaining telomeric integrity (90,91).
MITOCHONDRIAL GENOME G4 DNA
The mitochondrial genome is comprised of a guanine-rich DNA strand (the heavy ‘H’ strand) and a cytosine-rich strand (light ‘L’ strand). Therefore, the H strand has a pronounced potential for G-quadruplex formation. Applying an in silico approach, numerous sequences with G4-forming potential were identified in mitochondrial DNA. These sites map in close proximity to known mitochondrial deletion breakpoints (12). More recently JJ Lin's group provided evidence supporting the existence of mitochondrial DNA G4s in live cells employing a G4-specific fluorescent ligand (154)
The Twinkle enzyme, discovered in 2001, belongs to the helicase superfamily 4. Twinkle is a mitochondrial replicative DNA helicase encoded by the mitochondrial gene c10orf2 (155) that unwinds mitochondrial DNA substrates in a 5′ to 3′ direction in a NTP-dependent manner (156). Structurally, Twinkle contains two domains connected by a linker region (157). The C-terminal domain is responsible for the helicase activity. An in vitro test demonstrated that Twinkle alone was unable to resolve mitochondrial G4 structures, whereas it readily unwound duplex DNA substrates. It is conceivable that Twinkle or the mitochondrial replication machinery may act with other factors including auxiliary helicases (e.g. PIF1, SUV3) to resolve mitochondrial G4 DNA structures (158). In addition, it will be interesting to determine if Pif1 activity towards G4 is involved in its mitochondrial function. Interestingly, it has been estimated that there is a 10-fold enrichment in potential G4 forming sequences in the yeast mitochondrial DNA compared to nuclear DNA (159). In agreement with this observation, yeast cells lacking Pif1 experience mitochondrial DNA breakage at specific sites, even in the absence of genotoxic stress (160). Nevertheless, it seems reasonable to suggest that mitochondrial G4 motifs present a formidable barrier during the replication process and are likely to cause genome instability within the organelle (12).
METHODS USED TO STUDY G4 UNFOLDING BY HELICASES
Gel-based assays
Gel-based assays are the main biochemical technique employed to study the G4 unfolding activity of helicase enzymes. DNA substrates employed in gel assays are mainly based on tetra- and bi-molecular G-quadruplexes; thus the single-stranded products migrate faster than the dimeric or tetrameric substrates (Figure 4). Gel-based assays can be also used to measure helicase-catalyzed unwinding of uni-molecular G4 DNA substrates when a trap oligonucleotide complementary to the radiolabelled one is used (81,130).
Surface plasmon resonance
The first report on a helicase resolving an intramolecular G4 motif employed surface plasmon resonance (SPR). This real-time technique was used to study the SV40 T-ag helicase (161). Biotinylated human telomeric repeats, duplex DNA and single-stranded sequences were immobilized on the gold surface of the SPR sensor chip. Binding of the hexameric SV40 T-ag to the DNA substrate was found to be ATP dependent and, expectedly, the helicase activity also required ATP.
Single-molecule studies
Recently Budhathoki et al. studied the unwinding of intramolecular G-quadruplexes at the single-molecule level using FRET (smFRET; Figure 4) (105). In this study, performed under physiological salt concentrations, BLM enzyme was active even in the absence of ATP. In a more recent study, the same group reported an ATP-dependent unwinding activity for this protein (162). To explain this discrepancy, they suggested that BLM protein binds to a single-stranded region in the vicinity of a G4. This can lead to significant destabilization of the G-quadruplex structure.
The important addition of single-molecule techniques to the helicase-G4 field has aided mechanistic studies. The mechanism employed by BLM (106,110) and Pif1 (57,163) enzymes to unfold G-quadruplex structures were investigated using smFRET techniques.
Fluorescent-based bulk assays
The introduction of fluorescent-based techniques allowed monitoring of the G4 unfolding reaction in real-time and at high-throughput. Tan et al. designed a novel DNA system consisting of a hybrid duplex-quadruplex DNA to overcome the issue of rapid refolding of an intramolecular G4 (100). In this system an intermolecular DNA duplex motif is placed immediately downstream of a G-quadruplex structure. Therefore, the helicase enzyme unwinds the duplex DNA directly after having resolved the G-quadruplex structure (Figure 4). As this duplex is a bimolecular structure, its reformation is slower than and highly dependent on the strand concentration. Employing this G4-duplex DNA substrate, the helicase-promoted G4 unwinding can be monitored in real-time by labelling one of the DNA strands with an organic fluorophore.
In cellulo tools
A number of studies have demonstrated that various helicases resolve G-quadruplexes in vitro using purified enzymes on controlled substrates. However, the demonstration of their direct involvement in the resolution of G4 in cells has only been made in a limited number of cases. Early reports demonstrated that the inhibition or absence of the G4 helicases leaves a footprint in the genome that can be detected by bioinformatic studies and correlated with G4 propensity (49,72). More recently, G4-specific antibodies have been developed to detect G4 DNA structures in living cells, and the influence of G4 helicases in cells was studied in presence of these specific antibodies (164,165).The Lansdorp lab and their collaborators developed a murine monoclonal antibody specific for G4 structures (1H6), which exhibits strong nuclear staining in human and murine cells (165). Cells deficient in FANCJ show stronger nuclear staining than cells containing a normal level of this helicase. Therefore G4 DNA structures are less abundant in FANCJ-proficient cells, suggesting a specific G4-resolving role of this helicase. The same G4 DNA-specific antibody was used to show that G4 structures are present in heterochromatin, and that G4 staining in germline DNA was much stronger in somatic cells compared to stem cells in a variety of organisms including mammals (166), pointing to a role of G4 DNA in cell differentiation during development.
An antibody against γH2AX was used to identify DNA damage produced during cell replication in cells treated with a G4-specific ligand. Cells were exposed to pyridostatin (Figure 3) and an γH2AX antibody was used to precipitate genomic DNA flanking double-strand breaks (167). A chromatin immunoprecipitation sequencing analysis of these regions of DNA damage revealed that pyridostatin targeted genes containing sequences with potential to form G-quadruplexes. Thus, pyridostatin was able to regulate the expression of these genes.
There is also a growing field in which G4-specific molecular beacons or probes are being developed with the aim to have convenient G4 cellular imaging. There are several reviews on this topic (168–170). If such probes become readily available, then it would be of great interest to assess specific G4 DNA topologies that accumulate in certain helicase-deficient backgrounds.
DISCUSSION AND CONCLUSIONS
Common characteristics of G4 helicases
As it has been discussed in this review, helicases with G4-resolvase activity belong to different helicase families. All have certain features in common, however. All the reported G4-helicases require a single-stranded tail (either at the 3′ or 5′ end) in order to load onto the DNA substrate and act on the G4 structure. To the best of our knowledge, no helicase has been shown to act on blunt-ended G4 structures. In addition (with the exception of WRN and BLM acting on G4 structures surrounded by ssDNA) all the G4-helicases have been found to employ ATP hydrolysis as a source of energy to unfold the G4 structures.
Interestingly, most helicases are able to denature intermolecular G4 DNA structures more efficiently than duplex DNA. This is remarkable considering the high thermal stability of many G4 structures in comparison with duplexes. For many helicase proteins, the helicase activity can be modulated by presence of G4 ligands or DNA-binding proteins; although in some cases the reduction in helicase activity is not due to G4 stability but a direct effect on the helicase enzyme.
Not all G4 are born equal, neither are G4-helicases
G-quadruplexes should be considered as a family of structures rather than a single fold. Multiple topologies and strand stoichiometries have been observed. In addition, RNA, DNA, and hybrid G-quadruplexes can be formed, at least in vitro. Moreover, some stable G4 have one long single-stranded loop (possibly allowing helicase loading ‘within’ the G-quadruplex rather than at the 5′ or 3′ extremities. In addition to this structural heterogeneity, G4 structures exhibit important differences in thermal stability and in kinetics of folding. Mirroring this diversity, a number of helicases have been found to act on G4 structures, at least in vitro. At this stage, countless questions are still open: Why are so many helicases active on G4? Are their activities redundant or do they act on different G-quadruplex classes? Are helicases active at different stages of the cell cycle? Are there G4-resolving enzymes yet to be discovered? Do some proteins bind to G4 without unfolding these structures? Is a very high affinity for G4 a hindrance to G4 unfolding? How do helicases cooperate with other proteins?
When are G4-helicases required?
Helicases that act on G4 are required for certain cellular processes. That these helicases are necessary for normal development is demonstrated by the involvement of genes encoding helicases in a number of human diseases. It remains unclear whether the G4-resolving activities of these helicases are required. What are their roles during replication and transcription? Are they key components for genomic stability? How different are the leading and lagging strands? Are helicases involved during ribosome scanning? How conserved are these functions? Can one selectively inactivate G4-resolving activity while keeping duplex activity intact? Are there some pathologies associated with G4-resolving deficiency?
As more assays are being designed to study the mechanism of G4 DNA resolution by helicases in the cell, some of the remaining questions will surely be resolved in the near future.
Acknowledgments
O.M., A.B. and J.L.M. thank all lab members for helpful discussions.
FUNDING
Intramural Research program of the National Institutes of Health, NIA (in part, to R.M.B.); Conseil Régional d'Aquitaine and Agence Nationale de la Recherche (ANR grants ‘OligoSwitch’ [ANR-12-IS07-0001]; ‘Quarpdiem’ [ANR-12-BSV8-0008-01]; ‘VIBBnano’ [ANR-10-NANO-04-03 to J.L.M]. Funding for open access charge: Inserm.
Conflict of interest statement. None declared.
REFERENCES
- 1.Rhodes D., Lipps H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015;43:8627–8637. doi: 10.1093/nar/gkv862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Y., Patel D.J. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 3.Parkinson G.N., Lee M.P.H., Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 4.Todd A.K., Johnston M., Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huppert J.L., Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bedrat A., Lacroix L., Mergny J.-L. Re-evaluation of G-quadruplex propensity with G4Hunter. Nucleic Acids Res. 2016;44:1746–1759. doi: 10.1093/nar/gkw006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chambers V.S., Marsico G., Boutell J.M., Di Antonio M., Smith G.P., Balasubramanian S. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotech. 2015;33:877–881. doi: 10.1038/nbt.3295. [DOI] [PubMed] [Google Scholar]
- 8.Valton A., Hassan-Zadeh V., Lema I., Boggetto N., Alberti P., Saintomé C., Riou J., Prioleau M. G4 motifs affect origin positioning and efficiency in two vertebrate replicators. EMBO J. 2014;33:732–746. doi: 10.1002/embj.201387506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fragkos M., Ganier O., Coulombe P., Mechali M. DNA replication origin activation in space and time. Nat. Rev. Mol. Cell Biol. 2015;16:360–374. doi: 10.1038/nrm4002. [DOI] [PubMed] [Google Scholar]
- 10.Besnard E., Babled A., Lapasset L., Milhavet O., Parrinello H., Dantec C., Marin J.-M., Lemaitre J.-M. Unraveling cell type–specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat. Struct. Mol. Biol. 2012;19:837–844. doi: 10.1038/nsmb.2339. [DOI] [PubMed] [Google Scholar]
- 11.Fry M. Regulation of Gene Transcription by DNA G-quadruplexes. In: Spindler L, Fritzsche W, editors. Guanine Quartets. Cambridge: Royal Society of Chemistry; 2013. pp. 223–236. [Google Scholar]
- 12.Bharti S.K., Sommers J.A., Zhou J., Kaplan D.L., Spelbrink J.N., Mergny J.-L., Brosh R.M. DNA sequences proximal to human mitochondrial DNA deletion breakpoints prevalent in human disease form G-quadruplexes, a class of DNA structures inefficiently unwound by the mitochondrial replicative twinkle helicase. J. Biol. Chem. 2014;289:29975–29993. doi: 10.1074/jbc.M114.567073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Damas J., Carneiro J., Goncalves J., Stewart J.B., Samuels D.C., Amorim A., Pereira F. Mitochondrial DNA deletions are associated with non-B DNA conformations. Nucleic Acids Res. 2012;40:7606–7621. doi: 10.1093/nar/gks500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marusic M., Plavec J. The effect of DNA sequence directionality on G-quadruplex folding. Angew. Chem. Int. Ed. Engl. 2015;54:11716–11719. doi: 10.1002/anie.201505348. [DOI] [PubMed] [Google Scholar]
- 15.Zhang J., Zheng K., Xiao S., Hao Y., Tan Z. Mechanism and Manipulation of DNA:RNA Hybrid G-Quadruplex Formation in Transcription of G-Rich DNA. J. Am. Chem. Soc. 2014;136:1381–1390. doi: 10.1021/ja4085572. [DOI] [PubMed] [Google Scholar]
- 16.Bugaut A., Balasubramanian S. 5′-UTR RNA G-quadruplexes: translation regulation and targeting. Nucleic Acids Res. 2012;40:4727–4741. doi: 10.1093/nar/gks068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng K., Wu R., He Y., Xiao S., Zhang J., Liu J., Hao Y., Tan Z. A competitive formation of DNA:RNA hybrid G-quadruplex is responsible to the mitochondrial transcription termination at the DNA replication priming site. Nucleic Acids Res. 2014;42:10832–10844. doi: 10.1093/nar/gku764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Millevoi S., Moine H., Vagner S. G-quadruplexes in RNA biology. Wiley Interdiscip. Rev. RNA. 2012;3:495–507. doi: 10.1002/wrna.1113. [DOI] [PubMed] [Google Scholar]
- 19.Agarwala P., Pandey S., Maiti S. The tale of RNA G-quadruplex. Org. Biomol. Chem. 2015;13:5570–5585. doi: 10.1039/c4ob02681k. [DOI] [PubMed] [Google Scholar]
- 20.Li Q., Xiang J.-F., Yang Q.-F., Sun H.-X., Guan A.-J., Tang Y.-L. G4LDB: a database for discovering and studying G-quadruplex ligands. Nucleic Acids Res. 2013;41:D1115–D1123. doi: 10.1093/nar/gks1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patel S.S., Donmez I. Mechanisms of Helicases. J. Biol. Chem. 2006;281:18265–18268. doi: 10.1074/jbc.R600008200. [DOI] [PubMed] [Google Scholar]
- 22.Lohman T.M., Bjornson K.P. Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- 23.Singleton M.R., Dillingham M.S., Wigley D.B. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 24.Pyle A.M. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu. Rev. Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- 25.Bernstein K.A., Gangloff S., Rothstein R. The RecQ DNA helicases in DNA repair. Annu. Rev. Genet. 2010;44:393–417. doi: 10.1146/annurev-genet-102209-163602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dillingham M.S. Superfamily I helicases as modular components of DNA-processing machines. Biochem. Soc. Trans. 2011;39:413–423. doi: 10.1042/BST0390413. [DOI] [PubMed] [Google Scholar]
- 27.Jankowsky E. RNA helicases at work: Binding and rearranging. Trends Biochem. Sci. 2011;36:19–29. doi: 10.1016/j.tibs.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brosh R.M., Jr, Bohr V.A. Human premature aging, DNA repair and RecQ helicases. Nucleic Acids Res. 2007;35:7527–7544. doi: 10.1093/nar/gkm1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu Y. Unwinding and rewinding: double faces of helicase. J. Nucleic Acids. 2012;2012:1–14. doi: 10.1155/2012/140601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jain A., Bacolla A., Chakraborty P., Grosse F., Vasquez K.M. Human DHX9 Helicase unwinds triple-helical DNA structures. Biochemistry. 2010;49:6992–6999. doi: 10.1021/bi100795m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo M., Hundseth K., Ding H., Vidhyasagar V., Inoue A., Nguyen C.-H., Zain R., Lee J.S., Wu Y. A distinct triplex DNA unwinding activity of ChlR1 helicase. J. Biol. Chem. 2015;290:5174–5189. doi: 10.1074/jbc.M114.634923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsaneva I.R., Muller B., West S.C. RuvA and RuvB proteins of Escherichia coli exhibit DNA helicase activity in vitro. Proc. Natl. Acad. Sci. U.S.A. 1993;90:1315–1319. doi: 10.1073/pnas.90.4.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patel S.S., Donmez I. Mechanisms of Helicases. J. Biol. Chem. 2006;281:18265–18268. doi: 10.1074/jbc.R600008200. [DOI] [PubMed] [Google Scholar]
- 34.Suhasini A.N., Brosh R.M.J. Disease-causing missense mutations in human DNA helicase disorders. Mutat. Res. 2013;752:138–152. doi: 10.1016/j.mrrev.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Larsen N.B., Hickson I.D. RecQ helicases: conserved guardians of genomic integrity. Adv. Exp. Med. Biol. 2013;767:161–184. doi: 10.1007/978-1-4614-5037-5_8. [DOI] [PubMed] [Google Scholar]
- 36.Croteau D.L., Popuri V., Opresko P.L., Bohr V.A. Human RecQ helicases in DNA repair, recombination, and replication. Annu. Rev. Biochem. 2014;83:519–552. doi: 10.1146/annurev-biochem-060713-035428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brosh R.M. Jr. DNA helicases involved in DNA repair and their roles in cancer. Nat. Rev. Cancer. 2013;13:542–558. doi: 10.1038/nrc3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murat P., Balasubramanian S. Existence and consequences of G-quadruplex structures in DNA. Curr. Opin. Genet. Dev. 2014;25:22–29. doi: 10.1016/j.gde.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 39.Bochman M.L., Paeschke K., Zakian V.A. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012;13:770–780. doi: 10.1038/nrg3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baran N. The SV40 large T-antigen helicase can unwind four stranded DNA structures linked by G-quartets. Nucleic Acids Res. 1997;25:297–303. doi: 10.1093/nar/25.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harrington C., Lan Y., Akman S.A. The identification and characterization of a G4-DNA resolvase activity. J. Biol. Chem. 1997;272:24631–24636. doi: 10.1074/jbc.272.39.24631. [DOI] [PubMed] [Google Scholar]
- 42.Sun H., Karow J.K., Hickson I.D., Maizels N. The Bloom's syndrome helicase unwinds G4 DNA. J. Biol. Chem. 1998;273:27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
- 43.Sun H., Bennett R.J., Maizels N. The Saccharomyces cerevisiae Sgs1 helicase efficiently unwinds G-G paired DNAs. Nucleic Acids Res. 1999;27:1978–1984. doi: 10.1093/nar/27.9.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bessler J.B., Torres J.Z., Zakian V.A. The Pif1p subfamily of helicases: region-specific DNA helicases? Trends Cell Biol. 2001;11:60–65. doi: 10.1016/s0962-8924(00)01877-8. [DOI] [PubMed] [Google Scholar]
- 45.Lahaye A., Stahl H., Thines-Sempoux D., Foury F. PIF1: a DNA helicase in yeast mitochondria. EMBO J. 1991;10:997–1007. doi: 10.1002/j.1460-2075.1991.tb08034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boulé J.-B., Vega L.R., Zakian V.A. The yeast Pif1p helicase removes telomerase from telomeric DNA. Nature. 2005;438:57–61. doi: 10.1038/nature04091. [DOI] [PubMed] [Google Scholar]
- 47.Boulé J.-B., Zakian V.A. The yeast Pif1p DNA helicase preferentially unwinds RNA DNA substrates. Nucleic Acids Res. 2007;35:5809–5818. doi: 10.1093/nar/gkm613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Piazza A., Serero A., Boulé J.-B., Legoix-Né P., Lopes J., Nicolas A. Stimulation of gross chromosomal rearrangements by the human CEB1 and CEB25 minisatellites in Saccharomyces cerevisiae depends on G-quadruplexes or Cdc13. PLoS Genet. 2012;8:e1003033. doi: 10.1371/journal.pgen.1003033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ribeyre C., Lopes J., Boulé J.-B., Piazza A., Guédin A., Zakian V.A., Mergny J.-L., Nicolas A. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 2009;5:e1000475. doi: 10.1371/journal.pgen.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adrian M., Ang D.J., Lech C.J., Heddi B., Nicolas A., Phan A.T. Structure and conformational dynamics of a stacked dimeric G-quadruplex formed by the human CEB1 minisatellite. J. Am. Chem. Soc. 2014;136:6297–6305. doi: 10.1021/ja4125274. [DOI] [PubMed] [Google Scholar]
- 51.De Cian A., DeLemos E., Mergny J.-L., Teulade-Fichou M.-P., Monchaud D. Highly efficient G-quadruplex recognition by bisquinolinium compounds. J. Am. Chem. Soc. 2007;129:1856–1857. doi: 10.1021/ja067352b. [DOI] [PubMed] [Google Scholar]
- 52.Piazza A., Boulé J.-B., Lopes J., Mingo K., Largy E., Teulade-Fichou M.-P., Nicolas A. Genetic instability triggered by G-quadruplex interacting Phen-DC compounds in Saccharomyces cerevisiae. Nucleic Acids Res. 2010;38:4337–4348. doi: 10.1093/nar/gkq136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paeschke K., Capra J.A., Zakian V.A. DNA replication through G-Quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell. 2011;145:678–691. doi: 10.1016/j.cell.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paeschke K., Bochman M.L., Garcia P.D., Cejka P., Friedman K.L., Kowalczykowski S.C., Zakian V.A. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature. 2013;497:458–462. doi: 10.1038/nature12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lopes J., Piazza A., Bermejo R., Kriegsman B., Colosio A., Teulade-Fichou M.-P., Foiani M., Nicolas A. G-quadruplex-induced instability during leading-strand replication. EMBO J. 2011;30:4033–4046. doi: 10.1038/emboj.2011.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sanders C.M. Human Pif1 helicase is a G-quadruplex DNA-binding protein with G-quadruplex DNA-unwinding activity. Biochem. J. 2010;430:119–128. doi: 10.1042/BJ20100612. [DOI] [PubMed] [Google Scholar]
- 57.Zhou R., Zhang J., Bochman M.L., Zakian V.A., Ha T. Periodic DNA patrolling underlies diverse functions of Pif1 on R-loops and G-rich DNA. Elife. 2014;3:1–16. doi: 10.7554/eLife.02190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Byrd A.K., Raney K.D. A parallel quadruplex DNA is bound tightly but unfolded slowly by Pif1 helicase. J. Biol. Chem. 2015;290:6482–6494. doi: 10.1074/jbc.M114.630749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Duan X., Liu N., Yang Y., Li H.-H., Li M., Dou S., Xi X. G-quadruplexes significantly stimulate Pif1 helicase-catalyzed duplex DNA unwinding. J. Biol. Chem. 2015;290:7722–7735. doi: 10.1074/jbc.M114.628008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mendoza O., Gueddouda N.M., Boule J.-B., Bourdoncle A., Mergny J.-L. A fluorescence-based helicase assay: application to the screening of G-quadruplex ligands. Nucleic Acids Res. 2015;43:e71. doi: 10.1093/nar/gkv193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barranco-Medina S., Galletto R. DNA binding induces dimerization of Saccharomyces cerevisiae Pif1. Biochemistry. 2010;49:8445–8454. doi: 10.1021/bi100984j. [DOI] [PubMed] [Google Scholar]
- 62.Gao J., Zybailov B.L., Byrd A.K., Griffin W.C., Chib S., Mackintosh S.G., Tackett A.J., Raney K.D. Yeast transcription co-activator Sub1 and its human homolog PC4 preferentially bind to G-quadruplex DNA. Chem. Commun. (Camb). 2015;51:7242–7244. doi: 10.1039/c5cc00742a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hayashi N., Murakami S. STM1, a gene which encodes a guanine quadruplex binding protein, interacts with CDC13 in Saccharomyces cerevisiae. Mol. Genet. Genomics. 2002;267:806–813. doi: 10.1007/s00438-002-0712-3. [DOI] [PubMed] [Google Scholar]
- 64.Hereford L.M., Hartwell L.H. Defective DNA Synthesis in permeabilized yeast mutants. Nat. New Biol. 1971;234:171–172. doi: 10.1038/newbio234171a0. [DOI] [PubMed] [Google Scholar]
- 65.Bae S.H., Choi E., Lee K.H., Park J.S., Lee S.H., Seo Y.S. Dna2 of Saccharomyces cerevisiae possesses a single-stranded DNA-specific endonuclease activity that is able to act on double-stranded DNA in the presence of ATP. J. Biol. Chem. 1998;273:26880–26890. doi: 10.1074/jbc.273.41.26880. [DOI] [PubMed] [Google Scholar]
- 66.Budd M.E., Campbell J.L. A yeast gene required for DNA replication encodes a protein with homology to DNA helicases. Proc. Natl. Acad. Sci. U.S.A. 1995;92:7642–7646. doi: 10.1073/pnas.92.17.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kang Y.-H., Lee C.-H., Seo Y.-S. Dna2 on the road to Okazaki fragment processing and genome stability in eukaryotes. Crit. Rev. Biochem. Mol. Biol. 2010;45:71–96. doi: 10.3109/10409230903578593. [DOI] [PubMed] [Google Scholar]
- 68.Karanja K.K., Cox S.W., Duxin J.P., Stewart S.A., Campbell J.L. DNA2 and EXO1 in replication-coupled, homology-directed repair and in the interplay between HDR and the FA/BRCA network. Cell Cycle. 2012;11:3983–3996. doi: 10.4161/cc.22215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lin W., Sampathi S., Dai H., Liu C., Zhou M., Hu J., Huang Q., Campbell J., Shin-Ya K., Zheng L., et al. Mammalian DNA2 helicase/nuclease cleaves G-quadruplex DNA and is required for telomere integrity. EMBO J. 2013;32:1425–1439. doi: 10.1038/emboj.2013.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Imlay J.A. Iron-sulphur clusters and the problem with oxygen. Mol. Microbiol. 2006;59:1073–1082. doi: 10.1111/j.1365-2958.2006.05028.x. [DOI] [PubMed] [Google Scholar]
- 71.White M.F., Dillingham M.S. Iron–sulphur clusters in nucleic acid processing enzymes. Curr. Opin. Struct. Biol. 2012;22:94–100. doi: 10.1016/j.sbi.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 72.Wu Y., Shin-ya K., Brosh R.M. Jr. FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol. Cell. Biol. 2008;28:4116–4128. doi: 10.1128/MCB.02210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.London T.B.C., Barber L.J., Mosedale G., Kelly G.P., Balasubramanian S., Hickson I.D., Boulton S.J., Hiom K. FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts. J. Biol. Chem. 2008;283:36132–36139. doi: 10.1074/jbc.M808152200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Larson E.D., Duquette M.L., Cummings W.J., Streiff R.J., Maizels N. MutSalpha binds to and promotes synapsis of transcriptionally activated immunoglobulin switch regions. Curr. Biol. 2005;15:470–474. doi: 10.1016/j.cub.2004.12.077. [DOI] [PubMed] [Google Scholar]
- 75.Ray S., Qureshi M.H., Malcolm D.W., Budhathoki J.B., Çelik U., Balci H. RPA-mediated unfolding of systematically varying G-quadruplex structures. Biophys. J. 2013;104:2235–2245. doi: 10.1016/j.bpj.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Salas T.R., Petruseva I., Lavrik O., Bourdoncle A., Mergny J.-L., Favre A., Saintome C. Human replication protein A unfolds telomeric G-quadruplexes. Nucleic Acids Res. 2006;34:4857–4865. doi: 10.1093/nar/gkl564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nguyen B., Sokoloski J., Galletto R., Elson E.L., Wold M.S., Lohman T.M. Diffusion of human replication protein A along single-stranded DNA. J. Mol. Biol. 2014;426:3246–3261. doi: 10.1016/j.jmb.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Audry J., Maestroni L., Delagoutte E., Gauthier T., Nakamura T.M., Gachet Y., Saintome C., Geli V., Coulon S. RPA prevents G-rich structure formation at lagging-strand telomeres to allow maintenance of chromosome ends. EMBO J. 2015;34:1942–1958. doi: 10.15252/embj.201490773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cheung I., Schertzer M., Rose A., Lansdorp P.M. Disruption of dog-1 in Caenorhabditis elegans triggers deletions upstream of guanine-rich DNA. Nat. Genet. 2002;31:405–409. doi: 10.1038/ng928. [DOI] [PubMed] [Google Scholar]
- 80.Lemmens B., van Schendel R., Tijsterman M. Mutagenic consequences of a single G-quadruplex demonstrate mitotic inheritance of DNA replication fork barriers. Nat. Commun. 2015;6:8909. doi: 10.1038/ncomms9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bharti S.K., Sommers J.A., George F., Kuper J., Hamon F., Shin-ya K., Teulade-Fichou M.-P., Kisker C., Brosh R.M. Jr. Specialization among iron-sulfur cluster helicases to resolve G-quadruplex DNA structures that threaten genomic stability. J. Biol. Chem. 2013;288:28217–28229. doi: 10.1074/jbc.M113.496463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bridge W.L., Vandenberg C.J., Franklin R.J., Hiom K. The BRIP1 helicase functions independently of BRCA1 in the Fanconi anemia pathway for DNA crosslink repair. Nat. Genet. 2005;37:953–957. doi: 10.1038/ng1627. [DOI] [PubMed] [Google Scholar]
- 83.Wu Y., Sommers J.A., Loiland J.A., Kitao H., Kuper J., Kisker C., Brosh R.M., Jr The Q motif of Fanconi Anemia group J protein (FANCJ) DNA helicase regulates its dimerization, DNA binding, and DNA repair function. J. Biol. Chem. 2012;287:21699–21716. doi: 10.1074/jbc.M112.351338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sarkies P., Murat P., Phillips L.G., Patel K.J., Balasubramanian S., Sale J.E. FANCJ coordinates two pathways that maintain epigenetic stability at G-quadruplex DNA. Nucleic Acids Res. 2012;40:1485–1498. doi: 10.1093/nar/gkr868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schwab R.A., Nieminuszczy J., Shin-ya K., Niedzwiedz W. FANCJ couples replication past natural fork barriers with maintenance of chromatin structure. J. Cell Biol. 2013;201:33–48. doi: 10.1083/jcb.201208009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Henderson A., Wu Y., Huang Y.C., Chavez E.A., Platt J., Johnson F.B., Brosh R.M., Sen D., Lansdorp P.M. Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res. 2014;42:860–869. doi: 10.1093/nar/gkt957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wu Y., Sommers J.a., Khan I., De Winter J.P., Brosh R.M. Jr. Biochemical characterization of Warsaw breakage syndrome helicase. J. Biol. Chem. 2012;287:1007–1021. doi: 10.1074/jbc.M111.276022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bharti S.K., Khan I., Banerjee T., Sommers J.A., Wu Y., Brosh R.M. Jr. Molecular functions and cellular roles of the ChlR1 (DDX11) helicase defective in the rare cohesinopathy Warsaw breakage syndrome. Cell. Mol. Life Sci. 2014;71:2625–2639. doi: 10.1007/s00018-014-1569-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Calì F., Bharti S.K., Di Perna R., Brosh R.M., Pisani F.M. Tim/Timeless, a member of the replication fork protection complex, operates with the Warsaw breakage syndrome DNA helicase DDX11 in the same fork recovery pathway. Nucleic Acids Res. 2016;44:705–717. doi: 10.1093/nar/gkv1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vannier J.-B., Sandhu S., Petalcorin M.I., Wu X., Nabi Z., Ding H., Boulton S.J. RTEL1 is a replisome-associated helicase that promotes telomere and genome-wide replication. Science. 2013;342:239–242. doi: 10.1126/science.1241779. [DOI] [PubMed] [Google Scholar]
- 91.Vannier J.-B., Pavicic-Kaltenbrunner V., Petalcorin M.I.R., Ding H., Boulton S.J. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell. 2012;149:795–806. doi: 10.1016/j.cell.2012.03.030. [DOI] [PubMed] [Google Scholar]
- 92.Ballew B.J., Joseph V., De S., Sarek G., Vannier J.-B., Stracker T., Schrader K.A., Small T.N., O'Reilly R., Manschreck C., et al. A recessive founder mutation in regulator of telomere elongation helicase 1, RTEL1, underlies severe immunodeficiency and features of Hoyeraal Hreidarsson Syndrome. PLoS Genet. 2013;9:e1003695. doi: 10.1371/journal.pgen.1003695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Deng Z., Glousker G., Molczan A., Fox A.J., Lamm N., Dheekollu J., Weizman O.-E., Schertzer M., Wang Z., Vladimirova O., et al. Inherited mutations in the helicase RTEL1 cause telomere dysfunction and Hoyeraal–Hreidarsson syndrome. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E3408–E3416. doi: 10.1073/pnas.1300600110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ding H., Schertzer M., Wu X., Gertsenstein M., Selig S., Kammori M., Pourvali R., Poon S., Vulto I., Chavez E., et al. Regulation of murine telomere length by Rtel: an essential gene encoding a helicase-like protein. Cell. 2004;117:873–886. doi: 10.1016/j.cell.2004.05.026. [DOI] [PubMed] [Google Scholar]
- 95.Sfeir A., Kosiyatrakul S.T., Hockemeyer D., MacRae S.L., Karlseder J., Schildkraut C.L., de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chakraverty R.K., Hickson I.D. Defending genome integrity during DNA replication: a proposed role for RecQ family helicases. Bioessays. 1999;21:286–294. doi: 10.1002/(SICI)1521-1878(199904)21:4<286::AID-BIES4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 97.Karow J.K., Wu L., Hickson I.D. RecQ family helicases: roles in cancer and aging. Curr. Opin. Genet. Dev. 2000;10:32–38. doi: 10.1016/s0959-437x(99)00039-8. [DOI] [PubMed] [Google Scholar]
- 98.Park C.-J., Ko J., Ryu K.-S., Choi B.-S. Solution structure of the RecQ C-terminal domain of human Bloom syndrome protein. J. Biomol. NMR. 2014;58:141–147. doi: 10.1007/s10858-014-9812-8. [DOI] [PubMed] [Google Scholar]
- 99.Huber M.D., Duquette M.L., Shiels J.C., Maizels N. A conserved G4 DNA binding domain in RecQ family helicases. J. Mol. Biol. 2006;358:1071–1080. doi: 10.1016/j.jmb.2006.01.077. [DOI] [PubMed] [Google Scholar]
- 100.Liu J., Chen C., Xue Y., Hao Y., Tan Z. G-quadruplex hinders translocation of BLM helicase on DNA: a real-time fluorescence spectroscopic unwinding study and comparison with duplex substrates. J. Am. Chem. Soc. 2010;132:10521–10527. doi: 10.1021/ja1038165. [DOI] [PubMed] [Google Scholar]
- 101.Wang Q., Liu J.-Q., Chen Z., Zheng K.-W., Chen C.-Y., Hao Y.-H., Tan Z. G-quadruplex formation at the 3′ end of telomere DNA inhibits its extension by telomerase, polymerase and unwinding by helicase. Nucleic Acids Res. 2011;39:6229–6237. doi: 10.1093/nar/gkr164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ellis N.A., Groden J., Ye T.Z., Straughen J., Lennon D.J., Ciocci S., Proytcheva M., German J. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 103.Huber M.D., Lee D.C., Maizels N. G4 DNA unwinding by BLM and Sgs1p: substrate specificity and substrate-specific inhibition. Nucleic Acids Res. 2002;30:3954–3961. doi: 10.1093/nar/gkf530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li J.L., Harrison R.J., Reszka A.P., Brosh R.M., Bohr V.A., Neidle S., Hickson I.D. Inhibition of the Bloom's and Werner's syndrome helicases by G-quadruplex interacting ligands. Biochemistry. 2001;40:15194–15202. doi: 10.1021/bi011067h. [DOI] [PubMed] [Google Scholar]
- 105.Budhathoki J.B., Ray S., Urban V., Janscak P., Yodh J.G., Balci H. RecQ-core of BLM unfolds telomeric G-quadruplex in the absence of ATP. Nucleic Acids Res. 2014;42:11528–11545. doi: 10.1093/nar/gku856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chatterjee S., Zagelbaum J., Savitsky P., Sturzenegger A., Huttner D., Janscak P., Hickson I.D., Gileadi O., Rothenberg E. Mechanistic insight into the interaction of BLM helicase with intra-strand G-quadruplex structures. Nat. Commun. 2014;5:5556. doi: 10.1038/ncomms6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Janscak P., Garcia P.L., Hamburger F., Makuta Y., Shiraishi K., Imai Y., Ikeda H., Bickle T.A. Characterization and mutational analysis of the RecQ core of the bloom syndrome protein. J. Mol. Biol. 2003;330:29–42. doi: 10.1016/s0022-2836(03)00534-5. [DOI] [PubMed] [Google Scholar]
- 108.Ghosh A., Rossi M.L., Aulds J., Croteau D., Bohr V.A. Telomeric D-loops containing 8-Oxo-2′-deoxyguanosine are preferred substrates for Werner and Bloom Syndrome Helicases and are bound by POT1. J. Biol. Chem. 2009;284:31074–31084. doi: 10.1074/jbc.M109.027532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Garcia P.L., Liu Y., Jiricny J., West S.C., Janscak P. Human RECQ5β, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J. 2004;23:2882–2891. doi: 10.1038/sj.emboj.7600301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wu W.-Q., Hou X.-M., Li M., Dou S.-X., Xi X.-G. BLM unfolds G-quadruplexes in different structural environments through different mechanisms. Nucleic Acids Res. 2015;43:4614–4626. doi: 10.1093/nar/gkv361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yu C.E., Oshima J., Fu Y.H., Wijsman E.M., Hisama F., Alisch R., Matthews S., Nakura J., Miki T., Ouais S., et al. Positional cloning of the Werner's syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 112.Von Kobbe C., Karmakar P., Dawut L., Opresko P., Zeng X., Brosh R.M. Jr., Hickson I.D., Bohr V.A. Colocalization, physical, and functional interaction between Werner and Bloom syndrome proteins. J. Biol. Chem. 2002;277:22035–22044. doi: 10.1074/jbc.M200914200. [DOI] [PubMed] [Google Scholar]
- 113.Fry M., Loeb L.A. Structures of the fragile X syndrome repeat sequence d(CGG)n. Biochemistry. 1999;274:12797–12802. doi: 10.1074/jbc.274.18.12797. [DOI] [PubMed] [Google Scholar]
- 114.Mohaghegh P., Karow J.K., Brosh R.M., Jr, Bohr V.A., Hickson I.D. The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Johnson J.E., Cao K., Ryvkin P., Wang L.-S., Johnson F.B. Altered gene expression in the Werner and Bloom syndromes is associated with sequences having G-quadruplex forming potential. Nucleic Acids Res. 2010;38:1114–1122. doi: 10.1093/nar/gkp1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Damerla R.R., Knickelbein K.E., Strutt S., Liu F.-J., Wang H., Opresko P.L. Werner syndrome protein suppresses the formation of large deletions during the replication of human telomeric sequences. Cell Cycle. 2012;11:3036–3044. doi: 10.4161/cc.21399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Aggarwal M., Sommers J.A., Shoemaker R.H., Brosh R.M. Inhibition of helicase activity by a small molecule impairs Werner syndrome helicase (WRN) function in the cellular response to DNA damage or replication stress. Proc. Natl. Acad. Sci. U.S.A. 2011;108:1525–1530. doi: 10.1073/pnas.1006423108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Han H., Bennett R.J., Hurley L.H. Inhibition of unwinding of G-quadruplex structures by Sgs1 helicase in the presence of N,N’-Bis[2-(1-piperidino)ethyl]-3,4,9,10-perylenetetracarboxylic diimide, a G-quadruplex-interactive ligand. Biochemistry. 2000;39:9311–9316. doi: 10.1021/bi000482r. [DOI] [PubMed] [Google Scholar]
- 119.Hershman S.G., Chen Q., Lee J.Y., Kozak M.L., Yue P., Wang L.-S., Johnson F.B. Genomic distribution and functional analyses of potential G-quadruplex-forming sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 2007;36:144–156. doi: 10.1093/nar/gkm986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Smith J.S., Chen Q., Yatsunyk L.A., Nicoludis J.M., Garcia M.S., Kranaster R., Balasubramanian S., Monchaud D., Teulade-Fichou M.-P., Abramowitz L., et al. Rudimentary G-quadruplex–based telomere capping in Saccharomyces cerevisiae. Nat. Struct. Mol. Biol. 2011;18:478–485. doi: 10.1038/nsmb.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wu X., Maizels N. Substrate-specific inhibition of RecQ helicase. Nucleic Acids Res. 2001;29:1765–1771. doi: 10.1093/nar/29.8.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tuesuwan B., Kern J.T., Thomas P.W., Rodriguez M., Li J., David W.M., Kerwin S.M. Simian virus 40 large T-antigen G-Quadruplex DNA helicase inhibition by G-quadruplex DNA-interactive agents. Biochemistry. 2008;47:1896–1909. doi: 10.1021/bi701747d. [DOI] [PubMed] [Google Scholar]
- 123.Zhang S., Grosse F. Multiple functions of nuclear DNA Helicase II (RNA helicase A) in nucleic acid metabolism. Acta Biochim. Biophys. Sin. (Shanghai). 2004;36:177–183. doi: 10.1093/abbs/36.3.177. [DOI] [PubMed] [Google Scholar]
- 124.Chakraborty P., Grosse F. Human DHX9 helicase preferentially unwinds RNA-containing displacement loops (R-loops) and G-quadruplexes. DNA Repair (Amst). 2011;10:654–665. doi: 10.1016/j.dnarep.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 125.Jain A., Bacolla A., Chakraborty P., Grosse F., Vasquez K.M. Human DHX9 helicase unwinds triple-helical DNA structures. Biochemistry. 2010;49:6992–6999. doi: 10.1021/bi100795m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Akman S.A., Lingeman R.G., Doroshow J.H., Smith S.S. Quadruplex DNA formation in a region of the tRNA gene supF associated with hydrogen peroxide mediated mutations. Biochemistry. 1991;30:8648–8653. doi: 10.1021/bi00099a022. [DOI] [PubMed] [Google Scholar]
- 127.Vaughn J.P., Creacy S.D., Routh E.D., Joyner-Butt C., Jenkins G.S., Pauli S., Nagamine Y., Akman S.A. The DEXH protein product of the DHX36 gene is the major source of tetramolecular quadruplex G4-DNA resolving activity in HeLa cell lysates. J. Biol. Chem. 2005;280:38117–38120. doi: 10.1074/jbc.C500348200. [DOI] [PubMed] [Google Scholar]
- 128.Chen M.C., Murat P., Abecassis K., Ferre-D'Amare A.R., Balasubramanian S. Insights into the mechanism of a G-quadruplex-unwinding DEAH-box helicase. Nucleic Acids Res. 2015;43:2223–2231. doi: 10.1093/nar/gkv051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Creacy S.D., Routh E.D., Iwamoto F., Nagamine Y., Akman S.a., Vaughn J.P. G4 resolvase 1 binds both DNA and RNA tetramolecular quadruplex with high affinity and is the major source of tetramolecular quadruplex G4-DNA and G4-RNA resolving activity in HeLa cell lysates. J. Biol. Chem. 2008;283:34626–34634. doi: 10.1074/jbc.M806277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Giri B., Smaldino P.J., Thys R.G., Creacy S.D., Routh E.D., Hantgan R.R., Lattmann S., Nagamine Y., Akman S.A., Vaughn J.P. G4 Resolvase 1 tightly binds and unwinds unimolecular G4-DNA. Nucleic Acids Res. 2011;39:7161–7178. doi: 10.1093/nar/gkr234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lattmann S., Stadler M.B., Vaughn J.P., Akman S.A., Nagamine Y. The DEAH-box RNA helicase RHAU binds an intramolecular RNA Gquadruplex in TERC and associates with telomerase holoenzyme. Nucleic Acids Res. 2011;39:9390–9404. doi: 10.1093/nar/gkr630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Booy E.P., Meier M., Okun N., Novakowski S.K., Xiong S., Stetefeld J., McKenna S.A. The RNA helicase RHAU (DHX36) unwinds a G4-quadruplex in human telomerase RNA and promotes the formation of the P1 helix template boundary. Nucleic Acids Res. 2012;40:4110–4124. doi: 10.1093/nar/gkr1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lacroix L., Seosse A., Mergny J.-L. Fluorescence-based duplex-quadruplex competition test to screen for telomerase RNA quadruplex ligands. Nucleic Acids Res. 2011;39:e21. doi: 10.1093/nar/gkq1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lattmann S., Giri B., Vaughn J.P., Akman S.A., Nagamine Y. Role of the amino terminal RHAU-specific motif in the recognition and resolution of guanine quadruplex-RNA by the DEAH-box RNA helicase RHAU. Nucleic Acids Res. 2010;38:6219–6233. doi: 10.1093/nar/gkq372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sexton A.N., Collins K. The 5′ guanosine tracts of human telomerase RNA are recognized by the G-quadruplex binding domain of the RNA helicase DHX36 and function to increase RNA accumulation. Mol. Cell. Biol. 2011;31:736–743. doi: 10.1128/MCB.01033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Meier M., Patel T.R., Booy E.P., Marushchak O., Okun N., Deo S., Howard R., McEleney K., Harding S.E., Stetefeld J., et al. Binding of G-quadruplexes to the N-terminal recognition domain of the RNA helicase associated with AU-rich Element (RHAU) J. Biol. Chem. 2013;288:35014–35027. doi: 10.1074/jbc.M113.512970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Booy E.P., Howard R., Marushchak O., Ariyo E.O., Meier M., Novakowski S.K., Deo S.R., Dzananovic E., Stetefeld J., Mckenna S.A. The RNA helicase RHAU (DHX36) suppresses expression of the transcription factor PITX1. Nucleic Acids Res. 2014;42:3346–3361. doi: 10.1093/nar/gkt1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Huang W., Smaldino P.J., Zhang Q., Miller L.D., Cao P., Stadelman K., Wan M., Giri B., Lei M., Nagamine Y., et al. Yin Yang 1 contains G-quadruplex structures in its promoter and 5′-UTR and its expression is modulated by G4 resolvase 1. Nucleic Acids Res. 2012;40:1033–1049. doi: 10.1093/nar/gkr849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Smaldino P.J., Routh E.D., Kim J.H., Giri B., Creacy S.D., Hantgan R.R., Akman S.A., Vaughn J.P. Mutational dissection of telomeric DNA binding requirements of G4 resolvase 1 shows that G4-structure and certain 3′-tail sequences are sufficient for tight and complete binding. PLoS One. 2015;10:e0132668. doi: 10.1371/journal.pone.0132668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Thandapani P., Song J., Gandin V., Cai Y., Rouleau S.G., Garant J.-M., Boisvert F.-M., Yu Z., Perreault J.-P., Topisirovic I., et al. Aven recognition of RNA G-quadruplexes regulates translation of the mixed lineage leukemia protooncogenes. Elife. 2015;4:e06234. doi: 10.7554/eLife.06234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nguyen G.H., Tang W., Robles A.I., Beyer R.P., Gray L.T., Welsh J.A., Schetter A.J., Kumamoto K., Wang X.W., Hickson I.D., et al. Regulation of gene expression by the BLM helicase correlates with the presence of G-quadruplex DNA motifs. Proc. Natl. Acad. Sci. U.S.A. 2014;111:9905–9910. doi: 10.1073/pnas.1404807111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Smestad J.A., Maher L.J. Relationships between putative G-quadruplex-forming sequences, RecQ helicases, and transcription. BMC Med. Genet. 2015;16:91. doi: 10.1186/s12881-015-0236-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gray L.T., Vallur A.C., Eddy J., Maizels N. G quadruplexes are genomewide targets of transcriptional helicases XPB and XPD. Nat. Chem. Biol. 2014;10:313–318. doi: 10.1038/nchembio.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Li X.L., Lu X., Parvathaneni S., Bilke S., Zhang H., Thangavel S., Vindigni A., Hara T., Zhu Y., Meltzer P.S., et al. Identification of RECQ1-regulated transcriptome uncovers a role of RECQ1 in regulation of cancer cell migration and invasion. Cell Cycle. 2014;13:2431–2445. doi: 10.4161/cc.29419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sarkies P., Murat P., Phillips L.G., Patel K.J., Balasubramanian S., Sale J.E. FANCJ coordinates two pathways that maintain epigenetic stability at G-quadruplex DNA. Nucleic Acids Res. 2012;40:1485–1498. doi: 10.1093/nar/gkr868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Eddy S., Ketkar A., Zafar M.K., Maddukuri L., Choi J.-Y., Eoff R.L. Human Rev1 polymerase disrupts G-quadruplex DNA. Nucleic Acids Res. 2014;42:3272–3285. doi: 10.1093/nar/gkt1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Suhasini A.N., Brosh R.M. Fanconi anemia and Bloom's syndrome crosstalk through FANCJ-BLM helicase interaction. Trends Genet. 2012;28:7–13. doi: 10.1016/j.tig.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Castillo Bosch P., Segura-Bayona S., Koole W., van Heteren J.T., Dewar J.M., Tijsterman M., Knipscheer P. FANCJ promotes DNA synthesis through G-quadruplex structures. EMBO J. 2014;33:2521–2533. doi: 10.15252/embj.201488663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sabouri N., McDonald K.R., Webb C.J., Cristea I.M., Zakian V.A. DNA replication through hard-to-replicate sites, including both highly transcribed RNA Pol II and Pol III genes, requires the S. pombe Pfh1 helicase. Genes Dev. 2012;26:581–593. doi: 10.1101/gad.184697.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Smith J.S., Chen Q., Yatsunyk L.A., Nicoludis J.M., Garcia M.S., Kranaster R., Balasubramanian S., Monchaud D., Teulade-Fichou M.-P., Abramowitz L., et al. Rudimentary G-quadruplex-based telomere capping in Saccharomyces cerevisiae. Nat. Struct. Mol. Biol. 2011;18:478–485. doi: 10.1038/nsmb.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Crabbe L. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004;306:1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- 152.Edwards D.N., Machwe A., Chen L., Bohr V.A., Orren D.K. The DNA structure and sequence preferences of WRN underlie its function in telomeric recombination events. Nat. Commun. 2015;6:8331. doi: 10.1038/ncomms9331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Drosopoulos W.C., Kosiyatrakul S.T., Schildkraut C.L. BLM helicase facilitates telomere replication during leading strand synthesis of telomeres. J. Cell Biol. 2015;210:191–208. doi: 10.1083/jcb.201410061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Huang W.-C., Tseng T.-Y., Chen Y.-T., Chang C.-C., Wang Z.-F., Wang C.-L., Hsu T.-N., Li P.-T., Chen C.-T., Lin J.-J., et al. Direct evidence of mitochondrial G-quadruplex DNA by using fluorescent anti-cancer agents. Nucleic Acids Res. 2015;43:10102–10113. doi: 10.1093/nar/gkv1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Spelbrink J.N., Li F.-Y., Tiranti V., Nikali K., Yuan Q.-P., Tariq M., Wanrooij S., Garrido N., Comi G., Morandi L., et al. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat. Genet. 2001;28:223–231. doi: 10.1038/90058. [DOI] [PubMed] [Google Scholar]
- 156.Korhonen J.A., Gaspari M., Falkenberg M. TWINKLE has 5′ -> 3′ DNA helicase activity and is specifically stimulated by mitochondrial single-stranded DNA-binding protein. J. Biol. Chem. 2003;278:48627–48632. doi: 10.1074/jbc.M306981200. [DOI] [PubMed] [Google Scholar]
- 157.Ziebarth T.D., Farr C.L., Kaguni L.S. Modular architecture of the hexameric human mitochondrial DNA helicase. J. Mol. Biol. 2007;367:1382–1391. doi: 10.1016/j.jmb.2007.01.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ding L., Liu Y. Borrowing nuclear DNA helicases to protect mitochondrial DNA. Int. J. Mol. Sci. 2015;16:10870–10887. doi: 10.3390/ijms160510870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Capra J.A., Paeschke K., Singh M., Zakian V.A. G-quadruplex DNA sequences are evolutionarily conserved and associated with distinct genomic features in Saccharomyces cerevisiae. PLoS Comput. Biol. 2010;6:e1000861. doi: 10.1371/journal.pcbi.1000861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Cheng X., Dunaway S., Ivessa A.S. The role of Pif1p, a DNA helicase in Saccharomyces cerevisiae, in maintaining mitochondrial DNA. Mitochondrion. 2007;7:211–222. doi: 10.1016/j.mito.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 161.Plyler J., Jasheway K., Tuesuwan B., Karr J., Brennan J.S., Kerwin S.M., David W.M. Real-time investigation of SV40 Large T-antigen helicase activity using surface plasmon resonance. Cell Biochem. Biophys. 2009;53:43–52. doi: 10.1007/s12013-008-9038-z. [DOI] [PubMed] [Google Scholar]
- 162.Budhathoki J.B., Stafford E.J., Yodh J.G., Balci H. ATP-dependent G-quadruplex unfolding by Bloom helicase exhibits low processivity. Nucleic Acids Res. 2015;43:5961–5970. doi: 10.1093/nar/gkv531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hou X.-M., Wu W.-Q., Duan X.-L., Liu N.-N., Li H.-H., Fu J., Dou S.-X., Li M., Xi X.-G. Molecular mechanism of G-quadruplex unwinding helicase: sequential and repetitive unfolding of G-quadruplex by Pif1 helicase. Biochem. J. 2015;466:189–199. doi: 10.1042/BJ20140997. [DOI] [PubMed] [Google Scholar]
- 164.Biffi G., Tannahill D., McCafferty J., Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Henderson A., Wu Y., Huang Y.C., Chavez E.A., Platt J., Johnson F.B., Brosh R.M., Sen D., Lansdorp P.M. Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res. 2014;42:860–869. doi: 10.1093/nar/gkt957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hoffmann R.F., Moshkin Y.M., Mouton S., Grzeschik N.A., Kalicharan R.D., Kuipers J., Wolters A.H.G., Nishida K., Romashchenko A.V., Postberg J., et al. Guanine quadruplex structures localize to heterochromatin. Nucleic Acids Res. 2016;44:152–163. doi: 10.1093/nar/gkv900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Rodriguez R., Miller K.M., Forment J. V, Bradshaw C.R., Nikan M., Britton S., Oelschlaegel T., Xhemalce B., Balasubramanian S., Jackson S.P. Small-molecule–induced DNA damage identifies alternative DNA structures in human genes. Nat. Chem. Biol. 2012;8:301–310. doi: 10.1038/nchembio.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ma D.-L., Shiu-Hin Chan D., Yang H., He H.-Z., Leung C.-H. Luminescent G-quadruplex probes. Curr. Pharm. Des. 2012;18:2058–2075. doi: 10.2174/138161212799958314. [DOI] [PubMed] [Google Scholar]
- 169.Ma D.-L., Zhang Z., Wang M., Lu L., Zhong H.-J., Leung C.-H. Recent developments in G-quadruplex probes. Chem. Biol. 2015;22:812–828. doi: 10.1016/j.chembiol.2015.06.016. [DOI] [PubMed] [Google Scholar]
- 170.Vummidi B.R., Alzeer J., Luedtke N.W. Fluorescent probes for G-quadruplex structures. Chembiochem. 2013;14:540–558. doi: 10.1002/cbic.201200612. [DOI] [PubMed] [Google Scholar]
- 171.Wu Y., Sommers J.A., Suhasini A.N., Aggarwal M., Brosh R.M. Jr. Molecular analyses of DNA helicases involved in the replicational stress response. Methods. 2010;51:303–312. doi: 10.1016/j.ymeth.2010.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]