Abstract
An ATP-dependent RNA ligase from Methanobacterium thermoautotrophicum (MthRnl) catalyzes intramolecular ligation of single-stranded RNA to form a closed circular RNA via covalent ligase-AMP and RNA-adenylylate intermediate. Here, we report the X-ray crystal structures of an MthRnl•ATP complex as well as the covalent MthRnl–AMP intermediate. We also performed structure-guided mutational analysis to survey the functions of 36 residues in three component steps of the ligation pathway including ligase-adenylylation (step 1), RNA adenylylation (step 2) and phosphodiester bond synthesis (step 3). Kinetic analysis underscored the importance of motif 1a loop structure in promoting phosphodiester bond synthesis. Alanine substitutions of Thr117 or Arg118 favor the reverse step 2 reaction to deadenylate the 5′-AMP from the RNA-adenylate, thereby inhibiting step 3 reaction. Tyr159, Phe281 and Glu285, which are conserved among archaeal ATP-dependent RNA ligases and are situated on the surface of the enzyme, are required for RNA binding. We propose an RNA binding interface of the MthRnl based on the mutational studies and two sulfate ions that co-crystallized at the active site cleft in the MthRnl–AMP complex.
INTRODUCTION
RNA ligases participate in repair, splicing and editing pathways to repair breaks in RNA incurred during processing events. ATP-dependent RNA ligases catalyze the joining of RNA molecules via three nucleotidyltransfer steps similar to those used by ATP-dependent DNA ligases (1,2). In step 1, RNA ligase reacts with ATP to form a covalent ligase–AMP intermediate. In step 2, this AMP is transferred to the 5′-PO4 RNA end, to form RNA-adenylate (AppRNA). In step 3, AppRNA is attacked by the 3′-OH RNA to form a phosphodiester bond, an event that liberates AMP. RNA ligase can join two single-stranded RNA molecules with or without a complementary bridging polynucleotide. It can also catalyze intramolecular ligation, leading to the formation of a covalently-closed circular RNA (cRNA).
Five different families of ATP-dependent RNA ligases (Rnl1–Rnl5) have been classified based on structure and polynucleotide substrate specificity. Crystal structures have been solved for bacteriophage T4 RNA ligase 1 [Rnl1 family; (3)], bacteriophage T4 RNA ligase 2 and Trypanosoma brucei REL1 [Rnl2 family; (4–6)], Pyrococccus. abyssi Rnl [Rnl3 family; (7)], Clostridium thermocellum and Capnocytophaga gingivalis RNA ligases [Rnl4 family; (8,9)], and Naegleria gruberi RNA ligase [Rnl5 family; (10)]. All ATP-dependent ligases share a common adenylyltransferase domain with nucleotidyl transferase motifs (I, Ia, III, IIIa, IV and V) that make up the active site (11). Lys in motif 1[Kx(D/H/N)G] is the site of covalent AMP attachment and other conserved residues found in the nucleotidyltransferase motifs make direct contact with ATP and are required for step 1 covalent ligase–AMP intermediate formation.
The specificity for the polynucleotide substrate is dictated by an additional domain that is typically linked to the adenylyltransferase domain. In T4Rnl1, the C-terminal domain is responsible for recognition of the anticodon loop of tRNA (12). In T4Rnl2, the C-terminal domain was shown to be essential for step 2 RNA-adenylylation and binding to a nicked duplex RNA substrate (13). In contrast, Rnl5 lacks a C-terminal domain and recognition of a nick in the duplex RNA is dictated by a distinct N-terminal domain (14,15). Several bacterial species encode Rnl4, which consists of Pnkp–Hen1 heterotetramer or Pnkp–Rnl–Hen1 heterohexamer, to repair ribotoxin cleaved RNA molecules (16,17). CthPnkp and CgiRnl have the adenylyltransferase domain and are unable to catalyze the ligation unless they interact with Hen1, suggesting that Hen1 may contribute in substrate recognition (8,9).
Rnl3 is found in several archaea species and is unique among polynucleotide ligases in that it adopts a homodimeric quaternary structure (7,18). Each protomer consists of four structural domains: an N-terminal, the adenylyltransferase, a dimerization and a C-terminal domain. Although the physiological substrate and biological function of Rnl3 are not known, it can act to circularize single-stranded RNA and DNA (7,18). Methanobacterium thermoautotrophicum Rnl3 (MthRnl) can also convert 3′-phosphorylated RNA termini into 2′, 3′-cyclic phosphate (19). Under a non-optimal condition, MthRnl can remove the 5′-adenylate on AppRNA to generate 5′-PO4 RNA (18). While there is growing interest for the use of thermostable RNA ligases in constructing sequencing libraries of microRNAs and other small RNAs (20,21), how Rnl3 recognizes polynucleotide substrate is not clear. Deletion analysis of MthRnl suggests that the dimerization domain and/or C-terminal module likely contribute to strand-joining activity (18).
Here, we report the crystal structure of MthRnl with ATP bound and that of a covalent MthRnl–AMP intermediate at 2.5 Å resolution. A structure-guided mutational analysis was performed to survey the function of 36 residues on overall RNA ligation and step 3 AppRNA sealing activity. The effect on step 2 catalysis was assessed by formation of AppRNA during early time points in the composite pRNA ligation reaction, and by a reverse step 2 reaction to deadenylate AppRNA. We identified a structural component that promotes the forward ligation reaction in the direction of phosphodiester bond formation and propose an RNA binding interface based on the mutational studies.
MATERIALS AND METHODS
Cloning, expression and purification of WT and mutant MthRnl
Alanine mutations were introduced into pET–MthRnl by PCR-based methods (QuikChange; Stratagene) followed by sequencing to confirm desired mutations. Wild-type MthRnl and mutant proteins were expressed and purified by nickel-agarose chromatography as described previously (18). MthRnl mutants K73A, R76A, R118A and R275A were insoluble when induced by isopropyl-D-thiogalactoside (IPTG) at 37°C. Solubility was improved by varying the induction method as follows. The cultures were grown at 37°C until the A600 reached 0.4. Cells were placed on ice for 10 min, after which they were adjusted to 0.4 mM IPTG and 2% ethanol, then incubated at 17°C for 20 h with continuous shaking. Wild-type MthRnl was also produced by this protocol. The wild-type and mutant MthRnl preparations were stored at 4°C. Protein concentration was determined with the Bio-Rad dye reagent using bovine serum albumin as the standard.
Crystallization and X-ray structure determination
Crystals were obtained by using hanging drop vapor diffusion by mixing equal volumes of RNA ligase (∼10 mg/ml) and crystallization solution [0.1 M Tris-HCl (pH 8.5), 0.2 M lithium sulfate and 40% PEG400] and incubating at 22°C over the same crystallization solution. Crystals were harvested directly from a crystallization drop and frozen by liquid nitrogen. For preparing crystals of the ligase–AMP intermediate, crystals were soaked in the crystallization solution containing 20 mM magnesium sulfate overnight before frozen by liquid nitrogen. The crystals belong to the primitive monoclinic space group (Table 1) containing one RNA ligase homodimer per asymmetric unit.
Table 1. Data collections and refinement statistics of the Methanobacterium thermoautotrophicum (MthRnl) RNA ligase.
| Crystala | Form 1 | Form 2 |
|---|---|---|
| PDB code | 5D1O | 5D1P |
| Data collection | ||
| Space group | P21 | P21 |
| Cell dimensions | ||
| a (Å) | 50.25 | 50.50 |
| b (Å) | 114.87 | 115.30 |
| c (Å) | 91.29 | 91.57 |
| β (°) | 104.42 | 104.55 |
| Resolution (Å) | 30–2.65 | 30–2.20 |
| Total reflections | 56 562 | 159 151 |
| Unique reflections | 23 491 | 46 681 |
| Redundancy | 2.4 (2.2)* | 3.4 (2.5)* |
| Completeness (%) | 80.3 (76.2)* | 90.6 (56.2)* |
| I / σ | 7.47 (0.792)* | 12.4 (1.32)* |
| Rsym | 0.150 (>1.000)* | 0.100 (0.735)* |
| Rpim | 0.115 (0.810)* | 0.062 (0.565)* |
| CC1/2 | 0.661 | 0.514 |
| Refinement | ||
| Resolution (Å) | 30–2.65 | 30–2.2 |
| Rwork | 0.206 (0.287)* | 0.192 (0.283)* |
| Rfree | 0.265 (0.337)* | 0.241 (0.360)* |
| R.m.s deviations | ||
| Bond length (Å) | 0.021 | 0.022 |
| Bond angles (°) | 1.26 | 1.31 |
| Number of atoms | 6142 | 6470 |
| Protein | 5981 | 5972 |
| Ligands | 88 | 94 |
| Water | 73 | 404 |
| Average B factors (Å2) | 44.0 | 39.7 |
| Protein | 43.9 | 39.5 |
| Ligands | 47.6 | 41.0 |
| Water | 46.1 | 42.4 |
aForm 1: crystal was grown without Mg, Form 2: crystal was soaked in 20 mM MgAcetate.
*Highest resolution shells are shown in parenthesis.
The X-ray data sets were collected at the Pennsylvania State University X-Ray Crystallography Facility. The data were processed by HKL2000 (22). A crystal structure of Pyrococcus abyssi RNA ligase [PDB: 2VUG, (7)] without any ATP analog at active sites was used as a search model for the molecular replacement. The molecular replacement map allowed segments that were not present in the search model to be built manually by Coot (23). Positional refinement with non-crystallographic symmetry and secondary structure restraints was performed by the programs PHENIX (24). The Fo-Fc maps around active sties were carefully inspected and determined their reaction states before placing ATP, AMP, SO4 and Mg2+ at their active sites. Final coordinates and structure factors were submitted to the PDB under accession code 5D1O and 5D1P.
Step 1 adenylyltransferase assay
Reaction mixtures (20 μl) containing 50 mM Tris-HCl (pH 8.0), 5 mM dithiothreitol (DTT), 0.5 mM MgCl2, with various concentrations of [32P]α-ATP and wild-type (WT) or mutant MthRnl proteins were incubated for 15 min at 50°C. The reactions were quenched with SDS loading buffer, and the products were analyzed by SDS-PAGE. The [32P]ligase-AMP adduct was visualized by autoradiography of the dried gel and quantitated by scanning the gel with a Storm PhosphorImager.
Overall RNA ligation assay
Reaction mixtures (120 μl) containing 50 mM Tris-HCl (pH 6.5), 2 mM DTT, 2 mM MgCl2, 10 pmol of 32P-labeled 24-mer pRNA (5′-pAUUCCGAUAGUGCGUGUCGCCCUU) and 60 pmol of WT or mutant MthRnl protein were incubated at 55°C. Aliquots (10 μl) were withdrawn at indicated time points and quenched immediately with 90% formamide - 20 mM ethylene diamine tetraacetic acid (EDTA). Products were resolved by 18% PAGE containing 7 M urea. The extents of RNA circle (cRNA) and RNA adenylate (AppRNA) formation were quantified by phosphoimager analysis.
Step 3 sealing assay
Reaction mixtures (120 μl) containing 50 mM Tris-HCl (pH 6.5), 2 mM MgCl2, 2 mM DTT, 6 pmol 32P-labeled AppRNA and 60 pmol of WT or mutant protein were incubated at 55°C. Aliquots (10 μl) were withdrawn at each indicated time point and quenched immediately with 90% formamide 20 mM EDTA. Products were resolved on denaturing 18% PAGE containing 7 M urea. The extent of cRNA and pRNA was quantified by phosphoimager analysis.
RESULTS
Crystal structure of MthRnl in complex with ATP and a covalent AMP intermediate
We crystallized MthRnl and solved the structure at 2.65 Å resolution (see Materials and Methods, and Table 1, form 1 crystal). Consistent with sedimentation analysis (18), MthRnl crystallized as homodimer with two protomers per asymmetric unit (Figure 1A). The overall structure resembles the PabRnl structure (∼0.9 Å root mean square deviations) (7) and each protomer consists of four structural domains – an N-terminal catalytic domain that adopts a similar topology as T4Rnl1, a covalent nucleotidyltransferase domain that is found in ATP-dependent polynucleotide ligase and GTP-dependent mRNA capping enzyme, a dimerization domain and a C-terminal domain that protrudes into the ATP binding pocket within the partner molecule.
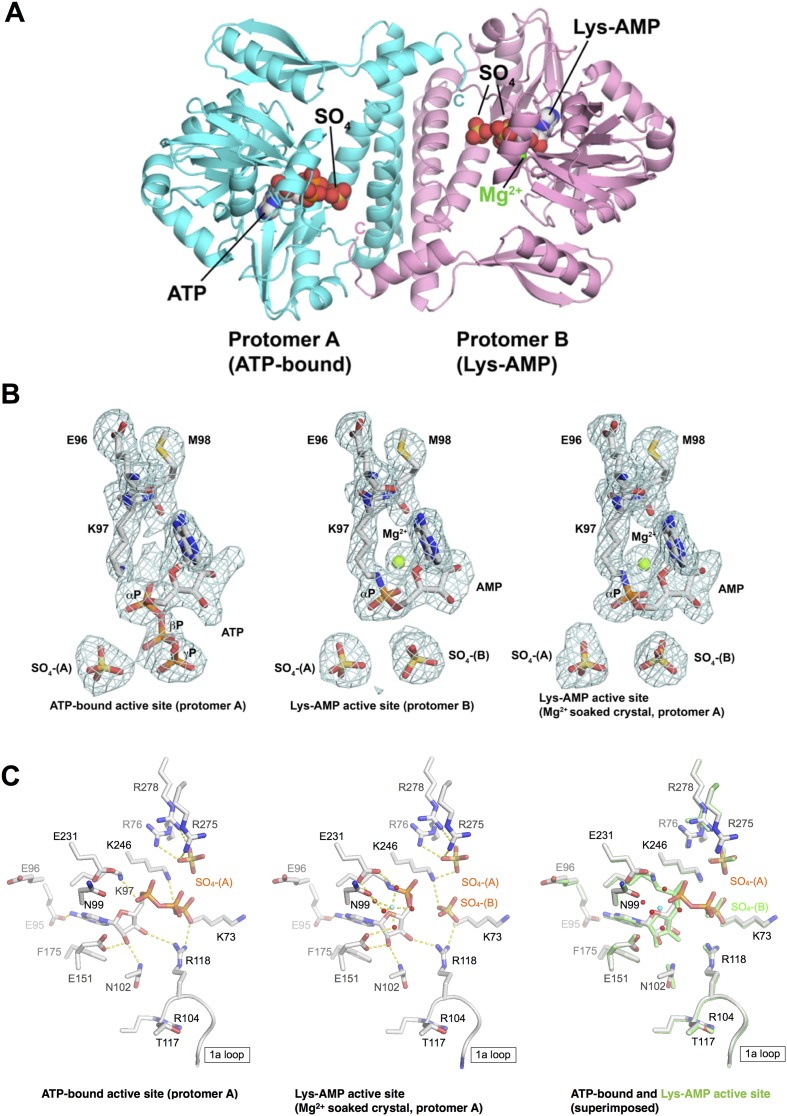
Figure 1.
(A) Structure of the MthRnl. Proteins are depicted as cartoon models and ATP, Lys-AMP, SO4 and Mg are depicted as spheres. A protomer A (cyan) contains ATP at active site while the promoter B (pink) contains adenylated Lys at active site. (B) Fo-Fc electron density maps (sigma cutoff = 3, likelihood-weighted omit maps) showing ATP-bound active site (left; protomer A from crystal 1), Lys-AMP active site (middle; protomer B from crystal 1) and Lys-AMP active site (right; promoter A form crystal 1). (C) ATP-binding pocket of protomer A of MthRnl•ATP bound complex (left), covalent MthRnl–AMP intermediate (middle), and superimposed structures of ATP-bound and covalent AMP intermediate (right). The covalent AMP intermediate structure on the right panel is shown in green. Amino acids, ATP/AMP and sulfate are shown as stick models. Atomic contacts between MthRnl and ATP/AMP are indicated by dashed lines. Mg is shown as cyan sphere. Waters in the metal coordination complex are shown in red spheres. The image was prepared with Pymol.
Recombinant MthRnl purified from bacteria comprises a mixture of apoenzyme and MthRnl–AMP intermediate (18). Interestingly, MthRnl is crystallized as ‘half-and-half’ containing one protomer having an ATP in the active site (protomer A), and the other protomer having an AMP covalently attached to the active site Lys-97 (protomer B) (Figure 1A and B). Soaking the MthRnl crystals in magnesium triggers the adenylylation reaction, leading to the formation of a covalent MthRnl-AMP intermediate and displacement of the pyrophosphate leaving group in protomer A (Figure 1B), and its crystal structure was determined at 2.2 Å resolution (see Materials and Methods and Table 1, form 2 crystal). The active site structure of protomer B remains the same before and after soaking Mg2+ into crystal. The electron density adjacent to the AMP phosphate was modeled as magnesium with respect to its octahedral coordination with surrounding water molecules. The overall structure and the nucleotide binding pocket of the ATP bound- and the AMP attached-MthRnl structures were nearly identical (∼0.48 Å root mean square deviations) without any specific local conformation difference, indicating that the MthRnl does not appear to undergo a conformational change during the step 1 reaction (Supplementary Figure S1A). The adenosine nucleoside adopts a syn conformation in both structures (Figure 1B).
The adenosine nucleotide binding pocket is located in the adenylyltransferase domain, which is composed of a cage of β-strands and intrastrand loops (Supplementary Figure S1). In Mg2+ free MthRnl crystal structure, Lys97 is ∼3.3 Å from the α-phosphate of ATP in protomer A, while the AMP moiety is covalently linked to Lys97 in protomer B. Other conserved side chains found in covalent nucleotidyltransferase motifs that make direct contact to the adenylate occupy equivalent positions in the MthRnl active site with respect to the position of the nucleotide (Figure 1C): (i) the aromatic ring of Phe175 makes stacking interactions with the adenine base; (ii) Glu151 coordinates the Mg2+ and engages with the 2′-O of ribose together with Asn102; (iii) Arg118 coordinates the γ-phosphate of ATP; (iv) Lys246 contacts the α- and γ-phosphates of the ATP. Glu95, which is conserved in archaeal ATP-dependent RNA ligase, makes contact with the 6-amino group of the adenine ring.
The instructive difference between the ATP- and AMP-bound structures is the position and the number of sulfates that co-crystallized with MthRnl. In the structure of the MthRnl-ATP (protomer A of Mg2+-free MthRnl crystal), a single well-ordered sulfate ion (SO4-A) was found within the nucleotide binding pocket coordinated by number of positive charged residues (Figure 1). In contrast, two sulfate ions (SO4-A and SO4-B) were present in the MthRnl–AMP structure (protomer B of Mg2+-free MthRnl crystal and both protomers A and B of Mg2+ soaked MthRnl crystal). SO4-A occupies the same position found in the ATP-bound form, while SO4-B is bound in the position previously occupied by the γ phosphate of the ATP. We infer that SO4-A and SO4-B found in the MthRnl structures mimic the position of the phosphate backbone of the RNA substrate during steps 2 and 3 reactions for the following reasons. First, the SO4-B position is identical to the 5′-PO4 at the DNA nick in the Chlorella virus DNA ligase structures (25,26). Second, SO4-B occupies an equivalent position to the β-phosphate of the AppN in the T4Rnl2-AppDNA complex (Supplementary Figure S1B). Third, the SO4-A, which is ∼6 Å away from the SO4-B, occupies the position of phosphate backbone between the first and second nucleotide of the polynucleotide substrate in the T4Rnl2-AppDNA complex. SO4-A is coordinated by Arg-76 and Lys-246, which also interact with the AMP phosphate and SO4-B, respectively, and Arg-275 and Arg-278, which protrude from the dimerization domain (Figure 1C). SO4-B is coordinated by Arg-118 located in motif Ia loop, Lys-246 in motif V and by Lys-73. Residues that make contacts to SO4-A and SO4-B identified from the MthRnl structures are conserved in other archaeal Rnl3-family ligases (Supplementary Figure S2) suggesting they play important roles in the ligation reaction.
Structure based mutagenesis of MthRnl
The crystal structure of MthRnl revealed 17 residues (Lys73, Arg76, Glu95, Glu96, Lys97, Asn99, Asn102, Arg104, T117, Arg118, Glu151, Phe175, Glu231, Lys246, Glu256, Arg275 and Arg278) in the nucleotide binding pocket (Figure 1C). We introduced alanine substitutions at each of these residues to assess their roles in the ligation reaction (Supplementary Figure S2). WT MthRnl and mutant MthRnl–Ala proteins were produced as His10-tagged derivatives in bacteria and then purified from soluble lysates by Ni-agarose chromatography (18). The 42-kDa MthRnl polypeptide was the predominant species detected by SDS-PAGE, and the extents of purification were comparable for mutant and WT enzymes (Supplementary Figure S3).
MthRnl purified from bacteria comprises a mixture of ligase apoenzyme and adenylated intermediate. Only the apoenzyme is available to react in vitro with ATP to form the ligase-AMP intermediate, which makes it difficult to evaluate the mutational effect on step 1 catalysis because the extent of reactive apoenzyme varied among the mutant enzyme preparations. For this reason, we measured the formation of the ligase–AMP intermediate for each mutant protein at various ATP concentrations (Table 2 and Supplementary Figure S4). As a criterion to evaluate the effect of mutations on step 1 catalysis, mutant proteins that failed to form any detectable ligase–AMP complex and do not form AppRNA in the ligation assay (described below), are classified as inert for step 1 adenylylation. Mutant proteins with decreased affinity for ATP of 20-fold compared to the WT are classified as severely defective. As expected, the K97A mutant failed to form ligase–AMP intermediate, consistent with the structural data showing that Lys97 is the site of covalent attachment. Similarly, N102A, E151A, E231A and K246A proteins were completely inert for step 1 adenylylation; no detectable complex was observed with up to 0.5 mM ATP (data not shown). R104A, R118A and F175A mutants formed traced amounts of ligase–AMP intermediates, with >40-fold decrease in affinity for ATP. R76A, E95A, E96A, N99A, E256A, R275A and R278A showed moderate effects on step 1 adenylylation (5- to 20-fold decrease in affinity for ATP) as compared to WT MthRnl, while K73A and T117A and had ATP affinity comparable to WT, consistent with the crystal structures showing that these residues do not directly interact with ATP.
Table 2. Effects of alanine mutations in the active site of MthRnl on adenylyltransferase activity.
| MthRnl Mutations | S50, ATP [μM] |
|---|---|
| WT | 0.21 ± 0.09 |
| K73A | 0.29 ± 0.04 |
| R76A | 3.65 ± 1.79 |
| E95A | 2.07 ± 0.59 |
| E96A | 2.16 ± 0.68 |
| K97A | ND |
| N99A | 2.00 ± 0.42 |
| N102A | ND |
| R104A | 12.24 ± 4.04 |
| T117A | 0.39 ± 0.09 |
| R118A | 8.46 ± 1.87 |
| E151A | ND |
| F175A | 11.68 ± 1.88 |
| E231A | ND |
| K246A | ND |
| E256A | 1.55 ± 0.23 |
| R275A | 1.65 ± 0.17 |
| R278A | 1.96 ± 0.32 |
The MthRnl1–Ala preparations were assayed for adenylyltransferase activity as described under Materials and Methods. Protein concentration used for the assay were 0.5 μM for wild-type, K73A, T117A and R278A proteins, and 2.0 μM for R76A, E95A, E96A, N99A, R104A, R118A, F175A, E256A and R275A proteins. Ligase-[32P]AMP formed was plotted as a function of ATP concentration and the S50 values were determined by plotting the data by non-linear regression to one-site binding model in Prism (Supplementary Figure S4). The data shown represent the average of three separate experiments with standard error. ND = not detected.
Mutational effects on RNA ligation
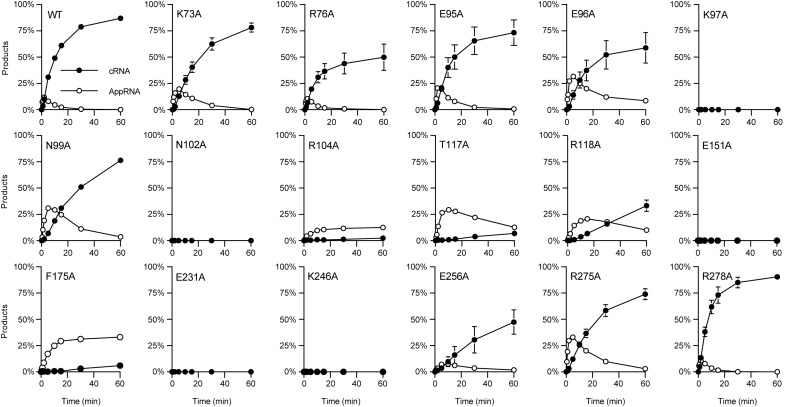
We subjected WT and mutant MthRnl to kinetic analysis of single-turnover RNA circularization in the absence of added ATP. The products were analyzed by denaturing PAGE, and the distributions of 32P-labeled RNAs [cRNA product, RNA-adenylate (AppRNA) intermediate, and residual pRNA substrate] were quantified by scanning the gels with a phosphorimager to determine the initial rate of pRNA ligation (Figure 2 and Supplementary Figure S5A). The initial rate of cRNA formation by the mutant proteins were compared to that for the WT protein. With WT MthRnl, AppRNA intermediate was detected at earlier time points (0.5–5 min) but the level of AppRNA declined to undetectable levels by 30 min, with concomitant decay of input 24-mer linear pRNA and formation of cRNA. Nearly 90% of input pRNA substrate was converted to cRNA by WT MthRnl in 60 min. The K97A, N102A, E151A, E231A and K246A mutants were completely inert in forming cRNA as well as AppRNA at all time points, consistent with their defect in step 1 adenylylation. The initial rate of RNA circularization by other mutant proteins were: R104A (<1% of compared to the wild-type enzyme), T117A (1%), R118A (5%) and F175A (1%). These mutant proteins also accumulated AppRNA during the course of the reaction, suggesting that they are likely defective in step 3 reaction. Mutations that showed moderate defects in circularizing pRNA include K73A (46%), E96A (45%), N99A (22%), E256A (16%) and R275A (40%). We note that the rate of RNA-adenylylation (sum of AppRNA plus cRNA formed over time) of K73A, E96A, N99A and R275A were similar to WT, implying that these mutations may specifically affect the step 3 sealing reaction. The R76A, E95A and R278A mutant proteins showed >60% activity of WT control.
Figure 2.
Effect of alanine mutations on RNA circularization. Overall RNA ligation assay with MthRnl specified was performed as described in Materials and Methods. The yield of cRNA product and AppRNA intermediate is plotted as a function of time. The data shown represent the average of three separate experiments. Standard error bars are included for each datum point.
Mutational effects on step 3 phosphodiester bond formation
A finer analysis to evaluate defects in the individual steps in the RNA ligation pathway is to assay for step 3 reaction in isolation using AppRNA as substrate. Reaction of WT MthRnl with AppRNA resulted in the appearance of a sealed cRNA (step 3 product) and a deadenylated pRNA generated by reversal of step 2 reaction (Figure 3 and Supplementary Figure S5). The cRNA formation is favored over AppRNA deadenylation (18). Approximately 25% of input AppRNA was deadenylated to form pRNA at early time points, but the level of pRNA declined after 5 min with a concomitant formation of cRNA under the conditions analyzed. The initial rate of AppRNA circularization in the step 3 isolation reaction was ∼2-fold faster than the initial rate for pRNA ligation reaction.
Figure 3.
Effect of alanine mutations on the phosphodiester bond formation on a preadenylated RNA. Step 3 sealing assay was performed with MthRnl specified as described in Materials and Methods. The yield of cRNA and deadenylated product (pRNA) is plotted as a function of time. The data shown represent the average of three separate experiments. Standard error bars are included for each datum point.
The E151A and E231A mutants, which were inert for step 1 and overall pRNA ligation, were unable to seal the AppRNA (Figure 3). Moreover, E151A and E231A were incapable of deadenylating AppRNA, suggesting that these glutamates also participate in step 2 catalysis. As noted previously, the K97A and K246A mutant proteins were as active for the step 3 reaction (20). However, the K97A mutant did not generate pRNA from AppRNA. A simple interpretation of this result is that the active site Lys97 plays a role in accepting AMP from AppRNA in the step 2 reverse reaction. In contrast, K246A was capable of deadenylating the AppRNA, albeit less efficiently then the WT, implying that Lys-246 may not be strictly required for the step 2 reaction. We also note that the initial rate of AppRNA circularization by K97A, K246A, as well as R278A, were slightly faster than the WT enzyme. The F175A and R104A proteins were severely defective for sealing AppRNA, consistent with the overall RNA ligation assay in which AppRNA accumulated. The N102A mutant protein, which was inert for step 1 adenylylation, can seal AppRNA, although the rate of phosphodiester bond synthesis was reduced to 7% of the wild-type enzyme. Similar to K246A, the N102A mutant protein could deadenylate the AppRNA.
Kinetic analysis of T117A and R118A in isolated step 3 reaction showed that these mutations cause MthRnl to preferentially deadenylate the AppRNA (Figure 3 and Supplementary Figure S5B). Remarkably, the rate of the reverse step 2 reaction by T117A was ∼250 times faster than the rate of phosphodiester bond formation. Nearly 70% of input AppRNA was converted to pRNA, while only 5% of input AppRNA was circularized in 60 min. The R118A substitution exerts a similar effect, as the initial rate of the reverse step 2 reaction was twice as fast as the rate of the step 3 reaction. These results suggest that the Thr117 and Arg118 side chains function in the step 3 reaction to promote phosphodiester bond synthesis.
Moderate defects in AppRNA ligation were observed for E95A, E96A, N99A and R275A. These mutant proteins appear to be selectively impaired in phosphodiester bond synthesis because the initial rate of AppRNA circularization in the isolated step 3 reaction was comparable to that for pRNA circularization (compare Figures 2 and 3). In contrast, the K73A and E256A substitutions did not significantly impact the step 3 reaction, which implies that the defect observed in the composite pRNA ligation assay is likely due to reduced step 2 activity. The R76A and R278A substitutions did not have a major effect on all three steps in the ligation reaction, which implies that these two arginines are dispensable for single-strand RNA circularization reaction. We conclude that Glu96, Asn99 and Arg275 side chains may contribute to step 3 phosphodiester bond formation, whereas Lys73 and Glu256 may participate in step 2 RNA adenylylation, consistent with the results obtained from the overall pRNA ligation assay.
To verify that pRNA generated during the isolated step 3 reaction was derived from deadenylation of AppRNA by MthRnl activity, and not by cleavage of AMP by a contaminating activity in the preparation or by an alternative mechanism, we incubated the WT and selected mutant MthRnl proteins with 32P-labeled AppRNA (radiolabel phosphate underlined), and visualized the covalent 32P protein–AMP formation (a product of step 1 reaction) by SDS-PAGE (Supplementary Figure S6). We verified that WT enzyme, as well as E95A, N102A, T117A, R118A and K246A mutant proteins, which were capable of generating pRNA from AppRNA, reacted with AppRNA to form a 32P-labeled protein–AMP complex. In contrast, K97A and E151A mutant proteins, which failed to generate pRNA in isolated step 3 reaction assay, did not form a 32P-labeled protein–AMP protein complex.
Defining the RNA binding surface of MthRnl
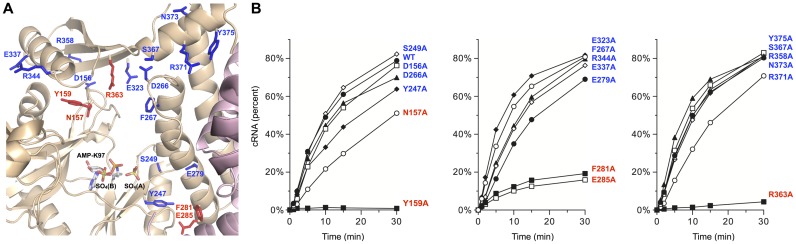
To probe for the nucleic acid binding surface on MthRnl, we conducted an alanine scan of 19 residues of the protein. We selected charged, hydrophilic or aromatic side chains located on the surface of the MthRnl as potential ligands for the phosphodiester backbone of single-stranded RNA (Figure 4A and Supplementary Figure S2). Mutant proteins were produced in Escherichia coli, purified as described above, and assayed for pRNA circularization activity (Supplementary Figure S3 and Figure 4B). Five mutant proteins displaying significant defects in ligation activity (as gauged by the initial rate of pRNA circularization respect to the WT initial rate) were: Y159A (<1%), R363A (2%), E285A (16%), F281A (20%) and N157A (35%). The D156A, E232A, S249A, Y247A, D266A, F267A, E279A, E337A, R344A, R358A, S367A, R371A, N373A and Y375A mutants displayed near WT ligation activity (within a factor of two).
Figure 4.
Structure-guided mutagenesis of MthRnl. (A) Residues on the surface of MthRnl shown were substituted by alanine. Position of active site lysine (K97) and sulfates are indicated. Residues that were important for RNA ligation activity are colored by red. (B) Kinetics of RNA circularization by WT and mutant MthRnl proteins. Kinetics analysis of single turnover pRNA ligation was performed as described in Materials and Methods. The yield of circular RNA product is plotted as a function of time. The data shown represent the average of two separate experiments.
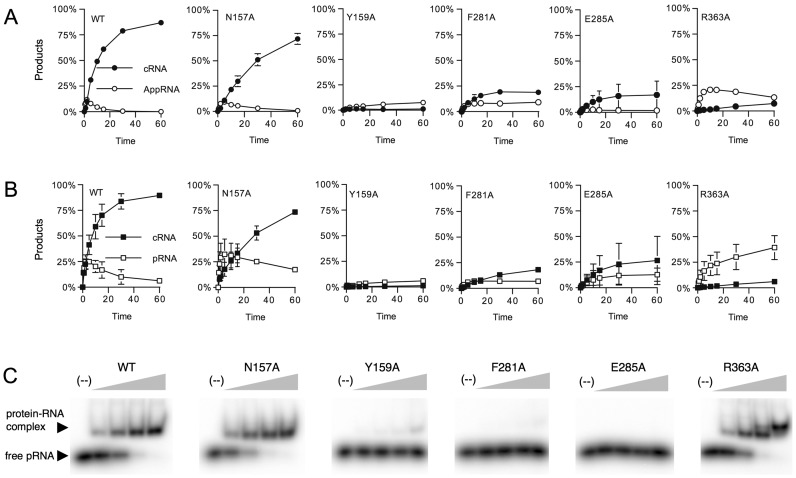
A finer kinetic analysis was performed for N157A, Y159A, F281A, E285A and R363A, to further evaluate their defects in ligation activity (Figure 5A and B). Y159A was virtually inert for step 2 and step 3 ligation reaction. This mutant protein failed to accumulate AppRNA in the overall pRNA ligation and did not circularize AppRNA in the isolated step 3 reaction. F281A and E285A appear to be defective in both step 2 and step 3 activities. R363A displayed WT rate for step 2 RNA adenylylation, but was specifically defective in step 3 reaction, as it accumulated high levels of AppRNA and slowly converted it to cRNA in the overall pRNA ligation. R363A preferentially deadenylates the input AppRNA, with nearly 40% of input AppRNA was converted to pRNA, while only 5% AppRNA was circularized. N157A shows a moderate defect in phosphodiester bond synthesis, as the initial rate of pRNA circularization was comparable to the initial rate of AppRNA circularization in the isolated step 3 reaction.
Figure 5.
Effect of N157A, Y159A, F281A, E285A and R363A on RNA ligation and RNA binding. (A) Overall RNA ligation assay was performed as described in Materials and Methods. The yield of circular RNA and AppRNA intermediate is plotted as a function of time. (B) Sealing of a preadenylated RNA. Step 3 sealing assay was performed as described in Materials and Methods. The yield of cRNA and pRNA is plotted as a function of time. The data shown represent the average of three separate experiments. Standard error bars are included for each datum point. (C) RNA binding. The extent of RNA-binding by wild-type and mutant MthRnl proteins was analyzed by gel mobility shift assay as described (18).
To further evaluate the defects in these mutant proteins, their binding affinity for single-stranded pRNA was measured by gel mobility shift assay (Figure 5C). The Y159A, F281A and E285A proteins showed significantly reduced binding, whereas N157A and R363A maintained strong binding to the pRNA forming RNA–protein complexes with properties comparable to WT MthRnl. We conclude that Tyr159, Phe281 and Glu285 likely participate in RNA recognition, which correlates with their defect in both step 2 and step 3 activities.
The quaternary structure of N157A, Y159A, F281A, E285A and R363A mutant proteins were assessed by sedimentation analysis through 15–30% glycerol gradients. Similar to the wild-type, all the above mutant proteins sediment between catalase and BSA marker proteins (Supplementary Figure S7). However, the Y159A (S value = 7.9), F281A (S value = 8.1) and E285A (S value = 7.9) proteins migrated slightly faster then the WT MthRnl (S value = 6.0). The observed sedimentation coefficient suggests that these mutant proteins may form a trimer or the substitution may have distorted the homodimeric structure.
DISCUSSION
The present study provides new insights to the structure and mechanism of archaeal ATP-dependent RNA ligase by capturing crystal structures of homodimeric MthRnl in complex with ATP and with AMP, which represents the state after step 1 catalysis and PPi product dissociation. The crystals were grown in the presence of ammonium sulfate, and the MthRnl•ATP complex contains one bound sulfate ion (SO4-A), whereas the MthRnl–AMP intermediate has additional sulfate ion (SO4-B) bound at the active site cleft. The distance between the two sulfur atoms is ∼6 Å, which is similar to the distance between two phosphorus atoms in single-stranded RNA. SO4-B probably occupies the binding site for the 5′-phosphate on pRNA (5′-pN1pN2) or AppRNA (AppN1pN2). We predict that SO4-A reflects the position of backbone phosphate on the RNA (5′-pN1pN2 or AppN1pN2) based on the structure of T4Rnl2 complex with adenylated nicked-duplex polynucleotide substrate (5)(Supplementary Figure S1).
To understand the mechanistic similarities with other members of RNA ligase families, and to define how Rnl3 recognizes its polynucleotide substrate, we employed kinetic analysis to assess the effects of alanine substitutions on individual steps in the ligation pathway. Within the active site pocket, alanine substitution at Lys97, Asn102, Arg104, Thr117, Arg118, Glu151, Phe175, Glu231 and Lys246 reduced the rate of ligation by at least an order of magnitude. Consistent with a previous report, the active site nucleophile Lys97 and the conserved motif V Lys246, are essential for step 1 ligase adenylylation, but not for step 3 sealing activity (20). MthRnl with alanine substituted at Lys97 also failed to deadenylate the AppRNA, which implies that Lys97 participates in the step 2 reaction. In contrast, K246A was capable of deadenylating the AppRNA to generate pRNA, which implies that this residue may not participate in the step 2 reaction. The sealing activity by K97A and K246A mutant proteins was slightly higher than the WT enzyme. Because the majority of the WT enzyme is pre-adenylated, which could preclude the binding of AppRNA adenylate to the active site and may suppress the formation of ligated circles. In contrast, the active site of K97A and K246A mutant proteins is free of AMP and may allow the adenylate on the AppRNA to be recognized more readily in the isolated step 3 reaction. Glu151 and Glu231, which coordinate the metal ion, are required for all three steps in the ligation reaction. Glu151 also makes contact with the adenylate ribose at the 2′-OH. Our results are also consistent with biochemical data indicating that divalent cation is required for all three ligation steps by ATP-dependent RNA ligases (9,14,27–30). Asn102, which contacts the 3′-OH of adenylate ribose in the MthRnl•ATP complex as well as in the MthRnl–AMP intermediate, is essential for step 1 ligase-adenylylation and showed reduced step 3 sealing activity, which suggests that it is important for adenylate recognition. Asn102 in motif 1 of archaeal ATP-dependent RNA ligases is shared between all Rnl2 members, but not with other family members of RNA ligases (as well as DNA ligases or mRNA capping enzymes). An equivalent alanine mutation in T4Rnl2 also showed reduced step 1 and step 3 activity (30), suggesting that Asn in motif 1 of MthRnl functions similarly as in T4Rnl2. Phe175 makes stacking interactions with the adenine base. Substitution of Phe175 with alanine reduced the affinity for ATP by 55-fold and was virtually inert for sealing AppRNA. The F175A mutant protein was capable for transferring AMP to pRNA to form AppRNA. Thus, this aromatic residue likely contributes to recognition of the adenine base to promote step 1 and step 3 catalysis, but is dispensable for step 2 reaction.
Arg118 and Thr117 are located between motifs I and III and form a loop structure designated as motif Ia, which is structurally conserved among all ATP-dependent RNA ligases. We show that motif Ia plays a critical role in promoting phosphodiester bond synthesis, as mutation within the loop structure favors reversal of step 2 reaction to deadenylate the input AppRNA. Arg118 makes contact with the 3′O-ribose of the adenylate and the γ-phosphate of ATP in the MthRnl•ATP structure. In the MthRnl–AMP complex, the later interaction is replaced with SO4-B, from which we infer that it represents the position of a 5′-PO4 on the polynucleotide substrate based on the structures of T4Rnl2 (contacted by Arg55), Chrolella virus DNA ligase–AMP intermediate (Arg42) and Human DNA ligase 1 (Arg589) (5,25–26,31). In the isolated step 3 ligation assay, the rate of AppRNA deadenylation was twice as fast as the rate of phosphodiester bond formation. Similar effects were observed for T4Rnl1, where substitution of the equivalent Arg54 predominantly results in deadenylation of AppRNA in the isolated step 3 reaction (28). We predict that Arg118 participates in releasing the pyrophosphate group from ATP during step 1 reaction, coordinates AMP on the enzyme and the 5′-PO4 of the RNA in the step 2 reaction, and assists in phosphodiester bond formation through interaction with the adenylate on the AppRNA, as proposed for Chrolella virus DNA ligase and T4Rnl2 (5,26). In contrast, Thr117 does not make direct contact with the ATP, which may explain why substitution of this residue does not affect step 1 adenylylation. Mutation of Thr117 exerts remarkable effects on the subsequent step in the ligation reaction; the rate of AppRNA deadenylation is ∼250-times faster than circularizing the AppRNA in the isolated step 3 reaction. It is conceivable that Thr117 contacts the 3′-OH end and may induce a conformational change on the RNA to align the reactive 3′-OH for phosphodiester bond formation. We hypothesize that lack of the Thr117 side chain may expose the 5′-adenylate to be deadenylated more readily than the WT because the reactive 3′-OH is not positioned correctly to promote the sealing reaction.
Arg104 in motif 1 is essential for all three steps in ligation. Arg104 does not contact the adenylate but forms a hydrogen bond with the main-chain carbonyl on Thr117. The defect observed for R104A could be attributed as an additive effect of T117A and R118A mutations: (i) The affinity of ATP for R104A was reduced to a similar extent as for R118A (reduced by 40- to 60-fold compared to the WT); (ii) The rates for RNA-adenylylation and phosphodiester bond formation by R104A were slower than for either T117A or R118A mutation alone. We speculate that the major contribution of Arg104 is to stabilize the motif Ia loop structure to maintain an active conformation of Thr117 and Arg118. These results are consistent with mutational analysis of T4Rnl1 (12). Lys117 of T4Rnl1 (equivalent of Arg104 in MthRnl) makes contacts with the main chain of Ser118 (equivalent of Thr117 in MthRnl) and Arg54 (equivalent of Arg 118 in MthRnl). The effect of K117A substitution on step 2 and step 3 reactions appeared to be much more severe than the S118A or R54A mutations in T4Rnl1 (12,28).
Tyr159 emerges from the mutational study as a residue required for interaction with one of the reactive RNA termini because Tyr159 makes no contacts with ATP, and alanine substitution abolished RNA binding. Tyr159 is situated on the surface of the protein in the vicinity of motif 1a loop (Figure 4A). Comparison of MthRnl and T4Rnl2 structures reveals that the position of Tyr159 in MthRnl is nearly identical to the position of Phe65-Phe66 dipeptide in T4Rnl2. The Phe65 of T4Rnl2 was shown to be essential for interaction with the 3′-OH acceptor strand (5). The Tyr159 side chain is ∼4 Å away from the Thr117 side chain. We speculate that Tyr159 and Thr117 may act together to coordinate the 3′-OH end of substrate. It is also conceivable that Tyr159 may stabilize the active conformation of the motif Ia loop, analogous to the role of Arg104, but from the opposite side of the loop.
Two conserved glutamates, Glu95 and Glu96, located N-terminal to the active site lysine, are highly conserved among the Rnl3 proteins. The Glu95 side chain makes contact with the 6-amino group of the adenine ring and elimination of this side chain shows minor defects in step 1 and step 3 activities. CthPnkp (Rnl4 family) also possesses glutamate at this position and makes an equivalent contact, but is not essential for adenylylation activity (9). Alanine substitution of the adjacent Glu96 resulted in reduced affinity for ATP and reduced sealing activity for AppRNA. The Glu96 side chain of MthRnl points away from the ATP and makes contact with the Arg204 and Arg230 side chains, which may allow the main-chain oxygen on Glu96 to hydrogen bond to N-6 of adenine base to stabilize the ATP and adenylate on AppRNA, as it is required for step1 and step 3 reactions. In T4Rnl2, an equivalent Glu has been proposed to stabilize the active site loop structure and is also required for step 1 and step 3 activity (30).
The Asn99 in the KxNG motif 1 is not essential for MthRnl ligase activity. An equivalent Asn in fungal tRNA ligase is also dispensable for tRNA splicing activity in vivo (32). In other RNA ligases, as well as DNA ligase and some members of the Rnl3 family, the equivalent position is occupied by either Asp (T4Rnl1 and DNA ligases, PabRnl3), His (T4Rnl2), Met (CthLig) or Ser (plant Trl1). Interestingly, in Chlorella virus DNA ligase, motif 1 Asp is essential for step 2 DNA adenylylation (33), while in T4Rnl2 motif 1 His is specifically required for the step 3 reaction (29,34).
Arg363, which is located near the C-terminal end of the protein, is required for the step 3 reaction. Elimination of the Arg363 side chain stimulated deadenylation of AppRNA, which phenocopies the effect observed in the Thr117 and Arg118 mutants. Arg363 is in close proximity of Asn157 and Tyr159, the later of which is required for RNA binding. We speculate that 3′-OH acceptor strand lies on the enzyme surface and is contacted by Thr117, Asn157, Tyr159 and Arg363 (Figures 1C and 4A). Similar to other RNA ligases, the specificity for the polynucleotide substrate for Rnl3 could be dictated by the C-terminal domain.
Conserved Phe281 and Glu285, situated at the edge of the dimerization domain, are also required for RNA binding and for efficient ligation (Figure 4A). Based on the position of these amino acids with respect to the active site and the SO4 molecules in the structures, we propose that Phe281 and Glu285 likely interact with the 5′-PO4 donor strand. Alternatively, these mutations may have distorted the dimeric structure of the enzyme and preclude RNA binding.
We and others have proposed that the dimer interface may serve as a platform for RNA binding because of its high positive charge potential on the dimer surface (7) and the deletion of dimerization domain abolish ligation and RNA binding (18). We attempted to produce a monomeric form of MthRnl by introducing a series of alanine substitutions within the dimer interface. However, the mutant protein aggregated in solution and therefore, we were unable to assess whether the dimeric structure contribute to the ligation activity (unpublished results).
Lys73, Arg76, Glu256, Arg 275 and Arg 278 are clustered near the sulfate molecules that co-crystallized in the active site pocket of the MthRnl (Figure 1C). These side chains may act to coordinate the ATP binding or 5′-PO4 donor strand of the RNA substrate. Replacement of Glu256 with Ala resulted in 5-fold decrease in step 2 RNA adenylylation, but had little effect on step 1 and step 3 catalysis. The Arg275 ala-substitution reduced the rate of the step 3 reaction by 4-fold without affecting step 2 RNA adenylylation. Glu256 may participate in recognition of 5′-PO4 donor strand in step 2 reaction, while Arg275 may assist in positioning the adenylate on the AppRNA prior to step 3 sealing reaction. Glu256 may also interact with Arg278, but this interaction does not appear to be important because replacing Arg278 with alanine did not significantly effect the ligation activity. Alanine substitution of Lys73 and Arg76 showed mild reduction of the rate of ligation. Lys73 and Arg76 are situated in close proximity to the SO4-A, but these residues may not directly recognize single-stranded RNA. However these residues may act on the physiological RNA substrate, which likely adopts a different structure than the RNA substrate used in our assay. The outstanding question concerning MthRnl is to determine the physiological RNA substrate and how homodimeric structure contributes to the ligation reaction.
ACCESSION NUMBERS
Coordinates for the MthRnl structures have been deposited in the Protein Data Bank under accession code 5D1O and 5D1P.
Supplementary Material
Acknowledgments
We thank Jinhee Kim and JeeEun Kim from SUNY Buffalo for their help with experiments.
Footnotes
Present address: Raka Ghosh, Department of Structural Biology and Bioinformatics, Indian Institute of Chemical Biology, Kolkata 32WB, India.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation [1050984 to C.K.H]; National Institutes of Health [R01GM087350 to K.S.M.]. Funding for open access charge: National Science Foundation. Funding for the Open Access publication charge was provided by National Science Foundation grant 1050984 [to C.K.H].
Conflict of interest statement. None declared.
REFERENCES
- 1.Uhlenbeck O.C., Gumport R.I. T4 RNA Ligase. In: Boyer PD, editor. The Enzymes. Vol. 15. NY: Academic Press; 1982. pp. 31–58. [Google Scholar]
- 2.Pascal J.M. DNA and RNA ligases: structural variations and shared mechanisms. Curr. Opin. Struct. Biol. 2008;18:96–105. doi: 10.1016/j.sbi.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 3.El Omari K., Ren J., Bird L.E., Bona M.K., Klarmann G., LeGrice S.F.J., Stammers D.K. Molecular architecture and ligand recognition determinants for T4 RNA ligase. J. Biol. Chem. 2006;281:1573–1579. doi: 10.1074/jbc.M509658200. [DOI] [PubMed] [Google Scholar]
- 4.Ho C.K., Wang L.K., Lima C.D., Shuman S. Structure and mechanism of RNA ligase. Structure. 2004;12:327–339. doi: 10.1016/j.str.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 5.Nandakumar J., Shuman S., Lima C.D. RNA ligase structures reveal the basis for RNA specificity and conformational changes that drive ligation forward. Cell. 2006;127:71–84. doi: 10.1016/j.cell.2006.08.038. [DOI] [PubMed] [Google Scholar]
- 6.Deng J., Schnaufer A., Salavati R., Stuart K.D., Hol W.G.J. High resolution crystal structure of a key editosome enzyme from Trypanosoma brucei: RNA editing ligase 1. J. Mol. Biol. 2004;343:601–613. doi: 10.1016/j.jmb.2004.08.041. [DOI] [PubMed] [Google Scholar]
- 7.Brooks M.A., Meslet-Cladiére L., Graille M., Kuhn J., Blondeau K., Myllykallio H., van Tilbeurgh H. The structure of an archaeal homodimeric ligase which has RNA circularization activity. Protein Sci. 2008;17:1336–1345. doi: 10.1110/ps.035493.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang P., Chan C.M., Christensen D., Zhang C., Selvadurai K., Huang R.H. Molecular basis of bacterial protein Hen1 activating the ligase activity of bacterial protein Pnkp for RNA repair. Proc. Natl. Acad. Sci. U.S.A. 2012;109:13248–13253. doi: 10.1073/pnas.1209805109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith P., Wang L.K., Nair P.A., Shuman S. The adenylyltransferase domain of bacterial Pnkp defines a unique RNA ligase family. Proc. Natl. Acad. Sci. U.S.A. 2012;109:2296–2301. doi: 10.1073/pnas.1116827109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Unciuleac M.C., Goldgur Y., Shuman S. Structure and two-metal mechanism of a eukaryal nick-sealing RNA ligase. Proc. Natl. Acad. Sci. U.S.A. 2015;112:13868–13873. doi: 10.1073/pnas.1516536112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shuman S., Lima C.D. The polynucleotide ligase and RNA capping enzyme superfamily of covalent nucleotidyltransferases. Curr. Opin. Struct. Biol. 2004;14:757–764. doi: 10.1016/j.sbi.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 12.Wang L.K., Nandakumar J., Schwer B., Shuman S. The C-terminal domain of T4 RNA ligase 1 confers specificity for tRNA repair. RNA. 2007;13:1235–1244. doi: 10.1261/rna.591807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nandakumar J., Ho C.K., Lima C.D., Shuman S. RNA substrate specificity and structure-guided mutational analysis of bacteriophage T4 RNA ligase 2. J. Biol. Chem. 2004;279:31337–31347. doi: 10.1074/jbc.M402394200. [DOI] [PubMed] [Google Scholar]
- 14.Raymond A., Shuman S. Deinococcus radiodurans RNA ligase exemplifies a novel ligase clade with a distinctive N-terminal module that is important for 5′-PO4 nick sealing and ligase adenylylation but dispensable for phosphodiester formation at an adenylylated nick. Nucleic Acids Res. 2007;35:839–849. doi: 10.1093/nar/gkl1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Unciuleac M.-C., Shuman S. Characterization of a novel eukaryal nick-sealing RNA ligase from Naegleria gruberi. RNA. 2015;21:824–832. doi: 10.1261/rna.049197.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan C.M., Zhou C., Huang R.H. Reconstituting bacterial RNA repair and modification in vitro. Science. 2009;326:247. doi: 10.1126/science.1179480. [DOI] [PubMed] [Google Scholar]
- 17.Jain R., Shuman S. Bacterial Hen1 is a 3′ terminal RNA ribose 2′-O-methyltransferase component of a bacterial RNA repair cassette. RNA. 2010;16:316–323. doi: 10.1261/rna.1926510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torchia C., Takagi Y., Ho C.K. Archaeal RNA ligase is a homodimeric protein that catalyzes intramolecular ligation of single-stranded RNA and DNA. Nucleic Acids Res. 2008;36:6218–6227. doi: 10.1093/nar/gkn602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhelkovsky A.M., McReynolds L.A. Polynucleotide 3′-terminal phosphate modifications by RNA and DNA ligases. J. Biol. Chem. 2014;289:33608–33616. doi: 10.1074/jbc.M114.612929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhelkovsky A.M., McReynolds L.A. Structure-function analysis of Methanobacterium thermoautotrophicum RNA ligase - engineering a thermostable ATP independent enzyme. BMC Mol. Biol. 2012;13:24. doi: 10.1186/1471-2199-13-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhelkovsky A.M., McReynolds L.A. Simple and efficient synthesis of 5′ pre-adenylated DNA using thermostable RNA ligase. Nucleic Acids Res. 2011;39:e117. doi: 10.1093/nar/gkr544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 23.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 24.Afonine P.V., Mustyakimov M., Grosse-Kunstleve R.W., Moriarty N.W., Langan P., Adams P.D. Joint X-ray and neutron refinement with phenix.refine. Acta Crystallogr. D. 2010;66:1153–1163. doi: 10.1107/S0907444910026582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nair P.A., Nandakumar J., Smith P., Odell M., Lima C.D., Shuman S. Structural basis for nick recognition by a minimal pluripotent DNA ligase. Nat. Struct. Mol. Biol. 2007;14:770–778. doi: 10.1038/nsmb1266. [DOI] [PubMed] [Google Scholar]
- 26.Odell M., Sriskanda V., Shuman S., Nikolov D.B. Crystal structure of eukaryotic DNA ligase-adenylate illuminates the mechanism of nick sensing and strand joining. Mol. Cell. 2000;6:1183–1193. doi: 10.1016/s1097-2765(00)00115-5. [DOI] [PubMed] [Google Scholar]
- 27.Wang L.K., Schwer B., Shuman S. Structure-guided mutational analysis of T4 RNA ligase 1. RNA. 2006;12:2126–2134. doi: 10.1261/rna.271706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang L.K., Ho C.K., Pei Y., Shuman S. Mutational analysis of bacteriophage T4 RNA ligase 1. Different functional groups are required for the nucleotidyl transfer and phosphodiester bond formation steps of the ligation reaction. J. Biol. Chem. 2003;278:29454–29462. doi: 10.1074/jbc.M304320200. [DOI] [PubMed] [Google Scholar]
- 29.Ho C.K., Shuman S. Bacteriophage T4 RNA ligase 2 (gp24.1) exemplifies a family of RNA ligases found in all phylogenetic domains. Proc. Natl. Acad. Sci. U.S.A. 2002;99:12709–12714. doi: 10.1073/pnas.192184699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yin S., Ho C.K., Shuman S. Structure-function analysis of T4 RNA ligase 2. J. Biol. Chem. 2003;278:17601–17608. doi: 10.1074/jbc.M300817200. [DOI] [PubMed] [Google Scholar]
- 31.Pascal J.M., O'Brien P.J., Tomkinson A.E., Ellenberger T. Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature. 2004;432:473–478. doi: 10.1038/nature03082. [DOI] [PubMed] [Google Scholar]
- 32.Sawaya R., Schwer B., Shuman S. Genetic and biochemical analysis of the functional domains of yeast tRNA ligase. J. Biol. Chem. 2003;278:43928–43938. doi: 10.1074/jbc.M307839200. [DOI] [PubMed] [Google Scholar]
- 33.Odell M., Shuman S. Footprinting of Chlorella virus DNA ligase bound at a nick in duplex DNA. J. Biol. Chem. 1999;274:14032–14039. doi: 10.1074/jbc.274.20.14032. [DOI] [PubMed] [Google Scholar]
- 34.Nandakumar J., Shuman S. Dual mechanisms whereby a broken RNA end assists the catalysis of its repair by T4 RNA ligase 2. J. Biol. Chem. 2005;280:23484–23489. doi: 10.1074/jbc.M500831200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.