Abstract
BACKGROUND
Peripartum cardiomyopathy shares some clinical features with idiopathic dilated cardiomyopathy, a disorder caused by mutations in more than 40 genes, including TTN, which encodes the sarcomere protein titin.
METHODS
In 172 women with peripartum cardiomyopathy, we sequenced 43 genes with variants that have been associated with dilated cardiomyopathy. We compared the prevalence of different variant types (nonsense, frameshift, and splicing) in these women with the prevalence of such variants in persons with dilated cardiomyopathy and with population controls.
RESULTS
We identified 26 distinct, rare truncating variants in eight genes among women with peripartum cardiomyopathy. The prevalence of truncating variants (26 in 172 [15%]) was significantly higher than that in a reference population of 60,706 persons (4.7%, P = 1.3×10−7) but was similar to that in a cohort of patients with dilated cardiomyopathy (55 of 332 patients [17%], P = 0.81). Two thirds of identified truncating variants were in TTN, as seen in 10% of the patients and in 1.4% of the reference population (P = 2.7×10−10); almost all TTN variants were located in the titin A-band. Seven of the TTN truncating variants were previously reported in patients with idiopathic dilated cardiomyopathy. In a clinically well-characterized cohort of 83 women with peripartum cardiomyopathy, the presence of TTN truncating variants was significantly correlated with a lower ejection fraction at 1-year follow-up (P = 0.005).
CONCLUSIONS
The distribution of truncating variants in a large series of women with peripartum cardiomyopathy was remarkably similar to that found in patients with idiopathic dilated cardiomyopathy. TTN truncating variants were the most prevalent genetic predisposition in each disorder.
Peripartum cardiomyopathy is marked by the development of maternal systolic heart failure late in pregnancy or early in the postpartum period.1,2 The incidence varies from 1 in 100 to 1 in 300 in geographic hot spots, including Nigeria and Haiti, to 1 in 1000 to 1 in 4000 in Europe and the United States. The strongest known risk factors are the presence of preeclampsia, twin gestation, and advanced maternal age. Among patients with peripartum cardiomyopathy, heart failure can resolve but often does not: rates of death of 5 to 10% are common, and 4% of cardiac transplantations in the United States among women are performed for the treatment of peripartum cardiomyopathy.
The cause of peripartum cardiomyopathy remains unknown. Hypotheses include fetal autoimmunity or microchimerism, myocarditis, and dietary excess of salt or deficiency of selenium.1–3 Previous studies have suggested that peripartum cardiomyopathy is largely a vascular disease, triggered by the hormonal milieu of late gestation and the early postpartum period.3–5 There are no clear explanations for why heart failure develops in only a small subgroup of women in these contexts.
Peripartum cardiomyopathy shares some clinical features with idiopathic dilated cardiomyopathy, including decreased systolic function, enlarged cardiac dimensions, and nonspecific histologic findings on biopsy. Mutations in a number of genes have been shown to cause idiopathic dilated cardiomyopathy. These genes include TTN, which encodes the sarcomere protein titin. Up to 25% of patients with familial dilated cardiomyopathy and 18% of those with sporadic dilated cardiomyopathy harbor deleterious truncating variants (i.e., variants that are predicted to result in the truncation of translation) in TTN.6
Some evidence supports the hypothesis that peripartum cardiomyopathy may have a hereditary or genetic component. Geographic hot spots of incidence, including Nigeria and Haiti, may reflect genetic factors, and a genome wide association study identified a locus near the gene encoding parathyroid hormone–like hormone (PTHLH).7 Although hot spots could be environmental, familial clustering of peripartum cardiomyopathy has been noted,8–11 and 15% of patients in a German cohort had a family history of cardiomyopathy (defined as the presence of peripartum cardiomyopathy, dilated cardiomyopathy, sudden death, or arrhythmias in a first-degree relative).12 Two groups of investigators who have studied rare pedigrees affected by these two types of cardiomyopathies have identified variants in genes encoding myofibrillar proteins, including TTN.13,14 However, very few cases of peripartum cardiomyopathy are clearly familial or associated with dilated cardiomyopathy. A genetic underpinning to the great majority of cases of peripartum cardiomyopathy remains uncertain.
We therefore sequenced DNA from 172 women with peripartum cardiomyopathy to determine the contribution of variants in 43 genes that have been previously associated with dilated cardiomyopathy.
METHODS
PATIENTS
Patients with peripartum cardiomyopathy were recruited from six independent groups. Group A was recruited from a cohort of patients who underwent either cardiac transplantation or placement of a left ventricular assist device (LVAD) at Temple University in Philadelphia. Group B was recruited from a cohort of patients that included some who had been referred for cardiac transplantation and who were being treated in a clinic at the University of Pennsylvania in Philadelphia. Group C was recruited from a cohort of patients who had been referred to a peripartum cardiomyopathy clinic at the University of Hannover in Hannover, Germany. Group D consisted of a subgroup of 9 women from a cohort of 100 Japanese women with peripartum cardiomyopathy from whom samples were obtained.15 Group E consisted of patients in the multicenter, prospective Intervention in Myocarditis and Acute Cardiomyopathy 2 (IMAC-2) study,16 which enrolled patients with acute nonischemic cardiomyopathy, including a subgroup with peripartum cardiomyopathy. Group F consisted of patients who were enrolled in the Investigations in Pregnancy Associated Cardiomyopathy (IPAC) trial, a multicenter, prospective study involving women with peripartum cardiomyopathy.17,18 The study was approved by the institutional review board at each study center. All the patients provided written informed consent.
Reference groups were taken from the Exome Aggregation Consortium (ExAC) in Cambridge, Massachusetts (http://exac.broadinstitute.org), which contains more than 60,000 exomes, and the Exome Variant Server (data release, ESP6500SI-V2; evs.gs.washington.edu/EVS), which contains exomic sequences from 6503 persons. For comparison, patients with dilated cardiomyopathy were recruited at the Royal Brompton and Harefield NHS Foundation Trust in London, as described previously.19
DNA SEQUENCING AND ANALYSES
We constructed bar-coded sequencing libraries from genomic DNA obtained from 172 patients with peripartum cardiomyopathy and 332 patients with dilated cardiomyopathy. The libraries were enriched for protein-coding portions of 43 genes that when mutated cause dilated cardiomyopathy and were then sequenced (Table S1 in the Supplementary Appendix, available with the full text of this article at NEJM.org). The 43 genes comprise the great majority of genes known to be associated with dilated cardiomyopathy and were present on the two platforms that were used to sequence patients with peripartum cardiomyopathy and those with dilated cardiomyopathy.
For the peripartum-cardiomyopathy libraries, genes were captured with the use of SureSelect Target Enrichment (Agilent Technologies) and sequenced on the HiSeq 2500 sequencing system (Illumina). Reads were aligned to hg19 (GRCh37) with the use of NovoAlign, version 3.02.04 (Novocraft), with measurements set for full Needleman– Wunsch alignments and a gap penalty of 10. Variants were identified with the use of the Genome Analysis Tool Kit (GATK) Haplotype-Caller tool on the basis of GATK Best Practices.20 Variants were annotated with the use of SnpEff and GRCh37.68.21 In samples obtained from patients with dilated cardiomyopathy, genes were captured with the use of in-solution hybridization and sequenced on the SOLiD 5500XL (Applied Biosystems). Reads were demultiplexed, optimized with the use of the SOLiD Accuracy Enhancement Tool, and aligned to hg19 with the use of the SOLiD targeted resequencing pipeline. Variants were identified with the use of Life-Scope genomic analysis software, version 2.5.1, with default measurements, and annotated with the use of the Ensembl Variant Effect Predictor (API version 75 on GRCh37).22
STATISTICAL ANALYSIS
We used either Fisher’s exact test (for two-tailed analyses) or Pearson’s chi-square test of association for cross-cohort and cross-group analyses. The frequency of truncating variations was compared between cohorts with the use of the binomial test. Analyses were performed with the use of the R statistical package.
RESULTS
STUDY PATIENTS
A total of 172 women with peripartum cardiomyopathy were recruited from the six cohorts (Table 1). Approximately one third of the patients were of African descent, consistent with the known increased prevalence of peripartum cardiomyopathy in this group. Ancestry was defined genetically by means of principle-component analysis of all common variants that were sequenced (Fig. S1 in the Supplementary Appendix). The ejection fraction was lowest in group A, which consisted of patients who were under evaluation for cardiac transplantation.
Table 1.
Demographic Characteristics of the Patients in Each Cohort and the Prevalence of Truncating Variants at Baseline.*
| Characteristic | Group A (N = 10) |
Group B (N = 26) |
Group C (N = 10) |
Group D (N = 9) |
Group E (N = 34) |
Group F (N = 83) |
All Patients (N = 172) |
|---|---|---|---|---|---|---|---|
| Temple University |
University of Pennsylvania |
University of Hannover, Germany |
Japan | IMAC-2 | IPAC | ||
| Age — yr | 34.2±7.6 | 34.1±7.4 | 34.3±6.7 | 30.8±3.4 | 31.2±6.8 | 29.8±6.3 | 31.3±6.7 |
| African descent — no. (%)† | 5 (50) | 16 (62) | 5 (50) | 0 | 11 (32) | 24 (29) | 61 (35) |
| Left ventricular ejection fraction — % | 10.0±3.9 | 30.1±13.5 | 27.6±10.8 | 29.1±10.1 | 27.2±7.4 | 29.8±9.7 | 28.6±10.4 |
| Patients with truncating variants — no. (%) | |||||||
| Any | 2 (20) | 1 (4) | 1 (10) | 1 (11) | 6 (18) | 15 (18) | 26 (15) |
| TTN | 0 | 0 | 1 (10) | 1 (11) | 4 (12) | 11 (13) | 17 (10) |
Plus–minus values are means ±SD. IMAC-2 denotes Intervention in Myocarditis and Acute Cardiomyopathy 2, and IPAC Investigations in Pregnancy Associated Cardiomyopathy.
Ancestry was defined genetically by means of principle-component analysis of all common variants that were sequenced.
DNA SEQUENCING
Next-generation sequencing was performed on 43 genes, including TTN (Table S1 in the Supplementary Appendix). More than 95% of targeted bases were sequenced to a read depth of more than 20 times (data not shown). Rare variants (ExAC frequency, <0.1%) were chosen for further analysis. We focused on truncating variants that included nonsense, frameshift, and splicing variants, because they are predicted to have a strong effect on protein structure and function. All truncating variants were confirmed by means of traditional Sanger sequencing.
GENETIC VARIATION IN PERIPARTUM CARDIOMYOPATHY
Among the 172 women with peripartum cardiomyopathy, we identified 26 who carried 26 distinct rare heterozygous truncating variants in eight different genes (Table S2 in the Supplementary Appendix). No homozygous or compound heterozygous truncating variants were observed. Eleven variants were nonsense, seven were frameshift, and eight affected canonical splicing sites. The prevalence of truncating variants did not differ significantly among the six cohorts (Table 1). The overall prevalence of truncating variants in 26 of 172 women with peripartum cardiomyopathy (15%) was significantly higher than that in the ExAC reference population of more than 60,000 samples (4.7%, P = 1.3×10−7) and similar to that in a cohort of patients with dilated cardiomyopathy (55 of 332 patients [17%], P = 0.81).
Of the 26 truncating variants, 17 (65%) affected TTN (in 10% of the cohort; P = 2.7×10−10 for the comparison with the reference population). TTN truncating variants were seen in 8 of 61 women of African descent (13%) and 8 of 102 women of European descent (8%). In the reference ExAC population, such variants were found in 2.1% of persons of African descent (P = 3.8×10−5) and in 1.1% of those of European descent (P = 1.4×10−5). Four of the TTN truncating variants (two nonsense and two splice-site donors) were identical to variants previously identified in 26 patients with dilated cardiomyopathy who were studied at Brigham and Women’s Hospital in Boston (Table S3 in the Supplementary Appendix). Three of these variants were absent from the more than 60,000 exomes in the ExAC database, and one was identified only once. Five of the TTN truncating variants (in 3% of the 172 women) are also identical to variants annotated as probably pathogenic for dilated cardiomyopathy in the ClinVar database (www.ncbi.nlm.nih.gov/clinvar), as compared with a frequency of 0.1% found in ExAC (P = 1.1×10−5).
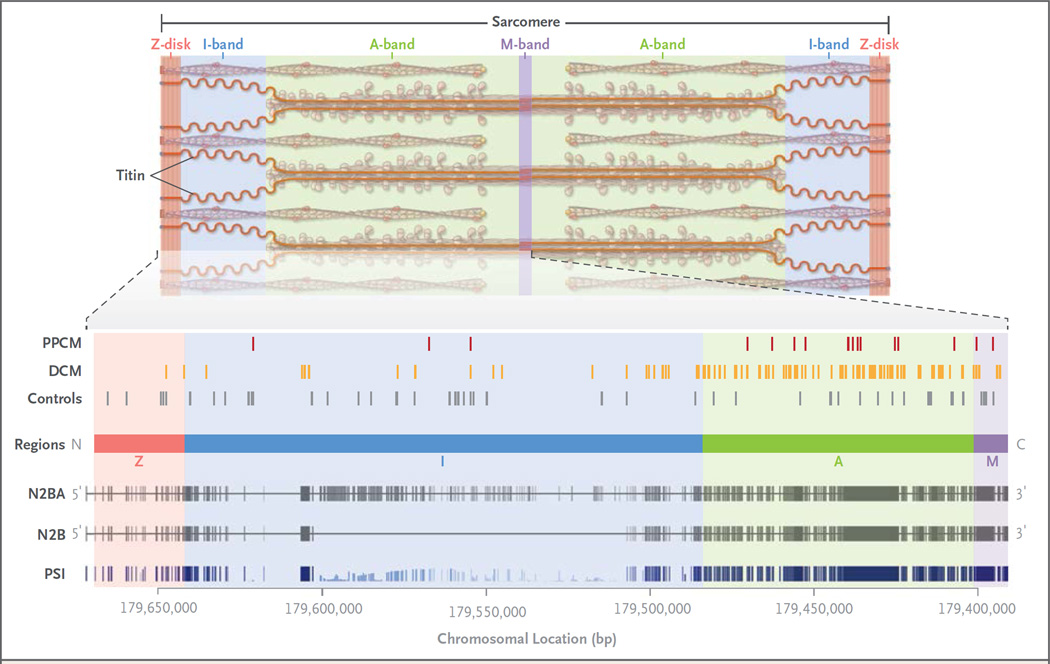
Of 17 truncating variants in TTN, 14 were located in a region that encodes the A-band portion of the protein (Fig. 1, and Table S3 in the Supplementary Appendix), as compared with 21 of 56 in a reference population19 (P = 0.002). (The A-band, named for its anisotropic properties on polarized microscopy, is the portion of the sarcomere that contains the myosin thick filament.) Truncating variants in patients with dilated cardiomyopathy cluster similarly in the region encoding the A-band.6,19 Sixteen of the 17 TTN truncating variants were in exons that are constitutively expressed in the heart (i.e., that appear in every messenger RNA isoform that is transcribed from the gene) (Fig. 1, and Table S3 in the Supplementary Appendix), as compared with 83 of 168 variants in the Exome Sequencing Project (P = 1.1×10−4). This finding again reflects the pattern seen in patients with dilated cardiomyopathy.19 Truncating variants were more likely to be positioned in the region of the gene encoding the C-terminal regions of the protein (Fig. 1). In persons with dilated cardiomyopathy, truncating variants that are found near the C-terminus are associated with more severe disease than are N-terminal truncating variants.19
Figure 1. Titin Protein and Spatial Distribution of Variants in the Protein.
Titin, a protein encoded by TTN, makes up one of the three major filaments of the cardiac sarcomere, the basic unit of striated muscle tissue. Regions of the sarcomere are designated as the Z-disk (red), I-band (named for its isotropic properties under a polarizing microscope, shown in blue), A-band (named for its anisotropic properties, shown in green), and M-band (from the German mittelscheibe, the disk in the middle of the sarcomere, shown in purple). The spatial distributions of the truncating variants that were found in samples obtained from patients with peripartum cardiomyopathy (PPCM) and dilated cardiomyopathy (DCM) are indicated, along with the distributions of such variants in healthy controls. At the bottom of the diagram, the genomic locus of TTN is shown. N2BA and N2B denote the exons (vertical lines) encoding the two main cardiac transcripts. For PSI (i.e., the proportion spliced in), the height of the vertical line indicates the proportion of cardiac transcripts obtained from patients with dilated cardiomyopathy that contain the exon. Images are adapted from Herman et al.6 and Roberts et al.19
The prevalence of missense variants among the 172 women with peripartum cardiomyopathy was high (1.06 per person among women of African descent and 1.25 per person among those of European descent) and was not significantly different from the burden found in the ExAc cohort (1.26 per person among women of African descent [P = 0.21] and 1.19 per person among those of European descent [P = 0.59]). No missense variants that were annotated in ClinVar as pathogenic or probably pathogenic for dilated cardiomyopathy were identified among the women with peripartum cardiomyopathy. Three missense variants that are associated in ClinVar with hypertrophic cardiomyopathy were identified in MYH7 (encoding a myosin heavy-chain isoform expressed mainly in the heart) in the 172 women with peripartum cardiomyopathy (P = 0.10 for the comparison with the reference population), and seven missense variants reported in association with the long-QT syndrome were identified in SCN5A (encoding a voltage-gated sodium-channel isoform) in these women (P = 0.02 for the comparison with the reference population).
CLINICAL CHARACTERISTICS
At baseline, there were no significant differences with respect to age, ancestry, or ejection fraction between the women with truncating variants and those without such variants (Table 2). In the IPAC study,17,18 enrollment took place within 13 weeks after delivery, which was followed by comprehensive clinical evaluation during 1-year follow-up. Of 83 women in the IPAC study, 15 (18%) carried truncating variants, including 11 (13%) who had such variants in TTN. During the index pregnancy, there were no significant differences between the women with truncating variants and those without such variants with respect to age, the number of previous pregnancies, or the number of previous births (Table 3). The reported prevalence of a family history of peripartum or dilated cardiomyopathy was similarly low in the two study groups (10%). Fewer patients with TTN truncating variants had hypertension than did those without such variants (P = 0.009). The burden of TTN truncating variants among the women without hypertension (10 of 43 [23%]) was significantly higher than that among those with hypertension (1 of 40 [2%]) (P = 0.005).
Table 2.
Age, Ancestry, and Ejection Fraction among 172 Patients with Peripartum Cardiomyopathy, According to Variant Status at Baseline.*
| Variable | No Variant (N = 146) |
TTN Variant (N = 17) |
P Value | Any Variant (N = 26) |
P Value |
|---|---|---|---|---|---|
| Age — yr | 31±7 | 29±6 | 0.23 | 30±7 | 0.42 |
| African descent — no. (%) | 48 (33) | 8 (47) | 0.28 | 13 (50) | 0.12 |
| Ejection fraction — % | 29±10 | 26±9 | 0.25 | 25±10 | 0.12 |
Plus–minus values are means ±SD.
Table 3.
Clinical Characteristics of the Patients with Peripartum Cardiomyopathy in the IPAC Study, According to Variant Status.*
| Characteristic | No Truncating Variant (N = 68) |
TTN Truncating Variant (N = 11) |
P Value |
|---|---|---|---|
| Age — yr | 30±6 | 28±6 | 0.25 |
| No. of pregnancies | 2.8±1.9 | 2.9±2.3 | 0.84 |
| No. of births | 2.1±1.2 | 2.1±1.5 | 0.92 |
| Family history of cardiomyopathy — no. (%) | 7 (10) | 1 (9) | 1.00 |
| Hypertension — no. (%) | 35 (51) | 1 (9) | 0.009 |
| Twin gestation — no. (%) | 15 (22) | 1 (9) | 0.45 |
| Ejection fraction — % | |||
| At enrollment | 35±9 | 30±12 | 0.14 |
| At 1 yr | 54±8 | 44±17 | 0.005† |
Plus–minus values are means ±SD.
P = 0.04 by the Wilcoxon rank-sum test.
In the IPAC study, cardiac function at presentation was not significantly different between the women with TTN truncating variants and those without such variants (mean [±SD] left ventricular ejection fraction, 30±12% and 35±9%, respectively; P = 0.14). However, a year after enrollment, the ejection fraction was significantly lower in the group with TTN truncating variants than in those without such variants (44±17 and 54±8%, P = 0.005). The majority of TTN truncating variants in the IPAC cohort (7 of 11) were found among women of African descent. At the 1-year follow-up, significant differences in the ejection fraction were observed within this subgroup of women (38±16% with TTN variants vs. 52±12% without TTN variants, P = 0.04) but not in the smaller number of women of European descent (56±17% vs. 55±10%, P = 0.75). There were no significant between-group differences in the incidence of LVAD placement, cardiac transplantation, or death, although the total number of such events was small (data not shown).
In the IMAC-2 study,16 4 of the women (3 of European descent) carried TTN truncating variants (Table 4). Within this smaller cohort, no significant differences were seen in the ejection fraction at enrollment or at 6-month follow-up. Follow-up echocardiography at 12 months was not available.
Table 4.
Clinical Characteristics of the Patients with Peripartum Cardiomyopathy in the IMAC-2 Study, According to Variant Status.*
| Characteristic | No Truncating Variant (N = 26) |
TTN Truncating Variant (N = 4) |
P Value |
|---|---|---|---|
| Age — yr | 31±7 | 31±5 | 0.94 |
| No. of pregnancies | 2.5±1.5 | 1.5±0.6 | 0.20 |
| No. of births | 2.0±0.9 | 1.5±0.6 | 0.31 |
| Family history of cardiomyopathy — no. (%) | 3 (12) | 2 (50) | 0.12 |
| Ejection fraction — % | |||
| At enrollment | 27±8 | 27±3 | 0.95 |
| At 6 mo | 45±14 | 51±11 | 0.51 |
Plus–minus values are means ±SD.
DISCUSSION
We identified truncating variants in eight genes among 26 of 172 women with peripartum cardiomyopathy. The prevalence of truncating variants in these genes was significantly higher among these women (15%) than in the reference group (P = 1.3×10−7), and most of the variants occurred in TTN. We conclude that many of these truncating variants lead to a strong genetic predisposition to peripartum cardiomyopathy. However, the presence of the more common variants probably is not a risk factor for penetrant peripartum cardiomyopathy in isolation, although such variants may be risk alleles for the disease.
The burden of genetic variants that were found in the women with peripartum cardiomyopathy resembled that found in patients with dilated cardiomyopathy. Most notably, 65% of variants occurred in TTN (in 10% of the patients, P = 2.7×10−10 for the comparison with the reference population). The great majority of these variants occurred in constitutively expressed exons and in the region encoding the A-band, akin to the distribution seen in patients with dilated cardiomyopathy.6,23 Seven of these variants have been found previously in association with dilated cardiomyopathy. Approximately 15% of patients with dilated cardiomyopathy and peripartum cardiomyopathy share the same types of truncating variants, so we propose that a shared mechanism is responsible for these cardiomyopathies. Since a gene-based diagnosis is clinically available for dilated cardiomyopathy, it is plausible that the same genetic diagnosis could be used for peripartum cardiomyopathy with similar sensitivity and specificity. However, further study is needed to understand the penetrance of variants identified in the context of peripartum cardiomyopathy.
Reliable prognostic indicators for peripartum cardiomyopathy are currently lacking. In a predefined, prospectively followed, and well-characterized subgroup of 83 women in the IPAC study, we found that the ejection fraction at 1-year follow-up was significantly lower among women with TTN truncating variants than among those without such truncating variants (P = 0.005). Dilated cardiomyopathy similarly carries a worse prognosis in the presence of truncating TTN variants.19 On the other hand, at 6-month follow-up of the 34 women in the IMAC-2 cohort, the ejection fraction was not significantly different among the 3 women with TTN truncating variants than among those without such variants. Although these data are suggestive, the value of genetic information in determining the prognosis for patients with peripartum cardiomyopathy will require further studies.
Truncating variants occurred equally among patients who did not report a family history of cardiomyopathy and among those who did. Similarly, dilated cardiomyopathy with causative TTN variants is frequently not familial.6 Of the 26 truncating variants, 13 (including 8 in TTN) were found in women of African heritage, which indicates that a genetic cause of peripartum cardiomyopathy is not exclusive to one ancestry. Peripartum cardiomyopathy occurs more frequently and has a poorer prognosis among women of African descent than among those of European descent,24 which may be due in part to the higher prevalence of TTN truncating variants among those of African descent.
Preeclampsia and gestational hypertension are strong risk factors for peripartum cardiomyopathy, but there has been controversy as to whether cardiomyopathy that is associated with hypertension may represent a separate disease from cardiomyopathy in the absence of hypertensive disorders. Among 15 women with truncating variants in the well-characterized IPAC cohort, only 4 had any form of hypertension (chronic or gestational), and among the 11 women with TTN truncating variants, only 1 had hypertension. This finding contrasts sharply with the overall 47% prevalence of hypertension in the IPAC cohort and most other cohorts.25 The prevalences of preeclampsia and hypertension are higher among women of African descent, and yet none of the 7 women of African descent with a TTN truncating variant had a hypertensive disorder, whereas 15 of the 17 women of African descent with peripartum cardiomyopathy who did not carry TTN variants had hypertension (P<0.001). Overall, the burden of truncating TTN variants among women without hypertension was 10 times as high as that among women with hypertension (23% vs. 2%, P = 0.005), a burden that approximates that found in familial dilated cardiomyopathy (25%).6 These post hoc analyses suggest that peripartum cardiomyopathy that is associated with hypertension may reflect a different pathophysiologic process than that in the absence of hypertension and that peripartum cardiomyopathy in the absence of risk factors such as hypertension may be of genetic origin.
Two women had truncating variants in DMD or LAMP, genes that lie on the X chromosome. Mutations in these genes cause Duchenne’s muscular dystrophy and Danon’s disease, respectively, in male patients and more rarely in female patients. Cardiomyopathy is a prominent feature of the two diseases.26 Peripartum cardiomyopathy may have occurred in these two women as a consequence of skewed X-chromosome inactivation27 or stresses of pregnancy superimposed on subclinical cardiac abnormalities. Two previous reports identified a DMD or LAMP2 mutation in a patient with peripartum cardiomyopathy.28,29
Previous studies have shown that peripartum hormonal changes can pose a vascular insult to the heart and trigger peripartum cardiomyopathy.4,5,30,31 What predisposes the development of this disease in only a small subgroup of women in this context remains unclear. We identified a potential genetic predisposition to peripartum cardiomyopathy in approximately 15% of women in our study, owing to truncating variants primarily affecting TTN. There must be additional environmental or genetic stimuli that explain why the average age at onset of dilated cardiomyopathy for women with TTN truncating variants is 65 years,6 whereas the age at onset of peripartum cardiomyopathy among women with TTN truncating variants is 28 years. In addition, women with peripartum cardiomyopathy recover systolic cardiac function more frequently than do women with dilated cardiomyopathy. Thus, defining the mechanistic interaction between truncations in TTN and late gestational antivascular insults will probably lead to further understanding of both peripartum cardiomyopathy and dilated cardiomyopathy.
We conclude that peripartum cardiomyopathy shares a genetic predisposition with both familial and sporadic idiopathic dilated cardiomyopathy. In addition, the presence of truncating variants in TTN is the most prevalent genetic predisposition for each of these disorders.
Supplementary Material
Acknowledgments
Supported by grants from the National Institutes of Health (HL094499, to Dr. Arany; HL080494, to Dr. J.G. Seidman; HL88577, to Dr. Cappola; and HL102429, to Dr. McNamara), the Fondation Leducq (to Drs. Ware, C.E. Seidman, and J.G. Seidman), the National Institute for Health Research Cardiovascular Biomedical Research Unit at Royal Brompton and Harefield NHS Foundation Trust and Imperial College London (to Dr. Ware), Regenerative Biology and Reconstructive Therapies (REBIRTH) 2 Cluster of Excellence and Bundesministerium für Bildung und Forschung (to Dr. Hilfiker-Kleiner), and Howard Hughes Medical Institute (to Dr. C.E. Seidman).
APPENDIX
The authors’ affiliations are as follows: the Department of Genetics, Harvard Medical School (J.S.W., E.M., C.M.Y., C.E.S., J.G.S.), the Howard Hughes Medical Institute (C.E.S.), and the Cardiovascular Division, Brigham and Women’s Hospital (J.S.W., E.M., C.E.S., J.G.S.) — all in Boston; the Cardiovascular Institute and the Department of Medicine, Perelman School of Medicine, University of Pennsylvania, Philadelphia (J.L., T.D., T.P.C., Z.A.), the Heart and Vascular Institute, University of Pittsburgh Medical Center, Pittsburgh (I.H., J.P., K.H.-Y., J.G., D.M.M.), and Penn State Hershey Medical Center, Hershey (J.B.) — all in Pennsylvania; the National Institute for Health Research Royal Brompton Cardiovascular Biomedical Research Unit (J.S.W., F.M., S.K.P.) and the National Heart and Lung Institute (J.S.W., F.M., S.A.C., S.K.P.), Imperial College London, London; the Division of Cardiology, Department of Medicine, College of Physicians and Surgeons, Columbia University, New York (E.J.T.), and the University of Rochester, Rochester (J.A.) — both in New York; the Department of Cardiology and Angiology, Hannover Medical School, Hannover, Germany (D.H.-K.); the Department of Perinatology and Gynecology, the National Cerebral and Cardiovascular Center, Osaka, Japan (C.A.K.); the National Heart Center and Duke–National University of Singapore, Singapore (S.A.C.); the Intermountain Medical Center, Murray, Utah (R.A.); Vanderbilt University, Nashville (J.D.); Cleveland Clinic, Cleveland (E.H.); University of Southern California, Los Angeles (U.E.); McGill University and Jewish General Hospital, Montreal (R.S.), University of Calgary, Calgary, AB (A.K.), and University of Toronto, Toronto (P.L.) — all in Canada; University of Maryland, College Park (G.R.), and Johns Hopkins Hospital, Baltimore (I.S.W.) — both in Maryland; Morristown Hospital, Morristown (J.S.), and Newark Beth Israel Medical Center, Newark (M.J.Z.) — both in New Jersey; Truman Medical Center, University of Missouri, Kansas City (D.F.P.); and Wake Forest University Baptist Medical Center, Winston-Salem, NC (V.T.).
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
REFERENCES
- 1.Elkayam U. Clinical characteristics of peripartum cardiomyopathy in the United States: diagnosis, prognosis, and management. J Am Coll Cardiol. 2011;58:659–670. doi: 10.1016/j.jacc.2011.03.047. [DOI] [PubMed] [Google Scholar]
- 2.Sliwa K, Fett J, Elkayam U. Peripartum cardiomyopathy. Lancet. 2006;368:687–693. doi: 10.1016/S0140-6736(06)69253-2. [DOI] [PubMed] [Google Scholar]
- 3.Hilfiker-Kleiner D, Sliwa K. Pathophysiology and epidemiology of peripartum cardiomyopathy. Nat Rev Cardiol. 2014;11:364–370. doi: 10.1038/nrcardio.2014.37. [DOI] [PubMed] [Google Scholar]
- 4.Patten IS, Rana S, Shahul S, et al. Cardiac angiogenic imbalance leads to peripartum cardiomyopathy. Nature. 2012;485:333–338. doi: 10.1038/nature11040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hilfiker-Kleiner D, Kaminski K, Podewski E, et al. A cathepsin D-cleaved 16 kDa form of prolactin mediates postpartum cardiomyopathy. Cell. 2007;128:589–600. doi: 10.1016/j.cell.2006.12.036. [DOI] [PubMed] [Google Scholar]
- 6.Herman DS, Lam L, Taylor MR, et al. Truncations of titin causing dilated cardiomyopathy. N Engl J Med. 2012;366:619–628. doi: 10.1056/NEJMoa1110186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horne BD, Rasmusson KD, Alharethi R, et al. Genome-wide significance and replication of the chromosome 12p11.22 locus near the PTHLH gene for peripartum cardiomyopathy. Circ Cardiovasc Genet. 2011;4:359–366. doi: 10.1161/CIRCGENETICS.110.959205. [DOI] [PubMed] [Google Scholar]
- 8.Ntusi NB, Wonkam A, Shaboodien G, Badri M, Mayosi BM. Frequency and clinical genetics of familial dilated cardiomyopathy in Cape Town: implications for the evaluation of patients with unexplained cardiomyopathy. S Afr Med J. 2011;101:394–398. [PubMed] [Google Scholar]
- 9.Massad LS, Reiss CK, Mutch DG, Haskel EJ. Familial peripartum cardiomyopathy after molar pregnancy. Obstet Gynecol. 1993;81:886–888. [PubMed] [Google Scholar]
- 10.Pearl W. Familial occurrence of peripartum cardiomyopathy. Am Heart J. 1995;129:421–422. doi: 10.1016/0002-8703(95)90032-2. [DOI] [PubMed] [Google Scholar]
- 11.Pierce JA, Price BO, Joyce JW. Familial occurrence of postpartal heart failure. Arch Intern Med. 1963;111:651–655. doi: 10.1001/archinte.1963.03620290117016. [DOI] [PubMed] [Google Scholar]
- 12.Haghikia A, Podewski E, Libhaber E, et al. Phenotyping and outcome on contemporary management in a German cohort of patients with peripartum cardiomyopathy. Basic Res Cardiol. 2013;108:366. doi: 10.1007/s00395-013-0366-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Spaendonck-Zwarts KY, Posafalvi A, van den Berg MP, et al. Titin gene mutations are common in families with both peripartum cardiomyopathy and dilated cardiomyopathy. Eur Heart J. 2014;35:2165–2173. doi: 10.1093/eurheartj/ehu050. [DOI] [PubMed] [Google Scholar]
- 14.Morales A, Painter T, Li R, et al. Rare variant mutations in pregnancy-associated or peripartum cardiomyopathy. Circulation. 2010;121:2176–2182. doi: 10.1161/CIRCULATIONAHA.109.931220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamiya CA, Kitakaze M, Ishibashi-Ueda H, et al. Different characteristics of peripartum cardiomyopathy between patients complicated with and without hypertensive disorders: results from the Japanese nationwide survey of peripartum cardiomyopathy. Circ J. 2011;75:1975–1981. doi: 10.1253/circj.cj-10-1214. [DOI] [PubMed] [Google Scholar]
- 16.McNamara DM, Starling RC, Cooper LT, et al. Clinical and demographic predictors of outcomes in recent onset dilated cardiomyopathy: results of the IMAC (Intervention in Myocarditis and Acute Cardiomyopathy)-2 study. J Am Coll Cardiol. 2011;58:1112–1118. doi: 10.1016/j.jacc.2011.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McNamara D, Damp J, Elkayam U, Hsich E, Ewald G, Cooper L. Myocardial recovery at six months in peripartum cardiomyopathy: results of the NHLBI multicenter IPAC study. Circulation. 2013;128:A12898. abstract. [Google Scholar]
- 18.McNamara DM, Elkayam U, Alharethi R, et al. Clinical outcomes for peripartum cardiomyopathy in North America: results of the Investigations of Pregnancy Associated Cardiomyopathy (IPAC) study. J Am Coll Cardiol. 2015;66:905–914. doi: 10.1016/j.jacc.2015.06.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roberts AM, Ware JS, Herman DS, et al. Integrated allelic, transcriptional, and phenomic dissection of the cardiac effects of titin truncations in health and disease. Sci Transl Med. 2015;7:270ra6. doi: 10.1126/scitranslmed.3010134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van der Auwera GA, Carneiro MO, Hartl C, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;11:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cingolani P, Platts A, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pugh TJ, Kelly MA, Gowrisankar S, et al. The landscape of genetic variation in dilated cardiomyopathy as surveyed by clinical DNA sequencing. Genet Med. 2014;16:601–608. doi: 10.1038/gim.2013.204. [DOI] [PubMed] [Google Scholar]
- 24.Goland S, Modi K, Hatamizadeh P, Elkayam U. Differences in clinical profile of African-American women with peripartum cardiomyopathy in the United States. J Card Fail. 2013;19:214–218. doi: 10.1016/j.cardfail.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 25.Bello N, Rendon IS, Arany Z. The relationship between pre-eclampsia and peripartum cardiomyopathy: a systematic review and meta-analysis. J Am Coll Cardiol. 2013;62:1715–1723. doi: 10.1016/j.jacc.2013.08.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arad M, Maron BJ, Gorham JM, et al. Glycogen storage diseases presenting as hypertrophic cardiomyopathy. N Engl J Med. 2005;352:362–372. doi: 10.1056/NEJMoa033349. [DOI] [PubMed] [Google Scholar]
- 27.Giliberto F, Radic CP, Luce L, Ferreiro V, de Brasi C, Szijan I. Symptomatic female carriers of Duchenne muscular dystrophy (DMD): genetic and clinical characterization. J Neurol Sci. 2014;336:36–41. doi: 10.1016/j.jns.2013.09.036. [DOI] [PubMed] [Google Scholar]
- 28.Cheng VE, Prior DL. Peripartum cardiomyopathy in a previously asymptomatic carrier of Duchenne muscular dystrophy. Heart Lung Circ. 2013;22:677–681. doi: 10.1016/j.hlc.2012.11.015. [DOI] [PubMed] [Google Scholar]
- 29.Toib A, Grange DK, Kozel BA, Ewald GA, White FV, Canter CE. Distinct clinical and histopathological presentations of Danon cardiomyopathy in young women. J Am Coll Cardiol. 2010;55:408–410. doi: 10.1016/j.jacc.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 30.Bello NA, Arany Z. Molecular mechanisms of peripartum cardiomyopathy: avascular/hormonal hypothesis. Trends Cardiovasc Med. 2015;25:499–504. doi: 10.1016/j.tcm.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Halkein J, Tabruyn SP, Ricke-Hoch M, et al. MicroRNA-146a is a therapeutic target and biomarker for peripartum cardiomyopathy. J Clin Invest. 2013;123:2143–2154. doi: 10.1172/JCI64365. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.