Abstract
Centromeres form specialized chromatin structures termed kinetochores which are required for accurate segregation of chromosomes. DNA lesions might disrupt protein-DNA interactions, thereby compromising segregation and genome stability. We show that yeast centromeres are heavily resistant to removal of UV-induced DNA lesions by two different repair systems, photolyase and nucleotide excision repair. Repair resistance persists in G1- and G2/M-arrested cells. Efficient repair was obtained only by disruption of the kinetochore structure in a ndc10-1 mutant, but not in cse4-1 and cbf1Δ mutants. Moreover, UV photofootprinting and DNA repair footprinting showed that centromere proteins cover about 120 bp of the centromere elements CDEII and CDEIII, including 20 bp of flanking CDEIII. Thus, DNA lesions do not appear to disrupt protein-DNA interactions in the centromere. Maintaining a stable kinetochore structure seems to be more important for the cell than immediate removal of DNA lesions. It is conceivable that centromeres are repaired by postreplication repair pathways.
Centromeres form specialized chromatin structures, termed kinetochores, which are required for accurate segregation of chromosomes during mitosis and meiosis. Mutations in yeast centromere sequences (CEN) and centromere-associated proteins result in chromosome instability and missegregation. Consequently, DNA lesions generated by intra- and extracellular DNA-damaging agents such as UV light might disrupt protein-DNA interactions at the centromere, thereby compromising segregation and genome stability.
Centromere sequences are not conserved among different organisms, but conservation of proteins, including a centromere-specific histone H3 variant (CENP-A; Cse4 in the yeast Saccharomyces cerevisiae), implies that centromere structure and function are epigenetically determined by formation of a specialized chromatin structure (8, 70).
The budding yeast centromere is by far the simplest and most dissected one (6, 30, 50). Centromere DNA is about 125 bp long and subdivided into three DNA elements: CDEI, an A+T-rich DNA CDEII (76 to 84 bp), and CDEIII (10, 17). Those elements associate with several protein complexes. Cbf1/Cpf1 binds to CDEI (31) and bends DNA (36). CDEIII is a binding site for the Cbf3 complex, which consists of Ndc10, Cep3, Ctf13, and Skp1 (20). Cse4, together with the histones H4, H2A, and H2B, forms a centromere-specific nucleosome (34). Cse4 has about 65% similarity to the histone H3 fold domain but contains a specialized N-terminal domain, responsible for kinetochore protein recruitment (7, 51). Mif2, a protein with similarity to metazoan CENP-C, interacts with Cbf1 (33). Beyond those complexes forming the inner kinetochore, additional components build up the central and outer kinetochores (6, 8, 30). Recent observations showed that the yeast heterochromatin protein Sir1p is also a component of centromeres and possibly attracts the histone deposition factor CAF-I to centromeric chromatin (46).
The yeast kinetochore complex produces nuclease footprints of about 150 to 250 bp (5, 11). Similar footprints were observed in cells arrested at different positions of the cell cycle, suggesting that a stable structure is present throughout the cell cycle (71). No high-resolution data that characterize the extension and actual size of the kinetochore footprint are available.
DNA is continuously damaged by intra- and extracellular DNA-damaging agents. DNA lesions may arrest the cell cycle, block transcription and replication, interfere with protein binding, and lead to mutations, cell death, and cancer (18). Cylcobutane pyrimidine dimers (CPDs) and pyrimidine-6-4-pyrimidone photoproducts (6-4PPs) are the two major classes of stable DNA lesions (pyrimidine dimers [PDs]) generated by UV light. In most organisms, CPDs and 6-4PPs are repaired by nucleotide excision repair (NER), which is a multistep pathway including damage recognition, excision of an oligonucleotide with the damage, and DNA repair synthesis (18, 39). In addition to NER, many organisms express CPD- or 6-4PP- specific photolyases, which bind to the photoproduct and revert the damage in a light-dependent reaction (photoreactivation [PR]) (43).
Both DNA repair mechanisms are modulated by chromatin structures at all levels, from individual protein-DNA interactions to nucleosomes and higher-order heterochromatin (23, 49, 58). Moreover, repair of UV lesions is modulated by transcription. NER preferentially repairs the transcribed strand of genes transcribed by RNA polymerase II, which is referred to as transcription-coupled repair (15, 18). Photolyase, on the other hand, is inhibited by RNA polymerases stalled at CPDs, which leads to slow repair of the transcribed strand (1, 24, 53). Hence, the modulation of repair allows conclusions to be drawn on the accessibility of DNA in chromatin (the repair footprint).
Mutations within centromere DNA can lead to chromosome loss (27). Therefore, UV lesions in centromere DNA represent a unique type of damage, as loss of function could result in catastrophic loss of the genetic material of an entire chromosome. Previous work reported that UV lesions were efficiently removed from the centromere of chromosome III by NER (41). In contrast to that report, we observed a strong inhibition of NER and PR in centromeres, which suggested that a stable kinetochore complex is more important than immediate repair. Moreover, UV photofootprinting and DNA repair footprinting provide novel information on the size and structure of the centromere complexes.
MATERIALS AND METHODS
Yeast strains and media.
We used the following strains: UCC510 (MATa ade2-101 his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ1 ura3-52 URA3 at telomere V), provided by D. Gottschling (40); RRY3 (same as UCC510, but rad1Δ::kanMX) (23); CCY2 (same as UCC510, but bar1Δ::LEU2); AHY6 (MATa ndc10-1ts leu2-del1 lys2-801 trp1-1 ura3-52) and AHY666 (MATa ade2-101 his3-del200 lys2-801 trp1-del63 ura3-53 leu2-3,112 cse4-1) provided by A. Hyman; T952 [W303.1a, but scc1::SCC1myc18(TRP1) cbf1Δ::HIS3sp], provided by T. Tanaka; and 7428 (MATa ura3 cdc20Δ::LEU2 his3 GAL-CDC20::TRP1), provided by K. Nasmyth (22). The following media were used: YPD (1% Bacto Yeast Extract, 2% Bacto Peptone, 2% dextrose), YPG (1% Bacto Yeast Extract, 2% Bacto Peptone, 2% galactose), and SD (synthetic minimal medium, 0.67% Bacto Yeast nitrogen base without amino acids 2% dextrose) (47).
Chromatin analysis with MNase.
Chromatin analysis with micrococcal nuclease (MNase) was done as previously described (59).
UV irradiation and repair.
The repair experiments were done as previously described (53, 54). Cells were grown in YPD to a density of ∼3 × 107 cells/ml, harvested, and resuspended in SD to a density of about 3 × 107 cells/ml. Aliquots were transferred to plastic trays and irradiated with UV light with germicidal lamps (Sylvania G15 T8 bulbs) at 1 mW/cm2 (measured with a radiometer; UVP Inc., San Gabriel, Calif.). After irradiation, the medium was supplemented with the appropriate amino acids. Aliquots were either photoreactivated using Sylvania F15 T8/BLB bulbs (emission peak, 366 nm) at 1.5 mW/cm2 for 7 to 120 min or incubated in the dark for NER. The cells were collected and chilled on ice. DNA was purified using QIAGEN Genomic Tips 500/G and dissolved in 10 mM Tris-HCl-1 mM EDTA (pH 8.0). All steps from UV irradiation to DNA extraction were done at the appropriate temperature and in yellow light (Sylvania GE Gold fluorescent light) to prevent undesired PR.
Repair experiments with G1-arrested cells.
CCY2 cells were incubated with α-factor (1.5 μg/ml) in YPD for 2 h, resuspended in SD containing 1.5 μg of α-factor/ml, and irradiated with 150 J of UV light/m2. For NER, the culture was supplemented with the appropriate amino acids and incubated in the dark. All steps were done at 30°C.
Repair experiments with G2/M-arrested cells.
G2/M arrest was achieved by synchronization of strain 7428 (Gal-Cdc20) with α-factor (2.6 μg/ml) in YPG for 3 h, followed by transfer of the cells to YPD for 4 h. Cells were harvested, resuspended in SD, irradiated with 150 J of UV light/m2, and incubated for NER as described above. All steps were done at 30°C. Flow cytometric analysis of the DNA content was done as described previously (35).
Analysis of CPDs by indirect end labeling.
CPDs were mapped by indirect end labeling and quantified as described previously (53, 54). Genomic DNA was cut with the appropriate restriction enzymes (legends to Fig. 1 and 3) and purified by phenol extractions. Aliquots were incubated for 2 h at 37°C with T4 endonuclease V (T4-Endo V; Epicentre) in 50 mM Tris-5 mM EDTA (pH 7.5) or mock treated with the same buffer. The DNA was electrophoresed on 1.5% alkaline agarose gels, blotted to Zeta GT nylon membranes (Bio-Rad), and hybridized with radioactively labeled strand-specific DNA probes. The membranes were analyzed and quantified with a PhosphorImager (Molecular Dynamics). The positions of DNA lesions were always determined using a size marker (256-bp intervals) and verified based on the sequence specificity of UV damage formation. The CPD content (CPDs per top strand and CPDs per bottom strand) was calculated using the Poisson expression, −ln(RFa/RFb), where RFa and RFb represent the signal intensities of the intact restriction fragment of the T4-Endo V- and mock-treated DNA, respectively (29). Region-specific damage was calculated as the signal of that region in the T4-Endo V-treated DNA and divided by the signal of the whole lane. The corresponding signal of the mock-treated DNA was subtracted as background. To generate repair curves, the values were normalized with respect to the initial damage (0 min, 100%).
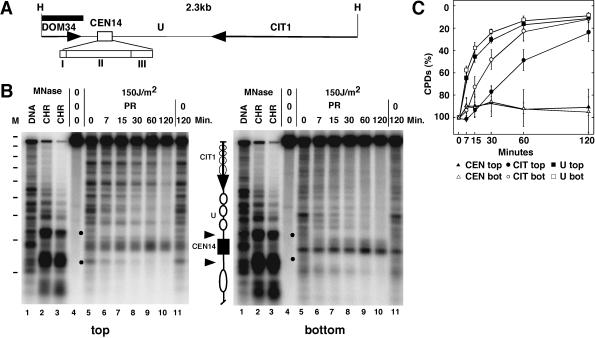
FIG. 1.
Inhibition of CPD repair by photolyase in the centromere of chromosome XIV. (A) Illustration of the 2.3-kb HindIII restriction fragment containing the centromere (CEN14) with the three sequence elements CDEI, CDEII, and CDEIII (inserts I, II, and III), a flanking region (U), parts of two genes (DOM34 and CIT1 [arrows]). (B) Comparison of chromatin structure (lanes 1 to 3) with repair of UV-induced CPDs by photolyase (lanes 4 to 11). For repair analysis, RRY3 (rad1Δ) cells were irradiated with 150 J/m2 of UV light (lanes 5 to 11), exposed to photoreactivating light for 0 to 120 min (lanes 5 to 10), or kept in the dark for NER (lanes 11). The DNA was purified, cut with HindIII, cut at CPDs with T4-Endo V, fractionated on alkaline agarose gels, blotted to nylon membranes, hybridized to strand-specific probes (350 nucleotides) (black bar in panel A), and exposed to PhosphorImager screens (top, top strand; bottom, bottom strand). For chromatin analysis, DNA (lanes 1) and chromatin (lanes 2 and 3) were digested with different amounts of MNase. The DNA was purified and cut with HindIII before being loaded on the agarose gel. Dashes in column M, positions of size markers (intervals of 256 bp). A schematic drawing illustrates the positions of CEN14 (black box), nuclease-hypersensitive regions (arrowheads), the nucleosomal region U with positioned nucleosomes (white ellipses), and CIT1 (arrow) with unknown nucleosome positions (faint circles). Dots, rapidly repaired CPDs. (C) Quantitative analysis of repair in the centromere (CEN), the nucleosomal region (U), and CIT1. The curves show the percentage of CPDs in a region at the indicated repair time in minutes. The bottom strand of CIT1 is the transcribed strand. The data represent averages ± standard deviations of results for four gels from two experiments.
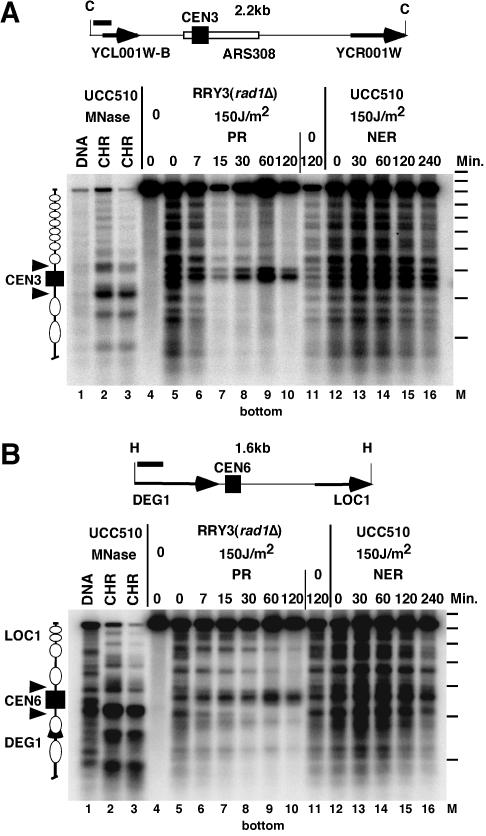
FIG. 3.
Inhibition of repair in CEN3 and CEN6. (A) Schematic drawing and repair data for CEN3. Indicated are the hybridization probe (260 nucleotides; black bar), the ClaI restriction fragment (C) containing the centromere (CEN3), ARS308, and parts of two open reading frames (YCL001W-B andYCR001W; arrows). (B) Schematic drawing and repair data for CEN6. Indicated are the hybridization probe (318 nucleotides; black bar), the HindIII restriction fragment (H) containing the centromere (CEN6), and parts of two genes (DEG1 and LOC1; arrows). Repair data (gels in panels A and B) were obtained as described in the legends to Fig. 1 and 2. Lanes 1 to 3, chromatin analysis; lanes 4 to 16, CPD analysis (cut with T4-Endo V). In the schematic drawing, white ellipses indicate positioned nucleosomes; a gap between nucleosomes indicates the presumed open promoter of LOC1 (panel B). Dashes in column M, positions of size markers (intervals of 256 bp).
Strand-specific probes were generated by primer extension with small DNA fragments as templates, strand-specific primers, [α-32P]CTP, and Taq polymerase (QIAGEN) for 30 cycles. DNA templates were generated by PCR from whole cells or genomic DNA. The oligonucleotides used for PCR and primer extension were CC/CEN14-A, 5′-ccgcGCTTGGTATGGTGAAAAAGAGG-3′; CC/CEN14-B, 5′-gctgGGAAGAAGTAAAGAGAATAATCC-3′; CC/CEN6-A, 5′-GCTTGATGGCTCAAAACAAAATTAC-3′; CC/CEN6-B, 5′-GCCTTATAGTCATCTACAGTAGG-3′; CC/CEN3-A, 5′-tatcgaattcGGTTGTAAGAAATCCTGAATAC-3′; and CC/CEN3-B, 5′-ccgcGCCATTGTTGTTTCCACTATTCT-3′. Lowercase letters indicate a sequence added for subcloning. Extension of an A primer generates a probe that hybridizes to the bottom strand; extension of a B primer generates a probe that hybridizes to the top strand.
Analysis of CPDs by primer extension.
Primer labeling and primer extension were done as described previously (1), with minor modifications. High-performance liquid chromatography-purified primers were 5′ end labeled with T4 nucleotide kinase (Biolabs) in the presence of [γ-32P]ATP. Primer extension was done with about 5 μg of HindIII-digested DNA, 0.6 pmol of labeled primer, 1 U of Taq DNA polymerase (QIAGEN) in 40 μl of 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 3 mM MgCl2, and 2% dimethyl sulfoxide for 30 cycles of repeated denaturation (95°C for 45 s), annealing (57°C for 4.5 min), and extension (72°C for 3 min). The primers were 5′-CGGATTATTCTCTTTACTTCTTCCC-3′ (hybridizes to the bottom strand) and 5′-TCCGTATTCATTCTTATCAGTCCAG-3′ (hybridizes to the top strand). The reaction products were ethanol precipitated, analyzed on 5 or 8% denaturing polyacrylamide gels, and quantified with a PhosphorImager (Molecular Dynamics) (56).
RESULTS
To investigate DNA repair of UV lesions in the centromere regions, yeast cells were irradiated with UV light and either exposed to photoreactivating light for DNA repair by photolyase or incubated in the dark for DNA repair by the NER pathway. The DNA was purified, and the distribution of CPDs was analyzed by gel electophoresis.
Inhibition of PR in CEN14.
Figure 1 shows a direct comparison of chromatin footprinting by MNase and repair of CPDs by photolyase in and around the centromere of chromosome XIV (CEN14). For chromatin analysis, nuclei and genomic DNA were digested with MNase. The DNA was purified, digested with HindIII, run on a denaturing agarose gel, blotted to a nylon membrane, and hybridized to short-strand-specific probes. CEN14 displays strong footprints flanked by sensitive regions and positioned nucleosomes, consistent with previous reports (Fig. 1B) (11). The footprints were observed on both strands, indicating that there was no substantial nicking by MNase within nucleosomes and the centromere.
To investigate PR in the absence of NER, we used S. cerevisiae strain RRY3 (rad1Δ), where NER was inactivated by deletion of RAD1. A growing culture was irradiated with UV light (150 J/m2) to generate about 0.35 CPD/kb. After damage induction, the cell suspension was exposed to photoreactivating light for up to 2 h at room temperature. DNA was purified, cleaved with HindIII, and cut at the CPDs with T4-Endo V. The cutting sites were compared with the MNase footprints in the same alkaline agarose gels. DNA of unirradiated cells showed the intact HindIII restriction fragment (Fig. 1B, lanes 4). DNA of irradiated cells revealed numerous bands and a weaker top band (lanes 5). The bands represent the yields and the distribution of CPDs along the chromosomal DNA. Top and bottom strands showed different patterns, demonstrating strand specificity of the probes and the strand-specific CPD distribution. Some CPD bands disappeared when cells were exposed to photoreactivating light (lanes 6 to 10), demonstrating that repair was done by photolyase. The CPDs remained when cells were kept in the dark (lanes 11), which confirmed that NER was inactivated (53).
Repair by photolyase was heterogeneous. Most striking was a strong inhibition of repair in a region which maps in the footprint of CEN14, while the flanking nucleosomal region was efficiently repaired. Quantification indicates that less than 20% of CPDs were removed from CEN14 in 2 h, but more than 80% were removed from the flanking regions (Fig. 1C). CPDs in the nuclease-sensitive region of CEN14 were readily accessible to photolyase and rapidly repaired (Fig. 1B). With respect to the accessibility of CPDs, the results suggest that the kinetochore complex was much more stable than the nucleosomes and remained stable during the time course of the experiment.
PR is inhibited by RNA polymerases stalled at CPDs (24, 53). The flanking region of CEN14 contains a gene coding for citrate synthase, CIT1 (Saccharomyces Genome Database). Figure 1 shows that repair was reduced in the top (transcribed) strand, but not in the bottom strand, suggesting that CIT1 was transcribed. Thus, the inhibition of centromere repair was also stronger than inhibition by stalled RNA polymerases in CIT1.
Inhibition of NER in CEN14.
To investigate NER and the combination of PR with NER, repair experiments were done with UCC510 (RAD1) at room temperature (Fig. 2A and C). NER was much slower than PR and NER together and slower than PR alone (see above). In 4 h, NER removed about 50% of CPDs from the nucleosomal regions and the bottom strand of CIT1 (nontranscribed strand). The top strand of CIT1 (transcribed strand) was more rapidly repaired, which is consistent with transcription-coupled repair. Thus, both the strand bias observed by PR and NER can be explained by transcription of CIT1. In contrast to efficient repair of the flanking regions, NER alone and both repair systems together were severely inhibited in CEN14.
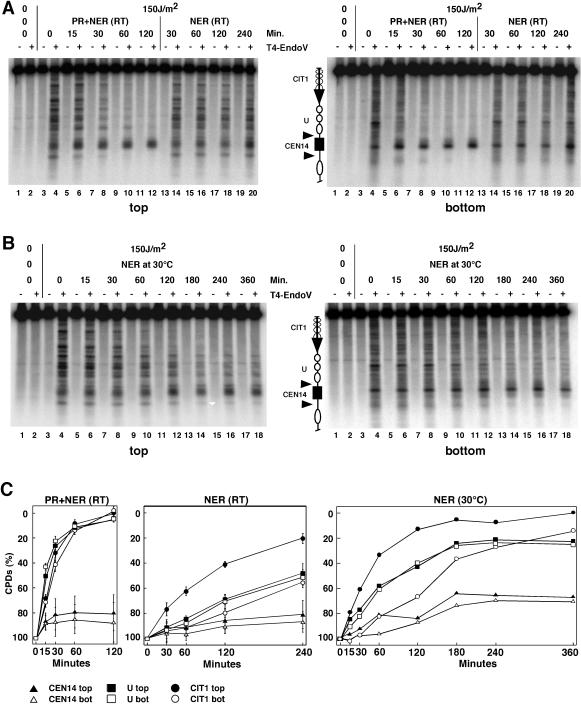
FIG. 2.
Inhibition of CPD repair by NER and photolyase (PR) in CEN14. (A) NER and PR at room temperature (RT). UCC510 cells were irradiated with 150 J of UV light/m2and exposed to photoreactivating light (lanes 3 to 12) or incubated in the dark (lanes 13 to 20). Lanes 1 and 2 contain DNA of nonirradiated cells. The DNA was cut with T4-Endo V (+) or was mock treated (−) and processed as described in the legend to Fig. 1. (B) UV irradiation and incubation in the dark were performed at 30°C. (C) Quantitative analysis of NER and PR at room temperature (RT) and NER at 30°C. The curves for experiments done at RT represent averages (± standard deviations) of results for four gels. For details about the schematic diagrams shown in panels A and B, see the legend to Fig. 1.
Since NER is slow at room temperature, the experiment was repeated at 30°C and repair times were extended to 6 h (Fig. 2B and C). While more than 70% of the lesions were repaired in the nucleosomal region and the CIT1 gene (including transcription-coupled repair of the transcribed strand), only about 30% of CPDs were removed from CEN14. Thus, NER remained inhibited in CEN14, and centromere protection remained stable at the higher temperature.
Inhibition of repair in CEN3 and CEN6.
It was previously reported that CPDs are efficiently repaired in the CEN3 and surrounding regions (41). Under our conditions, however, repair by NER and PR was inhibited in CEN3 and CEN6 (Fig. 3). Therefore, inhibition of repair appears to be a general property of yeast centromeres.
UV photofootprints and DNA repair footprints in CEN14.
Centromeres associate with numerous proteins to form a complex chromatin structure that generates a nuclease footprint of about 150 to 250 bp, but the precise dimensions are unknown (Fig. 1 and 2) (5, 11). Formation of DNA lesions by UV light is modulated by the DNA structure and the deformation of DNA in protein-DNA interactions (3). On the other hand, damage formation might disrupt protein-DNA complexes and facilitate repair at individual sites.
We therefore analyzed PDs on sequencing gels by using primer extension with radioactive primers and Taq polymerase (Fig. 4). CPDs and 6-4PPs efficiently block elongation of Taq polymerase (69). PDs were detected in CDEII and CDEIII and the flanking regions, but not in CDEI, which lacks thymidine clusters (Fig. 4B and C, lanes 7). To distinguish between CPDs and 6-4PPs, a fraction of in vivo UV-irradiated DNA was treated with Escherichia coli photolyase in vitro, which removes CPDs but leaves 6-4PPs (Fig. 4B and C, lanes 6). There were only a few sites (B4, T6, T9, and T12) which generated substantial amounts of 6-4PPs in chromatin (Fig. 4B).
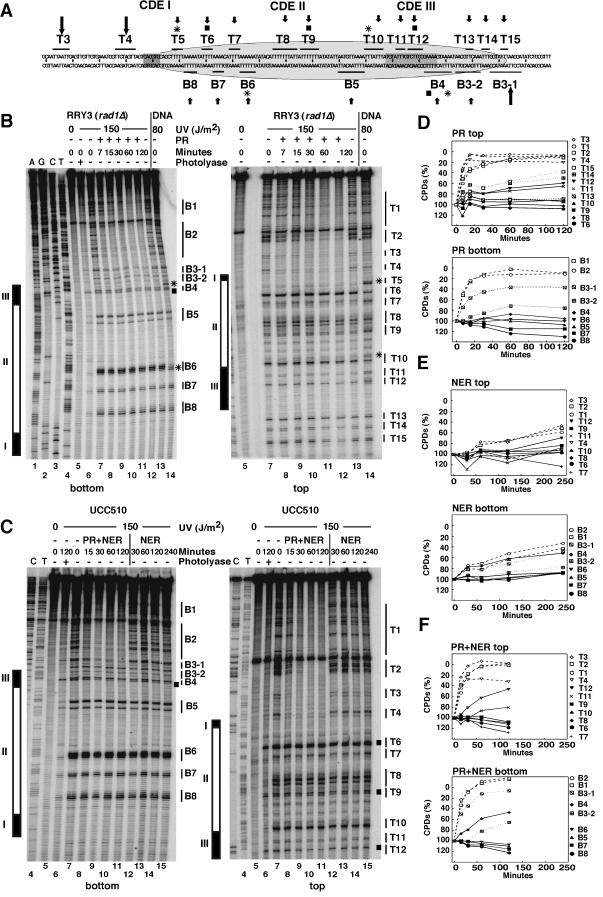
FIG.4.
UV photofootprints and repair footprints in CEN14. (A) Schematic summary. Indicated are the sequence of CEN14 and flanking regions; elements CDEI, CDEII, and CDEIII; pyrimidine clusters in the top (T3 to T15) and bottom (B3 to B8) strands; sites of significant UV photofootprints (stars) and 6-4PPs (squares); sites that are accessible and repaired (long arrows) or inaccessible (small arrows); and the size of the footprint (about 120 bp; ellipse). (B) Footprinting of PR was done by extension of a radioactively labeled primer by Taq polymerase, which is blocked by CPDs and 6-4PPs. DNA was from the same experiment as described in the legend to Fig. 1. Lanes 1 to 4, genomic sequencing lanes; lanes 5, unirradiated DNA; lanes 6, DNA of irradiated cells (lane 7) treated with photolyase in vitro to remove CPDs (the remaining lesions are presumably 6-4PPs); lanes 7, DNA of irradiated cells (initial damage, no repair); lanes 14, naked DNA irradiated with 80 J/m2. Different repair conditions are indicated for PR (panel B, lanes 8 to 13). (C) Footprinting of NER and PR. DNA was from experiments described in the legend to Fig. 2. Lanes 1 to 7 are as described in the legend to panel B. Lanes 8 to 11 and 12 to 15 show NER with PR and NER alone, respectively. CDEI, CDEII, and CDEIII (rectangles); sites of major UV footprints (asterisks); and 6-4PPs (squares) are indicated in panels B and C. (D to F) Repair curves obtained from data shown in panels B and C.
A distinct modulation of damage formation by chromatin (the UVphotofootprint) was observed in CDEII and CDEIII (compare chromatin and irradiated naked DNA) (Fig. 4B, lanes 7 and 14). Most obvious were one site (B6) with enhanced yields of lesions (Fig. 4A and B) and two sites, B4 and T5 (Fig. 4A and B), where damage formation was almost completely suppressed in chromatin compared with damage formation in naked DNA. Those UV footprints provide evidence for strong protein-DNA interactions, which alter DNA structure and modulate damage formation.
Complementary to the UV footprint, the PR results revealed a strong repair footprint. CPDs flanking the CDEs were very efficiently repaired (sites B1, B2 and T1 to T4), whereas CPDs within the CDEII, CDEIII, and the 20-bp flanking CDEIII were protected against photolyase activity. The photolyase footprint extended from T6 and B8 in CDEII to T15 and B3-2 outside of CDEIII. Site B3-1 was partially accessible (Fig. 4A).
Very similar repair footprints were obtained by photolyase and NER together (Fig. 4C and F). NER alone was slow at room temperature, but a repair footprint could be measured quantitatively (Fig. 4C and E). Interestingly, lesions in site B4 of CDEIII containing 6-4PPs were quite efficiently repaired by NER. This result suggests that DNA damage at this site might have destabilized the DNA-CBF3 complex and could thereby compromise chromosome stability. Similar repair footprints were obtained by NER at 30°C. Those footprints extended from sites T6 to B3, too (data not shown).
In conclusion, photolyase and NER were strongly inhibited in CDEII and CDEIII, and reduced inhibition was observed within 20 bp of CDEIII. The UV photofootprint and repair footprint covered about 120 bp (from sites T5 to B3 and T15) (Fig. 4A), a finding which correlates with the specialized structure of the centromere and indicates that the centromere complex remained stable despite UV damage (with the exception of the 6-4PPs in CDEIII).
Centromere chromatin remains stable in cse4-1 and cbf1Δ mutants, but not in ndc10-1.
Since repair experiments showed a strong inhibition of centromere repair, we were encouraged to test centromere stability and repair in strains with mutated kinetochore proteins.
Cbf1p binds to CDEI, bends DNA, interacts with CBF3, and may thereby clamp the centromere structure (8). Deletion of the gene (cbf1Δ) is not lethal but leads to a partial loss of the centromere function and an enhanced nuclease sensitivity at the centromere (31). UV photofootprinting at high resolution revealed damage formation in site T5 (data not shown), demonstrating that the suppression of damage formation observed in CBF1 wild-type cells was indeed due to binding of Cbf1p. Repair of the centromeres remained inhibited compared with the complete repair of the flanking regions (Fig. 5A). NER alone removed about 40% of CPDs in 4 h at 30°C, which was slightly more efficient than NER in CBF1 wild-type cells (Fig. 2). Thus, the centromere structure remained relatively stable in the absence of Cbf1p.
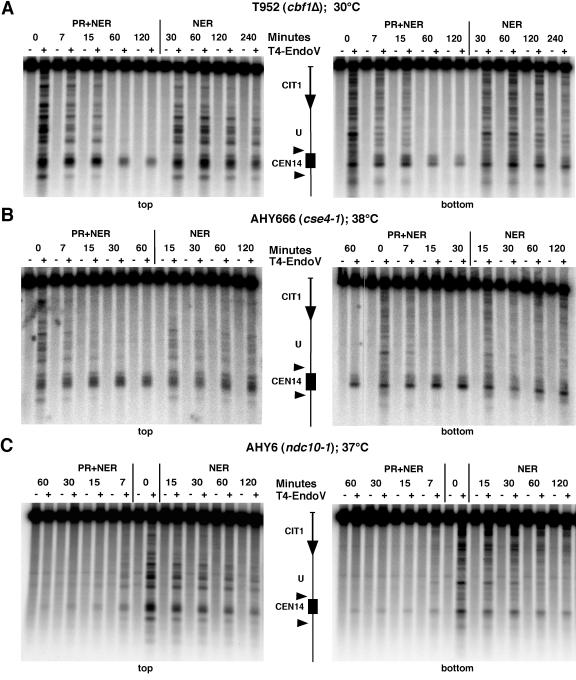
FIG. 5.
Mutations of centromere proteins differentially affect DNA repair. Repair analysis was done as described in the legends to Fig. 1 and 2. (A) T952 (cbf1Δ); (B) AHY666 (cse4-1); (C) AHY6 (ndc10-1).
Cse4p is a centromere-specific histone H3 variant required for formation of the centromere nucleosomes (34). Cse4-1 is a temperature-sensitive mutant and arrests in G2/M at restrictive temperatures (38°C) (51). In cse4-1 mutants, recruitment of Ctf3p and Ctf19p is severely impaired (28). AHY666 cells (cse4-1) were shifted for 6 h to 38°C, before the repair experiment was performed at the restrictive temperature. The rate of repair was below 30% (NER and PR; 1 h) and 20% (NER; 2 h), whereas damage outside of the centromere was completely removed (Fig. 5B). High-resolution analysis provided photofootprints and repair footprints similar to those for wild-type cells (data not shown). Hence, the results suggest that the kinetochore complex, including the centromeric nucleosome, remained stable despite a mutation in its histone H3 variant. The impaired recruitment of Ctf3p and Ctf19p did not destabilize the protein-DNA interactions of the complex.
Ndc10p belongs to the CBF3 complex, which binds CDEIII and recruits kinetochore proteins. The ndc10-1ts mutation has a defect in chromosome segregation, since the chromosomes remain at one pole of the anaphase spindle at restrictive temperatures (37°C) (14). In contrast to the wild type and the other mutants tested above, ndc10-1ts cells revealed very efficient repair of centromeres (Fig. 5C). CPDs were as rapidly removed from the centromeres as from the flanking nucleosomal region. This result supports a disruption of the centromere structure in the absence of a functional Ndc10p.
In summary, the repair experiments with the kinetochore mutants illustrate that only a severe disruption of the kinetochore structure leads to efficient repair of the underlying centromere sequence. Moreover, it is not the centromere sequence per se which inhibits repair.
Repair in cell cycle-arrested cells.
Despite the inhibition of repair, the CPD levels in centromeres appeared to be reduced after 6 h of incubation when experiments were started with exponentially growing cultures (Fig. 2). The high-resolution footprinting showed that this effect could not be accounted for by repair of individual sites within the centromere region (Fig. 4). UV irradiation leads to cell cycle arrest (37, 48, 67), but the cells eventually resume cycling and process the lesion by postreplication repair mechanisms (21, 38). Thus, if some cells resume DNA synthesis, the reduced CPD levels might reflect not only NER activity but also dilution by DNA synthesis and/or removal by postreplicative repair.
We therefore tested how centromeres are repaired when yeast cultures are arrested in G1 or in G2/M (Fig. 6). CCY2 (bar1Δ RAD1) cells were arrested in G1 by addition of α-factor in YPD for 2 h, transferred to SD containing α-factor, irradiated with 150 J/m2, and incubated for NER at 30°C. The initial damage was about 0.3 CPD/kb. In contrast to very efficient repair of the flanking regions, the G1-arrested cells showed less than 20% repair in CEN14 (Fig. 6A). Strain 7428 was used for analysis of DNA repair in G2/M. 7428 cells contain CDC20 under galactose control and arrest at G2/M when galactose is replaced by glucose (22). The cultures were grown in galactose, arrested with α-factor in G1 for 3 h, and then shifted to glucose for 4 h. The culture was irradiated with 150 J/m2 and incubated for repair at 30°C. Flow cytometric analysis of the DNA content showed that the cells remained arrested in G2/M (data not shown). The initial damage was about 0.1 CPD/kb. Repair was efficient in all regions, except in the centromeres. Less than 20% of CPDs were removed from CEN14 (Fig. 6B). Thus, repair of centromeres appears to be strongly inhibited both in G1- and G2/M-arrested cells.
FIG. 6.
Inhibition of centromere repair in G1- and G2/M-arrested cells. NER was done at 30°C in G1-arrested cells (A) and G2/M-arrested cells (B). Chromosomal regions U and CIT1 around CEN14 are shown (as in Fig. 1). bot, bottom strand; top, top strand.
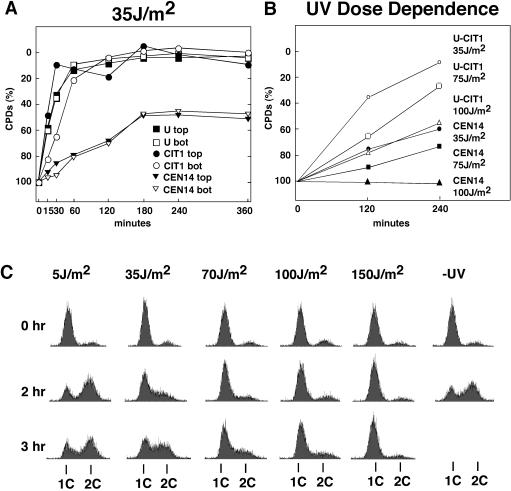
We further analyzed CPD levels in centromeres after irradiation with a lower dose (35, 75, or 100 J/m2) and incubation in the dark for 4 to 6 h (Fig. 7A and B). Under all conditions, the fraction of CPDs in CEN14 decreased much more slowly than that in the flanking regions, demonstrating that repair in centromeres was strongly inhibited. Moreover, the fraction of CPDs remained constant during the repair time in cultures irradiated with 100 J/m2 but decreased in cultures irradiated with 35 and 75 J/m2 (Fig. 7B). To evaluate whether the enhanced decrease of CPDs in centromeres is related to DNA synthesis of cells that restarted the cell cycle, synchronized cultures were released from α-factor arrest, irradiated with 5 to 150 J/m2, and incubated in the dark at room temperature. Flow cytometric analysis showed that a substantial fraction of cells irradiated with 5 and 35 J/m2 resumed DNA synthesis, in contrast to cells irradiated at higher doses (70, 100, and 150 J/m2) (Fig. 7C). Thus, the reduced CPD levels observed at 35 and 75 J/m2 (Fig. 7A and B) may reflect a dilution effect by DNA synthesis.
FIG. 7.
UV dose dependence of centromere repair and cell growth after UV damage. (A) Inhibition of CEN14 repair after irradiation with 35 J/m2. The fraction of CPDs in CEN14 and flanking regions is shown (U and CIT1) after exposure to a low dose of UV (35 J/m2) and incubation at room temperature in the dark for up to 360 min. Chromosomal regions U and CIT1 around CEN14 are shown (as in Fig. 1). bot, bottom strand; top, top strand. (B) Fraction of CPDs in CEN14 after irradiation at 35, 75, and 100 J/m2 and incubation in the dark at room temperature for 120 and 240 min. Data are for the bottom strand. (C) Cells arrested with α-factor in G1 were released by washing in SD without amino acids, irradiated at 5 to 150 J/m2 or left unirradiated (−UV), and after addition of the appropriate amino acids incubated in the dark at room temperature for up to 3 h. Cells were fixed, stained with propidium iodide, and analyzed by flow cytometry as described previously (35). 1C and 2C, DNA content of haploid cells before and after replication, respectively.
DISCUSSION
Centromeres form specialized chromatin structures required for accurate segregation of chromosomes. Since mutations in centromere sequences as well as mutations in centromere proteins result in chromosome instability and missegregation, a disruption of protein-DNA interactions by DNA lesions could be deleterious and result in the loss of entire chromosomes. Consequently, one would expect that centromeric lesions must be rapidly and efficiently repaired. Surprisingly, we found that repair of centromeres was strongly inhibited, indicating that the centromere-kinetochore complex was not disrupted by damage formation. Apparently, maintenance of a stable structure overrides the necessity for efficient DNA repair. These observations pose several questions on structural and dynamic properties of centromere chromatin and suggest that postreplication repair pathways repair centromeres.
A general inhibition of repair in yeast centromeres.
An earlier work described efficient repair of CEN3 by 3 h of NER in diploid yeast cells damaged with 100 J/m2; no cell divison was detected during that time, indicating that the cells were arrested in the cell cycle (41). In contrast to that work, we always observed an inhibition of repair in centromeres compared with efficient repair in other regions under all conditions, i.e., from 35 to 150 J/m2; in CEN3, CEN6, and CEN14; at room temperature, 30°C, and 38°C (cse4-1); in haploid and diploid strains (data not shown); and in G1- and in G2/M-arrested cells. Thus, we were unable to find efficient repair of centromeres except in the ndc10-1 mutant. The reason for the controversial results remains unknown, but it is unlikely that the strain used by Resnick et al. (41) has components that facilitate centromere repair which were absent in our strains derived from different sources.
Moreover, we found in our experiments that flanking nucleosomal regions, a transcribed gene (CIT1), and the inactive nucleosomal GAL10 gene (data not shown) were normally repaired by both pathways. The ndc10-1 mutant, which is deficient in recruitment of kinetochore proteins, showed efficient repair of centromeres, thereby demonstrating that the inhibition of repair was caused not by sequence specificity but by a stable kinetochore structure built on centromeres. Thus, the chromatin structure of the yeast centromeres remains stable after damage formation and prevents repair during repair incubation.
Centromeres are less dynamic than nucleosomes.
NER and PR are modulated by chromatin structures. Both reactions are inhibited in nucleosomes in vitro (12, 16, 45, 65). In yeast in vivo, however, repair is fast in linker DNA between nucleosomes and slow but complete in nucleosomes, suggesting that dynamic properties of nucleosomes facilitate repair, e.g., by transient disruption of nucleosomes or by moving the DNA lesion towards linker DNA, where it is readily accessible for damage recognition and repair (53, 56, 61, 68). Nucleosome positions in yeast cells are determined by DNA sequence, boundaries that limit nucleosome mobility, and chromatin folding (60). Compared with repair of nucleosomes, repair of centromeres was more strongly inhibited. Removal of Cbf1p only moderately enhanced repair. Thus, even in absence of Cbf1p, centromere chromatin is less dynamic than nucleosomes.
Yeast centromere chromatin covers 120 to 140 bp of DNA.
Structural models suggest that yeast centromere chromatin contains a nucleosome with a specialized histone H3 (Cse4p) and additional proteins bound to CDEI (Cbf1p), CDEII (Cse4p and Mif2p), and CDEIII (a CBF3 complex containing Ndc10p, Skp1p, Ctf13p, and Cep3p) (8, 32, 50). In vivo, these protein complexes and additional kinetochore components establish a nuclease-resistant structure. The reports vary on the dimensions of those footprints, citing lengths from 150 to 160 bp (11) to 220 to 250 bp (4). No high-resolution data have been published. An initial model proposed a nucleosome with CDEIII on the histone octamer surface. CDEI was close to CDEIII in location but was outside the nucleosome. This model accommodated the lower estimates of the nuclease-resistant core complex (34). Alternative models that were proposed on the basis of genetic interactions of CSE4 with CDEI and CDEII placed CDEIII outside a centromere-specific nucleosome, thus suggesting a nuclease protected region of 200 to 220 bp (19, 50).
Our in vivo UV photofootprint and repair footprint data provide novel information. First, UV photofootprinting identified a modulation of damage formation in chromatin at several sites in CDEII and CDEIII. Folding of DNA around histone octamers bends DNA (42) and affects CPD formation (13, 44, 45). Thus, the observation of a UV footprint is compatible with the existence of a nucleosome-like structure in the centromere. The striking inhibition of damage formation in sites T5 (close to CDEI) and B4 (close to CDEIII) showed that DNA was heavily constrained and not flexible enough for damage formation. The inhibition of damage formation in T5 was dependent on Cbf1, since it was not observed in cbf1Δ cells (data not shown). Second, both NER and photolyase revealed similar and strong repair footprints. Thus, damage recognition and processing were inhibited in both pathways. The footprints have a minimal length of about 120 bp extending from B8 and T5 at the left end of CDEII to B3 and T15, which is about 20 bp outside of CDEIII. The footprints closely match the in vitro data obtained with the CBF3 complex. Those proteins generated a 56-bp DNase I footprint which included CDEIII (20). An additional extension of about 20 bp as observed by cross-linking (9) was not detected by repair. Recent chromatin immunoprecipitation data also support extended contacts (34). Taken together, the minimal footprint (inhibition of repair) and maximal footprints (distance between rapidly repaired sites) are about 120 and 140 bp, respectively, and include the CDEIII-flanking region. The repair data support the initial model and not a structure which extends outside of CDEI.
Consequences of DNA damage in centromeres.
UV-induced DNA lesions can interfere with binding of transcription factors (63). Moreover, repair heterogeneity observed in promoter and regulatory regions implies that some factors are released while others remain bound after damage induction (2, 55, 57, 64). Therefore, the putative consequences of damage formation in centromeres are twofold. (i) If DNA damage disrupts protein-DNA interactions, the functional activity of that complex might be destroyed. A disruption, however, could make the lesion accessible for repair, thereby making the DNA available for reformation of the functional complex. We observed one damage site which qualifies for that interpretation, a presumed 6-4PP in CDEIII (site B4) was repaired, while CPDs close by were not removed by NER. Whether damage formation indeed disrupts the complex and whether complex formation occurs after repair cannot be addressed in vivo, since it is not possible to generate UV lesions at one specific site only.
(ii) If DNA lesions do not disrupt or even stabilize the complex, repair enzymes may be unable to remove the damage, which may preserve the function of the complex but could eventually lead to mutations. Most of the lesions in the centromeres were not repaired for several hours and hence were inaccessible by repair enzymes. The conclusion must be that the kinetochore complex remains stable, tolerates the lesions, and is likely to maintain its function in chromosome segregation. In this view, a correct segregation of chromosomes appears to be more important than DNA repair. Additional studies will be required to investigate whether chromosomes with one site-specific damage in the centromere are compromised in segregation. Again, the experimental problems are that UV lesions cannot be selectively induced in centromeres and that each centromere has numerous sites for CPD formation (Fig. 4), each of which might react in different ways.
DNA repair in centromeres.
What happens to DNA lesions that are not repaired? The fate of damaged cells is controlled by checkpoints, a surveillance system that delays progression through the cell cycle when cellular components such as chromosomes or the mitotic spindle are either damaged or incompletely assembled. The concerted action of DNA repair and checkpoint mechanisms prevents accumulation of mutations and ensures genetic integrity (25, 66). Cells eventually override the checkpoint and continue through the cell cycle, even if they are unable to repair the damage (62). The pathways by which cells tolerate UV damage during DNA replication have been termed postreplication repair pathways, although the damage remains in the DNA and is gradually eliminated in successive generations (38). One mechanism is translesion synthesis and appears to be error free or error prone, depending on whether DNA polymerase η (Rad30) or DNA polymerase ζ (Rev3), respectively, is involved in DNA synthesis opposite to the damage. In the other pathway, the lesion is bypassed by recombination with the sister chromatid or by DNA synthesis after template switching with the sister chromatid (21, 37).
Despite the strong inhibition, we observed a time-dependent decrease of centromeric CPDs by about 30 to 50%, starting with growing cultures irradiated at 150 and 35 J/m2, respectively (Fig. 2 and 7). On the other hand, no big change was obvious in cells that were arrested in G1 or G2/M (Fig. 6). How can this reduction of CPDs be explained? First, different NER activities in arrested and growing cells can be excluded, since DNA outside of centromeres was always efficiently repaired. However, we do not know whether kinetochores in G1- and G2/M-arrested cells are more stable than those in the exponentially growing cells. It is also conceivable that microtubule attachment modulates kinetochore stability and thereby regulates repair repression of centromeric DNA. Second, it is also conceivable that the replication process itself may lead to repair (replication-coupled repair). When replication forks move through chromatin templates, nucleosomes are disrupted in front of the fork and reassembled with new and old histones about 200 to 300 bp behind the fork (26). Thus, there is a short window of DNA accessibility, which could eventually be used for damage recognition and repair. Whether replication of centromeres and the assembly of the kinetochore behind the replication fork follow kinetics similar to that of the replication of nucleosomes is not known. The most likely explanation is, however, that in experiments with growing cultures, a fraction of cells resumes the cell cycle after most of the lesions (except those in the centromeres) are repaired, as illustrated by flow cytometric analysis of synchronized cells (Fig. 7). This mechanism leads to a dilution of centromere damage by translesion synthesis, by the other postreplication repair mechanisms described above, or by the growing fraction of cells that never had damage in the centromere.
Centromere repair in other eukaryotes.
It is interesting that some centromere proteins are conserved in evolution but that centromere DNA sequences are not. Moreover, centromeres of higher eukaryotes are much larger and more complex than the point centromeres of the budding yeast (52). On one side, it is conceivable that repair in those complexes could be as strongly inhibited as in yeast. Those centromere regions contain repetitive sequences. If irreparable DNA lesions are removed by postreplicative recombination (37), the high copy number of repetitive DNA might facilitate recombination events. Alternatively, the lack of repair in centromeres as observed in yeast cells might be necessary when no redundant attachment sites for microtubules exist. In higher eukaryotes, the larger centromeres may allow the cells to move proteins out of the way to allow repair while still maintaining sufficient contacts of the spindle with the centromere for appropriate chromosome segregation.
Acknowledgments
We thank U. Suter for continuous support, D. Gottschling, A. Hyman, K. Nasmyth, and T. Tanaka for yeast strains, and E. Niederer (Institut für Biomedizinische Technik ETH and UNI Zürich) for help with flow cytometry.
This work was supported by the Swiss National Science Foundation, the Swiss Federal Institute of Technology (ETH), and the Roche Research Foundation.
REFERENCES
- 1.Aboussekhra, A., and F. Thoma. 1998. Nucleotide excision repair and photolyase preferentially repair the nontranscribed strand of RNA polymerase III-transcribed genes in Saccharomyces cerevisiae. Genes Dev. 12:411-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aboussekhra, A., and F. Thoma. 1999. TATA-binding protein promotes the selective formation of UV-induced (6-4)-photoproducts and modulates DNA repair in the TATA box. EMBO J. 18:433-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker, M. M., and J. C. Wang. 1984. Use of light for footprinting DNA in vivo. Nature 309:682-687. [DOI] [PubMed] [Google Scholar]
- 4.Bloom, K. S., E. Amaya, J. Carbon, L. Clarke, A. Hill, and E. Yeh. 1984. Chromatin conformation of yeast centromeres. J. Cell Biol. 99:1559-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bloom, K. S., and J. Carbon. 1982. Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell 29:305-317. [DOI] [PubMed] [Google Scholar]
- 6.Cheeseman, I. M., D. G. Drubin, and G. Barnes. 2002. Simple centromere, complex kinetochore: linking spindle microtubules and centromeric DNA in budding yeast. J. Cell Biol. 157:199-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, Y., R. E. Baker, K. C. Keith, K. Harris, S. Stoler, and M. Fitzgerald-Hayes. 2000. The N terminus of the centromere H3-like protein Cse4p performs an essential function distinct from that of the histone fold domain. Mol. Cell. Biol. 20:7037-7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cleveland, D. W., Y. Mao, and K. F. Sullivan. 2003. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112:407-421. [DOI] [PubMed] [Google Scholar]
- 9.Espelin, C. W., K. B. Kaplan, and P. K. Sorger. 1997. Probing the architecture of a simple kinetochore using DNA-protein crosslinking. J. Cell Biol. 139:1383-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitzgerald-Hayes, M., L. Clarke, and J. Carbon. 1982. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell 29:235-244. [DOI] [PubMed] [Google Scholar]
- 11.Funk, M., J. H. Hegemann, and P. Philippsen. 1989. Chromatin digestion with restriction endonucleases reveals 150-160 bp of protected DNA in the centromere of chromosome XIV in Saccharomyces cerevisiae. Mol. Gen. Genet. 219:153-160. [DOI] [PubMed] [Google Scholar]
- 12.Gaillard, H., D. J. Fitzgerald, C. L. Smith, C. L. Peterson, T. J. Richmond, and F. Thoma. 2003. Chromatin remodeling activities act on UV-damaged nucleosomes and modulate DNA damage accessibility to photolyase. J. Biol. Chem. 278:17655-17663. [DOI] [PubMed] [Google Scholar]
- 13.Gale, J. M., K. A. Nissen, and M. J. Smerdon. 1987. UV-induced formation of pyrimidine dimers in nucleosome core DNA is strongly modulated with a period of 10.3 bases. Proc. Natl. Acad. Sci. USA 84:6644-6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goh, P. Y., and J. V. Kilmartin. 1993. NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol. 121:503-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanawalt, P. C. 2001. Controlling the efficiency of excision repair. Mutat. Res. 485:3-13. [DOI] [PubMed] [Google Scholar]
- 16.Hara, R., J. Mo, and A. Sancar. 2000. DNA damage in the nucleosome core is refractory to repair by human excision nuclease. Mol. Cell. Biol. 20:9173-9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hieter, P., D. Pridmore, J. H. Hegemann, M. Thomas, R. W. Davis, and P. Philippsen. 1985. Functional selection and analysis of yeast centromeric DNA. Cell 42:913-921. [DOI] [PubMed] [Google Scholar]
- 18.Hoeijmakers, J. H. 2001. Genome maintenance mechanisms for preventing cancer. Nature 411:366-374. [DOI] [PubMed] [Google Scholar]
- 19.Keith, K. C., and M. Fitzgerald-Hayes. 2000. CSE4 genetically interacts with the Saccharomyces cerevisiae centromere DNA elements CDE I and CDE II but not CDE III. Implications for the path of the centromere DNA around a cse4p variant nucleosome. Genetics 156:973-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lechner, J., and J. Carbon. 1991. A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell 64:717-725. [DOI] [PubMed] [Google Scholar]
- 21.Lehmann, A. R. 2000. Replication of UV-damaged DNA: new insights into links between DNA polymerases, mutagenesis and human disease. Gene 253:1-12. [DOI] [PubMed] [Google Scholar]
- 22.Lim, H. H., P. Y. Goh, and U. Surana. 1998. Cdc20 is essential for the cyclosome-mediated proteolysis of both Pds1 and Clb2 during M phase in budding yeast. Curr. Biol. 8:231-234. [DOI] [PubMed] [Google Scholar]
- 23.Livingstone-Zatchej, M., R. Marcionelli, K. Moller, R. De Pril, and F. Thoma. 2003. Repair of UV lesions in silenced chromatin provides in vivo evidence for a compact chromatin structure. J. Biol. Chem. 278:37471-37479. [DOI] [PubMed] [Google Scholar]
- 24.Livingstone-Zatchej, M., A. Meier, B. Suter, and F. Thoma. 1997. RNA polymerase II transcription inhibits DNA repair by photolyase in the transcribed strand of active yeast genes. Nucleic Acids Res. 25:3795-3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lowndes, N. F., and J. R. Murguia. 2000. Sensing and responding to DNA damage. Curr. Opin. Genet. Dev. 10:17-25. [DOI] [PubMed] [Google Scholar]
- 26.Lucchini, R., and J. M. Sogo. 1995. Replication of transcriptionally active chromatin. Nature 374:276-280. [DOI] [PubMed] [Google Scholar]
- 27.McGrew, J., B. Diehl, and M. Fitzgerald-Hayes. 1986. Single base-pair mutations in centromere element III cause aberrant chromosome segregation in Saccharomyces cerevisiae. Mol. Cell. Biol. 6:530-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Measday, V., D. W. Hailey, I. Pot, S. A. Givan, K. M. Hyland, G. Cagney, S. Fields, T. N. Davis, and P. Hieter. 2002. Ctf3p, the Mis6 budding yeast homolog, interacts with Mcm22p and Mcm16p at the yeast outer kinetochore. Genes Dev. 16:101-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mellon, I., G. Spivak, and P. C. Hanawalt. 1987. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell 51:241-249. [DOI] [PubMed] [Google Scholar]
- 30.Mellone, B. G., and R. C. Allshire. 2003. Stretching it: putting the CEN(P-A) in centromere. Curr. Opin. Genet. Dev. 13:191-198. [DOI] [PubMed] [Google Scholar]
- 31.Mellor, J., W. Jiang, M. Funk, J. Rathjen, C. A. Barnes, T. Hinz, J. H. Hegemann, and P. Philippsen. 1990. CPF1, a yeast protein which functions in centromeres and promoters. EMBO J. 9:4017-4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meluh, P. B., and D. Koshland. 1997. Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes Dev. 11:3401-3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meluh, P. B., and D. Koshland. 1995. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol. Biol. Cell 6:793-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meluh, P. B., P. Yang, L. Glowczewski, D. Koshland, and M. M. Smith. 1998. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell 94:607-613. [DOI] [PubMed] [Google Scholar]
- 35.Nash, R., G. Tokiwa, S. Anand, K. Erickson, and A. B. Futcher. 1988. The WHI1+ gene of Saccharomyces cerevisiae tethers cell division to cell size and is a cyclin homolog. EMBO J. 7:4335-4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niedenthal, R. K., M. Sen-Gupta, A. Wilmen, and J. H. Hegemann. 1993. Cpf1 protein induced bending of yeast centromere DNA element I. Nucleic Acids Res. 21:4726-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paulovich, A. G., C. D. Armour, and L. H. Hartwell. 1998. The Saccharomyces cerevisiae RAD9, RAD17, RAD24 and MEC3 genes are required for tolerating irreparable, ultraviolet-induced DNA damage. Genetics 150:75-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prakash, L. 1981. Characterization of postreplication repair in Saccharomyces cerevisiae and effects of rad6, rad18, rev3 and rad52 mutations. Mol. Gen. Genet. 184:471-478. [DOI] [PubMed] [Google Scholar]
- 39.Prakash, S., and L. Prakash. 2000. Nucleotide excision repair in yeast. Mutat. Res. 451:13-24. [DOI] [PubMed] [Google Scholar]
- 40.Renauld, H., O. M. Aparicio, P. D. Zierath, B. L. Billington, S. K. Chhablani, and D. E. Gottschling. 1993. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 7:1133-1145. [DOI] [PubMed] [Google Scholar]
- 41.Resnick, M. A., J. Westmoreland, E. Amaya, and K. Bloom. 1987. UV-induced damage and repair in centromere DNA of yeast. Mol. Gen. Genet. 210:16-22. [DOI] [PubMed] [Google Scholar]
- 42.Richmond, T. J., and C. A. Davey. 2003. The structure of DNA in the nucleosome core. Nature 423:145-150. [DOI] [PubMed] [Google Scholar]
- 43.Sancar, A. 2003. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem. Rev. 103:2203-2238. [DOI] [PubMed] [Google Scholar]
- 44.Schieferstein, U., and F. Thoma. 1996. Modulation of cyclobutane pyrimidine dimer formation in a positioned nucleosome containing poly(dA.dT) tracts. Biochemistry 35:7705-7714. [DOI] [PubMed] [Google Scholar]
- 45.Schieferstein, U., and F. Thoma. 1998. Site-specific repair of cyclobutane pyrimidine dimers in a positioned nucleosome by photolyase and T4 endonuclease V in vitro. EMBO J. 17:306-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharp, J. A., D. C. Krawitz, K. A. Gardner, C. A. Fox, and P. D. Kaufman. 2003. The budding yeast silencing protein Sir1 is a functional component of centromeric chromatin. Genes Dev. 17:2356-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 48.Siede, W., A. S. Friedberg, I. Dianova, and E. C. Friedberg. 1994. Characterization of G1 checkpoint control in the yeast Saccharomyces cerevisiae following exposure to DNA-damaging agents. Genetics 138:271-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smerdon, M. J., and A. Conconi. 1999. Modulation of DNA damage and DNA repair in chromatin. Prog. Nucleic Acid Res. Mol. Biol. 62:227-255. [DOI] [PubMed] [Google Scholar]
- 50.Smith, M. M. 2002. Centromeres and variant histones: what, where, when and why? Curr. Opin. Cell Biol. 14:279-285. [DOI] [PubMed] [Google Scholar]
- 51.Stoler, S., K. C. Keith, K. E. Curnick, and M. Fitzgerald-Hayes. 1995. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 9:573-586. [DOI] [PubMed] [Google Scholar]
- 52.Sullivan, B. A., M. D. Blower, and G. H. Karpen. 2001. Determining centromere identity: cyclical stories and forking paths. Nat. Rev. Genet. 2:584-596. [DOI] [PubMed] [Google Scholar]
- 53.Suter, B., M. Livingstone-Zatchej, and F. Thoma. 1997. Chromatin structure modulates DNA repair by photolyase in vivo. EMBO J. 16:2150-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suter, B., M. Livingstone-Zatchej, and F. Thoma. 1999. Mapping cyclobutane-pyrimidine dimers in DNA and using DNA-repair by photolyase for chromatin analysis in yeast. Methods Enzymol. 304:447-461. [DOI] [PubMed] [Google Scholar]
- 55.Suter, B., G. Schnappauf, and F. Thoma. 2000. Poly(dA.dT) sequences exist as rigid DNA structures in nucleosome-free yeast promoters in vivo. Nucleic Acids Res. 28:4083-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suter, B., and F. Thoma. 2002. DNA-repair by photolyase reveals dynamic properties of nucleosome positioning in vivo. J. Mol. Biol. 319:395-406. [DOI] [PubMed] [Google Scholar]
- 57.Suter, B., R. E. Wellinger, and F. Thoma. 2000. DNA repair in a yeast origin of replication: contributions of photolyase and nucleotide excision repair. Nucleic Acids Res. 28:2060-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thoma, F. 1999. Light and dark in chromatin repair: repair of UV-induced DNA lesions by photolyase and nucleotide excision repair. EMBO J. 18:6585-6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thoma, F. 1996. Mapping of nucleosome positions. Methods Enzymol. 274:197-214. [DOI] [PubMed] [Google Scholar]
- 60.Thoma, F. 1992. Nucleosome positioning. Biochim. Biophys. Acta 1130:1-19. [DOI] [PubMed] [Google Scholar]
- 61.Tijsterman, M., R. de Pril, J. G. Tasseron-de Jong, and J. Brouwer. 1999. RNA polymerase II transcription suppresses nucleosomal modulation of UV-induced (6-4) photoproduct and cyclobutane pyrimidine dimer repair in yeast. Mol. Cell. Biol. 19:934-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Toczyski, D. P., D. J. Galgoczy, and L. H. Hartwell. 1997. CDC5 and CKII control adaptation to the yeast DNA damage checkpoint. Cell 90:1097-1106. [DOI] [PubMed] [Google Scholar]
- 63.Tommasi, S., P. M. Swiderski, Y. Tu, B. E. Kaplan, and G. P. Pfeifer. 1996. Inhibition of transcription factor binding by ultraviolet-induced pyrimidine dimers. Biochemistry 35:15693-15703. [DOI] [PubMed] [Google Scholar]
- 64.Tu, Y., S. Tornaletti, and G. P. Pfeifer. 1996. DNA repair domains within a human gene: selective repair of sequences near the transcription initiation site. EMBO J. 15:675-683. [PMC free article] [PubMed] [Google Scholar]
- 65.Ura, K., M. Araki, H. Saeki, C. Masutani, T. Ito, S. Iwai, T. Mizukoshi, Y. Kaneda, and F. Hanaoka. 2001. ATP-dependent chromatin remodeling facilitates nucleotide excision repair of UV-induced DNA lesions in synthetic dinucleosomes. EMBO J. 20:2004-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weinert, T. 1998. DNA damage checkpoints update: getting molecular. Curr. Opin. Genet. Dev. 8:185-193. [DOI] [PubMed] [Google Scholar]
- 67.Weinert, T. A., and L. H. Hartwell. 1988. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science 241:317-322. [DOI] [PubMed] [Google Scholar]
- 68.Wellinger, R. E., and F. Thoma. 1997. Nucleosome structure and positioning modulate nucleotide excision repair in the non-transcribed strand of an active gene. EMBO J. 16:5046-5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wellinger, R. E., and F. Thoma. 1996. Taq DNA polymerase blockage at pyrimidine dimers. Nucleic Acids Res. 24:1578-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wiens, G. R., and P. K. Sorger. 1998. Centromeric chromatin and epigenetic effects in kinetochore assembly. Cell 93:313-316. [DOI] [PubMed] [Google Scholar]
- 71.Wilmen, A., and J. H. Hegemann.n 1996. The chromatin of the Saccharomyces cerevisiae centromere shows cell-type specific changes. Chromosoma 104:489-503. [DOI] [PubMed] [Google Scholar]