Abstract
Transcription factor Nrf2 is a major regulator of genes encoding phase 2 detoxifying enzymes and antioxidant stress proteins in response to electrophilic agents and oxidative stress. In the absence of such stimuli, Nrf2 is inactive owing to its cytoplasmic retention by Keap1 and rapid degradation through the proteasome system. We examined the contribution of Keap1 to the rapid turnover of Nrf2 (half-life of less than 20 min) and found that a direct association between Keap1 and Nrf2 is required for Nrf2 degradation. In a series of domain function analyses of Keap1, we found that both the BTB and intervening-region (IVR) domains are crucial for Nrf2 degradation, implying that these two domains act to recruit ubiquitin-proteasome factors. Indeed, Cullin 3 (Cul3), a subunit of the E3 ligase complex, was found to interact specifically with Keap1 in vivo. Keap1 associates with the N-terminal region of Cul3 through the IVR domain and promotes the ubiquitination of Nrf2 in cooperation with the Cul3-Roc1 complex. These results thus provide solid evidence that Keap1 functions as an adaptor of Cul3-based E3 ligase. To our knowledge, Nrf2 and Keap1 are the first reported mammalian substrate and adaptor, respectively, of the Cul3-based E3 ligase system.
Biological responses to toxic environmental stresses are regulated by several coordinated functions of cellular factors, providing animals with a means of cellular protection. The cellular factors usually involve a component that senses a stress and transmits the information as a cellular signal to transcription factors. Transcription factors then induce or regulate the expression of genes encoding cytoprotective enzymes and proteins (16, 23). It has been shown that repression of stress-responsive transcription factors is crucial for the maintenance of cellular homeostasis. This repression is especially important for avoiding unnecessary gene activation in the absence of stress stimuli. Several prototype mechanisms of such inhibitory action have been identified. For instance, von Hippel-Lindau protein (pVHL) acts as a repressor of transcription factor Hif-1α by accelerating protein degradation under conditions of normoxia (23). pVHL works to ubiquitinate Hif-1α, leading to degradation of the protein in a proteasome-dependent manner. The transcriptional activity of Hif-1α is thus repressed during normoxia (11).
Oxidative and xenobiotic stresses are known to cause many diseases, such as cancer, diabetes, and arteriosclerosis. Recent progress in this field has provided solid evidence for the contention that these stresses are sensed by the Nrf2-Keap1 system, which in response achieves cytoprotection by regulating the expression of phase 2 drug-metabolizing enzymes and antioxidant response proteins (6, 10, 16). In the absence of stress stimuli, the cytoplasmic protein Keap1 binds Nrf2 and prevents its translocation to the nucleus (10). This cytoplasmic sequestration of Nrf2 requires at least two cysteine residues in the intervening region (IVR) of Keap1. In experiments conducted in vitro, four cysteine residues in the IVR appeared to be modified by electrophiles (3, 14, 27), suggesting that Keap1 functions as a sensor for oxidative and xenobiotic stimuli through these cysteines.
Extensive studies have been executed to elucidate the molecular mechanisms governing Nrf2 activity. These studies revealed that Nrf2 is degraded rapidly by the ubiquitin-proteasome pathway (9, 15, 17, 25, 31). The rapid turnover of Nrf2 was reproducible throughout these studies and proven in vivo in experiments with a gene-manipulated mouse (9). However, the molecular mechanisms regarding Nrf2 degradation are still controversial. One group reported that ubiquitination of Nrf2 is carried out in a Keap1-independent manner, whereas another group claimed Keap1-dependent degradation of Nrf2 (15, 31). We also found that Nrf2 is degraded through two distinct pathways: a proteasome-dependent rapid turnover and a relatively slow turnover in the nucleus (9).
The ubiquitin-dependent proteolysis system regulates the abundance of proteins and serves a central regulatory function in many biological processes, including cell cycle progression, signal transduction, and transcription (7). Ubiquitin conjugation to substrate proteins is carried out by the sequential reaction of three enzymes (19): the ubiquitin-activating enzymes (E1), the ubiquitin-conjugating enzymes (E2), and the ubiquitin ligases (E3). E3 ligases provide two distinct functions; one is to target substrate protein, and the other is to catalyze isopeptide bond formation between the substrate protein and ubiquitin.
It has been shown that there are several types of E3 ligase. Cullin (Cul)-based E3 ligases regulate the turnover of important transcription factors and are composed of several subunits. Cul is a scaffold protein in the E3 ligase complex and forms a catalytic core complex with Roc1/Rbx1/Hrt1, with Roc1 recruiting a cognate E2 enzyme. Six Cul protein members have been identified in mammals: Cul1, Cul2, Cul3, Cul4A, Cul4B, and Cul5 (29). To target substrate proteins specifically, the Cul-Roc1 complex requires an adaptor molecule. Cul1 preferentially binds the adaptor molecules Skp1 and F-box protein, while Cul2 binds pVHL and elongin C. These complexes are referred to as the SCF (Skp1-Cul1-F-box) and ECS (elongin-Cul2-SOCS) E3 ligases, respectively. Recently, in Caenorhabditis elegans and Schizosaccharomyces pombe, a subset of proteins containing a BTB domain was reported to function as a distinct group of substrate-specific adaptors which preferentially bind to Cul3. The BTB proteins seem to target certain substrates into the E3 ligase complex by virtue of their protein-interaction domains, the Kelch motifs and MATH domain (4, 5, 21, 30). However, a specific substrate for the mammalian Cul3 system has not yet been identified.
Two distinct molecular mechanisms for the contribution of Keap1 to the rapid turnover of Nrf2 have been assumed (9). Keap1 may contribute to the turnover of Nrf2 by merely retaining Nrf2 in the cytoplasm such that Nrf2 is kept in close proximity to the proteasome system. Alternatively, Keap1 may promote Nrf2 degradation more effectively through the active recruitment of E3 ligase and proteasome subunits. To examine which case is actually operating, we established a system that analyzes Nrf2 degradation in vivo. This paper describes the molecular mechanisms of Nrf2 degradation involving the ubiquitin-proteasome system. Our results provide conclusive evidence that Keap1 is a stress sensor protein that functions directly as an adaptor molecule in the Cul3-based E3 ligase system in the rapid degradation of Nrf2. Thus, Keap1 plays essential roles in the Nrf2-Keap1 stress response system, not only as a sensor of oxidative and electrophilic stresses but also as a regulator of Nrf2 degradation.
MATERIALS AND METHODS
Chemical reagents.
MG132 and the calpain inhibitor E64 were purchased from Peptide Institute Inc. Clasto-lactacystin β-lactone was from Calbiochem.
Plasmid construction.
Cul1, Cul2, Cul4A, and Cul5 cDNAs were cloned by reverse transcription-PCR using mouse brain and testis cDNAs and inserted into the Asp718 and XhoI sites of pcDNA3-Myc (Invitrogen). Cul3 cDNA was cloned by similar reverse transcription-PCR and inserted into the EcoRI and XbaI sites of p3XFLAG-CMV-10 (Sigma). Primers used for the amplification are available on request. pCMVNrf2ΔETGE was generated by substituting the KpnI-EcoRV fragment of pCMVmNrf2 with two pieces of PCR fragments digested with KpnI and EcoRI and with EcoRI and EcoRV, respectively. pCMVNrfaΔC and pCMVNrfaΔC/ETGE were constructed by inserting the XbaI linker (amber mutation) into the blunt-ended HindIII sites of pCMVNrf2 and pCMVNrf2ΔETGE, respectively. Expression plasmids of Cul3 deletion mutants, p3Xflag Cul3N280 and p3Xflag Cul3 ΔN280, were generated by inserting PCR products into the EcoRI and XbaI sites of p3XFLAG-CMV-10 (Sigma). Primers used for the amplification are available on request. Keap1 deletion mutants and Cys mutants in the IVR were generated as described previously (12, 27). All constructs were verified by sequencing.
Cell culture and transfection.
Cos7 cells and 293T cells were cultured in Dulbecco modified Eagle medium (Sigma) supplemented with 10% fetal calf serum (Gibco), 4500 mg of glucose per liter, 40 μg of streptomycin per ml, and 40 U of penicillin per ml. The DNA transfection was performed with Fugen6 (Roche) and Lipofectamine Plus (Invitrogen).
Turnover of Nrf2 in the presence of Keap1 and mutants.
Full-length Keap1 or Keap1 mutants were expressed in Cos7 cells along with enhanced green fluorescent protein (EGFP). At 36 h after transfection, the cells were treated with cyclohexamide (final concentration, 10 μM) to stop de novo protein synthesis and harvested by scraping. The cells were boiled in Laemmli sample buffer supplemented with β-mercaptoethanol (final concentration, 2%) (Wako Chemicals) at several time points, as described in the legend to Fig. 1. Cell extracts were subjected to immunoblot analysis with an anti-Nrf2 antibody (C4) against the C-terminal end of Nrf2 and anti-EGFP antibody (Santa Cruz). Experiments were performed twice in duplicate.
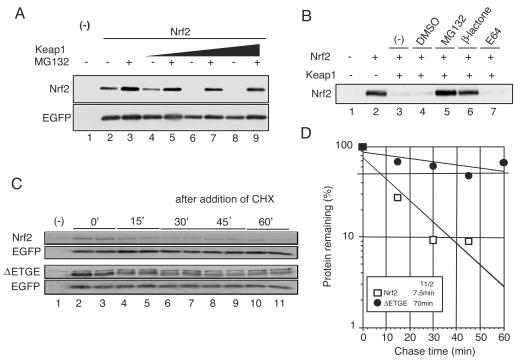
FIG. 1.
Assay system to examine the degradation mechanism of Nrf2. (A) Keap1 promotes Nrf2 degradation in the in vivo degradation system. An Nrf2 expression vector (2 μg) was transfected into Cos7 cells (90% confluent) with or without the Keap1 expression vector (1.5 μg). At 24 h after transfection, the cells were treated with dimethyl sulfoxide (DMSO) (lanes 1, 2, 4, 6, and 8) and 2 μM MG132 (lanes 3, 5, 7, and 9) for 12 h and directly lysed in sodium dodecyl sulfate sample buffer. (Upper panel) Whole-cell extracts were subjected to immunoblot analysis with an anti-Nrf2 antibody. (Lower panel). The expression level of cotransfected EGFP was used as an internal control. (B) Proteasome-specific inhibitors stabilize the Nrf2 protein. Transfected cells were treated with DMSO (lane 4), 2 μM MG132 (lane 5), 2 μM clasto-lactacystin β-lactone (lane 6), and E64 (lane 7) for 12 h. Immunoblot analysis was performed as described above. (C and D) The Nrf2 expressed in this system was rapidly degraded in a Keap1-dependent manner. Nrf2 and ΔETGE mutant were transfected into cells along with Keap1. At 36 h after transfection, the cells were treated with 10 μM cycloheximide (CHX) per ml for the periods indicated. (Upper panel) Whole-cell extracts were subjected to immunoblot analysis with an anti-Nrf2 antibody. (Lower panel). The expression level of EGFP was used as an internal control. The averages of the relative band intensities of Nrf2 (open squares) and ΔETGE mutant (closed circles) represent two independent experiments performed in duplicate.
Immunohistochemical staining.
Cos7 cells expressing Nrf2 and deletion mutants were grown on cultured dishes. At 36 h after transfection, the cells were fixed with 4% paraformaldehyde and acetone, blocked with 2% goat serum and 5% skim milk for 1 h, and incubated with an anti-Nrf2 antibody (100-fold dilution). The cells were incubated with anti-rabbit antibody conjugated with fluorescein isothiocyanate (100-fold dilution) (Zymed) and washed with phosphate-buffered saline. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). The cells were visualized by fluorescence microscopy (Leica DMIRB).
In vivo ubiquitination assay.
293T cells were transfected with several combinations of plasmids as described in the legend to Fig. 6, along with His-tagged Ub vector (26). At 24 h following transfection, the cells were treated with MG132 (final concentration, 2 μM) for 12 h to inhibit the proteasome function. Whole-cell extracts were prepared in lysis buffer I (20 mM Tris-HCl [pH 7.5], 0.5 M NaCl, 8 M urea, 5 mM imidazole) and incubated overnight with Ni2+ affinity beads (Probond resin; Invitrogen). After being washed three times with lysis buffer, the beads were boiled in the sample buffer and the eluate was subjected to immunoblot analysis with an anti-Nrf2 antibody recognizing the Neh2 domain.
FIG. 6.
The BTB domain of Bach1 does not bind Cul3. Whole-cell extracts prepared from 293T cells transfected with expression plasmids (3 μg) in the combinations indicated were subjected to immunoprecipitation (IP) with anti-Flag antibody-conjugated beads followed by immunoblot (IB) analysis with an anti-Myc antibody (top). The expression level of each protein was monitored by immunoblot analysis with anti-Myc (middle) and anti-Flag (bottom) antibodies, respectively.
Immunoprecipitation.
To determine the interaction of endogenous Keap1 with Cul3 in culture cells, an immunoprecipitation experiment using anti-Keap1 antibody was performed. Whole-cell extracts of 293T cells were prepared in lysis buffer II (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 0.1% Nonidet P-40, protease inhibitor cocktail [Roche Diagnostic], 10 μM MG132) and treated overnight with anti-Keap1 antibody against the N-terminal peptide (12). Immunocomplexes were precipitated with protein G (Pierce), washed three times with lysis buffer II, and subjected to immunoblot analysis using anti-Cul3 antibody (Santa Cruz), the ABC kit (Vector Laboratory), and the ECL kit (Amersham).
To analyze the interaction between Keap1 and several Cul proteins, expression plasmids for these factors were transfected into 293T cells by using Lipofectamine Plus. At 36 h after transfection, cytoplasmic extracts were prepared in buffer A (10 mM HEPES-KOH [pH 7.9], 10 mM KCl, 0.1 mM EDTA, 1 mM MgCl2, 0.5 mM dithiothreitol, protease inhibitor cocktail), and NaCl was added to a final concentration of 70 mM. Cell extracts were incubated with anti-Flag M2 beads (Sigma) by generous rocking at 4°C for 4 h. The immunocomplexes were washed three times with buffer (10 mM Tris-HCl [pH 7.5], 100 mM NaCl, 0.1 mM EDTA, 1 mM MgCl2, 0.1% Nonidet P-40) and subjected to immunoblot analysis with anti-Myc, anti-HA (Santa Cruz), and anti-Keap1 antibodies, separately.
RESULTS
Degradation of Nrf2 requires association with Keap1.
To examine the contribution of Keap1, we established a system enabling analysis of the mechanisms involved in Nrf2 degradation. In this system, Nrf2 was transiently expressed in Cos7 cells by transfection in the presence or absence of Keap1, and a whole-cell extract was prepared to determine the stability of Nrf2 by immunoblot analysis with an anti-Nrf2 antibody. As shown in Fig. 1A, Nrf2 accumulated in Cos7 cells after transfection (lanes 1 and 2). The accumulation of Nrf2 was augmented by the addition of a proteasome inhibitor, MG132 (lane 3), indicating that Nrf2 is otherwise rapidly degraded through the proteasome pathway. Important observations are that coexpression of Keap1 significantly reduced the amounts of Nrf2 (lanes 4, 6, and 8) and that this Keap1-dependent reduction of Nrf2 was inhibited by the addition of MG132 (lanes 5, 7, and 9).
We also challenged a couple of other protease inhibitors. Of the protease inhibitors, clasto-lactacystin β-lactone (β-lactone) is known to be more specific to the proteasome pathway than MG132 whereas E64 is a specific inhibitor of calpain proteases. As shown in Fig. 1B, degradation of Nrf2 was inhibited by β-lactone and MG132 but not by E64 (lanes 5 to 7). These results indicate that Nrf2 is degraded in this Cos7 system in a proteasome-dependent manner.
To further examine how closely this system recapitulates the endogenous Nrf2 degradation machinery, we determined the half-life of Nrf2 in this system and compared it with that of endogenous Nrf2 (Fig. 1C and D). The half-life of Nrf2 was determined by cotransfecting Nrf2 and Keap1 expression plasmids into Cos7 cells and then treating them with cycloheximide for specific periods to inhibit de novo protein synthesis. Whole-cell extracts were then prepared from these cells and subjected to immunoblot analysis. The half-life of Nrf2 was determined to be 7.5 min in the present Cos7 system (Fig. 1D). This is in very good agreement with the half-life of 18.5 min determined for endogenous Nrf2 in our previous analysis using peritoneal macrophages (9).
The ETGE motif of Nrf2 is indispensable in the association of Nrf2 with Keap1 (13); therefore, our ETGE deletion mutant will not interact with Keap1 (data not shown). Thus, we would expect significant stabilization of this Nrf2 mutant if Keap1 is truly involved in the active degradation of Nrf2 in vivo. We measured the half-life of the ΔETGE mutant of Nrf2 and found that it was indeed stabilized markedly (Fig. 1D, t1/2 = 70 min). These results thus demonstrate that Nrf2 is degraded in a Keap1-dependent manner in this system. Collectively, these data support our contention that the present degradation system of Nrf2 mimics the endogenous degradation of Nrf2 fairly well.
Keap1 directly promotes degradation of Nrf2.
To address the question whether Keap1 actively contributes to the rapid degradation of Nrf2, we measured the turnover of Nrf2 localized exclusively in the cytoplasm. To this end, we generated two deletion mutants of Nrf2. One is a ΔC mutant, which lacks the C-terminal region including the nuclear localization signal, while the other is a ΔC/ETGE mutant, which lacks the ETGE motif as well as the C-terminal region (Fig. 2A, ΔC and ΔC/ETGE, respectively). Keap1 associated with the ΔC mutant, but not with the ΔC/ETGE mutant (data not shown). Importantly, immunohistochemical analysis showed that both mutants were localized in the cytoplasm, even in the absence of Keap1 (Fig. 2B).
FIG. 2.
Rapid turnover of Nrf2 requires its association with Keap1. (A) Schematic presentation of Nrf2 deletion mutants. ΔC lacks the C terminus including the NLS of wild-type Nrf2. ΔC/ETGE lacks both this C terminus and the ETGE motif, which is crucial for association with Keap1. (B) Cytoplasmic localization of ΔC and ΔC/ETGE mutants in Cos7 cells (b and c). These mutant proteins were stained by an immunohistochemical method with anti-Nrf2 (C4) antibody. Nuclei were stained with DAPI (d to f). (C and D) Deletion of the ETGE motif abolished the degradation of Nrf2 by Keap1. This suggests that Keap1 positively regulates the degradation of Nrf2 through its association. The experimental procedure was described in the legend to Fig. 1. The averages of the band intensities of ΔC and ΔC/ETGE mutants represent two independent experiments done in duplicate.
Exploiting the Cos7 system, we again examined the half-life of Nrf2. The hypothesis behind this study is that if rapid degradation of Nrf2 requires a physical interaction with Keap1, the ΔC mutant should degrade more rapidly than the ΔC/ETGE mutant. After cotransfection of expression plasmids of these Nrf2 mutants and Keap1 into Cos7 cells, the cells were treated with cycloheximide for 36 h and harvested at several time points (Fig. 2C and D). We determined the half-life of these Nrf2 mutants by normalizing the amount of Nrf2 protein with that of coexpressed EGFP. Deletion of the C-terminal region slightly stabilized Nrf2 (t1/2 = 25 min) compared to full-length Nrf2. In contrast, simultaneous deletion of the ETGE motif with the C-terminal region significantly stabilized Nrf2 (t1/2 = 150 min). These data suggest that Keap1 actively promotes Nrf2 degradation whereas the mere presence of Nrf2 in the cytoplasm does not lead to its active degradation.
BTB and IVR domains of Keap1 contribute to the Nrf2 degradation.
The next important study was to decipher the molecular mechanisms of how Keap1 regulates Nrf2 degradation. We executed a series of domain functional analyses of Keap1 to identify the domains in Keap1 crucial for Nrf2 degradation. Keap1 consists mainly of the BTB/POZ, IVR, and DGR domains (Fig. 3A). We therefore expressed deletion mutants of each of these domains in Cos7 cells together with Nrf2 and monitored the stability of Nrf2 by immunoblot analysis using whole-cell extracts (Fig. 3B). Deletion of the DGR domain completely abolished the Nrf2 degradation activity of Keap1 since the DGR domain is essential for the association of Keap1 with Nrf2 (lane 6). Surprisingly, deletion of either the BTB or IVR domain also impaired the Keap1 activity that leads to Nrf2 degradation (lanes 4 and 5). The expression levels of the deletion mutants were verified to be comparable by immunoblot analysis using anti-Keap1 antibodies (Fig. 3C). The BTB and IVR domain mutants could interact with Nrf2 and localize the Nrf2 reporter protein (Neh2-GFP) exclusively in the cytoplasm (12). Taken together, these results suggest that Keap1 promotes Nrf2 degradation through interaction between certain regulatory factors with the BTB or IVR domain.
FIG. 3.
BTB and IVR domains contribute to degradation of Nrf2 by Keap1. (A) Schematic presentation of Keap1 deletion mutants. Keap1 contains mainly three characteristic domains, the BTB, IVR, and DGR domains. (B) Deletion of the BTB and IVR domains abolished Nrf2 degradation in the in vivo degradation assay (top). The experimental procedure is described in the legend to Fig. 1. The expression of cotransfected EGFP was used as an internal control (bottom). (C) The expression of Keap1 deletion mutants was also monitored by immunoblot analysis with two anti-Keap1 antibodies against the C-terminal and N-terminal ends (lanes 1 to 4 and lanes 5 to 7, respectively). wt, wild type.
Keap1 associates with Cul3 in vivo.
To identify regulatory factors associated with the degradation property of Keap1, we performed several rounds of yeast two-hybrid screens using full-length Keap1 or the BTB domain as bait. However, in spite of these extensive analyses, we could not isolate factors regulating Nrf2 degradation (data not shown). Switching to a candidate approach showed that a subset of BTB proteins function as an adaptor for the Cul3-type E3 ligase complex (4, 5, 21, 30). We hypothesized that Keap1 might interact with Cul3 through the BTB domain and promote the ubiquitination of Nrf2, thereby resulting in the rapid degradation of Nrf2.
To test this hypothesis, we tested whether Keap1 interacts with Cul3 in vivo through an immunoprecipitation analysis. We immunoprecipitated endogenous Keap1 in 293T cells with an anti-Keap1 antibody and performed an immunoblot analysis with an anti-Cul3 antibody (Fig. 4A). Cul3 was observed in the Keap1 immunocomplex (lane 2), indicating that Keap1 associates with Cul3.
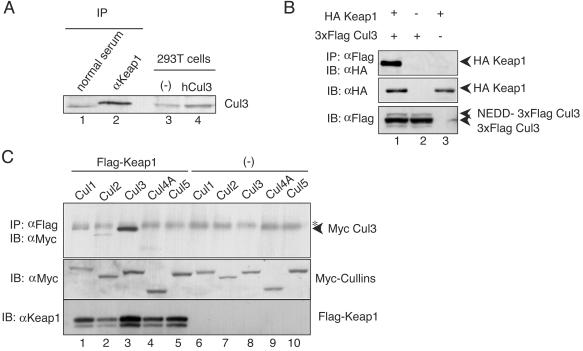
FIG. 4.
Keap1 associates specifically with Cul3 in vivo and in vitro. (A) Complex formation of Keap1 and Cul3 in 293T cells. Endogenous Keap1 was precipitated with anti-Keap1 antibody and protein G beads (IP). The immunocomplex was subjected to immunoblot analysis with anti-Cul3 antibody. Whole-cell extracts of 293T cells expressing human Cul3 were used as a control (lanes 3 and 4). (B) Association between Keap1 and Cul3 in a transient-expression system. Whole-cell extracts prepared from 293T cells transfected with expression plasmids of HA-tagged Keap1 (1 μg) and 3xFlag Cul3 (1 μg) were subjected to immunoprecipitation (IP) with anti-Flag (M2) beads and immunoblot analysis with anti-HA antibody (IB). Analyses of cells expressing 3xFlag Cul3 with or without HA-Keap1 (lanes 1 and 2) are shown. Lane 3 is loaded with cell extracts expressing HA-Keap1 alone. (C) Among the Cul family proteins, Cul3 specifically interacts with Keap1. Expression plasmids (1 μg each) of Cul1 (lanes 1 and 6), Cul2 (lanes 2 and 7), Cul3 (lanes 3 and 8), Cul4A (lanes 4 and 9), and Cul5 (lanes 5 and 10) were transfected into 293T cells in the presence (lanes 1 to 5) or absence (lanes 6 to 10) of Flag-fused Keap1. Immunoprecipitation and immunoblot analyses were performed as described above (top). The asterisk indicates a nonspecific band. The expression levels of Cul proteins and Flag-Keap1 were verified by immunoblot analysis with anti-Myc and anti-Keap1 antibodies (middle and bottom, respectively)
To further clarify the mechanisms of the association of Keap1 with Cul3, we carried out a similar immunoprecipitation analysis but with a DNA transfection system. 293T cells were transfected with Flag-tagged Cul3 and HA-tagged Keap1 expression vectors. Cytoplasmic extracts were prepared from the cells 36 h after transfection and subjected to immunoprecipitation with anti-Flag antibody-conjugated beads. In an immunoblot analysis with anti-HA antibody, HA-tagged Keap1 was clearly visualized in the immunoprecipitates (Fig. 4B), indicating that Keap1 coimmunoprecipitated with Cul3. We also examined the specificity of the association between Keap1 and Cul3 by examining the association of Keap1 with other Cul protein family members by using immunoprecipitation analyses. For this purpose, we expressed Myc-tagged Cul1, Cul2, Cul4A, and Cul5 in 293T cells along with Flag-tagged Keap1. We found that Cul3 specifically interacts with Keap1 whereas the other Cul proteins do not (Fig. 4C). These data thus suggest that Keap1 associates specifically with Cul3 and forms an E3 ligase complex.
The IVR domain of Keap1 associates with the N-terminal region of Cul3.
To identify the association interface of Keap1 and Cul3, we performed an immunoprecipitation analysis with 293T cells expressing three Keap1 deletion mutants independently. Since Cul3 was reported to interact with BTB domains, we anticipated that the BTB domain might comprise the interacting interface of Keap1. Surprisingly, however, we could see a positive band to anti-HA antibody in the immunoprecipitates of cell extracts expressing ΔBTB mutant with an anti-Flag antibody (Fig. 5A, lane 3), indicating that deletion of the BTB domain does not abolish the association between Keap1 and Cul3. Similarly, the ΔDGR mutant interacts with Cul3 (lane 5). In contrast, the ΔIVR mutant did not give any positive signals for the anti-HA antibody, indicating that this deletion completely abolishes the interaction with Cul3 (lane 4).
FIG. 5.
The IVR domain of Keap1 associates with the N-terminal end of Cul3. (A) Whole-cell extracts prepared from 293T cells transfected with various expression plasmids of HA-tagged Keap1 deletion mutants (1 μg) and 3xFlag tagged Cul3 (1 μg) were subjected to an immunoprecipitation (IP) assay with anti-Flag antibody beads and immunoblot (IB) analysis with anti-HA antibody (top). The expression levels of Keap1 deletion mutants and 3xFlag Cul3 were verified by immunoblot analysis with anti-HA and anti-Flag antibodies (middle and bottom, respectively). Analyses of cell lysates coexpressing 3xFlag Cul3 and HA-Keap1 (lane 2), ΔBTB (lane 3), ΔIVR (lane 4), or ΔDGR (lane 5) are shown. Lane 1 is loaded with cell extract expressing only 3xFlag Cul3. (B) Cysteine residues in the IVR domain are not crucial for the association between Keap1 and Cul3. The IVR contains seven cysteine residues (from Cys226 to Cys297), renumbered arbitrarily from 1 to 7. The association between Cul3 and Keap1 Cys point mutants (27) was examined by immunoprecipitation (upper panel) as described above. The expression levels of Cys mutants were verified by immunoblot analysis with anti-Keap1 antibody (lower panel). (C) Schematic presentation of Cul3 deletion mutants. (D) The N-terminal sequence of Cul3 is crucial for its association with Keap1. Whole-cell extracts of 293T cells transfected with expression plasmids of Cul3 deletion mutants (1 μg) and Keap1 (1 μg) were prepared and subjected to immunoprecipitation with anti-Flag (M2) beads and immunoblotting with anti-Keap1 antibody. Analyses of cell lysates expressing 3xFlag Cul3 (lanes 1 and 3), N280 (lanes 4 and 5), or ΔN280 (lanes 6 and 7) in the presence (lanes 2, 3, 5, and 7) or absence (lanes 1, 4, and 6) of Keap1 are shown (top). The expression levels of Keap1 and Cul3 deletion mutants were verified by immunoblot analysis with anti-Keap1 and anti-Flag antibodies (middle and bottom, respectively)
We previously showed that the IVR domain possesses four cysteine residues that bind to the electrophilic agent, dexamethasone 21-mesylate (Dex-mes), (3). Of these four cysteine residues, we further identified Cys273 and Cys288 as playing essential roles in sensing oxidative stress (27). Recently, it was reported that mutation of Cys273 to Ser reduced Keap1-dependent ubiquitination of Nrf2, resulting in the stabilization of Nrf2 (31). Considering these lines of evidence, we next examined the contribution of these cysteine residues to the formation of the Cul3 complex by exploiting cysteine point mutants of Keap1 (Fig. 5B) (27). However, mutation of these cysteine residues within the IVR domain did not significantly affect the association of Keap1 with Cul3, indicating that Cul3 recognizes the interface that is formed by other residues in the IVR domain.
As a reverse strategy, we generated two Cul3 deletion mutants, N280 and ΔN280 (Fig. 5C), and examined their interaction with Keap1. Crystal structure analysis of Cul1 showed that the N-terminal end region of Cul1, which is conserved among the Cul proteins, is the surface of Cul1 that directly associates with the adaptor protein Skp1 (32). Consistent with this observation, Keap1 coimmunoprecipitated with N280 (Fig. 5D, lane 5) but did not recognize the C terminus of Cul3 (lane 7, ΔN280). These results indicate that the association between Keap1 and Cul3 is achieved in an N280 domain-specific manner.
The BTB domain of Bach1 does not bind to Cul3.
Although Cul3 was reported to interact with BTB domains, we found that Cul3 recognizes the IVR domain but not the BTB domain of Keap1. To examine whether this nature of Cul3 is specific for Keap1 or can be seen with other proteins, we performed a similar immunoprecipitation analysis with 293T cells expressing Bach1. Bach1 is known to be an Nrf2-related transcription factor and contains a BTB domain (18). We did not detect any positive bands that specifically interacted with an anti-Myc antibody in the immunoprecipitates with an anti-Flag antibody, indicating that Bach1 cannot interact with Cul3 (Fig. 6, lane 4). Since we could clearly reproduce an association between Cul3 and a control BTB protein under the same conditions (data not shown), these data indicate that Cul3 preferentially interacts with certain types of BTB protein as a substrate-specific adaptor.
In vivo ubiquitination of Nrf2 by the Cul3-Keap1 complex.
To investigate whether the Keap1-Cul3 complex contributes to the ubiquitination process of Nrf2, we performed an in vivo ubiquitination assay. For this purpose, we transfected into 293T cells the expression plasmids for Nrf2, Keap1, Cul3, and Roc1 (a subunit of the Cul3 complex). We also simultaneously expressed histidine-tagged ubiquitin (26) in the cells. The latter experiment enabled us to purify ubiquitinated Nrf2 by using nickel affinity beads. At 24 h after transfection, the cells were treated with MG132 for 12 h to inhibit the proteasomal degradation of Nrf2. Nrf2 was examined by immunoblot analysis with an anti-Nrf2 antibody.
In the immunoblot analysis, we first examined Nrf2 in the nickel bead precipitates after purification of ubiquitinated Nrf2 from whole-cell extracts (Fig. 7). For the cells expressing wild-type Nrf2, the immunoblot analysis detected multiple bands and showed a smear migration pattern, suggesting that Nrf2 is conjugated with ubiquitin chains in various patterns. Importantly, while the smearing pattern was weak for Nrf2 expressed alone (Fig. 7, upper panel, lane 2), concomitant expression of Keap1 promoted Nrf2 ubiquitination (lane 3). Coexpression of Cul3 and Roc1 enhanced Nrf2 ubiquitination, albeit weakly (lane 5) (see Discussion). In the absence of Keap1, Cul3 plus Roc1 did not provoke ubiquitination of Nrf2 (lane 4). Furthermore, the ΔETGE mutant of Nrf2 did not show this pattern of smearing and ubiquitination (lanes 6 to 9). These results thus demonstrate that the Nrf2 ubiquitination process is strictly dependent on the presence of and interaction with Keap1.
FIG. 7.
Ubiquitination of Nrf2 by Keap1 and Cul3 in vivo. Nrf2 (1 μg) was expressed in 293T cells, along with several combinations of Keap1 (0.5 μg) and Cul3 (1.5 μg)-Roc1 (1 μg), as indicated in the figure, in the presence of His-tagged ubiquitin (HisUb; 1 μg). As a control, the ΔETGE mutant was also transfected. Whole-cell extracts were prepared and subjected to affinity purification with Ni2+ resin. Precipitates (ppt) were eluted by boiling in sodium dodecyl sulfate sample buffer and subjected to immunoblot (IB) analysis (upper panel) with anti-Nrf2 antibody. The expression level of Nrf2 in the whole-cell extracts was also verified by immunoblot analysis with anti-Nrf2 antibody (lower panel).
We observed enhancement of Keap1-based Nrf2 ubiquitination by Cul3 and Roc1 more clearly by immunoblot analysis using whole-cell extracts (Fig. 7, lower panel, lanes 2 to 5) than when using nickel affinity column-purified immunoprecipitates. Again, the ΔETGE mutant of Nrf2 that lacks the ability to bind to Keap1 did not effectively promote the ubiquitination of Nrf2 (lanes 6 to 9). Taken together, these data demonstrate that the Keap1-Cul3 complex ubiquitinates Nrf2 and regulates the turnover of Nrf2.
DISCUSSION
In this study, we aimed to clarify the molecular mechanisms governing rapid Nrf2 turnover and the contribution of Keap1 to this degradation pathway. We revealed that the rapid degradation of Nrf2 requires direct association with Keap1. We also found that the IVR domain of Keap1 specifically interacts with Cul3, a component of the E3 ligase complex. These results provide the first convincing evidence for proteasomal degradation of Nrf2 and the function of Keap1 as an adaptor for Cul3-based E3 ligase (Fig. 8). The critical stress-responsive transcription factors IκB, Hif-1α, and Nrf2 have now been shown to share the ubiquitin-proteasome system in their rapid turnover and to use specific Cul-type E3 ligases, with IκB, Hif-1α, and Nrf2 using Cul1, Cul2, and Cul3, respectively. To our knowledge, Nrf2 and Keap1 are the first mammalian substrate and adaptor reported for the Cul3-based E3 ligase system.
FIG. 8.
Schematic model of the Keap1-Cul3 complex function as an E3 ligase. The cytoplasmic factor Keap1, bound on actin filaments, acts as a sensor for oxidative and electrophilic stress through two cysteine residues in the IVR domain. In the absence of stimuli, Keap1 sequesters the transcription factor Nrf2, a major regulator of the oxidative stress response, in the cytoplasm. In addition, Keap1 functions as an adaptor of the Cul3-based E3 ligase. This E3 ligase conjugates ubiquitin to Nrf2 and promotes rapid degradation of Nrf2 by proteasome in order to inhibit the expression of oxidative stress response genes under normal conditions.
While Cul3 in Caenorhabditis elegans was reported to ubiquitinate the substrate MEI-1/katanin through MEL26 as an adaptor (20, 21), the molecular mechanisms of Cul3 activity remain to be elucidated. A subset of proteins harboring the BTB domain were recently reported to serve as an adaptor in the Cul3-based E3 ligase system (4, 5, 21, 30). Our present data indicate that the BTB domain of Keap1 is necessary for Keap1 to function as an accelerator of Nrf2 degradation. However, one unexpected finding in this study is that deletion of the BTB domain does not significantly affect the association of Keap1 with Cul3. Rather, Keap1 effectively associates with Cul3 through the IVR domain. The BTB domain of Bach1 does not bind to Cul3 either (Fig. 6), demonstrating that not all BTB domains interact with Cul3.
The molecular mechanism by which the IVR domain interacts with Cul3 is unclear at present. Zhang and Hannink recently reported that mutation of the reactive cysteines Cys273 and Cys288 in the IVR domain repressed Nrf2 degradation due to an impaired ubiquitin pathway (31). Based on their observation, they proposed the hypothesis that these two cysteines in the IVR domain are crucial for the complex formation between Keap1 and an unknown E3 ligase. Although our present data indicate that Cul3 recognizes the IVR domain, our data clearly show that Cul3 binds to Keap1 despite mutations in the two reactive cysteines, indicating that Cul3 recognizes an alternative motif in the IVR domain. Furthermore, alanine substitutions of both Cys273 and Cys288 did not affect the ubiquitination of Nrf2 (data not shown). Thus, the present results disagree with their report, so this point remains to be clarified.
We envisage that two lines of evidence may be pertinent in this regard. First, the electrophilic reagent Dex-mes binds to the two reactive cysteines in the IVR domain (3) and liberates Nrf2 from Keap1. Second, Keap1 containing alanine substitutions of both Cys273 and Cys288 did not effectively repress the transactivation activity of Nrf2 in a cell culture system (27). Based on these observations, we propose that the two reactive cysteine residues in the IVR regulate the association of Keap1 with Nrf2.
Our results in Fig. 3 suggest that the BTB domain of Keap1 contributes to the turnover of the Nrf2 protein. Consistent with these data Zipper and Mulcahy (33) showed that the BTB domain of Keap1 is crucial for its dimerization and negative regulation of Nrf2. Furthermore, Zhang and Hannink (31) have recently reported that a C151S Keap1 mutation in the BTB domain significantly reduced Nrf2 activation in response to oxidants (31). Thus, while our analysis indicates that Cys151 is not a direct target of Dex-mes conjugation (3), it seems likely that this residue contributes to the function of Keap1. Further work to clarify the role of BTB in the Keap1/Nrf2 pathway is required.
Stresses on the endoplasmic reticulum were recently reported to activate the Nrf2-Keap1 system through direct phosphorylation of Nrf2 by pancreatic endoplasmic reticulum eukaryotic initiation factor 2α kinase (2). This observation suggests that the Nrf2-Keap1 system can be activated by signals other than oxidative stress. The ubiquitin-like protein NEDD8 regulates the E3 ligase activity of Cul3 by covalent modification, which is essential for the association of Cul3 with E2 enzyme (8, 20). The Cul3 complex contains a COP9 signalosome that functions as a deneddylation enzyme (1, 29). Hence, one emerging hypothesis is that down-regulation of Cul3 activity by deneddylation may enable Nrf2 stabilization, by which signals can induce the expression of cytoprotective target genes. Supporting this notion, the addition of a proteasome inhibitor was reported to induce the expression of the GCL gene, which encodes the catalytic subunit of γ-glutamylcysteine synthetase (22). Under our present experimental conditions, however, we could not detect the promotion of NEDD8 modification of Cul3 by oxidative stress (data not shown). Nonetheless, we feel that it is still of interest to explore this hypothesis.
The Neh2 domain of Nrf2 is ubiquitinated in a Keap1-dependent manner and possesses seven lysine residues, some of which might be conjugated with a polyubiquitin chain (Y. Kato, K. Itoh, and M. Yamamoto, submitted for publication). In addition, deletion of the C-terminal Neh1, Neh3, and Neh6 domains of Nrf2 renders the protein more stable than full-length Nrf2 (Fig. 1 and 2, compare 7.5 and 25 min, respectively), suggesting that Nrf2 contains an alternative degradation signal in this C-terminal portion. We previously showed two modes of Nrf2 degradation, namely, Keap1-dependent degradation in the cytoplasm and Keap1-independent degradation in the nucleus (9). The turnover of Nrf2 by the latter pathway is slower than that by the former. We surmise that Nrf2 degradation in the nucleus also uses the proteasome pathway but must be Cul3 independent. Therefore, the next important task would be to identify an E3 ligase complex that is essential for the nuclear degradation of Nrf2.
We exploited Cul3 double-stranded RNA (dsRNA) in a preliminary examination to assess whether endogenous Cul3 regulates the degradation of Nrf2 in collaboration with Keap1. Cul3 dsRNA was transfected into HeLa cells, and cyclin E was first analyzed as a control, since Cul3 was reported to determine the stability of cyclin E in the ubiquitin-proteasome system (24). However, although transfection of Cul3 dsRNA significantly affected the expression of Cul3 mRNA, only a marginal accumulation of cyclin E was found (data not shown). Similarly, down-regulation of Cul3 did not affect the stability of endogenous Nrf2 (data not shown). One plausible explanation for this observation is that because the dsRNA did not completely abrogate the Cul3 protein, the residual small amount of Cul3 might have been enough for rapid degradation of Nrf2 to take place. Supporting this hypothesis, we found that expression of Keap1 alone significantly enhanced the ubiquitination of Nrf2 in an in vivo ubiquitin assay whereas further expression of Cul3 and Roc1 enhanced ubiquitination only marginally (Fig. 7). These results suggest that Cul3 and Roc1 are abundant within living cells. An alternative explanation is that Keap1-independent degradation, as described above, might compensate for the loss of Cul3 function.
The function of Keap1 as an E3 ligase adaptor has similarity to the function of pVHL. It has been well documented that multiple human diseases are provoked by mutations in pVHL. These mutations usually affect the function of pVHL as an adaptor and cause aberrant stabilization of Hif-1α (11). Thus, cellular homeostasis requires not only inducible activation of transcription factors in response to stress stimuli but also continuous inactivation of the transcription factors through their rapid degradation or subcellular compartmentalization during unstressed conditions. Interestingly, a subset of Cul proteins requires WD40 repeat proteins as an adaptor to target substrates. This WD40 repeat domain is known to form a β-barrel structure (28). The DGR domain of Keap1 also forms a β-barrel structure. These observations further support our contention that Cul-based E3 ligases have common properties in both function and structure.
Acknowledgments
We are grateful to Tomohiko Ohta, Dirk Bohmann, and Tae Yamamoto for their generous donation of plasmids. We also thank Ken Itoh, Hozumi Motohashi, Makoto Kobayashi, Yasutake Kato, and Tania O'Connor for discussion and advice.
This work was supported in part by grants-in-aid from JST-ERATO (M.Y.), Ministry of Education, Sports, Science and Technology (A.K. and M.Y.), Ministry of Health, Labor and Welfare (M.Y.), Atherosclerosis Foundation (M.Y.), and Naito Foundation (M.Y.).
REFERENCES
- 1.Cope, G. A., and R. J. Deshaies. 2003. COP9 signalosome: a multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell 114:663-671. [DOI] [PubMed] [Google Scholar]
- 2.Cullinan, S. B., D. Zhang, M. Hannink, E. Arvisais, R. J. Kaufman, and J. A. Diehl. 2003. Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival. Mol. Cell. Biol. 23:7198-7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dinkova-Kostova, A. T., W. D. Holtzclaw, R. N. Cole, K. Itoh, N. Wakabayashi, Y. Katoh, M. Yamamoto, and P. Talalay. 2002. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc. Natl. Acad. Sci. USA 99:11908-11913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Furukawa, M., Y. J. He, C. Borchers, and Y. Xiong. 2003. Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat. Cell Biol. 5:1001-1007. [DOI] [PubMed] [Google Scholar]
- 5.Geyer, R., S. Wee, S. Anderson, J. Yates, and D. A. Wolf. 2003. BTB/POZ domain proteins are putative substrate adaptors for cullin 3 ubiquitin ligases. Mol. Cell 12:783-790. [DOI] [PubMed] [Google Scholar]
- 6.Hayes, J. D., S. A. Chanas, C. J. Henderson, M. McMahon, C. Sun, G. J. Moffat, C. R. Wolf, and M. Yamamoto. 2000. The Nrf2 transcription factor contributes both to the basal expression of glutathione S-transferases in mouse liver and to their induction by the chemopreventive synthetic antioxidants, butylated hydroxyanisole and ethoxyquin. Biochem. Soc. Trans. 28:33-41. [DOI] [PubMed] [Google Scholar]
- 7.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 8.Hori, T., F. Osaka, T. Chiba, C. Miyamoto, K. Okabayashi, N. Shimbara, S. Kato, and K. Tanaka. 1999. Covalent modification of all members of human cullin family proteins by NEDD8. Oncogene 18:6829-6834. [DOI] [PubMed] [Google Scholar]
- 9.Itoh, K., N. Wakabayashi, Y. Katoh, T. Ishii, T. O'Connor, and M. Yamamoto. 2003. Keap1 regulates both cytoplasmic-nuclear shuttling and degradation of Nrf2 in response to electrophiles. Genes Cells 8:379-391. [DOI] [PubMed] [Google Scholar]
- 10.Itoh, K., T. Ishii, N. Wakabayashi, and M. Yamamoto. 1999. Regulatory mechanisms of cellular response to oxidative stress. Free Radic. Res. 31:319-324. [DOI] [PubMed] [Google Scholar]
- 11.Kaelin, W. G., Jr. 2002. Molecular basis of the VHL hereditary cancer syndrome. Nat. Rev. Cancer 2:673-682. [DOI] [PubMed] [Google Scholar]
- 12.Kang, M.-I., A. Kobayashi, N. Wakabayashi, S. G. Kim, and M. Yamamoto. 2004. Scaffolding of Keap1 to the actin cytoskeleton controls the function of Nrf2 as key regulator of cytoprotective phase 2 genes. Proc. Natl. Acad. Sci. USA 101:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi, M., K. Itoh, T. Suzuki, H. Osanai, K. Nishikawa, Y. Katoh, Y. Takagi, and M. Yamamoto. 2002. Identification of the interactive interface and phylogenic conservation of the Nrf2-Keap1 system. Genes Cells 7:807-820. [DOI] [PubMed] [Google Scholar]
- 14.Levonen, A. L., A. Landar, A. Ramachandran, E. K. Ceaser, D. A. Dickinson, G. Zanoni, J. D. Morrow, and V. M. Darley-Usmar. 2004. Cellular mechanisms of redox cell signaling: role of cysteine modification in controlling antioxidant defenses in response to electrophilic lipid oxidation products. Biochem. J. 378:373-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McMahon, M., K. Itoh, M. Yamamoto, and J. D. Hayes. 2003. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J. Biol. Chem. 278:21592-21600. [DOI] [PubMed] [Google Scholar]
- 16.Nguyen, T., P. J. Sherratt, and C. B. Pickett. 2003. Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu. Rev. Pharmacol. Toxicol. 43:233-260. [DOI] [PubMed] [Google Scholar]
- 17.Nguyen, T., P. J. Sherratt, H. C. Huang, C. S. Yang, and C. B. Pickett. 2003. Increased protein stability as a mechanism that enhances Nrf2-mediated transcriptional activation of the antioxidant response element. Degradation of Nrf2 by the 26 S proteasome. J. Biol. Chem. 278:4536-4541. [DOI] [PubMed] [Google Scholar]
- 18.Oyake, T., K. Itoh, H. Motohashi, N. Hayashi, H. Hoshino, M. Nishizawa, M. Yamamoto, and K. Igarashi. 1996. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol. Cell. Biol. 16:6083-6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pickart, C. M. 2001. Ubiquitin enters the new millennium. Mol. Cell 8:499-504. [DOI] [PubMed] [Google Scholar]
- 20.Pintard, L., T. Kurz, S. Glaser, J. H. Willis, M. Peter, and B. Bowerman. 2003. Neddylation and deneddylation of CUL-3 is required to target MEI-1/katanin for degradation at the meiosis-to-mitosis transition in C. elegans. Curr. Biol. 13:911-921. [DOI] [PubMed] [Google Scholar]
- 21.Pintard, L., J. H. Willis, A. Willems, J. L. Johnson, M. Srayko, T. Kurz, S. Glaser, P. E. Mains, M. Tyers, B. Bowerman, and M. Peter. 2003. The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature 425:311-316. [DOI] [PubMed] [Google Scholar]
- 22.Sekhar, K. R., S. R. Soltaninassab, M. J. Borrelli, Z. Q. Xu, M. J. Meredith, F. E. Domann, and M. L. Freeman. 2000. Inhibition of the 26S proteasome induces expression of GLCLC, the catalytic subunit for gamma-glutamylcysteine synthetase. Biochem. Biophys. Res. Commun. 270:311-317. [DOI] [PubMed] [Google Scholar]
- 23.Semenza, G. L. 2001. HIF-1 and mechanisms of hypoxia sensing. Curr. Opin. Cell Biol. 13:167-171. [DOI] [PubMed] [Google Scholar]
- 24.Singer, J. D., M. Gurian-West, B. Clurman, and J. M. Roberts. 1999. Cullin-3 targets cyclin E for ubiquitination and controls S phase in mammalian cells. Genes Dev. 13:2375-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stewart, D., E. Killeen, R. Naquin, S. Alam, and J. Alam. 2003. Degradation of transcription factor Nrf2 via the ubiquitin-proteasome pathway and stabilization by cadmium. J. Biol. Chem. 278:2396-2402. [DOI] [PubMed] [Google Scholar]
- 26.Treier, M., L. M. Staszewski, and D. Bohmann. 1994. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell 78:787-798. [DOI] [PubMed] [Google Scholar]
- 27.Wakabayashi, N., A. T. Dinkova-Kostova, W. D. Holtzclaw, M-I. Kang, A. Kobayashi, M. Yamamoto, T. W. Kensler, and P. Talalay. 2004. Protection against electrophile and oxidant stress by induction of the phase 2 response: Fate of cysteines of the Keap1 sensor modified by inducers. Proc. Natl. Acad. Sci. USA 101:2040-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wall, M. A., D. E. Coleman, E. Lee, J. A. Iniguez-Lluhi, B. A. Posner, A. G. Gilman, and S. R. Sprang. 1995. The structure of the G protein heterotrimer Gi alpha 1 beta 1 gamma 2. Cell 83:1047-1058. [DOI] [PubMed] [Google Scholar]
- 29.Wolf, D. A., C. Zhou, and S. Wee. 2003. The COP9 signalosome: an assembly and maintenance platform for cullin ubiquitin ligases? Nat. Cell Biol. 5:1029-1033. [DOI] [PubMed] [Google Scholar]
- 30.Xu, L., Y. Wei, J. Reboul, P. Vaglio, T. H. Shin, M. Vidal, S. J. Elledge, and J. W. Harper. 2003. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature 425:316-321. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, D. D., and M. Hannink. 2003. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol. Cell. Biol. 23:8137-8151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng, N., B. A. Schulman, L. Song, J. J. Miller, P. D. Jeffrey, P. Wang, C. Chu, D. M. Koepp, S. J. Elledge, M. Pagano, R. C. Conaway, J. W. Conaway, J. W. Harper, and N. P. Pavletich. 2002. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416:703-709. [DOI] [PubMed] [Google Scholar]
- 33.Zipper, L. M., and R. T. Mulcahy. 2002. The Keap1 BTB/POZ dimerization function is required to sequester Nrf2 in cytoplasm. J. Biol. Chem. 277:36544-36552. [DOI] [PubMed] [Google Scholar]