Abstract
When co-overexpressed, the epidermal growth factor receptor (EGFR) and c-Src cooperate to cause synergistic increases in EGF-induced DNA synthesis, soft agar colony growth, and tumor formation in nude mice. This synergy is dependent upon c-Src-mediated phosphorylation of a unique tyrosine on the EGFR, namely, tyrosine 845 (Y845). Phenylalanine substitution of Y845 (Y845F) was found to inhibit EGF-induced DNA synthesis without affecting the catalytic activity of the receptor or its ability to phosphorylate Shc or activate mitogen-activated protein kinase. These results suggest that synergism may occur through alternate signaling pathways mediated by phosphorylated Y845 (pY845). One such pathway involves the transcription factor Stat5b. Here we describe another pathway that involves cytochrome c oxidase subunit II (CoxII). CoxII was identified as a specific binding partner of a pY845-containing peptide in a phage display screen. EGF-dependent binding of CoxII to the wild type but not to the mutant Y845F-EGFR was confirmed by coimmunoprecipitation experiments. This association also required the kinase activity of c-Src. Confocal microscopy, as well as biochemical fractionation, indicated that the EGFR translocates to the mitochondria after EGF stimulation, where it colocalizes with CoxII. Such translocation required the catalytic activity of the receptor but not phosphorylation of Y845. However, ectopic expression of the Y845F-EGFR prevented the EGF from protecting MDA-MB-231 breast cancer cells from adriamycin-induced apoptosis, whereas two mutants of Stat5b, a dominant-interfering mutant (DNstat5b) and a tyrosine mutation at 699 (Y699F-Stat5b) did not. Taken together, these data suggest that, through the ability of EGFR to translocate to the mitochondria, the binding of proteins such as CoxII to pY845 on the EGFR may positively regulate survival pathways that contribute to oncogenesis.
The epidermal growth factor receptor (EGFR) is a critical regulator of many normal cellular processes, including cell growth, differentiation, survival, and migration. The EGFR also is implicated in pathological processes of cellular transformation and oncogenesis due to its overexpression in many types of cancers (including 20 to 30% of all breast cancers) (2) and its ability to induce morphological transformation of cultured cells and tumor formation in nude mice (11, 37). Many different signaling pathways have been discovered to mediate the effects of EGF, the most notable being Ras-mitogen-activated protein kinase (MAPK) and the phosphatidylinositol 3-kinase pathways (reviewed in reference 9). In these pathways EGF binds to the EGFR to induce dimerization, catalytic activation, and autophosphorylation of tyrosines in the C-terminal tail of the EGFR. These phosphorylated tyrosines provide docking sites for adapter proteins such as Grb2, Shc, and Gab2 to link the receptor to the Ras-MAPK, as well as phosphatidylinositol 3-kinase pathways. Interestingly, activation of both pathways has been implicated in both normal and pathological processes involving the EGFR.
Co-overexpression of the EGFR and the nonreceptor tyrosine kinase c-Src has been observed in a number of breast cancer tissues and cell lines (reviewed in reference 5), and those exhibiting such co-overexpression display more aggressive growth in nude mice than those overexpressing only one of the pair (3). In addition, in a murine fibroblast model system engineered to overexpress both the EGFR and c-Src, synergistic increases in DNA synthesis, anchorage-independent cell growth, and tumorigenesis in nude mice have been observed, compared to the expression of the EGFR or c-Src alone (22). In an attempt to understand the molecular mechanism of this synergism, a unique site of tyrosine phosphorylation was identified within the activation loop of the kinase domain on the EGFR, tyrosine 845 (Y845) (4). Phosphorylation of this highly conserved tyrosine is mediated by c-Src and dependent on EGF stimulation (4, 35, 44). Of particular interest, a mutant receptor harboring a phenylalanine substitution for Y845 (Y845F) acts in a dominant-interfering fashion and inhibits the ability of EGF to induce DNA synthesis without affecting the kinase activity of the receptor or its ability to recruit and phosphorylate substrates such as Shc or to activate MAPK (35). These data suggest that signals emanating from phosphorylated Y845 (pY845) on the EGFR may involve effector molecules other than Shc and MAPK.
One such effector molecule is the transcription factor Stat5b. Functionally, Stat5b, as well as Stat1, -3, and -5a, has been shown to be important not only for cytokine-regulated events but also for processes regulated by G protein-coupled agonists and peptide growth factors, including EGF (33). However, studies by Silva and coworkers (20) have specifically demonstrated that EGF stimulates the tyrosine phosphorylation and subsequent transcriptional activation of Stat5b in a pY845-dependent manner. In addition, dominant-negative Stat5b abolishes EGF-induced DNA synthesis (20). Together, these findings suggest that pY845 mediates its effects on EGF-induced cell proliferation at least in part through Stat5b. However, it is possible that pY845 has additional downstream effectors that could cooperate with Stat5b or function in any one of the multitude of cellular processes influenced by EGF.
We identify here a novel binding partner for pY845, namely, cytochrome c oxidase subunit II (CoxII). CoxII is a mitochondrion-encoded protein that is a critical component of the oxidative phosphorylation pathway (8). It traverses the inner membrane of the mitochondria twice, with its N and C termini located in the inner-membrane space (see Fig. 1B). CoxII is encoded by mitochondrial DNA, synthesized in the mitochondria, and inserted into the inner membrane by mitochondrion-dependent pathways (13). It also has been shown to bind directly to cytochrome c and is speculated to regulate apoptosis through this affinity for cytochrome c (7, 45). We demonstrate in the present study that when the EGFR associates with CoxII it localizes at or near the mitochondria. Furthermore, transient ectopic expression of the Y845F mutant receptor reduces EGF-dependent survival of breast cancer cells that overexpress wild-type (wt) EGFR, suggesting that the interaction between the EGFR and CoxII may play a role in the regulation of apoptosis.
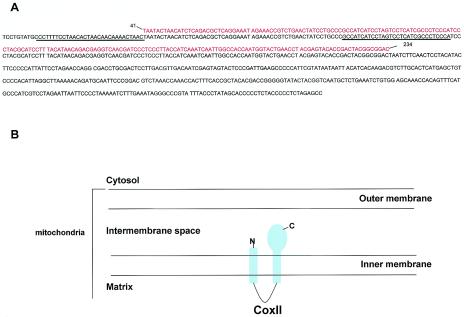
FIG. 1.
Identification of CoxII clones and structure of CoxII. (A) Using phage display, clones 6, 12, 14, 16, and 19 (red text) were identified as being identical to the human CoxII mitochondrial protein (black text). The sequence identified in the screen correlates with the matrix-embedded loop formed by the two transmembrane domains (underlined) the C-terminal transmembrane domain, and a portion of the C-terminal tail. (B) Model of CoxII localization in the mitochondria.
MATERIALS AND METHODS
Reagents.
Peptides corresponding to sequences surrounding Y845 of the EGFR were synthesized in the University of Virginia Biomolecular Research Facility. The sequence of the unphosphorylated 845 peptide (Y845) is LLGAEEKEYHAEGGKVPIKW, and the sequence of the phosphorylated 845 peptide (pY845) is LLGAEEKEpYHAEGGKVPIKW.
DNA constructs used in the present study included pcDNA-EGFR, pcDNA-c-Src, pcDNA Y845F-EGFR, pcDNA K721A-EGFR (kinase defective), pcDNA K−c-Src, hemagglutinin (HA)-tagged wt-Stat5b, Y699F-Stat5b, and DN-Stat5b.
Phage display.
The technique of phage display was utilized to identify proteins that bind pY845. Reagents and procedures supplied by the manufacturer (Novagen, Madison, Wis.) were modified slightly, as described previously (46). Specifically, the peptides described above were biotinylated for use with streptavidin-conjugated magnetic beads. Biotinylation was performed as follows, First, 2 mg of peptide was biotinylated by incubation with 2.5 mg of biotin for 2 h at 4°C according to the manufacturer's recommendation (Pierce, Rockford, Ill.). A rabbit anti-Y845 peptide antibody (made in collaboration with BioSource, Inc., Camarillo, Calif.) was then incubated with the biotin-plus-peptide solution for an additional 2 h at 4°C. To separate biotin-conjugated peptide from free biotin, a slurry of protein A-agarose beads (50 μl; UBI, Waltham, Mass.) was then added for 30 min at 4°C. The slurry was washed three times with phosphate-buffered saline (PBS) and once with 10 mM sodium phosphate (pH 6.8). The peptide-biotin conjugate was eluted from the antibody with 50 μl of 100 mM glycine (pH 1.8) for 5 min at room temperature. This eluant was then incubated with 500 μl of streptavidin MagneSphere Paramagnetic Particlues (Promega, Madison, Wis.) for 30 min at room temperature to absorb biotin-conjugated peptide from the solution. The beads were washed three times with PBS and stored in 500 μl of PBS plus 0.1% NP-40 (PBS-NH) until use.
The biotinylated pY845 peptide linked to streptavidin beads was incubated with an amplified breast tumor tissue phage library (Novagen) for 30 min at room temperature, and beads were washed four times with PBS-NH. Phage expressing specific phosphotyrosine-binding proteins were eluted by incubation with 100 mM phenylphosphate for 15 min at room temperature. The eluant was added to BLT5614 bacteria growing at log phase and induced with IPTG (isopropyl-β-d-thiogalactopyranoside) to express phage-binding proteins. This incubation continued until the eluted phage lysed the bacteria, at which time the solution was centrifuged at 10,000 × g for 10 min. The supernatant was saved, and the procedure described above was repeated to select for higher-affinity binding interactions.
Positive phage clones were screened against the nonphosphorylated Y845 peptide to eliminate proteins that failed to bind in a pY845-specific manner. Clones deemed positive were used to infect a lawn of log-phase bacteria, from which plaques were picked and subjected to DNA extraction. DNA was amplified by using PCR primers supplied by Novagen. PCR products were separated by agarose gel electrophoresis, purified by using gel extraction (Qiagen, Valencia, Calif.), and sequenced by the University of Virginia Biomolecular Research Facility. Identities of the sequences were determined by comparison with multiple databases by using the basic local alignment search tool (BLAST) search engine.
Cell lines and transient-transfection assays.
Stable, clonal cell lines of C3H10T1/2 murine fibroblasts overexpressing c-Src alone or EGFR and c-Src together (22, 41) were cultured in Dulbecco modified Eagle medium (DMEM; Gibco-BRL, Carlsbad, Calif.) containing 10% fetal bovine serum (FBS) and 1 mg of G418 (Gibco-BRL)/ml. Cos-7 cells were cultured in the same medium lacking G418. The breast cancer cell lines, MDA-MB-468 and MDA-MB-231, were cultured in DMEM containing 10% FBS and 1 mM sodium pyruvate.
Cos-7 cells were transfected by using the Effectene transfection reagents as directed by the manufacturer for 100-mm plates (Qiagen). MDA-MB-231 were transfected by using FuGENE6 (Roche) or by electroporation, as indicated. Briefly, cells were treated with trypsin, washed three times in PBS, and resuspended in PBS to a concentration of 5 × 106 cells/ml. Electroporation was carried out in 0.4-μm cuvettes by using the Bio-Rad Gene-Pulser electroporator (Hercules, Calif.) at 250 μF and 400 V in the presence of 10 μg of DNA. Transfected cells were plated on coverslips in DMEM plus 10% FBS for 3 h to recover and then placed in the culture conditions described for each experiment. Control electroporation experiments with green fluorescent protein demonstrated a >60% transfection efficiency.
Immunoprecipitation and immunoblotting.
Fibroblasts, Cos7 cells, and breast cancer cell lines were cultured in the medium described above for 24 h. Cells were placed in serum-free (SF) medium for 18 to 24 h and then stimulated with 50 ng of EGF (Sigma, St. Louis, Mo.)/ml for the indicated time period. EGF was removed by three washes with PBS. Cells were then lysed on ice in CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate} lysis buffer (10 mM CHAPS, 50 mM Tris [pH 8.0], 150 mM NaCl, 2 mM EDTA) containing protease and tyrosine phosphatase inhibitors (1 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM sodium orthovanadate, and 17 μg of aprotinin/ml), and lysates were clarified by centrifugation at 13,000 rpm for 10 min at 4°C. Protein was quantitated by using the Bradford assay (Bio-Rad). Then, 500 μg of lysate protein was precleared with 40 μl of protein A-agarose beads for 30 min at 4°C and incubated with EGFR antibody (monoclonal antibody [MAb] 108) (43) or a negative antibody control for 1 h at 4°C, with constant rocking. Protein A-agarose beads were then added (40 μl), followed by incubation for an additional 30 min at 4°C. Immunoprecipitates were washed three times with CHAPS or radioimmunoprecipitation assay lysis buffer (1% NP-40, 1% deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 0.15 M NaCl, 0.01 sodium phosphate dibasic, 2 mM EDTA, 50 mM NaF) and three times with TBS (Tris-buffered saline [pH 8.0]). Precipitated proteins were resolved by SDS-polyacrylamide gel electrophoresis on 7.5 or 10% acrylamide gels and electrophoretically transferred to polyvinylidene difluoride (PVDF) membranes. For direct immunoblotting of cellular lysates or subcellular fractions, the indicated amounts of lysate or equal volumes of subcellular fractions were subjected to SDS-7.5 or 10% PAGE and transferred to PVDF membranes. Immunoblotting was carried out with the following antibodies at the indicated dilutions: EGFR (rabbit Ab12 at 1:250; NeoMarkers, Fremont, Calif.), CoxII (mouse MAb at 1:250; Molecular Probes, Eugene, Oreg.), EEA1 (mouse MAb at 1:250; Transduction Labs, San Jose, Calif.), Lamp1 (mouse MAb at 1:1,000; Transduction Labs), nucleoporin (mouse MAb at 1:1,000; Transduction Labs), pY845 EGFR (rabbit polyclonal at 1:500; Biosource), phosphotyrosine (RC20H at 1:2,000 [Transduction Labs] or PY99 mouse MAb at 1:1,000 [Santa Cruz]), or cytochrome c (mouse MAb at 1:1,000; Transduction Labs). Briefly, PVDF membranes were blocked in a solution of 5% nonfat dry milk or 5% bovine serum albumin (where indicated) in TBS-T (TBS containing 0.1% Tween 20) for 1 h at room temperature. The membrane was then incubated with the primary antibodies listed above for 1 to 2 h, with the indicated concentrations in 5% nonfat dry milk or bovine serum albumin. Membranes were washed three times for 5 min with TBS-T, incubated at a 1:5,000 dilution of anti-mouse or -rabbit horseradish peroxidase (Amersham, Piscataway, N.J.) for 1 h, and washed three times with TBS-T. Antigen-antibody complexes were visualized with enhanced chemiluminescence reagent (Amersham), followed by autoradiography.
Confocal microscopy.
C3H10T1/2 murine fibroblasts overexpressing the EGFR and c-Src- or Src-overexpressing fibroblasts transiently transfected with Y845F-EGFR or K721A-EGFR were plated on coverslips in six-well tissue culture dishes (Corning, Corning, N.Y.) at a density of 100,000 cells per coverslip. Cells were serum starved for 24 h and stimulated with 50 ng of EGF/ml for 0 to 30 min at 37°C. MitoTracker Red (MTR; Molecular Probes) was added at 500 nM for 30 min (prior to and during ligand stimulation) to allow for incorporation into mitochondria. Cells were then fixed in 3% paraformaldehyde for 20 min and permeabilized with 0.1% Triton X-100 and 0.1% sodium citrate for 2 min on ice. Nonspecific antibody binding sites were blocked by incubation with 20% goat serum for 30 min at 37°C, and blocked cells were probed with EGFR antibody (mouse MAb at 1:1,000 or 1:250 for time course studies [Transduction Labs] and sheep antibody at 1:250 for triple staining [UBI]) and/or CoxII antibody (mouse MAb at 1:500; Molecular Probes) for 1 h at 37°C. Cells were washed three times with PBS and incubated with fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse, FITC-conjugated donkey anti-sheep, or Alexa Fluor 647-conjugated goat anti-mouse antibody (Molecular Probes) at 1:1,000 for 1 h at room temperature in the dark. Again, cells were washed three times with PBS and once with water and then mounted onto microscope slides with Vectashield mounting medium and Cytoseal. Images were captured with a Zeiss PASCAL confocal microscope.
Cellular fractionation.
Cells were seeded into 20 100-mm plates, grown to 50% confluency, serum starved for 24 h, and stimulated with 50 ng of EGF/ml for 15 min. Washed monolayers were then scraped from the plate in PBS and pelleted. Cells were swollen in homogenization buffer (250 mM sucrose, 1 mM EDTA, 50 mM Tris-HCl [pH 7.4], and 1 mM dithiothreitol) plus inhibitors (1 mM PMSF, 9 mM benzamidine, 0.28 U of aprotinin/ml, 50 μg of leupeptin/ml, 7 μg of pepstatin A/ml, and 100 μM sodium orthovanadate) for 1 h and lysed by Dounce homogenization (50 strokes) on ice. Unbroken cells were pelleted by centrifugation at 14,000 × g for 10 s. The nuclear fraction was isolated by centrifugation of the supernatant at 1,000 × g for 10 min at 4°C. The supernatant from the nuclear fraction was then pelleted at 3,000 × g for 10 min at 4°C to isolate the heavy membrane (mitochondrial) pellet or at 14,000 × g for 10 min at 4°C to isolate the light membrane fraction. The heavy mitochondrial pellet was washed three times with wash buffer (250 mM sucrose, 1 mM EGTA, 10 mM Tris [pH 7.4]) to remove any contaminating lysosomes and endosomes. The remaining supernatant was concentrated by trichloroacetic acid precipitation, and all pellets were resuspended in 300 μl of CHAPS or radioimmunoprecipitation assay buffer with protease and phosphatase inhibitors (1 mM PMSF, 9 mM benzamidine, 0.28 U of aprotinin/ml, 50 μg of leupeptin/ml, 7 μg of pepstatin A/ml, and 100 μM sodium orthovanadate) to maintain cell equivalent volumes for all of the fractions. Alternatively, the supernatant was centrifuged at 100,000 × g for 45 min to isolate the microsomal fraction. In separate experiments, this centrifugation step was also applied prior to mitochondrial isolation to prepare microsomal fractions that were enhanced in yield and purity (after multiple washes with wash buffer). Fractions were then immunoprecipitated and/or immunoblotted, as described above.
Activated caspase 3 assay.
Caspase 3 activity was measured in an immunofluorescence assay with an antibody that recognizes the cleaved (active) form of caspase 3. MDA-MB-231 cells were seeded onto 100-mm tissue culture plates at a density of 106 cells/plate (four plates/assay point) in DMEM containing 10% FBS and allowed to equilibrate for 24 h. Where indicated, cells were then washed three times with PBS and incubated with 2 μg of adriamycin/ml for 1 h at 37°C. After incubation, the adriamycin was removed by washing the cells three times with PBS, and the cells were subsequently placed in SF DMEM or SF DMEM containing 20 ng of EGF/ml for 24 h at 37°C. The cells were then electroporated with EGFR constructs, as described above. Approximately 4 × 106 electroporated cells were immediately seeded onto glass coverslips in a six-well tissue culture dish and allowed to recover for 3 h in DMEM plus 10% FBS. Cells were then returned to their appropriate medium and incubated for an additional 48 h. When Stat5b constructs were used, cells were transiently transfected with FuGENE6 (Roche) after adriamycin treatment and allowed to incubate for 48 h. All cells were subsequently fixed in 3% paraformaldehyde, permeabilized in TBS-T, and blocked in 5% goat serum in TBS-T. To detect activated caspase 3, cells were incubated with rabbit anti-caspase 3 (Cell Signaling Technologies, Waltham, Mass.) at a 1:50 dilution and mouse anti-HA antibody (for the Stat5b experiments) overnight at 4°C, washed three times with TBS-T, followed by incubation with Texas red-conjugated goat anti-rabbit antibody (Molecular Probes) at 1:1,000 and FITC-conjugated goat anti-mouse antibody (Molecular Probes) at 1:1,000 (for the Stat5b experiment) for 1 h at room temperature in the dark, washed three times with TBS-T, and mounted on microscope slides. Cells containing activated caspase 3 and/or expressing Stat5b were visualized by immunofluorescence microscopy, as described above.
RESULTS
Cytochrome c oxidase subunit II is a binding partner of the EGFR.
To extend our understanding of the mechanisms by which EGFR and c-Src synergize in oncogenesis, the technique of phage display was utilized to identify proteins that specifically bind to pY845. A 20-mer peptide corresponding to the region of the human EGFR containing pY845 was incubated with the phage display library, and binding partners were identified as described in Materials and Methods. One interacting protein was identified in 5 of 20 independent clones as having 100% homology to the mitochondrial protein, cytochrome c oxidase subunit II (CoxII) (Fig. 1A). To verify the specificity of this interaction, positive phage clones were incubated with biotinylated peptides corresponding to the sequence surrounding unphosphorylated Y845, as well as to streptavidin-linked magnetic beads alone. No binding occurred to either of these negative controls, suggesting that the phosphorylation at Y845 on the peptide is required for this coassociation. These results are consistent with the ability of the phosphorylated, but not the unphosphorylated Y845-containing peptide to inhibit EGF-induced S-phase entry in breast cancer cell (J. L. Boerner, J. S. Biscardi, D. Sproles, C. M. Silva, and S. J. Parsons, unpublished data).
CoxII is a mitochondrion-encoded protein that is a subunit of complex IV of the respiratory chain. Complex IV is one of five mitchondrial complexes that mediate the oxidation of cytochrome c and the generation of ATP (8). Structurally, CoxII is a two-pass transmembrane protein with the N and C termini of the protein located in the intermembrane space of the mitochondria (8, 36) (Fig. 1B). The region of CoxII encoded by all five of the phage clones (indicated in red above the complete sequence [Fig. 1A]) included the loop created by the transmembrane domains, the C-terminal transmembrane domain, as well as a small portion of the extensive C-terminal tail of the molecule. Studies are currently under way to narrow down this large region of the protein to identify the precise location of pY845 association with CoxII.
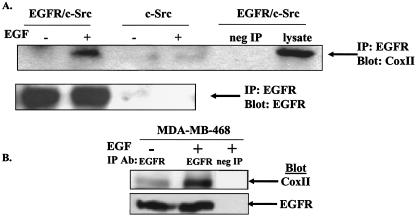
To determine whether the EGFR and CoxII interact in cells, murine fibroblasts (10T1/2 cells) stably co-overexpressing the EGFR and c-Src or c-Src alone were stimulated with 50 ng of EGF/ml for 20 min, and the ability of CoxII to coimmunoprecipitate with the EGFR was tested. Figure 2A demonstrates that endogenously expressed CoxII can coimmunoprecipitate with EGFR from EGFR/c-Src- but not from c-Src-overexpressing cells. In addition, the interaction appears to be induced or increased when cells are stimulated with EGF. Furthermore, as depicted in Fig. 2B, EGFR and CoxII were found to coimmunoprecipitate from extracts of the MDA-MB-468 breast cancer cell line, with an enhancement in the amount of coimmunoprecipitation seen upon EGF stimulation. This cell line contains elevated levels of EGFR and c-Src compared to nontumorigenic mammary epithelial cells (3). Taken together, these data support the phage display identification of CoxII as a binding partner of the EGFR and further show that the binding is dependent on EGF stimulation.
FIG. 2.
Coimmunoprecipitation of the EGFR and CoxII. (A and B) C3H10T1/2 murine fibroblasts overexpressing the EGFR and c-Src or c-Src alone (A) or MDA-MB-468 breast cancer cells (B) were grown in the absence of serum and stimulated with (or without) 50 ng of EGF/ml for 20 min. A total of 500 μg of cell lysate was immunoprecipitated with EGFR-specific MAb 108 or paxillin antibody as a negative control, and precipitated proteins were immunoblotted with CoxII or EGFR (Neomarkers) antibodies.
The binding of CoxII to the EGFR is dependent on the phosphorylation of Y845 on the EGFR.
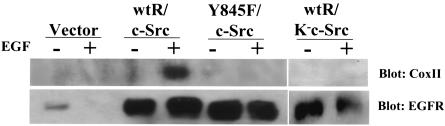
The phage display analysis described above identified CoxII as a binding partner of the phosphorylated but not the unphosphorylated Y845 peptide. It was, therefore, important to determine in intact cells whether coimmunoprecipitation of CoxII with the EGFR was dependent on the phosphorylation of Y845. To address this question, Cos-7 cells were transiently transfected with plasmids encoding wt-EGFR or a mutant receptor bearing a phenylalanine substitution at Y845 (Y845F), together with wt-or kinase-defective c-Src, and stimulated with EGF. Cell lysates were immunoprecipitated with EGFR antibodies and immunoblotted with CoxII or EGFR antibodies. As shown in Fig. 3, the association of CoxII with the EGFR was inhibited not only by mutation of Y845 on the EGFR but also by introduction of a catalytically inactive c-Src (K−c-Src). Treatment of cells with PP2 (a Src family kinase inhibitor) gave similar results (data not shown). These data are consistent with previous data from our laboratory and others, which demonstrated that c-Src mediates tyrosine phosphorylation of Y845 (4, 35, 44) and hence the association of the EGFR with CoxII. Together, these data confirm the specificity of the in vivo interaction between the EGFR and CoxII and demonstrate by mutation (Y845F) and lack of phosphorylation (when K−c-Src is expressed) that phosphorylation of Y845 is required for this event.
FIG. 3.
CoxII fails to coimmunoprecipitate with Y845F-EGFR. Cos-7 cells were transfected with vector only or plasmids encoding c-Src, K−c-Src, the wt EGFR, or the mutant Y845F-EGFR, serum starved for 24 h, left unstimulated, or stimulated with 50 ng of EGF/ml for 20 min, and lysed. Then, 500 μg of cell lysate was immunoprecipitated and immunoblotted as in Fig. 2.
The EGFR localizes to the mitochondria after EGF stimulation.
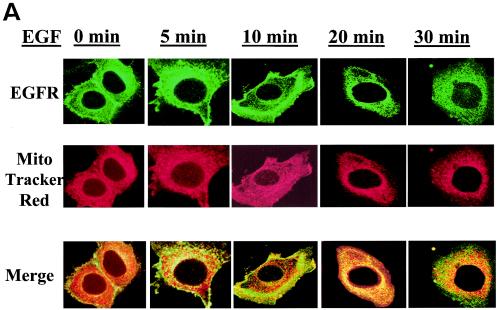
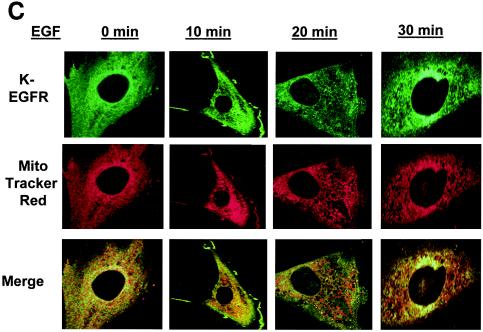
Because CoxII is a mitochondrial protein, we sought to determine whether EGFR could localize at or near the mitochondria and, if so, whether this localization was regulated by EGF. To address this question, several approaches were taken. First, 10T1/2 murine fibroblasts overexpressing the EGFR and c-Src were starved of serum, stimulated with EGF for various lengths of time, and examined by fluorescence confocal microscopy for the subcellular localization of EGFR. Mitochondria were localized with MTR, a dye that becomes incorporated into intact organelles. Figure 4A reveals that at 0 min EGF stimulation the EGFR is localized primarily at or near the plasma membrane (green). After 10 to 20 min of EGF stimulation the EGFR can clearly be seen translocating to intracellular compartments. Most importantly, merged images of MTR and EGFR (green) stained cells demonstrate that at 20 min of EGF treatment, the red and green staining colocalize to produce a yellow color. By 30 min after the addition of EGF, colocalization is less apparent, and by 60 min it returns to the unstimulated state (data not shown). More than 60% of the cells displayed similar kinetics of translocation of EGFR colocalization with the mitochondria. These data suggest that the EGFR can translocate to the mitochondria where it is in a position to interact with CoxII and that the movement to the mitochondria is EGF dependent and transient.
FIG. 4.
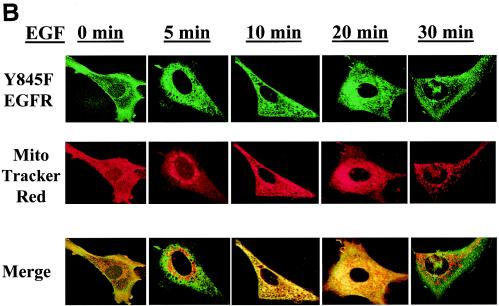
Colocalization of the EGFR to the mitochondria. 10T1/2 murine fibroblasts overexpressing the EGFR and c-Src (A) and 10T1/2 murine fibroblasts overexpressing c-Src and transiently expressing Y845F-EGFR (B) or K721A-EGFR (C) were cultured in SF medium for 24 h and stimulated with 50 ng of EGF/ml for 0 to 30 min. Just prior to EGF addition, MTR (a mitochondrial dye) was added at 500 nM for 30 min at 37°C. After stimulation, cells were prepared for immunofluorescent confocal microscopy by incubation with EGFR antibodies (Transduction Labs) and FITC-conjugated goat anti-mouse immunoglobulin G. Colors: green, EGFR; red, mitochondria; yellow, overlap.
Of interest, this translocation of the EGFR to the mitochondria appears to occur independently of its ability to be phosphorylated on Y845 but does require its kinase activity. Figure 4B demonstrates that the mutant Y845F-EGFR (when transiently transfected into 10T1/2 cells overexpressing c-Src) retains its ability to translocate to the mitochondria with the same time course and frequency as cells overexpressing the wt receptor (∼60%). To determine whether the overall kinase activity of the receptor is required for its translocation to the mitochondria, a kinase inactive mutant of the EGFR (K721A-EGFR) was transiently transfected into c-Src-overexpressing fibroblasts and monitored by confocal microscopy after EGF treatment. Figure 4C demonstrates that translocation of this EGFR mutant to the mitochondria is impaired in the absence of kinase activity. Therefore, although tyrosine phosphorylation of 845 is required for its molecular interaction with CoxII, the catalytic activity of the EGFR appears to be required for translocation of the receptor to the mitochondria.
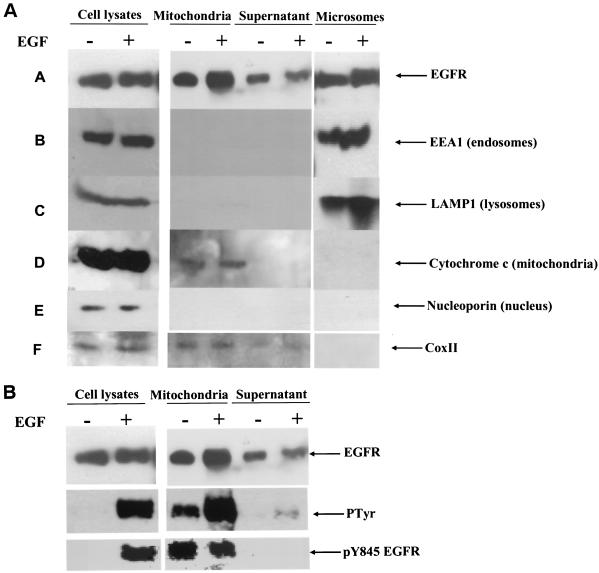
To further investigate this colocalization, biochemical fractionation was undertaken. 10T1/2 murine fibroblasts overexpressing the EGFR and c-Src were stimulated with EGF after serum deprivation and fractionated by using variable-speed centrifugation as described in Materials and Methods. Fractions were analyzed by immunoprecipitation and immunoblotting for the presence of the EGFR and for mitochondrial, endosomal, nuclear, and lysosomal markers. Figure 5A demonstrates that the EGFR is found in the mitochondrial fraction of the cell, with elevated amounts observed in cells stimulated with EGF. However, we found a portion of the EGFR to be constitutively present in the mitochondrial fraction, perhaps as a result of overexpression and spontaneous activation and translocation of the receptor. This phenomenon was also seen in breast cancer cells in which the EGFR and c-Src are endogenously overexpressed (data not shown). In addition, it can be observed that the mitochondrial fraction is relatively free of lysosomal and endosomal markers, suggesting a minimal lysosomal and endosomal contamination in the mitochondrial fraction. The supernatant (or cytoplasmic) fraction represents the remainder of the homogenate after the nuclear, microsomal, and mitochondrial fractions have been removed. These data are consistent with the findings of the immunofluorescence analysis, namely, that after EGF treatment a portion of the plasma membrane-associated EGFR migrates to the mitochondria.
FIG. 5.
Cofractionation of the EGFR with mitochondria. 10T1/2 murine fibroblasts overexpressing the EGFR and c-Src were cultured in SF medium for 24 h and either left unstimulated or stimulated with 50 ng of EGF/ml for 20 min. (A) Cells were biochemically fractionated as described in Materials and Methods, and fractions (except for the beginning lysate) representing cell equivalents were either immunoprecipitated with EGFR-specific MAb 108 and immunoblotted with EGFR antibody (Neomarkers) or directly immunoblotted with EEA1 (endosomal marker), LAMP1 (lysosomal marker), cytochrome c (mitochondrial marker), nucleoporin 62 (nuclear marker), or CoxII antibodies. (B) Lysates were immunoblotted with EGFR-, phosphotyrosine-, or pY845-specific antibodies as indicated.
Figure 5B shows that the EGFR found in the mitochondrial fraction is activated and tyrosine phosphorylated, with greater levels of tyrosine phosphorylation detected after EGF stimulation. These results are consistent with the amount of EGFR seen in the mitochondrial fraction before and after growth factor stimulation. Phosphorylation of Y845 was also observed in the mitochondrial fraction, but the level was unaltered by EGF treatment. At the present time, we do not know the reason for our inability to observe an EGF-inducible phosphorylation of Y845 in the mitochondrial fraction. This phenomenon could reflect increased phosphorylation of Y845 during purification of the mitochondria, where c-Src is known to reside (24, 32), or it could reflect an inactivation or separation from phosphatases that dephosphorylate pY845 in unstimulated cells. Alternatively, it could indicate that a separate pool of EGFR that is constitutively phosphorylated on Y845 resides in the mitochondria and is insensitive or less so to EGF than the plasma membrane-associated receptor.
EGFR and CoxII colocalization at the mitochondria.
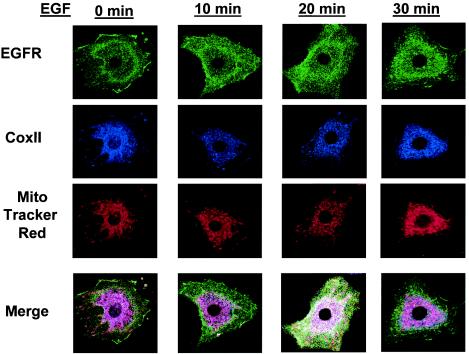
Although the data presented above demonstrate that the EGFR localizes at or near the mitochondria, it was not definitively determined whether the EGFR and CoxII interaction occurred at this site or in the cytoplasm. To determine whether the EGFR interacts with CoxII at or near the mitochondria, 10T1/2 fibroblasts overexpressing the EGFR and c-Src were serum starved, stimulated with EGF for various lengths of time, and examined for EGFR, mitochondrial, and CoxII subcellular localization by confocal microscopy. Figure 6 depicts the localization of CoxII and the EGFR in relationship to the mitochondria at a resting state and shows that CoxII and the mitochondria overlap (magenta), whereas the EGFR (green) is predominantly localized to the plasma membrane. However, at 20 min EGF stimulation, the EGFR can be seen to colocalize with CoxII at the mitochondria (white areas in Fig. 6). Costaining of the EGFR and CoxII (that does not include the mitochondrial stain) also can be observed in the perinuclear region (orange areas in Fig. 6). Together, these data suggest that CoxII and the EGFR may interact both independently of and at and/or within the mitochondria, although the conclusion that they associate outside of the mitochondria should take into consideration the fact that MTR may not stain all of the mitochondria in the cell due to organelle breakdown during cell fixation, permeabilization, and so forth.
FIG. 6.
Colocalization of the EGFR with CoxII and mitochondria. 10T1/2 murine fibroblasts overexpressing the EGFR and c-Src were serum starved overnight and stimulated with 50 ng of EGF/ml for 0 to 30 min. Cells were fluorescently stained with MTR (for mitochondria) or with CoxII and EGFR antibodies as described in Materials and Methods and then examined for colocalization by using confocal microscopy.
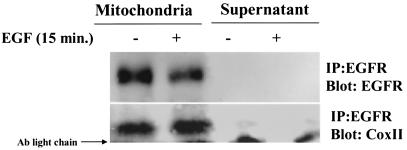
To further assess colocalization of the EGFR and CoxII at or near the mitochondria, biochemical fractionation techniques were again used, this time with MDA-MB-231 breast cancer cells that express elevated levels of EGFR and c-Src (3). After differential centrifugation, EGFR was immunoprecipitated from the various fractions, and the precipitates were immunoblotted with CoxII antibodies. Figure 7 shows that in the mitochondrial fraction, the EGFR and CoxII coimmunoprecipitated. In the cytoplasmic (supernatant) fraction, however, neither CoxII nor the EGFR were detected. Similar to the MDA-MB-468 cells (Fig. 2), MDA-MB-231 cells did not show an EGF-inducible association of the EGFR and CoxII in the mitochondria, possibly due to constitutive overexpression and translocation of the EGFR. Nevertheless, these data provide further evidence that the EGFR and CoxII colocalize at or near the mitochondria.
FIG. 7.
Coimmunoprecipitation of the EGFR and CoxII from the mitochondrial fraction. MDA-MB-231 breast cancer cells were cultured in SF medium for 24 h, left unstimulated, or stimulated with 50 ng of EGF/ml for 20 min and then subjected to biochemical fractionation as described in Materials and Methods. The indicated fractions were immunoprecipitated with EGFR-specific MAb 108, and precipitated proteins were immunoblotted with EGFR (top panel; Neomarkers) or CoxII (bottom panel) antibodies.
Expression of Y845F-EGFR induces apoptosis in MDA-MB-231 breast cancer cells.
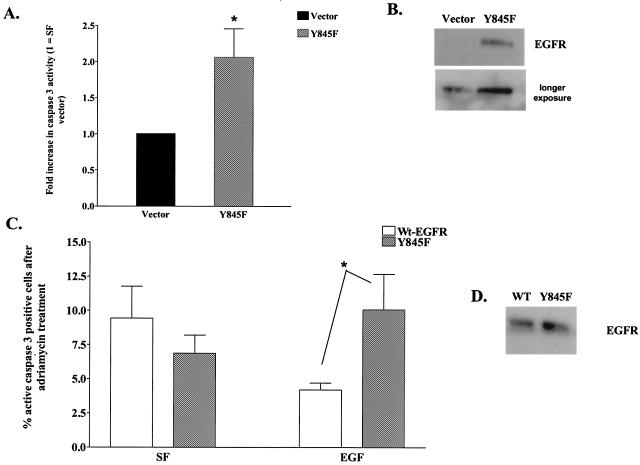
The association of EGFR with CoxII raises the question of whether the interaction between these two proteins could influence apoptotic or survival pathways. Much evidence suggests that both proteins are involved in such processes (12, 14, 15, 38). To test whether phosphorylation of Y845 on the EGFR was required for the ability of the EGFR to function as a survival factor in SF conditions, MDA-MB-231 cells (which express EGFR ∼6-fold above normal breast epithelium) (3) were transiently transfected with cDNA encoding the Y845F-EGFR or vector only and examined for the number of apoptotic cells in response to serum deprivation. Figure 8B demonstrates by immunoblotting that Y845F-EGFR was expressed approximately two- to threefold over endogenous EGFR in MDA-MB-231 cells. If phosphorylation of Y845 were required for survival during serum deprivation, the dominant-interfering effect of the Y845F mutant receptor would be expected to further decrease survival or increase apoptosis under such conditions. Apoptosis was measured by counting cells that stained positively in an immunofluorescence assay, with antibodies specific for activated caspase 3. Figure 8A shows that the number of apoptotic cells increased >2-fold in the presence of Y845F-EGFR when the cells were deprived of serum. Similar results were observed by using a TUNEL (terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling) assay (data not shown). These results demonstrated that phosphorylation of Y845 on the EGFR plays a role in protecting cells from apoptosis induced by serum starvation.
FIG. 8.
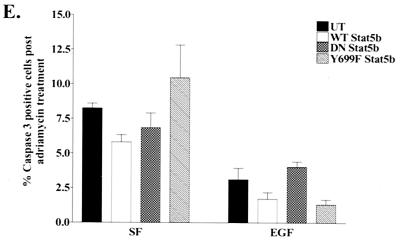
Caspase 3 activity in serum-deprived or adriamycin-treated MDA-MB-231 cells. (A) MDA-MB-231 cells were electroporated with plasmids encoding vector only (▪) or Y845F EGFR (░⃞), incubated in SF DMEM, and prepared for immunofluorescence microscopy to assess caspase 3 activity as described in Materials and Methods. The percentages of active caspase 3-positive cells, normalized to the percent positive for SF, vector-transfected cells (∼15%), are graphed for the Y845F-EGFR-expressing cells. The results are expressed as the mean ± the SEM for four experiments. ✽, P < 0.05. (B) Representative Western blot showing expression of the Y845F EGFR after electroporation with the indicated construct. (C) Electroporated MDA-MB-231 cells expressing wt (□) or Y845F EGFR (░⃞) were treated with adriamycin for 1 h prior to incubation in SF DMEM or EGF and assessment of caspase 3 activation. The results from three experiments are graphed as the percentage of cells positive for active caspase 3. Significantly greater numbers of positive cells were observed in the mutant EGFR-expressing population than in the wt EGFR population. ✽, P = 0.03. (D) Representative immunoblot demonstrating similar amounts of EGFR and Y845F-EGFR protein expressed after transfection. (E) MDA-MB-231 cells were transiently transfected with HA-tagged wt Stat5b, DNstat5b, or Y699F-Stat5b constructs and treated with adriamycin, followed by EGF stimulation as described above. The percentages of cells expressing the Stat5b constructs that were positive for active caspase 3 were determined by immunofluorescence. Significantly fewer positive cells were observed in the EGF-treated cells compared to each of the transfected counterparts in the SF group. ✽, P < 0.05; n = 4. None of the members of the SF group were significantly different from one another; this was also true of the EGF-treated group.
In another test of the role of pY845 in survival signaling, we took advantage of the finding that MDA-MB-231 breast cancer cells are sensitive to apoptosis induced by adriamycin, with EGF acting as a survival factor (14). Figure 8C shows that, in the absence of EGF or serum (i.e., SF), MDA-MB-231 cells treated with adriamycin exhibited apoptosis in approximately 7 to 10% of the cells (as measured by active caspase 3 immunofluorescence), whether they overexpressed wt or mutant EGFR. (Fig. 8D shows that approximately equal levels of wt and mutant EGFR are overexpressed). However, treatment with EGF partially protected wt-EGFR overexpressing MDA-MB-231 cells from apoptosis, whereas it had little to no affect on Y845F-EGFR-overexpressing cells. These data are consistent with the interpretation that the inability of the Y845F-EGFR mutant to bind to CoxII abolishes the protective effect of EGF on MDA-MB-231 breast cancer cells induced to undergo apoptosis in response to adriamycin.
Lastly, it has been reported that the transcription factor Stat5b is a mediator of pY845 proliferative signaling by the EGFR (20). To determine whether activation of Stat5b by the EGFR is required for EGF protection from adriamycin-induced apoptosis, we transiently transfected MDA-MB-231 cells with wt Stat5b, a dominant-interfering Stat5b mutant (DNstat5b), or a tyrosine-to-phenylalanine mutation at the tyrosine required for transcriptional activity (Y699F-Stat5b) and analyzed the cells for EGF protection from adriamycin-induced apoptosis. Figure 8E demonstrates that even in the presence of Y699F-Stat5b or DNAStat5b, EGF still protects MDA-MB-231 cells from adriamycin-induced apoptosis, indicating that Stat5b tyrosine phosphorylation (and subsequent activation) downstream of Y845 phosphorylation is not required for this phenotype.
DISCUSSION
Co-overexpression of the EGFR and c-Src in murine fibroblasts leads to synergistic increases in DNA synthesis, anchorage-independent cell growth, and tumor growth in nude mice (22). These synergisms are reproduced in breast cancer cells that endogenously co-overexpress the two proteins (3). The mechanisms of this synergy are not completely understood, but we do know that Y845 of the EGFR is phosphorylated under these conditions in a c-Src-dependent manner and that this phosphorylation is required for entrance into S phase in response to EGF (4, 35). Because of this requirement, we speculate that pY845 activates downstream effectors that are critical regulators of cell proliferation. One pY845 effector, Stat5b, has been identified and shown to be required for EGF-induced DNA synthesis (20). The results described in the present study demonstrate that Y845 phosphorylation also promotes the association of the EGFR with another potential effector, CoxII, and that this association occurs at or near the mitochondria. In addition, we show that when Y845 is mutated to the nonphosphorylatable phenylalanine or when Y845 phosphorylation is prevented by kinase-defective c-Src, CoxII and EGFR coassociation fails to occur and apoptosis, brought about by adriamycin, is no longer reduced by EGF treatment. That Stat5b is not a mediator of this survival signal is evidenced by the inability of a dominant-negative form of Stat5b to inhibit EGF rescue of adriamycin-induced apoptosis. Together, these findings suggest another possible mechanism for the biological synergisms observed when EGFR and c-Src are co-overexpressed, namely, promotion of a survival signal.
The association of the EGFR with CoxII appears to occur through a mechanism independent of phosphotyrosine-SH2 domain interactions. The sequence surrounding tyrosine 845 does not match any prototypical SH2 or PTB-binding domain motifs, and CoxII does not contain these motifs (34). We were unable to identify another protein containing the sequence surrounding Y845. Although the location of this tyrosine within the activation loop of the kinase domain is conserved in other tyrosine kinase receptors and nonreceptor tyrosine kinases, the sequence surrounding the tyrosine is not, nor are there any additional residues besides the Y845 homologues that are conserved throughout the panel of tyrosine kinases examined (17). The nature of the molecular interaction between pY845 and CoxII is now under investigation.
Traditionally, the EGFR has been thought to regulate various physiological processes of the cell (i.e., proliferation, survival, migration) by recruiting signaling proteins to the cellular membrane (where it is primarily located during quiescence) and, from there, emanating signals throughout the cell. However, recent evidence has suggested that membrane receptors, such as the EGFR, may continue to emit important signals after they are internalized (39, 40). In addition, Lin et al. (21) have demonstrated that the EGFR itself transits from the plasma membrane to the nucleus of cells, where it acts as a transcription factor. Our observations suggesting that the EGFR localizes to the mitochondria, where it may regulate cell survival, add to this increasing evidence of alternate subcellular localizations of the EGFR.
In addition to alternate subcellular localizations of the EGFR, studies of the EGFR in human tumorigenesis have revealed nontraditional signaling pathways emanating from the receptor. A mutant of the EGFR, i.e., EGFRvIII, has been identified in human gliomas (42), and expression of EGFRvIII in NIH 3T3 cells results in cellular transformation (26). The signaling pathway through which this transformation and potential tumorigenesis occurs appears to be distinct from classical EGFR signaling pathways (26). In this regard, a naturally occurring avian viral homolog of EGFRvIII, v-ErbB, was found to activate the small GTP-binding protein Rho (6). Furthermore, the activation of Rho was found to be required for transformation mediated by v-ErbB (6). Taken together, these two examples support the notion that nonclassical EGFR signaling proteins have the ability to be activated in response to EGFR activation.
Our observation that the EGFR migrates to the mitochondria after EGF stimulation is supported by the apparent localization of other receptors (as well as signaling molecules) to the mitochondria. For example, it has long been suggested that steroid receptors such as the androgen and estrogen receptors are located within the mitochondria, since they appear to regulate mitochondrial gene transcription (28, 30, 31). Recently, the β isoform of the estrogen receptor was found to localize predominantly to the mitochondrial and microsomal fractions of rat uterine cells, where it binds the majority of estrogen in the cell (25). Of added interest and in support of these observations, tamoxifen, an antagonist of estrogen, can prevent mitochondrial membrane depolarization, as well as affect other mitochondrial functions, such as calcium release (10). In addition, cellular signaling proteins known to be activated by EGF, such as Grb10 and Raf-1, are found in mitochondrial fractions (27). The localization of Raf-1 to the mitochondria is dependent on the insulin-like growth factor-I receptor and leads to antiapoptotic cellular signaling (29). In all, these findings support the observations reported here, namely, that the EGFR translocates to the mitochondria following EGF stimulation, where it interacts with CoxII and is appropriately positioned to regulate cell survival.
In addition to their localization to the mitochondria, signaling molecules have been shown to regulate or associate with members of the oxidative respiratory chain. In osteoclasts, c-Src recently was shown to localize within the mitochondria, where it tyrosine phosphorylates CoxII (24, 32). Introduction of a kinase-inactive Src has the ability to reduce CoxII activity to ca. 80% of normal, linking tyrosine phosphorylation to the regulation of CoxII activity (24). In addition, a mutant of the androgen receptor that contains an expanse of polyglutamine repeats recently was demonstrated to bind CoxVb, another subunit of cytochrome c oxidase (1). It was hypothesized that this interaction sequesters CoxVb away from its normal function and leads to mitochondrial impairments (1). Lastly, a constitutively active regulatory subunit of protein kinase A was shown to interact with CoxVb (45). This interaction is regulated by cyclic AMP treatment (45). Furthermore, loss of this interaction (via cyclic AMP treatment) led to a decrease in cytochrome c oxidase activity and an increase in cytochrome c release from the mitochondria (45). Taken together, these data support the hypothesis that nuclearly encoded cellular proteins that interact with members of the mitochondrial respiratory chain can regulate mitochondrial activity.
The mechanism by which the EGFR translocates to the mitochondria is currently unknown. However, in the case of the androgen receptor mutant binding to CoxVb, it was shown that Hsp70 stimulates this association (1). In a similar fashion, chaperone proteins may facilitate translocation of the EGFR from the plasma membrane to the mitochondria. The Y845F mutant of the EGFR lacks the ability to be associated with CoxII; however, it still retains the ability to translocate to the mitochondria, suggesting that Y845 itself does not target the molecule to the mitochondria and raising the question of whether other sequences within the EGFR may contain this activity. Indeed, the EGFR does contain a putative mitochondrial targeting sequence (18, 23), residues 645 to 666, located within the juxtamembrane domain of the molecule, suggesting that a chaperone protein or organelle may facilitate its intracellular movement. Indeed, the finding that receptor kinase activity is required for mitochondrial localization is consistent with the idea that endocytosis plays a critical role in translocation to the mitochondria, since the catalytic activity of the EGFR is known to be required for its endocytosis (16, 19). Whatever the mechanism of translocation may turn out to be, the observation that expression of Y845F-EGFR increases serum deprivation- and adriamycin-induced apoptosis suggests that it is the association of the EGFR with CoxII and not simply the translocation of the receptor to the mitochondria that enhances cellular survival.
Acknowledgments
We thank the PWP research group for their helpful discussions.
This study was generously supported by NRSA F32-CA93028 (J.L.B.) and DHHS R01-CA71449 (S.J.P.).
REFERENCES
- 1.Beauchemin, A. M. J., B. Gottlieb, L. K. Beitel, Y. A. Elhaji, L. Pinsky, and M. A. U. Trifiro. 2001. Cytochrome c oxidase subunit Vb interacts with human androgen receptor: a potential mechanism for neurotoxicity in spinobulbar muscular atrophy. Brain Res. Bull. 56:285-297. [DOI] [PubMed] [Google Scholar]
- 2.Biscardi, J., R. Ishizawar, C. Silva, and S. Parsons. 2000. Tyrosine kinase signalling in breast cancer: epidermal growth factor receptor and c-Src interactions in breast cancer. Breast Cancer Res. Com. 2:203-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biscardi, J. S., A. P. Belsches, and S. J. Parsons. 1998. Characterization of human epidermal growth factor receptor and c-Src interactions in human breast tumor cells. Mol. Carcinog. 21:261-272. [DOI] [PubMed] [Google Scholar]
- 4.Biscardi, J. S., M. C. Maa, D. A. Tice, M. E. Cox, T. H. Leu, and S. J. Parsons. 1999. c-Src-mediated phosphorylation of the epidermal growth factor receptor on Tyr845 and Tyr1101 is associated with modulation of receptor function. J. Biol. Chem. 274:8335-8343. [DOI] [PubMed] [Google Scholar]
- 5.Biscardi, J. S., D. A. Tice, and S. J. Parsons. 1999. c-Src, receptor tyrosine kinases, and human cancer. Adv. Cancer Res. 76:61-119. [DOI] [PubMed] [Google Scholar]
- 6.Boerner, J. L., A. J. Danielsen, M. J. McManus, and N. J. Maihle. 2001. Activation of Rho is required for ligand-independent oncogenic signaling by a mutant EGF receptor. J. Biol. Chem. 276:3691-3695. [DOI] [PubMed] [Google Scholar]
- 7.Brown, G. C., and V. Borutaite. 1999. Nitric oxide, cytochrome c and mitochondria. Biochem. Soc. Symp. 66:17-25. [DOI] [PubMed] [Google Scholar]
- 8.Cabral, F., M. Solioz, Y. Rudin, G. Schatz, L. Clavilier, and P. P. Slonimski. 1978. Identification of the structural gene for yeast cytochrome c oxidase subunit II on mitochondrial DNA. J. Biol. Chem. 253:297-304. [PubMed] [Google Scholar]
- 9.Carpenter, G. 2000. The EGF receptor: a nexus for trafficking and signaling. Bioessays 22:697-707. [DOI] [PubMed] [Google Scholar]
- 10.Custodio, J. B., A. J. Moreno, and K. B. Wallace. 1998. Tamoxifen inhibits induction of the mitochondrial permeability transition by Ca2+ and inorganic phosphate. Toxicol. Appl. Pharmacol. 152:10-17. [DOI] [PubMed] [Google Scholar]
- 11.Di Fiore, P. P., J. H. Pierce, T. P. Fleming, R. Hazan, A. Ullrich, C. R. King, J. Schlessinger, and S. A. Aaronson. 1987. Overexpression of the human EGF receptor confers an EGF-dependent transformed phenotype to NIH 3T3 cells. Cell 51:1063-1070. [DOI] [PubMed] [Google Scholar]
- 12.Di Giovanni, S., M. Mirabella, M. Papacci, F. Odoardi, G. Silvestri, and S. Servidei. 2001. Apoptosis and ROS detoxification enzymes correlate with cytochrome c oxidase deficiency in mitochondrial encephalomyopathies. Mol. Cell Neurosci. 17:696-705. [DOI] [PubMed] [Google Scholar]
- 13.Forsha, D., C. Church, P. Wazny, and R. O. Poyton. 2001. Structure and function of Pet100p, a molecular chaperone required for the assembly of cytochrome c oxidase in Saccharomyces cerevisiae. Biochem. Soc. Trans. 29:436-441. [DOI] [PubMed] [Google Scholar]
- 14.Geier, A., R. Beery, M. Haimsohn, R. Hemi, Z. Malik, and A. Karasik. 1994. Epidermal growth factor, phorbol esters, and aurintricarboxylic acid are survival factors for MDA-231 cells exposed to adriamycin. In Vitro Cell Dev. Biol. 30A:867-874. [DOI] [PubMed] [Google Scholar]
- 15.Gibson, S., S. Tu, R. Oyer, S. M. Anderson, and G. L. Johnson. 1999. Epidermal growth factor protects epithelial cells against Fas-induced apoptosis: requirement for Akt activation. J. Biol. Chem. 274:17612-17618. [DOI] [PubMed] [Google Scholar]
- 16.Glenney, J. R., Jr., W. S. Chen, C. S. Lazar, G. M. Walton, L. M. Zokas, M. G. Rosenfeld, and G. N. Gill. 1988. Ligand-induced endocytosis of the EGF receptor is blocked by mutational inactivation and by microinjection of anti-phosphotyrosine antibodies. Cell 52:675-684. [DOI] [PubMed] [Google Scholar]
- 17.Hanks, S. K., A. M. Quinn, and T. Hunter. 1988. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science 241:42-52. [DOI] [PubMed] [Google Scholar]
- 18.Hobert, M. E., S. J. Kil, M. E. Medof, and C. R. Carlin. 1997. The cytoplasmic juxtamembrane domain of the epidermal growth factor receptor contains a novel autonomous basolateral sorting determinant. J. Biol. Chem. 272:32901-32909. [DOI] [PubMed] [Google Scholar]
- 19.Honegger, A. M., T. J. Dull, S. Felder, E. Van Obberghen, F. Bellot, D. Szapary, A. Schmidt, A. Ullrich, and J. Schlessinger. 1987. Point mutation at the ATP binding site of EGF receptor abolishes protein-tyrosine kinase activity and alters cellular routing. Cell 51:199-209. [DOI] [PubMed] [Google Scholar]
- 20.Kloth, M. T., K. K. Laughlin, J. S. Biscardi, J. L. Boerner, S. J. Parsons, and C. M. Silva. 2003. STAT5b, a mediator of synergism between c-Src and the epidermal growth factor receptor. J. Biol. Chem. 278:1671-1679. [DOI] [PubMed] [Google Scholar]
- 21.Lin, S. Y., K. Makino, W. Xia, A. Matin, Y. Wen, K. Y. Kwong, L. Bourguignon, and M. C. Hung. 2001. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat. Cell Biol. 3:802-808. [DOI] [PubMed] [Google Scholar]
- 22.Maa, M. C., T. H. Leu, D. J. McCarley, R. C. Schatzman, and S. J. Parsons. 1995. Potentiation of epidermal growth factor receptor-mediated oncogenesis by c-Src: implications for the etiology of multiple human cancers. Proc. Natl. Acad. Sci. USA 92:6981-6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin-Nieto, J., and A. Villalobo. 1998. The human epidermal growth factor receptor contains a juxtamembrane calmodulin-binding site. Biochemistry 37:227-236. [DOI] [PubMed] [Google Scholar]
- 24.Miyazaki, T., L. Neff, S. Tanaka, W. C. Horne, and R. Baron. 2003. Regulation of cytochrome c oxidase activity by c-Src in osteoclasts. J. Cell Biol. 160:709-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monje, P., and R. Boland. 2001. Subcellular distribution of native estrogen receptor alpha and beta isoforms in rabbit uterus and ovary. J. Cell Biochem. 82:467-479. [DOI] [PubMed] [Google Scholar]
- 26.Moscatello, D. K., R. B. Montgomery, P. Sundareshan, H. McDanel, M. Y. Wong, and A. J. Wong. 1996. Transformational and altered signal transduction by a naturally occurring mutant EGF receptor. Oncogene 13:85-96. [PubMed] [Google Scholar]
- 27.Nantel, A., M. Huber, and D. Y. Thomas. 1999. Localization of endogenous Grb10 to the mitochondria and its interaction with the mitochondrial-associated Raf-1 pool. J. Biol. Chem. 274:35719-35724. [DOI] [PubMed] [Google Scholar]
- 28.Noteboom, W. D., and J. Gorski. 1965. Stereospecific binding of estrogens in the rat uterus. Arch. Biochem. Biophys. 111:559-568. [DOI] [PubMed] [Google Scholar]
- 29.Peruzzi, F., M. Prisco, A. Morrione, B. Valentinis, and R. Baserga. 2001. Anti-apoptotic signaling of the insulin-like growth factor-I receptor through mitochondrial translocation of c-Raf and Nedd4. J. Biol. Chem. 276:25990-25996. [DOI] [PubMed] [Google Scholar]
- 30.Pietras, R. J., and C. M. Szego. 1979. Estrogen receptors in uterine plasma membrane. J. Steroid Biochem. 11:1471-1483. [DOI] [PubMed] [Google Scholar]
- 31.Pietras, R. J., and C. M. Szego. 1980. Partial purification and characterization of estrogen receptors in subfractions of hepatocyte plasma membranes. Biochem. J. 191:743-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salvi, M., A. M. Brunati, L. Bordin, N. La Rocca, G. Clari, and A. Toninello. 2002. Characterization and location of Src-dependent tyrosine phosphorylation in rat brain mitochondria. Biochim. Biophys. Acta 1589:181-195. [DOI] [PubMed] [Google Scholar]
- 33.Schindler, C., and J. E. Darnell, Jr. 1995. Transcriptional responses to polypeptide ligands: the JAK-STAT pathway. Annu. Rev. Biochem. 64:621-651. [DOI] [PubMed] [Google Scholar]
- 34.Songyang, Z., S. E. Shoelson, J. McGlade, P. Olivier, T. Pawson, X. R. Bustelo, M. Barbacid, H. Sabe, H. Hanafusa, T. Yi, et al. 1994. Specific motifs recognized by the SH2 domains of Csk, 3BP2, fps/fes, GRB-2, HCP, SHC, Syk, and Vav. Mol. Cell. Biol. 14:2777-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tice, D. A., J. S. Biscardi, A. L. Nickles, and S. J. Parsons. 1999. Mechanism of biological synergy between cellular Src and epidermal growth factor receptor. Proc. Natl. Acad. Sci. USA 96:1415-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsukihara, T., H. Aoyama, E. Yamashita, T. Tomizaki, H. Yamaguchi, K. Shinzawa-Itoh, R. Nakashima, R. Yaono, and S. Yoshikawa. 1995. Structures of metal sites of oxidized bovine heart cytochrome c oxidase at 2.8 Å. Science 269:1069-1074. [DOI] [PubMed] [Google Scholar]
- 37.Velu, T. J., L. Beguinot, W. C. Vass, M. C. Willingham, G. T. Merlino, I. Pastan, and D. R. Lowy. 1987. Epidermal-growth-factor-dependent transformation by a human EGF receptor proto-oncogene. Science 238:1408-1410. [DOI] [PubMed] [Google Scholar]
- 38.Walker, F., A. Kato, L. J. Gonez, M. L. Hibbs, N. Pouliot, A. Levitzki, and A. W. Burgess. 1998. Activation of the Ras/mitogen-activated protein kinase pathway by kinase-defective epidermal growth factor receptors results in cell survival but not proliferation. Mol. Cell. Biol. 18:7192-7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, Y., S. Pennock, X. Chen, and Z. Wang. 2002. Endosomal signaling of epidermal growth factor receptor stimulates signal transduction pathways leading to cell survival. Mol. Cell. Biol. 22:7279-7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ware, M. F., D. A. Tice, S. J. Parsons, and D. A. Lauffenburger. 1997. Overexpression of cellular Src in fibroblasts enhances endocytic internalization of epidermal growth factor receptor. J. Biol. Chem. 272:30185-30190. [DOI] [PubMed] [Google Scholar]
- 41.Wilson, L. K., D. K. Luttrell, J. T. Parsons, and S. J. Parsons. 1989. pp60c-src tyrosine kinase, myristylation, and modulatory domains are required for enhanced mitogenic responsiveness to epidermal growth factor seen in cells overexpressing c-Src. Mol. Cell. Biol. 9:1536-1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong, A. J., J. M. Ruppert, S. H. Bigner, C. H. Grzeschik, P. A. Humphrey, D. S. Bigner, and B. Vogelstein. 1992. Structural alterations of the epidermal growth factor receptor gene in human gliomas. Proc. Natl. Acad. Sci. USA 89:2965-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wright, J. D., C. W. Reuter, and M. J. Weber. 1996. Identification of sites on epidermal growth factor receptors which are phosphorylated by pp60src in vitro. Biochim. Biophys. Acta 1312:85-93. [DOI] [PubMed] [Google Scholar]
- 44.Wu, W., L. M. Graves, G. N. Gill, S. J. Parsons, and J. M. Samet. 2002. Src-dependent phosphorylation of the epidermal growth factor receptor on tyrosine 845 is required for zinc-induced Ras activation. J. Biol. Chem. 277:24252-24257. [DOI] [PubMed] [Google Scholar]
- 45.Yang, W. L., L. Iacono, W. M. Tang, and K. V. Chin. 1998. Novel function of the regulatory subunit of protein kinase A: regulation of cytochrome c oxidase activity and cytochrome c release. Biochemistry 37:14175-14180. [DOI] [PubMed] [Google Scholar]
- 46.Zozulya, S., M. Lioubin, R. J. Hill, C. Abram, and M. L. Gishizky. 1999. Mapping signal transduction pathways by phage display. Nat. Biotechnol. 17:1193-1198. [DOI] [PubMed] [Google Scholar]