Abstract
Members of the Spalt gene family encode putative transcription factors characterized by seven to nine C2H2 zinc finger motifs. Four genes have been identified in mice—Spalt1 to Spalt4 (Sall1 to Sall4). Spalt homologues are widely expressed in neural and mesodermal tissues during early embryogenesis. Sall3 is normally expressed in mice from embryonic day 7 (E7) in the neural ectoderm and primitive streak and subsequently in the brain, peripheral nerves, spinal cord, limb buds, palate, heart, and otic vesicles. We have generated a targeted disruption of Sall3 in mice. Homozygous mutant animals die on the first postnatal day and fail to feed. Examination of the oral structures of these animals revealed that abnormalities were present in the palate and epiglottis from E16.5. In E10.5 embryos, deficiencies in cranial nerves that normally innervate oral structures, particularly the glossopharyngeal nerve (IX), were observed. These studies indicate that Sall3 is required for the development of nerves that are derived from the hindbrain and for the formation of adjacent branchial arch derivatives.
The Spalt genes were originally identified as important regulators of terminal system development in Drosophila. Spalt proteins have subsequently been shown to be required for appropriate migration, differentiation, and pattern formation in the development of the Drosophila wing, trachea, antenna, oenocytes-chordotonal organs, and photoreceptors (10, 11, 12, 17, 31, 32, 39, 48). Homologues have been isolated from a wide range of organisms—from Caenorhabditis elegans to humans (3, 5, 14, 15, 24, 26, 27, 28, 29, 31, 43, 44). In vertebrates, the Spalt or Sall genes are expressed early in development and in a diversity of embryonic and adult structures (5, 6, 13, 14, 27, 30, 42, 44, 45, 49). Four murine Sall genes have been identified to date (6, 27, 28, 29, 44). The patterns of expression of Sall1 (previously mSal-3) (45) and Sall3 (previously mSal-1) (44) during murine development (45) were previously reported. Both genes are expressed from embryonic day 7 (E7) in the mesoderm and ectoderm. Sall3 is subsequently expressed in the developing central nervous system, heart, limb buds, kidneys, ears, and palate.
Mutations in at least two of the human Spalt homologues are known to result in autosomal dominant human syndromes. Townes-Brocks syndrome, characterized by imperforate anus, pinna malformations, sensorineural hearing loss, polydactyly, kidney defects, and occasionally mental retardation, is associated with point mutations in SALL1 (7, 16, 25). Mutations in the SALL4 gene have been linked to Okihiro/acro-renal-ocular syndrome, in which patients exhibit forearm malformations and eye movement deficits (1, 28, 29). Although no diseases are yet directly linked to SALL3, this gene is located on chromosome 18 in a region commonly deleted in 18q deletion syndrome. Patients with this deletion exhibit hearing loss, cardiac problems, mental retardation, midfacial hypoplasia, delayed growth, and limb abnormalities (26, 50). No known disorders are associated with mutations in SALL2. It is becoming clear that Sall genes play a crucial role during the development of a number of systems in humans.
The function of Sall proteins in normal cellular and developmental processes is not known. The proteins are predicted to function as transcription factors. The first two zinc fingers of the Drosophila Spalt-related gene product have been shown to possess DNA binding activity, and potent transcriptional repression activity has been demonstrated for the N-terminal regions of human SALL1, murine Sall1, and chicken cSal-1 and cSal-3 proteins (2, 20, 41, 52). Studies of Sall1 have indicated that the transcriptional repression mediated by Spalt transcription factors likely involves histone deacetylase recruitment (20). Although the downstream effects of Spalt expression in these systems remain unclear, recent studies have suggested that Spalt genes may be involved in the regulation of the cell cycle and/or cell adhesion (8, 34, 37).
In an effort to elucidate the role of Sall molecules in vivo, targeted disruptions of these transcription factors were generated in mice. Targeted disruption of Sall1, the murine homologue of human SALL1, supports a role for this gene in kidney development, as these structures fail to form normally in mice that lack Sall1 (42). On the other hand, mice lacking Sall2 fail to demonstrate any abnormal phenotype, a finding which correlates with the fact that no diseases have been associated with its human homologue, SALL2 (49). In order to investigate the function of Sall3, we generated a targeted disruption in the Sall3 gene. The resulting animals survive until birth and die on the first postnatal day. This report describes the phenotype of animals from E9.5 until birth and identifies the structures targeted by loss of the Sall3 gene.
MATERIALS AND METHODS
Gene targeting of the Sall3 locus.
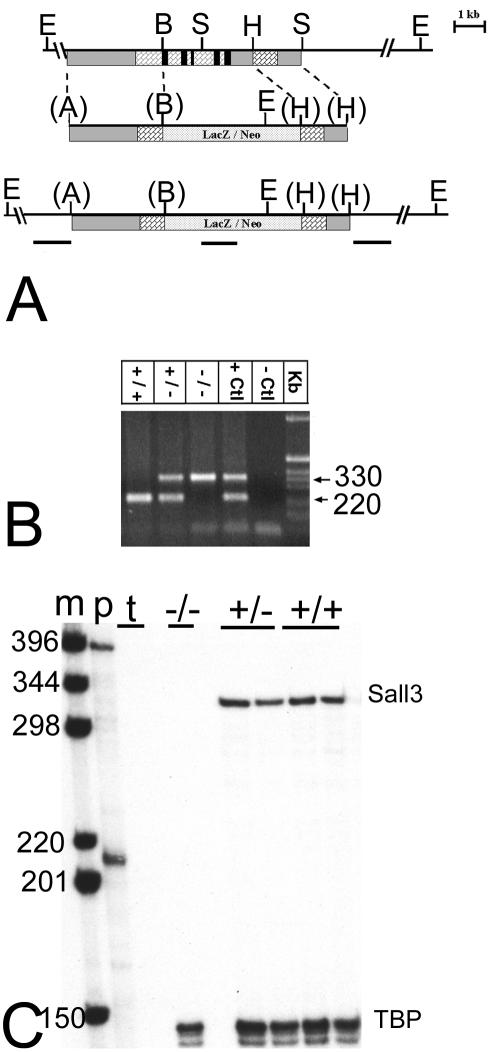
The targeting strategy for the Sall3 locus is indicated in Fig. 1A. To generate a null allele of the sall3 gene, a 3.1-kb ApaI/BamHI 5′ arm of homology from exon 2 was fused in frame to the β-galactosidase-promoterless neomycin resistance cassette in vector PHM3 (18). A 1.7-kb HindIII fragment was used as the 3′ arm of homology. The correctly targeted allele replaced 95% of the coding region, including all zinc fingers, thus producing a null allele. This construct was linearized, electroporated into 129SVJ embryonic stem cells, and selected with 315 μg of G418/ml for 7 days. One hundred positive clones were selected and screened by Southern hybridization with the 5′ and 3′ probes indicated in Fig. 1A to identify correctly targeted alleles. To ensure that selected clones had only one integration site, an internal probe for the lacZ gene was also used for Southern hybridization (Fig. 1A). Three positive clones were selected and injected into blastocysts, which were implanted in pseudopregnant females. Only one chimera gave rise to germ line transmission, and all heterozygous animals were screened by PCR (Fig. 1B). Southern blot and RNA analyses were performed as described previously (40).
FIG. 1.
Targeted disruption of Sall3. (A) (Top) Genomic structure of a portion of the Sall3 locus. (Middle) Targeting vector. (Bottom) Targeted locus. The majority of the coding sequence, including all zinc fingers, was removed and replaced with a LacZ-neomycin resistance cassette (LacZ/Neo). Grey bars indicate noncoding regions, brick-pattern bars indicate exons, stipple-pattern bars indicate LacZ/Neo, and black bars indicate locations of zinc finger motifs. E, B, S, H, and A, EcoRI, BamHI, SalI, HindIII, and ApaI restriction sites; parentheses indicate restriction sites that no longer exist. The locations of probes used for Southern hybridization (5′, LacZ, and 3′) are shown as black horizontal lines below the targeted locus. (B) Genotyping of Sall3 mutant animals. A 330-bp mutant band and the 220-bp wild-type band are shown. Samples from heterozygote animals yielded both bands. A positive control (+Ctl) and a negative control (−Ctl) are also shown. (C) RNase protection assay of a probe covering zinc fingers 4 and 5 indicating the absence of a Sall3 transcript in mutant animals. Lane m, labeled 1-kb marker; lane p, sizes of the unprotected Sall3 probe (394 bp) and the internal control TATA binding protein (TBP, which indicates that RNA was present in all samples) (210 bp); lane t, negative tRNA control. Other lanes show that mutant animals do not have a protected Sall3 band (330 bp), whereas wild-type and heterozygous animals do have a protected band.
PCR.
Collected tissues were incubated overnight at 56°C in 50 mM Tris (pH 8)-100 mM EDTA-0.5% sodium dodecyl sulfate-0.5 μg of proteinase K/μl. Proteinase K then was inactivated by incubation at 95°C for 10 min. PCR conditions were as follows: 1 cycle at 94°C for 10 min; 30 cycles at 94°C for 45 s, 69°C for 45 s, and 72°C for 45 s; and a final elongation cycle at 72°C for 10 min. PCR products were 330 bp for the mutant allele and 220 bp for the wild-type band and were visualized on a 1% agarose gel. The primers used were as follows: LacZ 2513 to 2492, 5′-CGCCATTCGCCATTCAGGCTGC-3′; 3′ 1240 to 1219, 5′-CAACGCACTGTCACTGCCGAAG-3′; and 5′ 1027 to 1048, 5′-TCAGACTTCATCCAGCAGTGTC-3′.
Mating and staging.
Heterozygous Sall3 mutant animals were mated to produce +/+, +/−, and −/− animals. Embryos were collected via cesarian section at different developmental stages ranging from E9.5 to E18.5. Embryonic ages were defined as days postcoitum, and the day of discovery of the vaginal plug was designated E0.5. Embryos were staged by embryonic age and/or physical characteristics. For postnatal ages, the first postnatal day was designated P0.
Behavioral tests.
Behavioral tests were performed on newborn pups in order to obtain a general measure of their coordination and response to external stimuli. Pups were collected and kept warm during experiments. They were weighed, and their reaction to pain was tested by a foot pinch. Any squeak elicited in response to the foot pinch was recorded as a positive response. As a general measure of coordination, animals were placed in a lying position on their fore and hind limbs. “Support” was assessed as the ability to support their weight and not fall onto their backs for 3 s. Any animal that did not role onto its back within that 3-s period was scored as +; all others were scored as −. Righting reflexes in these animals were also examined. Animals were rolled onto their backs to elicit a righting response. A positive response was recorded for any animal that lifted its hind limbs and put its forepaws down to right itself. A negative response was recorded for any animal that could not right itself or that lifted its forepaws into the air after a righting response was elicited. Righting responses were limited to these two opposite behaviors.
Histologic analysis.
Embryos ranging in age from E9.5 to E18.5 were preserved in Carnoy's fixative and embedded in paraffin. Serial sections of 17 μm were collected for each stage and stained with either hematoxylin-eosin or cresyl violet. Sections were examined and visualized with a Nikon fluorescent microscope and photographed with a Photometrics Cool Snap digital camera and IP Lab software.
In situ hybridization.
Selected 17-μm sections from embryos at E9.5 to E18.5 were processed for in situ hybridization. In situ prehybridization, hybridization, and probe synthesis were carried out as described previously (55). The probes used were Shh (a gift from Andy McMahon), HNF3α (40), HNF3β (40), Pax6 (a gift from Bob Hill), Sall1 (45), Sall3 (44), and Myf5, Myf6, MyoD, and myogenin (gifts from Marcia Ontell). Probes were hydrolyzed to 200-bp fragments, dissolved at 60 ng/ml in hybridization buffer, and hybridized overnight at 63°C (55). Slides were dipped in Kodak NTB2 emulsion diluted 1:1 with water, exposed at 4°C for 5 to 10 days, and developed in Kodak D19 and Kodakfix at 15°C for 4 min. Sections then were counterstained with hematoxylin-eosin or cresyl violet.
Whole-mount neurofilament staining.
E10.5 embryos were collected in phosphate-buffered saline (PBS), extraembryonic membranes were dissected away, and a small piece of tail or foot was retained for PCR genotyping. Embryos then were fixed overnight at 4°C in Dent's fixative (methanol-dimethyl sulfoxide [4:1]). Whole-mount neurofilament staining was performed as previously described (54). Mouse anti-neurofilament 160 primary antibody (Sigma clone NN18) was diluted 1/40 in PBS-2% Carnation milk powder-0.1% Triton X-100. Sall3−/− embryos were matched with a +/+ littermate. Occasionally, mutant embryos outnumbered +/+ littermates. In these instances, a +/− littermate match was selected (n = 26; 26 −/− versus 20 +/+ and 6 +/−). Excluding +/− animals and their −/− littermate matches from this analysis had no consistent effect on the outcome.
β-Galactosidase staining.
Newborn animals were sacrificed and decapitated. The skull was opened and immersion fixed in 4% paraformaldehyde-PBS at 4°C for 1 h. Newborn heads were rinsed in cold PBS, then immersed in PBS containing 4 mM potassium ferricyanide, 4 mM potassium ferrocyanide, 0.01% sodium deoxycholate, 0.02% NP-40, 2 mM magnesium chloride, and 1 mg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal)/ml, and gently agitated at 37°C overnight.
RESULTS
Gene targeting of the Sall3 locus.
The isolation of the Sall3 genomic locus and its organization have been previously described (44). The targeting strategy for the Sall3 locus replaced the majority of the coding region, including the DNA binding zinc fingers predicted to be required for transcription factor function (Fig. 1A). One hundred homologous recombinant ES cell clones were produced by electroporation and screened by Southern analysis. One chimera achieved germ line transmission of the targeted allele, and heterozygous progeny were identified initially by Southern analysis and subsequently by PCR. PCR of digested tail snips produced a 220-bp wild-type band and/or a 330-bp mutant band (Fig. 1B). Disruption of the Sall3 locus was confirmed by Southern analysis. The absence of the Sall3 transcript was confirmed by RNase protection analysis (Fig. 1C), since a Sall3-specific antibody is not available for Western analysis.
Sall3 is required for perinatal survival.
Heterozygous animals were fertile, indistinguishable from wild-type animals, and were intercrossed to produce homozygous progeny. Sall3−/− animals were produced at the expected Mendelian ratio and were alive at the time of birth (Table 1) but did not survive. These animals were of normal weight and length and were usually indistinguishable from littermates (Table 1 and data not shown). All but one Sall3 mutant animal died or was cannibalized within 12 h after birth.
TABLE 1.
Status of 285 Sall3 animals at birtha
| Animals | % (no. of animals) at P0 | Mean ± SD wt (g) | % of animals showing
|
n1 | % of animals showing Rt Rfx | n2 | |||
|---|---|---|---|---|---|---|---|---|---|
| Pain | Milk | Support | Pale pallor | ||||||
| +/+ | 28.1 (80) | 1.36 ± 0.08 | 100 | 85.3 | 100 | 5.9 | 17 | 100.0 | 15 |
| +/− | 48.1 (137) | 1.36 ± 0.13 | 100 | 78.0 | 97.6 | 0.0 | 41 | 100.0 | 27 |
| −/− | 23.9 (68) | 1.36 ± 0.09 | 100 | 42.9 | 42.9 | 21.4 | 14 | 72.7 | 11 |
Pain, reaction to foot pinch; milk, presence of milk in the stomach; pale pallor, presence of a pale complexion at birth; n1, numbers of animals used for weight measurement and the first four tests; Rt Rfx, presence of a righting reflex at birth; n2, number of animals used for the righting reflex test.
To investigate the cause of death, animals were collected promptly after birth. To determine whether animals could feed normally, the presence of milk in the stomach was assessed visually. Animals were scored as + or − for the presence or absence of milk. Animals with only small amounts of visible milk were scored as ±. Scores were tabulated as 1 for +, 0.5 for ±, and 0 for − (Table 1). Unlike littermates, few Sall3 mutant animals had observable milk in their stomachs (Table 1). To examine feeding behavior, dams were anesthetized, and each pup was placed alone with her to feed. Wild-type and heterozygous animals were able to identify nipples on the prone dam and suckle successfully. Homozygous Sall3 mutant animals were able to locate nipples and made an effort to suckle, indicating at least nominal functionality of olfactory, sensory, and motor systems. However, the mutant animals were unable to bring the nipples fully into their mouths and suckle normally. To determine whether dehydration was likely to be the immediate cause of death, Sall3+/+, Sall3+/−, and Sall3−/− pups were isolated at birth and given intraperitoneal injections of 20% sucrose-PBS every 2 h for 24 h. Homozygous mutant animals were indistinguishable from wild-type littermates and remained alive for the duration of the experiment, suggesting that dehydration was a likely cause of death.
We performed additional behavioral tests on mutant animals to assess the basic functionality of sensory and motor systems in Sall3−/− animals at birth. All mutant animals responded to painful stimuli (foot pinch); however, more than half of the mutants assessed were less capable of supporting their weight than their littermates (Table 1). A few (3 of 11; Table 1) Sall3 mutant animals also had altered righting reflexes (Table 1). These mutant pups moved their limbs in directions opposite those of wild-type and heterozygous animals when a righting response was elicited. These findings indicate that gross motor and sensory systems are intact but that subtle abnormalities may exist in motor coordination in Sall3−/− animals.
The morphology of the oral cavity is abnormal in Sall3−/− animals.
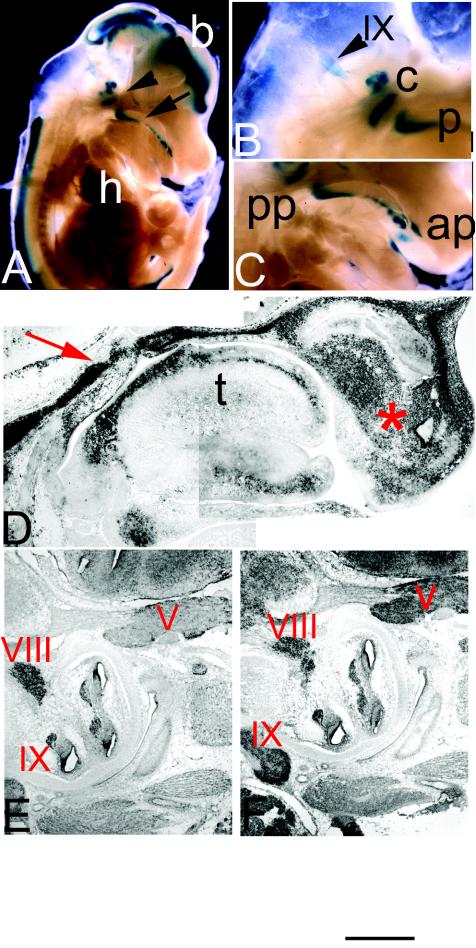
Ott et al. (44, 45) previously reported the patterns of expression of Sall1 and Sall3 in developing craniofacial regions in early development (E7.5 to E12.5). These studies indicated that transcripts for Sall1 and Sall3 are transiently found in tissues that give rise to structures of the palate and tongue and in regions of the central nervous system that innervate these structures (44, 45). In light of these findings and the observed suckling deficits in mutant animals, we assessed the normal distribution of Sall3 transcripts in developing craniofacial regions at later developmental stages (E14.5 until birth). From E14.5 on, Sall3 expression is up-regulated in the developing palate and tongue (Fig. 2A to D). Sall3 is found in a prominent band parallel to the dorsal surface of the tongue, with moderate expression in several other developing muscle groups of the tongue (Fig. 2D). Sall3 is also strongly expressed in the frontonasal process (Fig. 2D). In the palate, it is expressed in the mesenchyme of the rugae. Anteriorly, expression is found at the point of fusion of the primary and secondary palates (Fig. 2C); posteriorly, expression is found in areas surrounding the epiglottis and the opening of the nasopharynx and oropharynx (Fig. 2C). At P0, Sall3 is highly expressed in the rugae and in the primary and secondary palates. Sall3 transcripts are also found in developing cranial ganglia from E14.5 until birth (Fig. 2A, B, and E). In particular, the glossopharyngeal (IX), trigeminal (V), and vestibulocochlear (VIII) ganglia express robust levels of Sall3 at E16.5 (Fig. 2E). The related family member Sall1 is expressed in overlapping regions in the developing brain and palate at early developmental stages (45). At E16.5, Sall1 is also highly expressed in the glossopharyngeal (IX), trigeminal (V), and vestibulocochlear (VIII) ganglia (Fig. 2F). Differences were not observed in the expression of Sall3 transcripts detected by in situ hybridization and β-galactosidase staining in heterozygous and mutant animals, indicating that Sall3 does not regulate its own expression in developing craniofacial regions.
FIG. 2.
Expression of Sall3 in developing craniofacial regions from E13.5 to E16.5. (A) Whole-mount β-galactosidase staining of a parasagittal section through a mutant E14 embryo. Staining is observed in the brain (b), heart (h), palate (arrow), and cranial nerves (arrowhead). (B and C) Magnified views of the hindbrain (B) and palate (C) regions from E14 embryos. Sall3 transcripts are detected in cochlear ganglia (c), in the glossopharyngeal ganglion (IX), in the posterior palate (pp), and in the anterior palate (ap). (D to F) Digoxigenin-labeled in situ hybridization in sagittal sections of E13.5 (D) and E16.5 (E and F) embryos demonstrating the patterns of expression of Sall3 (D and E) and Sall1 (F). Expression is evident in the posterior palate (red arrow), the frontonasal process (asterisk), and the developing tongue (t). Both Sall3 and Sall1 are expressed in the trigeminal (V), the vestibulocochlear (VIII), and the glosspharyngeal (IX) ganglia. In all sections, rostral is to the right and dorsal is to the left. The scale bar represents 2 mm for panel A, 750 μm for panels B and C, 350 μm for panel D, and 500 μm for panels E and F.
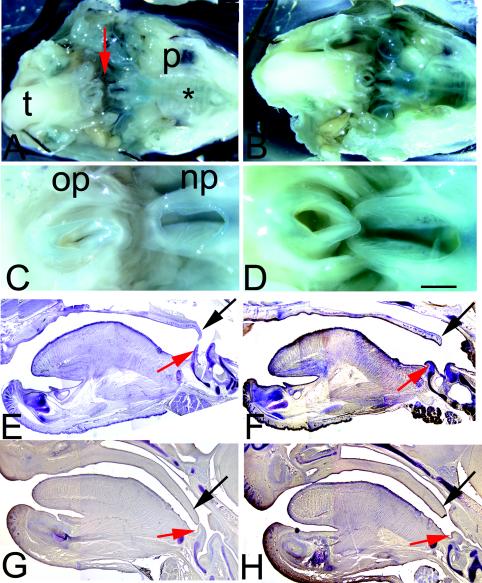
Malformations of the palate are one of the most common craniofacial abnormalities associated with feeding deficits. The palate and surrounding oral structures of Sall3 mutant animals were therefore examined from E12.5 until birth. In mutant animals at P0, palate fusion appears to have occurred normally between the shelves of the secondary palate and between the primary and secondary palates anteriorly. A normal pattern of rugae was also present (Fig. 3A and B). We used whole-mount alizarin red and alcian blue staining to examine the skull, mandible, and maxilla in Sall3−/− animals at P0. Analysis of craniofacial regions failed to show any consistent gross abnormalities in Sall3−/− animals at P0 (n = 6) (data not shown). Although the majority of the oral cavity appeared to be normal, some deficits were evident even at the gross level at P0. Examination of whole-mount specimens stained for β-galactosidase showed that the nasopharyngeal opening was much larger in Sall3 mutant animals than in heterzygous control animals (n = 4) (Fig. 3A to D).
FIG. 3.
Oral structures are abnormal in Sall3−/− animals. (A and B) Open-mouth views of whole-mount β-galactosidase staining of heterozygote (A) and mutant (B) embryos at P0. t, tongue; p, palate; asterisk, rugae. (C and D) Higher magnifications of the nasopharyngeal (np) and oropharyngeal (op) openings indicated by a red arrow in panel A. β-Galactosidase staining indicates that Sall3 is expressed throughout much of the palate, around the nasopharyngeal opening, and in the inner ear. In mutant animals (D), the nasopharyngeal opening is enlarged. (E to H) Cresyl violet staining of sagittal sections of tongues and palates of Sall3+/+ (E and G) and Sall3−/− (F and H) animals at P0 (E and F) and E16.6 (G and H). An abnormally large gap between the palate (velum) (black arrow) and the epiglottis (red arrow) is present in Sall3−/− animals. The velum and the epiglottis are smaller in Sall3 mutant animals at both P0 (F) and E16.5 (H). The scale bar represents 3 mm for panels A and B, 1.3 mm for panels C to F, and 1 mm for panels G and H.
To assess the morphology of structures surrounding the nasopharyngeal opening, we examined serial sagittal sections of intact P0 heads stained with cresyl violet (n = 6). In Sall3+/+ animals, the posterior end of the soft palate (velum) tapered and flexed ventrally toward the epiglottis (Fig. 3E). In mutant animals, the velum terminated prematurely, often ending as a rounded or bulbous structure. The ventral extension of the velum was also reduced or absent in mutant animals (Fig. 3F). Additionally, the epiglottis was abnormal in Sall3−/− animals. It was reduced in size and often had a sagging or wilted appearance (Fig. 3F). Serial sagittal sections of palatal structures were also analyzed at E16.5, just after palatal fusion is normally complete, in wild-type and mutant animals. At E16.5, like those of wild-type animals, the palates of Sall3 mutant animals had fused centrally, and fusion had extended rostrally and caudally, indicating that there was not a substantial delay in palate fusion. However, as at P0, the palates of Sall3 mutant animals at E16.5 were shorter, resulting in a substantial gap between the epiglottis and the posterior limit of the secondary palate (n = 2) (Fig. 3G and H). During normal suckling, the velum and the epiglottis overlap, separating the airway and the pharynx and permitting simultaneous breathing and swallowing. Our findings suggest that Sall3−/− animals are unable to exclude milk from the trachea or breathe while feeding because the velum and the epiglottis do not overlap.
Deficits in the tongue could also contribute to the abnormal feeding behavior observed in Sall3−/− animals at birth. We first examined the structure and the musculature of the tongues of Sall3+/+ and Sall3−/− animals in whole mounts and in serial coronal and sagittal sections stained with hematoxylin-eosin. At P0, multinucleate striated muscle of the tongue was organized into the typical intrinsic and extrinsic muscle groups in both wild-type and mutant littermates (n = 5). Taste buds were similar in size and number on the superior surface of the tongue in both wild-type and mutant animals. Measurements of the width of tongues revealed that the anterior portion of mutant tongues was, on average, 8% wider than that of wild-type littermate tongues (n = 5) (P < 0.05). To determine whether the deficits observed in the tongues at birth had a developmental origin, histological examination of the development of wild-type and mutant tongues and the surrounding mandible, maxilla, and dentition was performed with serial coronal and sagittal sections from E10 to E16.5. Differences in the size or organization of the tongues were not observed at these stages. Furthermore, the expression of the myogenic regulatory genes MRF-4, MRF-5, myogenin, and Myo-D was analyzed in Sall3−/− animals by in situ hybridization at E14.5, near the peak of their expression (57). All regulatory factors investigated were expressed at E14.5 in mutant tongues at levels and locations indistinguishable from those observed in wild-type littermate tongues (data not shown).
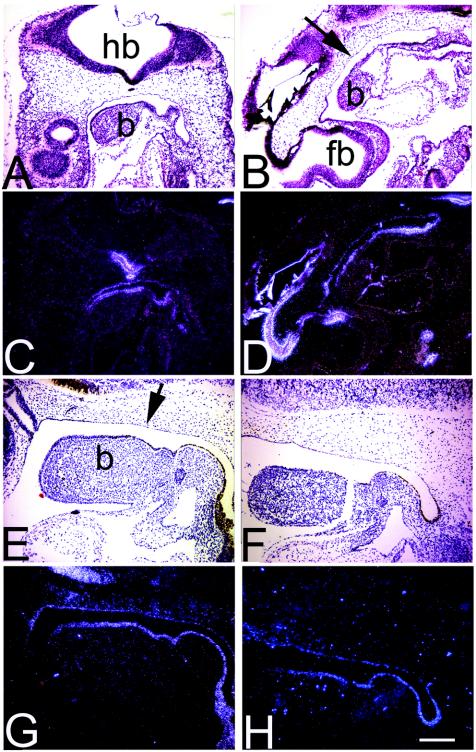
Sall3 is expressed not only at late developmental stages (E14 on) in oral structures but also earlier in the tissues and cells that give rise to these elements. The observed abnormalities in tissues surrounding the oral cavity could therefore reflect an earlier abnormality in patterning. To begin to answer this question, we examined the expression of a number of markers that are expressed in either the epithelium or the mesenchyme by in situ hybridization. Sonic hedgehog (Shh) (Fig. 4A to D), HNF3α (Fig. 4E to H), and Pax1 (data not shown) were used as markers of the epithelium of the first branchial arch, tongue, and palate. Msx2 was used as a mesenchyme marker for the first branchial arch and facial mesenchyme (data not shown). From E9.5 to E12.5, no differences in the patterns of expression of these markers between wild-type and mutant animals were observed.
FIG. 4.
Shh expression and HNF3α expression in the first branchial arch and tongue are normal in Sall3−/− animals. (A to H) Bright-field (A, B, E, and F) and dark-field (C, D, G, and H) photomicrographs of in situ hybridization of midsagittal sections from Sall3+/+ and −/− animals for Shh at E9.5 (A to D) and HNF3α at E10.5 (E to H). The epithelium of the palate (arrows), the first branchial arch (b), and the esophagus are labeled with Shh (A to D) and HNF3α (E to H) in a similar manner in both wild-type and mutant animals. Rostral is to the left, caudal is to the right, dorsal is to the top, and ventral is to the bottom in each panel. fb, forebrain; hb, hindbrain. Scale bar, 200 μm.
Sall3 influences cranial nerve development.
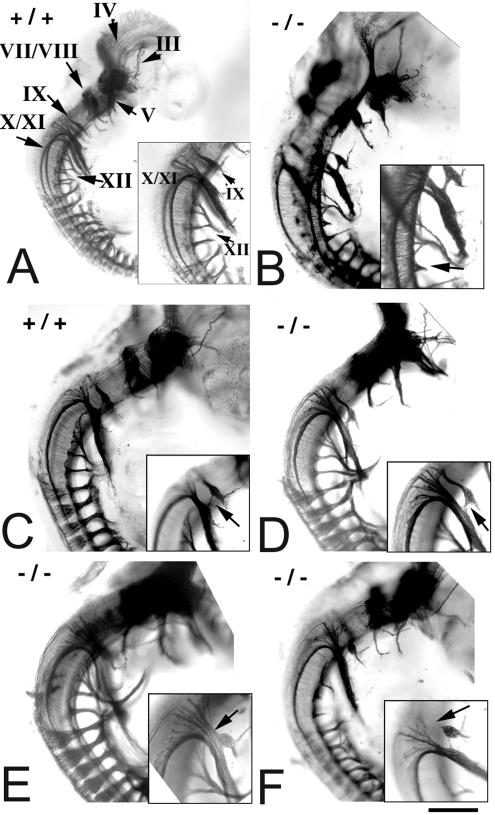
Sall3 is expressed in several cranial motor nuclei and nerves during development, including those that project to branchial arch derivatives (45) (Fig. 2). A lack of Sall3 in these nerves in mutant animals might result in cranial nerve abnormalities that would contribute to the failure of mutant animals to feed. To address this issue, we examined cranial nerve morphology by whole-mount neurofilament staining of a series of embryos taken from E10.5 to E11.5. To account for small within-litter variations in developmental age, each stained embryo was staged based on the overall extent of nerve outgrowth. The morphology and extent of outgrowth of most cranial and all spinal nerves were similar in wild-type and mutant embryos. Differences were observed, however, in the glossopharyngeal (cranial nerve IX), hypoglossal (cranial nerve XII), and anterior cervical ganglia when mutant (−/− [n = 26]) and control (+/+ [n = 20]; ± [n = 6]) groups were compared.
In the majority of control embryos (77%), a clearly defined glossopharyngeal nerve (IX) arose from the lateral surface of the hindbrain, just rostral to the vagal (X) and spinal (XI) accessory nerves (Table 2 and Fig. 5A). In a minority of wild-type embryos, either the proximal glossopharyngeal nerve was indivisible from the vagal or spinal accessory nerves or small connections existed between the two trunks (Fig. 5C). The frequency of these fusions or connections in Sall3−/− animals was doubled (46 versus 23%; Table 2) and was more severe than that in wild-type animals (Fig. 5C, D, and E). In addition, in 8% of mutant embryos (2 of 26), the inferior glossopharyngeal ganglion lacked a connection to the hindbrain (Fig. 5F and Table 2). This type of abnormality was never observed in control embryos. Abnormalities in the glossopharyngeal nerve, which mediates visceral sensations of the posterior oral cavity important in swallowing, also may have been partially responsible for the feeding deficits exhibited by Sall3−/− animals.
TABLE 2.
Cranial nerve deficits in Sall3−/− animals
| Animals (no. tested) | % of animals showing
|
||||
|---|---|---|---|---|---|
| Stunted IX | Connections between IX and X/XI | Total IX abnormalities | Persistent cervical DRGa | XII truncations | |
| +/+ (20) and +/− (6) | 0.0 | 23.1 | 23.1 | 7.7 | 30.8 |
| −/− (26) | 11.5 | 46.2 | 53.8 | 15.4 | 42.3 |
DRG, dorsal root-like ganglia.
FIG. 5.
Cranial nerve development is abnormal in Sall3−/− animals at E10.5 to E11.5. (A to F) Whole-mount neurofilament staining of wild-type (A and C) and Sall3−/− (B, D, E, and F) animals at E10.5 to E11.5. Rostral is to the top, caudal is to the bottom, dorsal is to the left, and ventral is to the right in each panel. Insets show higher magnifications of the areas of interest in each panel. (A) Wild-type embryo demonstrating the locations and normal morphology of cranial nerves (III, oculomotor; IV, trochlear; V, trigeminal; VII/VIII, facial/vestibulocochlear; IX, glossopharyngeal; X/XI, vagal/spinal accessory; XII, hypoglossal). (B and B inset) Sall3−/− animal in which cervical roots that normally contribute to the hypoglossal (XII) nerve are truncated (arrow in inset). (C, D, C inset, and D inset) Bundles of fibers connecting the glossopharyngeal (IX) nerve with the trunks of the vagal and spinal accessory (X and XI) nerves in a wild-type animal (C) and, more conspicuously, in a Sall3−/− animal (D) (arrows in insets). (E and E inset) Complete fusion of the glossopharyngeal (IX) and vagal/spinal accessory (X/XI) trunks in a Sall3−/− animal (arrow in inset). (F and F inset) Example in which the glossopharyngeal inferior (IX) ganglion lacked a connection to the brain stem (arrow in inset). The scale bar represents 800 μm for panel A and 500 μm for panels B to F.
At the stages examined, the hypoglossal (XII) nerve can be observed as multiple roots that originate from the ventral hindbrain and that converge deep in relation to the spinal accessory nerve (Fig. 5A). More distally, the hypoglossal nerve curves rostrally and projects into the tongue. Forty-two percent of Sall3 mutant animals showed unilateral or bilateral truncations of the most posterior roots that contribute to the hypoglossal nerve (Fig. 5B and Table 2). In contrast, only 31% of control animals showed similar reductions (Table 2). Such deficits were often unilateral and were not accompanied by reductions in adjacent nerve outgrowth.
Finally, dorsal root-like ganglia persisted at the anterior cervical levels in 15% of mutant embryos, while these structures were observed in only 8% of control embryos (Table 2). Prior studies suggested that such transient ganglia can occur normally in vertebrates (33); however, these abnormalities were observed more often in Sall3−/− animals. In half of the affected mutant animals, these supernumerary ganglia were also bilateral, while in the control animals, these structures were always unilateral (data not shown).
The glossopharyngeal and hypoglossal nerves innervate sensory and motor aspects of the tongue, respectively. To determine whether the abnormalities observed between E10.5 and E11.5 in these nerves led to an altered pattern of innervation of the tongue, serial sagittal sections of the tongue stained with anti-neurofilament 160 antibody were examined from E16 to birth (n = 2). Tongues of wild-type and mutant animals were both densely innervated. No gross abnormalities were observed in the innervation patterns of the tongues of Sall3 mutant animals.
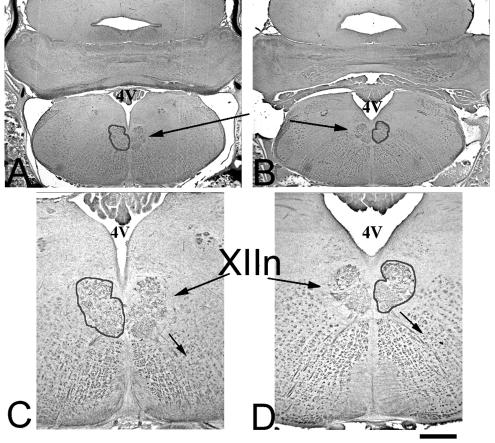
To determine whether the hypoglossal nucleus in the brain stem was normal in Sall3 mutant animals, we examined the expression of neurofilaments in serial coronal sections of the brain stem at P0. Neurons in the nucleus appeared to be similar in size and location, and their axons could be visualized projecting ventrolaterally from the nucleus to the ventral surface of the hindbrain and into the periphery (Fig. 6A to D).
FIG. 6.
The hypoglossal nucleus is normal in Sall3−/− animals. (A to D) Neurofilament staining of coronal sections demonstrates the normal location and appearance of the hypoglossal nucleus (XIIn) (perimeter highlighted) in Sall3+/+ (A and C) and Sall3−/− (B and D) animals. 4V, fourth ventricle; small arrows, axons. Dorsal is to the top and ventral is to the bottom in each panel. The scale bar represents 500 μm for panels A and B and 200 μm for panels C and D.
The ear and the spinal cord are normal in Sall3 mutant animals.
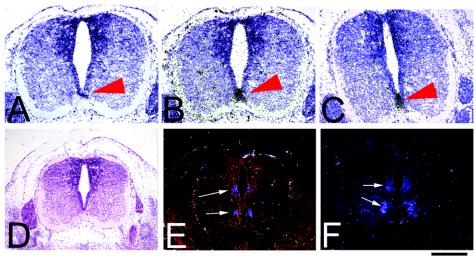
Behavior analyses indicated that some mutant animals had uncoordinated movements (Table 1). Feeding difficulties could result not only from deficits in craniofacial structure but also from abnormalities in the inner ear, affecting balance, or in the spinal cord itself, affecting the ability of the animals to produce coordinated movements. Sall3 is normally highly expressed in the inner ear, in the dorsal horn of the spinal cord, and in some motor neurons of the ventral horn. We therefore examined the structure and development of the inner ear and spinal cord. Histologically, at P0, no gross abnormalities were observed in the inner ear or in the organization and lamination of the neuronal cell bodies and fiber tracts in the spinal cord in mutant animals (n = 3). To determine whether the dorsal and ventral patterning of the spinal cord was normal, we examined the expression of HNF3α (Fig. 7B and C), Shh (data not shown), and HNF3β (data not shown) as markers of the notochord and floor plate and Pax6 as a regional spinal cord marker by in situ hybridization from E10.5 to E12.5 (Fig. 7E and F). No differences in the expression domains for these markers were observed (n = 3). Furthermore, alizarin red and alcian blue studies verified that skeletal and cartilaginous elements of the ribs, vertebrae, and limbs had developed normally (n = 6).
FIG. 7.
Early expression of transcription factors in the spinal cord is normal in Sall3−/− animals. (B, C, E, and F) Coronal sections of the spinal cord at E12.5 show the expression of HNF3α RNA in the floor plate (B and C; black grains; red arrowheads) and Pax6 RNA (E and F; white arrows; dark-field images) in Sall3+/+ (B and E) and Sall3−/− (C and F) animals. The gross patterning of the spinal cord in Sall3−/− animals (C and F) is similar to that in wild-type animals (B and E). (A and D) Bright-field images of Sall3+/+ spinal cords are included for reference adjacent to the experimental sections (B, C, E, and F). Dorsal is to the top and ventral is to the bottom in each panel. The scale bar represents 300 μm for all panels.
Sall3 mutants do not have kidney or heart deficits.
Sall3 is expressed in a number of other organs that, if perturbed, could result in perinatal lethality. Sall3 is expressed in the cushion pads of the heart and in differentiating glomeruli of the kidneys (45). We examined the structure of these organs to eliminate the possibility that the perinatal lethality seen in Sall3 mutant animals was due to organ failure rather than to dehydration. Gross histological analysis of serial sections stained with hematoxylin-eosin revealed no perturbations in the structure of any of these organs in Sall3−/− animals at P0 (n = 6) (data not shown).
DISCUSSION
To investigate the role of Sall3 during development we have generated a line of mice that lacks this putative transcription factor. Sall3−/− animals survived until birth, but died on the first postnatal day. Gross abnormalities were not detected in the kidneys, heart, skeleton, limbs or spinal cord. However, mutants appeared unable to suckle properly and occasionally showed an abnormal righting reflex. In an effort to determine the underlying cause of the suckling defect, we assessed the morphology and development of oral and craniofacial components required for feeding behavior. Sall3-deficient animals exhibit malformations of the palate, epiglottis, and tongue, all of which may inhibit normal feeding behavior. The morphology of the cranial nerves supplying these structures, especially the glossopharyngeal, was also found to be abnormal in mutant animals during early embryonic stages. These findings suggest that death of Sall3-deficient animals is caused at least in part by an inability to feed properly as a consequence of abnormalities in oral structures.
Sall3 is required for the terminal development of several oral structures.
Sall3 is expressed in the mesenchyme of the branchial arches early in development (from E8.5 on). It is subsequently down-regulated in oral structures derived from arch tissue until E13.5. At this time, intense expression of Sall3 appears within the palate at sites of midline and anterior fusion, in rugae, and in tongue musculature. At birth, expression is evident in the palate, epiglottis, nasopharynx, and oropharynx. The dynamic nature of these expression patterns suggests that Sall3 plays multiple roles in craniofacial development.
In Sall3 mutant animals, abnormalities in craniofacial structures were evident at mid-gestation and at birth. From E16 to P0, the palates of Sall3−/− animals terminate prematurely, and the velum was reduced or absent. Additionally, the epiglottis of mutant animals was reduced in size, and the tongue was widened anteriorly. These abnormalities would lead to an inability of the palate and pharynx to close and allow air to escape during swallowing and speech. Closure of the palate and pharynx during feeding is necessary to produce the appropriate suction to extract milk from the mother and subsequently to separate air from food to prevent aspiration and allow normal feeding and breathing. Interestingly, in addition to poor sucking or swallowing reflexes, humans with velopharyngeal insufficiency, characterized by a failure of the palate and pharynx to close, also exhibit hypernasality as air leaks from the nose during speech. Sall3−/− animals may also have abnormal vocalizations as a consequence of these palate deficiencies.
In spite of the early and widespread pattern of expression of Sall3 in branchial mesenchyme, the majority of palatal development and fusion occurs normally. In the absence of Sall3, alternate Spalt family members may compensate during early development in mutant animals. Sall1 is a likely candidate as it is also found in the 1st branchial arch and in head mesenchyme from E8 until E13.5 (45). At later stages (E14 on), Sall1 is not robustly expressed in the palate (M. Parrish and A. P. Monaghan, unpublished data) and is therefore unlikely to compensate for Sall3. Two additional Sall genes, Sall2 and Sall4, have been identified, and it will be interesting to define their expression patterns in the developing craniofacial regions.
Spalt homologues have been shown to be downstream targets of sonic hedgehog (Shh) signaling in vertebrates (30, 51). Humans and mice with mutations in Sall1 (SALL1; Townes-Brocks disorder) exhibit renal, ear, limb, anal, and cardiac abnormalities (21, 25, 42, 53). Patients with 18q deletion syndrome, that encompasses the SALL3 locus among other genes (26) also exhibit ear, limb, and cardiac as well as face and palatal deficiencies (56). Many of these abnormalities are also found in patients with alterations in Shh signaling. For example, patients with mutations in Gli3 (Pallister-Hall [PH] syndrome), a downstream target of Shh, exhibit, renal, ear, limb, anal, and cardiac abnormalities. Most strikingly, patients with PH syndrome can have laryngeal cleft and hypoplasia or absence of the epiglottis, similar to our findings for Sall3-deficient animals (4, 19, 38). Also, the VACTERL association that has been hypothesized to be a consequence of altered Shh signaling, includes tracheal-esophageal fistula, renal, limb, anal, and cardiac deficiencies (9, 22, 23, 35, 36, 47). Our observations on the Sall3 phenotype strengthens the link between Sall and Shh signaling and raises the possibility that reduced Sall expression or signaling could contribute to the phenotypes observed in syndromes such as PH syndrome.
Sall3 is important for cranial nerve development.
Several minor cranial nerve abnormalities were observed in Sall3 mutant animals. Although occasionally present in wild-type animals, the deficits observed were more frequent and of greater severity in Sall3−/− animals. The glossopharyngeal nerve (cranial nerve IX) was most consistently affected. This nerve mediates visceral sensory input from the posterior aspect of the tongue and palate in addition to other functions important in swallowing. Deficits in the function of this nerve would likely compound feeding difficulties resulting from the abnormal structure of the tongue, palate and epiglottis and contribute to the perinatal lethality observed in these animals. The hypoglossal nerve (XII) that innervates the genioglossal, hypoglossal, styloglossal and intrinsic muscles of the tongue was also variably altered in the absence of Sall3. Subtle alterations in the innervation pattern could lead to uncoordinated movements of the tongue during mastication and swallowing. The absence of overt abnormalities in the innervation pattern of the tongue at birth could be misleading. These subtle abnormalities could be difficult to detect due to the variable and unilateral nature of the defects observed during early embryonic development. It is therefore possible that more quantitative methods could reveal subtle abnormalities at birth. In light of this possibility, it is interesting that some mutants show an anterior enlargement of the tip of the tongue. It is also possible that the cranial nerve alterations observed at E10.5 are compensated for at later developmental stages. It is interesting that the deficits in the glossopharyngeal nerve described here are identical to those seen in animals in which mCoup-TFI has been disrupted (46). Similarities between Sall3 and mCoup-TFI in their function or the downstream effectors they impinge upon have yet to be fully described. It remains unclear if these deficits are a direct result of altered expression of Sall3 in the hindbrain or a secondary effect of alterations in branchial arch development resulting from the absence of Sall3 in these structures.
Role of Sall3 in spinal cord development.
Although Sall3 mutant animals responded to painful stimuli and demonstrated functionality of a number of sensory and motor systems, they were less capable of supporting their weight than their littermates and occasionally exhibited altered righting reflexes. General structure and development of the spinal cord, however, appeared normal. It is possible that defects exist in spinal cords of Sall3 mutant animals and would be revealed by electrophysiological or ultrastructural analysis. These behaviors, however, might be more simply explained by alterations in balance information conveyed by the inner ear or possibly attributed to the effects of dehydration. Sall3 is highly expressed in the inner ear during development and, although we observed no gross perturbations in this structure, changes in physiological function were not explored in this study.
The phenotype of Sall3 mutant animals is surprisingly mild in comparison to its expression pattern. Sall3 is expressed from E7 throughout the rest of development and also in the adult (45; Parrish and Monaghan, unpublished). The Sall1 and Sall3 gene products have similar structures, and their expression patterns differ only slightly in timing and location. Further studies will determine whether other Sall family members can compensate for Sall3 during development. If functional compensation can occur between members of the Spalt transcription factor family, compound mutations will be necessary to fully explore the role of Spalt genes in murine development.
Acknowledgments
We thank Dagmar Bock, Kristin Bond, Emily Sours, and Kirsten Boyer-Mauro for excellent technical assistance. We are grateful to Kathleen Kuznicki for commenting on the manuscript. We thank the following people for providing probes: Marsha Ontell (myogenic regulatory factors) and Andy McMahon (Shh).
This study was supported by NIMH grant no. 5RO1MH060774-03, MOD Basil O'Connor grant no. S-FY98-756, and NIH grant no. T32 MH18273.
REFERENCES
- 1.Al-Baradie, R., K. Yamada, C. St Hilaire, W. M. Chan, C. Andrews, N. McIntosh, M. Nakano, E. J. Martonyi, W. R. Raymond, S. Okumura, M. M. Okihiro, and E. C. Engle. 2002. Duane radial ray syndrome (Okihiro syndrome) maps to 20q13 and results from mutations in SALL4, a new member of the SAL family. Am. J. Hum. Genet. 71:1195-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrio, R., M. J. Shea, J. Carulli, K. Lipkow, U. Gaul, G. Frommer, R. Schuh, H. Jäckle, and F. C. Kafatos. 1996. The spalt-related gene of Drosophila melanogaster is a member of an ancient gene family, defined by the adjacent, region-specific homeotic gene spalt. Dev. Genes Evol. 206:315-325. [DOI] [PubMed] [Google Scholar]
- 3.Basson, M., and H. R. Horvitz. 1996. The Caenorhabditis elegans gene sem-4 controls neuronal and mesodermal cell development and encodes a zinc finger protein. Genes Dev. 10:1953-1965. [DOI] [PubMed] [Google Scholar]
- 4.Bose, J., L. Grotewold, and U. Ruther. 2002. Pallister-Hall syndrome phenotype in mice mutant for Gli3. Hum. Mol. Genet. 11:1129-1135. [DOI] [PubMed] [Google Scholar]
- 5.Buck, A., L. Archangelo, C. Dixkens, and J. Kohlhase. 2000. Molecular cloning, chromosomal localization, and expression of the murine SALL1 ortholog Sall1. Cytogenet. Cell Genet. 89:150-153. [DOI] [PubMed] [Google Scholar]
- 6.Buck, A., A. Kispert, and J. Kohlhase. 2001. Embryonic expression of the murine homologue of SALL1, the gene mutated in Townes-Brocks syndrome. Mech. Dev. 104:143-146. [DOI] [PubMed] [Google Scholar]
- 7.Cameron, T. H., A. M. Lachiewicz, and A. S. Aylsworth. 1991. Townes-Brocks syndrome in two mentally retarded youngsters. Am. J. Med. Genet. 41:1-4. [DOI] [PubMed] [Google Scholar]
- 8.Cantera, R., K. Luer, T. E. Rusten, R. Barrio, F. C. Kafatos, and G. M. Technau. 2002. Mutations in spalt cause a severe but reversible neurodegenerative phenotype in the embryonic central nervous system of Drosophila melanogaster. Development 129:5577-5586. [DOI] [PubMed] [Google Scholar]
- 9.Clarren, S. K., E. C. Alvord, Jr., and J. G. Hall. 1980. Congenital hypothalamic hamartoblastoma, hypopituitarism, imperforate anus, and postaxial polydactyly—a new syndrome? Part II. Neuropathological considerations. Am. J. Med. Genet. 7:75-83. [DOI] [PubMed] [Google Scholar]
- 10.de Celis, J. F., R. Barrio, and F. C. Kafatos. 1996. A gene complex acting downstream of dpp in Drosophila wing morphogenesis. Nature 381:421-424. [DOI] [PubMed] [Google Scholar]
- 11.de Celis, J. F., R. Barrio, and F. C. Kafatos. 1999. Regulation of the spalt/spalt-related gene complex and its function during sensory organ development in the Drosophila thorax. Development 126:2653-2662. [DOI] [PubMed] [Google Scholar]
- 12.Dong, P. D., J. S. Dicks, and G. Panganiban. 2002. Distal-less and homothorax regulate multiple targets to pattern the Drosophila antenna. Development 129:1967-1974. [DOI] [PubMed] [Google Scholar]
- 13.Farrell, E. R., and A. E. Munsterberg. 2000. csal1 is controlled by a combination of FGF and Wnt signals in developing limb buds. Dev. Biol. 225:447-458. [DOI] [PubMed] [Google Scholar]
- 14.Farrell, E. R., G. Tosh, E. Church, and A. E. Munsterberg. 2001. Cloning and expression of CSAL2, a new member of the spalt gene family in chick. Mech. Dev. 102:227-230. [DOI] [PubMed] [Google Scholar]
- 15.Hollemann, T., R. Schuh, T. Pieler, and R. Stick. 1996. Xenopus Xsal-1, a vertebrate homolog of the region specific homeotic gene spalt of Drosophila. Mech. Dev. 55:19-32. [DOI] [PubMed] [Google Scholar]
- 16.Ishikiriyama, S., F. Kudoh, N. Shimojo, J. Iwai, and T. Inoue. 1996. Townes-Brocks syndrome associated with mental retardation. Am. J. Med. Genet. 61:191-192. [DOI] [PubMed] [Google Scholar]
- 17.Jurgens, G. 1988. Head and tail development of the Drosophila embryo involves spalt, a novel homeotic gene. EMBO J. 7:189-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaestner, K. H., L. Montoliu, H. Kern, M. Thulke, and G. Schutz. 1994. Universal beta-galactosidase cloning vectors for promoter analysis and gene targeting. Gene 148:67-70. [DOI] [PubMed] [Google Scholar]
- 19.Kang, S., J. M. Graham, Jr., A. H. Olney, and L. G. Biesecker. 1997. GLI3 frameshift mutations cause autosomal dominant Pallister-Hall syndrome. Nat. Genet. 15:266-268. [DOI] [PubMed] [Google Scholar]
- 20.Kiefer, S. M., B. W. McDill, J. Yang, and M. Rauchman. 2002. Murine Sall1 represses transcription by recruiting a histone deacetylase complex. J. Biol. Chem. 277:14869-14876. [DOI] [PubMed] [Google Scholar]
- 21.Kiefer, S. M., K. K. Ohlemiller, J. Yang, B. W. McDill, J. Kohlhase, and M. Rauchman. 2003. Expression of a truncated Sall1 transcriptional repressor is responsible for Townes-Brocks syndrome birth defects. Hum. Mol. Genet. 12:2221-2227. [DOI] [PubMed] [Google Scholar]
- 22.Kim, J., P. Kim, and C. C. Hui. 2001. The VACTERL association: lessons from the Sonic hedgehog pathway. Clin. Genet. 59:306-315. [DOI] [PubMed] [Google Scholar]
- 23.Kim, P. C., R. Mo, and C. C. Hui. 2001. Murine models of VACTERL syndrome: role of sonic hedgehog signaling pathway. J. Pediatr. Surg. 36:381-384. [DOI] [PubMed] [Google Scholar]
- 24.Kohlhase, J., R. Schuh, G. Dowe, R. P. Kuhnlein, H. Jackle, B. Schroeder, W. Schulz-Schaeffer, H. A. Kretzschmar, A. Kohler, U. Muller, M. Raab-Vetter, E. Burkhardt, W. Engel, and R. Stick. 1996. Isolation, characterization, and organ-specific expression of two novel human zinc finger genes related to the Drosophila gene spalt. Genomics 38:291-298. [DOI] [PubMed] [Google Scholar]
- 25.Kohlhase, J., A. Wischermann, Reichenbach, H., U. Froster, and W. Engel. 1998. Mutations in the SALL1 putative transcription factor gene cause Townes-Brocks syndrome. Nat. Genet. 18:81-83. [DOI] [PubMed] [Google Scholar]
- 26.Kohlhase, J., S. Hausmann, G. Stojmenovic, C. Dixkens, K. Bink, W. Schulz-Schaeffer, M. Altmann, and W. Engel. 1999. SALL3, a new member of the human spalt-like gene family, maps to 18q23. Genomics 62:216-222. [DOI] [PubMed] [Google Scholar]
- 27.Kohlhase, J., M. Altmann, L. Archangelo, C. Dixkens, and W. Engel. 2000. Genomic cloning, chromosomal mapping, and expression analysis of msal-2. Mamm. Genome 11:64-68. [DOI] [PubMed] [Google Scholar]
- 28.Kohlhase, J., M. Heinrich, M. Liebers, L. Frohlich Archangelo, W. Reardon, and A. Kispert. 2002. Cloning and expression analysis of SALL4, the murine homologue of the gene mutated in Okihiro syndrome. Cytogenet. Genome Res. 98:274-277. [DOI] [PubMed] [Google Scholar]
- 29.Kohlhase, J., M. Heinrich, L. Schubert, M. Liebers, A. Kispert, F. Laccone, P. Turnpenny, R. M. Winter, and W. Reardon. 2002. Okihiro syndrome is caused by SALL4 mutations. Hum. Mol. Genet. 11:2979-2987. [DOI] [PubMed] [Google Scholar]
- 30.Koster, R., R. Stick, F. Loosli, and J. Wittbrodt. 1997. Medaka spalt acts as a target gene of hedgehog signaling. Development 124:3147-3156. [DOI] [PubMed] [Google Scholar]
- 31.Kuhnlein, R. P., G. Frommer, M. Friedrich, M. Gonzalez-Gaitan, A. Weber, J. F. Wagner-Bernholz, W. J. Gehring, H. Jackle, and R. Schuh. 1994. spalt encodes an evolutionarily conserved zinc finger protein of novel structure which provides homeotic gene function in the head and tail region of the Drosophila embryo. EMBO J. 13:168-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuhnlein, R. P., and R. Schuh. 1996. Dual function of the region-specific homeotic gene spalt during Drosophila tracheal system development. Development 122:2215-2223. [DOI] [PubMed] [Google Scholar]
- 33.Kuratani, S., S. Tanaka, Y. Ishikawa, and C. Zukeran. 1988. Early development of the hypoglossal nerve in the chick embryo as observed by the whole-mount nerve staining method. Am. J. Anat. 182:155-168. [DOI] [PubMed] [Google Scholar]
- 34.Li, D., Y. Tian, Y. Ma, and T. Benjamin. 2004. p150(Sal2) is a p53-independent regulator of p21(WAF1/CIP). Mol. Cell. Biol. 24:3885-3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martinez-Frias, M. L., and J. L. Frias. 1999. VACTERL as primary, polytopic developmental field defects. Am. J. Med. Genet. 83:13-16. [DOI] [PubMed] [Google Scholar]
- 36.Martinez-Frias, M. L., E. Bermejo, and J. L. Frias. 2001. The VACTERL association: lessons from the Sonic hedgehog pathway. Clin. Genet. 60:397-398. [DOI] [PubMed] [Google Scholar]
- 37.Milan, M., U. Weihe, L. Perez, and S. M. Cohen. 2001. The LRR proteins capricious and Tartan mediate cell interactions during DV boundary formation in the Drosophila wing. Cell 106:785-794. [DOI] [PubMed] [Google Scholar]
- 38.Ming, J. E., E. Roessler, and M. Muenke. 1998. Human developmental disorders and the Sonic hedgehog pathway. Mol. Med. Today 4:343-349. [DOI] [PubMed] [Google Scholar]
- 39.Mollereau, B., M. Dominguez, R. Webel, N. J. Colley, B. Keung, J. F. de Celis, and C. Desplan. 2001. Two-step process for photoreceptor formation in Drosophila. Nature 412:911-913. [DOI] [PubMed] [Google Scholar]
- 40.Monaghan, A. P., K. H. Kaestner, E. Grau, and G. Schutz. 1993. Postimplantation expression patterns indicate a role for the mouse forkhead/HNF-3 alpha, beta and gamma genes in determination of the definitive endoderm, chordamesoderm and neuroectoderm. Development 119:567-578. [DOI] [PubMed] [Google Scholar]
- 41.Netzer, C., L. Rieger, A. Brero, C. D. Zhang, M. Hinzke, J. Kohlhase, and S. K. Bohlander. 2001. SALL1, the gene mutated in Townes-Brocks syndrome, encodes a transcriptional repressor which interacts with TRF1/PIN2 and localizes to pericentromeric heterochromatin. Hum. Mol. Genet. 10:3017-3024. [DOI] [PubMed] [Google Scholar]
- 42.Nishinakamura, R., Y. Matsumoto, K. Nakao, K. Nakamura, A. Sato, N. G. Copeland, D. J. Gilbert, N. A. Jenkins, S. Scully, D. L. Lacey, M. Katsuki, M. Asashima, and T. Yokota. 2001. Murine homolog of SALL1 is essential for ureteric bud invasion in kidney development. Development 128:3105-3115. [DOI] [PubMed] [Google Scholar]
- 43.Onuma, Y., R. Nishinakamura, S. Takahashi, T. Yokota, and M. Asashima. 1999. Molecular cloning of a novel Xenopus spalt gene (Xsal-3). Biochem. Biophys. Res. Commun. 264:151-156. [DOI] [PubMed] [Google Scholar]
- 44.Ott, T., K. H. Kaestner, A. P. Monaghan, and G. Schutz. 1996. The mouse homolog of the region specific homeotic gene spalt of Drosophila is expressed in the developing nervous system and in mesoderm-derived structures. Mech. Dev. 56:117-128. [DOI] [PubMed] [Google Scholar]
- 45.Ott, T., M. Parrish, K. Bond, Schwaeger-Nickolenko, A., and A. P. Monaghan. 2001. A new member of the spalt like zinc finger protein family, Msal-3, is expressed in the CNS and sites of epithelial/mesenchymal interaction. Mech. Dev. 101:203-207. [DOI] [PubMed] [Google Scholar]
- 46.Qiu, Y., F. A. Pereira, F. J. DeMayo, J. P. Lydon, S. Y. Tsai, and M. J. Tsai. 1997. Null mutation of mCOUP-TFI results in defects in morphogenesis of the glossopharyngeal ganglion, axonal projection, and arborization. Genes Dev. 11:1925-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rittler, M., J. E. Paz, and E. E. Castilla. 1996. VACTERL association, epidemiologic definition and delineation. Am. J. Med. Genet. 63:529-536. [DOI] [PubMed] [Google Scholar]
- 48.Rusten, T. E., R. Cantera, J. Urban, G. Technau, F. C. Kafatos, and R. Barrio. 2001. Spalt modifies EGFR-mediated induction of chordotonal precursors in the embryonic PNS of Drosophila promoting the development of oenocytes. Development 128:711-722. [DOI] [PubMed] [Google Scholar]
- 49.Sato, A., Y. Matsumoto, U. Koide, Y. Kataoka, N. Yoshida, T. Yokota, M. Asashima, and R. Nishinakamura. 2003. Zinc finger protein sall2 is not essential for embryonic and kidney development. Mol. Cell. Biol. 23:62-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Strathdee, G., R. Sutherland, J. J. Jonsson, R. Sataloff, M. Kohonen-Corish, D. Grady, and J. Overhauser. 1997. Molecular characterization of patients with 18q23 deletions. Am. J. Hum. Genet. 60:860-868. [PMC free article] [PubMed] [Google Scholar]
- 51.Sturtevant, M. A., B. Biehs, E. Marin, and E. Bier. 1997. The spalt gene links the A/P compartment boundary to a linear adult structure in the Drosophila wing. Development 124:21-32. [DOI] [PubMed] [Google Scholar]
- 52.Sweetman, D., T. Smith, E. R. Farrell, A. Chantry, and A. Munsterberg. 2003. The conserved glutamine-rich region of chick csal1 and csal3 mediates protein interactions with other spalt family members. Implications for Townes-Brocks syndrome. J. Biol. Chem. 278:6560-6566. [DOI] [PubMed] [Google Scholar]
- 53.Townes, P. L., and E. R. Brocks. 1972. Hereditary syndrome of imperforate anus with hand, foot, and ear anomalies. J. Pediatr. 81:321-326. [DOI] [PubMed] [Google Scholar]
- 54.Wall, N. A., C. M. Jones, B. L. Hogan, and C. V. Wright. 1992. Expression and modification of Hox 2.1 protein in mouse embryos. Mech. Dev. 37:111-120. [DOI] [PubMed] [Google Scholar]
- 55.Wilkinson, D. G. 1992. Whole mount in situ hybridization of vertebrate embryos, p. 75-83. In D. G. Wilkinson (ed.), In situ hybridization: a practical approach. Oxford University Press, New York, N.Y.
- 56.Wilson, M. G., J. W. Towner, I. Forsman, and E. Siris. 1979. Syndromes associated with deletion of the long arm of chromosome 18[del(18q)]. Am. J. Med. Genet. 3:155-174. [DOI] [PubMed] [Google Scholar]
- 57.Yamane, A., M. Mayo, C. Shuler, D. Crowe, Y. Ohnuki, K. Dalrymple, and Y. Saeki. 2000. Expression of myogenic regulatory factors during the development of mouse tongue striated muscle. Arch. Oral Biol. 45:71-78. [DOI] [PubMed] [Google Scholar]