Abstract
We have programmed human cells to express physiological levels of recombinant RNA polymerase II (RNAPII) subunits carrying tandem affinity purification (TAP) tags. Double-affinity chromatography allowed for the simple and efficient isolation of a complex containing all 12 RNAPII subunits, the general transcription factors TFIIB and TFIIF, the RNAPII phosphatase Fcp1, and a novel 153-kDa polypeptide of unknown function that we named RNAPII-associated protein 1 (RPAP1). The TAP-tagged RNAPII complex is functionally active both in vitro and in vivo. A role for RPAP1 in RNAPII transcription was established by shutting off the synthesis of Ydr527wp, a Saccharomyces cerevisiae protein homologous to RPAP1, and demonstrating that changes in global gene expression were similar to those caused by the loss of the yeast RNAPII subunit Rpb11. We also used TAP-tagged Rpb2 with mutations in fork loop 1 and switch 3, two structural elements located strategically within the active center, to start addressing the roles of these elements in the interaction of the enzyme with the template DNA during the transcription reaction.
RNA polymerase II (RNAPII) is the multisubunit enzyme that synthesizes all mRNA precursors in eukaryotes. RNAPII is highly conserved among species, and in humans, RNAPII consists of 12 subunits, named Rpb1 to Rpb12 (16, 88). The two largest subunits, Rpb1 (220 kDa) and Rpb2 (140 kDa), form the enzyme's catalytic center and are homologous to the β′ and β subunits of bacterial RNAP, respectively. Five subunits, Rpb5, Rpb6, Rpb8, Rpb10, and Rpb12, are also found in RNAPI and RNAPIII. Rpb3 and Rpb11 are homologous to the α2 homodimer involved in bacterial RNAP assembly. Rpb9 was attributed a role in elongation through its action at DNA arrest sites (3). In Saccharomyces cerevisiae, Rpb4 and Rpb7 form a subcomplex that can dissociate from the enzyme upon changes in environmental conditions. Under active growth conditions, most yeast RNAPII molecules do not contain the Rpb4-Rpb7 dimer, which primarily associates during the stationary phase or following stress (11). Functional studies of human RNAPII have been limited due to the lack of appropriate systems for purifying variant forms of the human enzyme.
The availability of crystal structures of both yeast (17, 18, 26, 30) and bacterial (8, 57, 58, 84, 89) RNAPs has been invaluable for understanding many of the molecular features of the transcription reaction. For example, the structure of elongating RNAPII has revealed the positioning of the RNA-DNA duplex during the transcription reaction (30). The structures available support a model in which the DNA enters the enzyme through a channel formed by a pair of “jaws” before accessing a deep cleft, at the bottom of which is buried the Mg2+-ion-containing active site; the DNA then turns by about 90° along a wall where the upstream end exits the enzyme (30). Many loops and helices either directly contact or closely approach the RNA-DNA duplex, thus suggesting putative functions for these structural elements in the transcription reaction. Again, the lack of an efficient method for purifying variant forms of RNAPII with mutations within specific structural elements has precluded any detailed structure-function analysis of human RNAPII.
The activity of RNAPII at the various stages of the transcription reaction is regulated by several proteins, including the general transcription factors (14, 32, 65). TATA-binding protein (TBP), the TATA box-binding subunit of TFIID, recognizes the TATA box of promoters, and its binding induces a bend of approximately 80° in the DNA (45, 46). TFIIB associates with the TBP-promoter complex and participates in RNAPII recruitment (54). TFIIF, composed of subunits RAP74 and RAP30, binds directly to RNAPII and also helps to recruit the enzyme to the promoter (15, 80). TFIIF can induce further bending and wrapping of the promoter DNA around RNAPII during preinitiation complex formation (72). Both TFIIE and TFIIH were shown to participate in promoter melting at the transcription initiation site (38, 66). TFIIE, which is also composed of two subunits, TFIIE56/α and TFIIE34/β, stimulates the helicase and kinase activities of TFIIH (53, 62, 63). The helicase activity of TFIIH is required to fully open the promoter DNA between nucleotides (nt) −9 and +2 (37, 39, 44, 83), and its kinase activity phosphorylates the carboxy-terminal domain (CTD) of Rpb1 (21, 53, 75-77). The phosphorylation of the CTD on Ser5 by TFIIH has been shown to be required for the transition from initiation to early elongation and allows for the recruitment of the capping enzyme (35, 73). In yeast, the Ctk1 kinase phosphorylates the CTD on Ser2 at a later stage of the elongation phase (10), leading to the recruitment of mRNA processing factors (78). The CTD phosphatase Fcp1 is able to dephosphorylate the Rpb1 CTD on Ser2 in order to recycle RNAPII for reinitiation (10, 48, 49).
Unlike the case for all of the general transcription factors and the prokaryotic RNAP, it has been impossible to date to purify functionally active forms of mammalian RNAPII reconstituted from its recombinant subunits. We now report the purification of a functionally active human RNAPII by the use of doubly tagged subunits expressed in mammalian cells. Using the TAP procedure (71), we isolated a human RNAPII-containing complex which comprises the 12 subunits of core RNAPII,TFIIB, TFIIF, Fcp1, and RPAP1, a novel polypeptide of unknown function. The tagged RNAPII complex is functionally active both in vitro and in vivo. We capitalized on this purification procedure to generate two variant forms of human RNAPII by using TAP-tagged Rpb2. The first mutant has a deletion in fork loop 1, a structure that is expected to participate in maintaining the open state of the transcription bubble in elongating RNAPII (18, 30) or during RNA-DNA strand separation (85). The second mutant carries a triple alanine substitution in switch 3, a loop that directly contacts the DNA template within the DNA-RNA hybrid. Our results allowed us to detail the roles of fork loop 1 and switch 3 in transcriptional mechanisms both in vitro and in vivo.
MATERIALS AND METHODS
Protein factors and antibodies.
Recombinant yeast TBP (42), human TFIIB (31), RAP74 and RAP30 (22), TFIIE56/α and TFIIE34/β (64, 67, 81), highly purified TFIIH (20, 27), and calf thymus RNAPII (36) were purified as previously described. Antibodies raised against Rpb1 (N-20) were obtained from Santa Cruz Biotechnology.
TAP of human RNAPII-containing complex.
The cDNAs encoding human Rpb2 (kindly provided by Marc Vigneron, IGBMC, Strasbourg, France) (1), Rpb4 (Invitrogen; accession number BE883306), Rpb7 (Invitrogen; accession number BE256210), Rpb11 (Invitrogen; accession number AA814184), TFIIB (31), and RAP30 (22) were cloned into the mammalian expression vector pMZI (61) in such a way that each polypeptide carries a tandem affinity purification (TAP) tag at its C terminus (71). Stable cell lines carrying these plasmids in EcR-293 cells (Invitrogen) were produced by transfection, using the calcium phosphate method. Induction for 24 h with 1 to 3 μM ponasterone A, an ecdysone analog, was used to express the TAP-tagged subunits of RNAPII, TFIIB, and RAP30 at near-physiological levels. The TAP-tagged complexes were then purified as previously described (71). The TAP eluates were concentrated and stored at −80°C until use.
Protein identification.
The TAP eluates were run in sodium dodecyl sulfate (SDS) gels and silver stained. The protein bands were excised from the gels, reduced, alkylated, and subjected to in-gel tryptic digestion as previously described (50). The resulting tryptic peptides were purified and identified by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (50).
DNA microarray analysis of Tet-promoter mutants.
Tet-promoter alleles were constructed as previously described (86). Mutant and isogenic wild-type cultures were grown in parallel in SC medium with 10 μg of doxycycline/ml for a total of 22 h. Dilutions were made at 16 h to ensure that mutant and wild-type optical densities matched as closely as possible to 107 cells/ml at harvest. The cells were pelleted by a 2-min centrifugation in a room-temperature table-top centrifuge and then frozen immediately in liquid nitrogen. Total RNAs were prepared by hot acidic phenol-chloroform extraction followed by ethanol precipitation. Poly(A)+ RNA purification and cDNA labeling were performed as described previously (55). Microarrays were composed of 70-mer oligonucleotides, each specific to a different yeast gene (Operon Technologies), which were spotted onto poly-l-lysine-coated slides. Spotting buffer, blocking, hybridization, and washing were according to a procedure described by Hegde and collaborators (34). Each array was normalized by using grid-by-grid lowess smoothing (87). All measurements were taken in fluor-reversed pairs [i.e., each time a mutant was analyzed, it was hybridized to two arrays, and data were combined by averaging the normalized log(ratio)].
ChIP.
Chromatin immunoprecipitation (ChIP) assays were performed as previously described (60, 82), with the following minor modifications. Approximately 2 × 108 cells were cross-linked by the use of formaldehyde, and chromatin fragments of an average size of 0.3 to 0.5 kbp were prepared by four sonication treatments (Fisher Sonic Dismembrator; 20-s burst at 50% maximum power). For the immunoprecipitation step, immunoglobulin G (IgG)-Sepharose beads (Amersham Biosciences) were first blocked with a 5-mg/ml solution of bovine serum albumin in phosphate-buffered saline and then incubated with the sample for 1 h. After reversal of the cross-links, PCR analyses were performed with both the immunoprecipitated and input DNAs. Primer sets designed for amplifying both the promoter and the 5′ untranslated region of the CCNA1 gene were used.
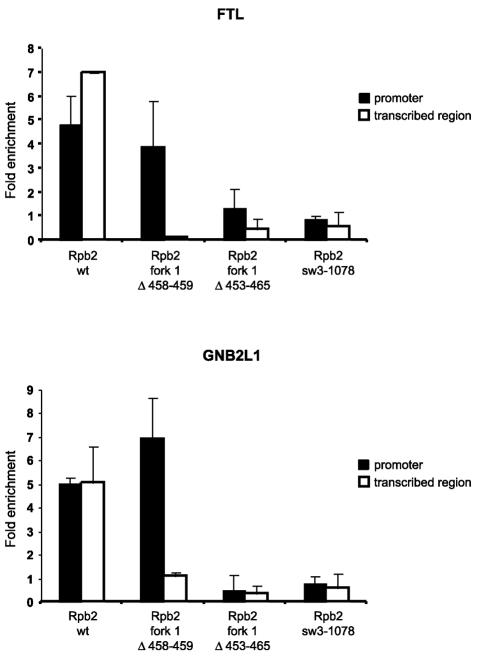
For the experiments shown in Fig. 8, relative occupancy was evaluated by quantitative PCR (Q-PCR) as previously described (5), with minor modifications. The quantification of the target DNA in the immunoprecipitated sample was carried out by generating a standard curve with a fivefold dilution series of the nonimmunoprecipitated sample (input DNA) for each couple of primer sets and DNA. The apparent immunoprecipitation efficiency was calculated by dividing the amount of the DNA fragments spanning the promoter or the coding region of the specified genes by the amount of the DNA fragment from the control region in the immunoprecipitated sample. The following cell lines were analyzed: Rpb2 wt, Rpb2 fork1 Δ458-459, Rpb2 sw3-1078, and Rpb2 fork1 Δ453-465. Q-PCR was performed by using the Mx4000 Multiplex Quantitative PCR system (Stratagene). The PCR protocol was applied according to the manufacturer's specifications (Qiagen). Primers were designed for both the promoter and transcribed regions of the GNB2L1 (positions −200 to +55 and +998 to +1165, respectively) and FTL (positions −157 to +33 and +955 to +1176, respectively) genes and also for an internal sequence of the expressed sequence tag located at chr17:49926700-49927999 as a control. Oligonucleotide sequences are available upon request. All amplified DNA fragments were unique, as determined by a BLAST search on the April 2003 assembly version of the UCSC Genome Browser (http://www.genome.ucsc.edu), by a melting curve analysis, and by agarose gel electrophoresis (data not shown).
FIG. 8.
Association of mutant forms of the RNAPII complex with chromatin in vivo. The fold enrichment over the control region of both the promoter and the transcribed region of the FTL and GNB2L1 genes after the immunoprecipitation of chromatin fragments was quantified by Q-PCR. Enrichment was normalized to an intergenic region of chromosome 17 (see Materials and Methods). Data points represent the means of four experiments, including two independent ChIP assays.
Gel mobility shift assays.
Gel mobility shift assays were performed as previously described (52). Complexes were assembled by using highly purified TBP (80 ng), TFIIB (60 ng), RAP30 (320 ng), RAP74 (640 ng), TFIIE34 (160 ng), TFIIE56 (320 ng), and calf thymus RNAPII or the TAP-tagged human RNAPII complex (the amounts of RNAPII used in the various experiments are detailed in the figure legends).
Histone tail peptide binding assay.
Histone tail peptide binding reactions were performed to determine whether the complex binds specifically to acetylated histone tails. For each reaction, 10 ng of biotinylated H3 peptides was bound to streptavidin-conjugated magnetic beads at 25°C for 30 min. The beads were then incubated with 1 nM RNAPII complex (Rpb11 TAP purified or calf thymus) for 1 h at 30°C. The beads were pelleted with a magnetic particle concentrator (MPC-S; Dynal) and the supernatant was removed. After being washed twice, the beads were resuspended in SDS-polyacrylamide gel electrophoresis (SDS-PAGE) lysis buffer and subjected to SDS-PAGE analysis. The complex was detected by immunoblotting with an anti-Rpb1 antibody (N-20; Santa Cruz).
Transcription assay.
Transcription reactions were performed as described previously (6). TBP (30 ng), TFIIB (30 ng), RAP30 (30 ng), RAP74 (65 ng), TFIIE34 (40 ng), TFIIE56 (60 ng), TFIIH (4 ng), and calf thymus RNAPII or the TAP-tagged human RNAPII complex (the amounts of RNAPII used for the various experiments are detailed in the figure legends) were incubated with 500 ng of linearized DNA template containing the adenovirus major late (AdML) promoter from nt −50 to +10 fused to a G-less cassette. Under these conditions, a 391-nt runoff transcript was produced.
Abortive initiation assay.
Abortive initiation assays were performed as described previously (52), with the following modifications. The DNA template (12 ng), either premelted in the −9 to +2 region or fully double-stranded, was incubated with TBP (60 ng), TFIIB (30 ng), RAP30 (30 ng), RAP74 (65 ng), TFIIE34 (40 ng), TFIIE56 (60 ng), and the TAP-tagged human RNAPII complex (the amounts of RNAPII used for the various experiments are detailed in the figure legends) in a 20-μl reaction mixture containing 750 μM ATP, 750 μM CTP, 10 μM UTP, 2.5 μCi of [α-32P]UTP, 12.5 mM MgCl2, 3 mM EGTA, and 0.84 U of RNase inhibitor/ml. The transcripts were analyzed in a 23% polyacrylamide denaturing gel containing 7 M urea.
Elongation assay.
The template was prepared by annealing two complementary oligonucleotides of random sequences, one of which carried an extension of 15 CMP as described previously (66). Typically, the template (9.3 ng) was incubated with 88 ng of TAP-tagged human RNAPII Rpb2 wt or 440 ng of TAP-tagged human RNAPII Rpb2 fork1 (Δ458-459)-TAP, and the reactions were performed as described previously (66). The transcripts were analyzed in 18% polyacrylamide-9% urea denaturing gels.
Nucleotide sequence accession number.
The RPAP1 name and sequence were registered in the HUGO database under accession number NP_056355.
RESULTS
TAP of human RNAPII-containing complex.
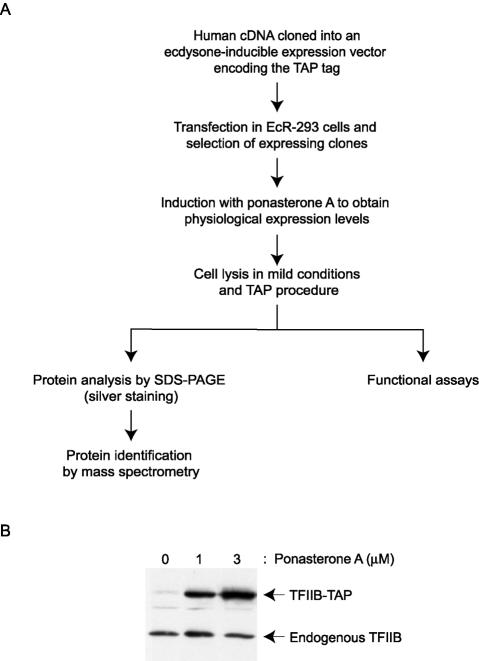
We have adapted the TAP procedure (69, 71), originally developed to isolate protein complexes from yeast, for the purification of human RNAPII and general transcription factors. Native conditions were maintained during the TAP procedure through the use of buffers with a low salt concentration (0.1 M Na+) and a neutral pH. As shown in Fig. 1A, which summarizes the TAP procedure, full-length cDNAs encoding polypeptides of interest were cloned in an ecdysone-inducible mammalian expression vector (61). The vector was engineered to encode polypeptides carrying the following at the C terminus: a TAP tag consisting of two copies of the IgG-binding domain of Staphylococcus aureus protein A, a cleavage site for the tobacco etch virus protease, and a calmodulin-binding peptide (CBP) (71). The tagged polypeptides were expressed in EcR-293 cells at near-physiological levels after a 24-h induction with the ecdysone analog ponasterone A. The concentration of ponasterone A required to obtain near-physiological levels of expression of the tagged polypeptides was determined in a pilot experiment in which we compared the levels of TAP-tagged polypeptides with the endogenous polypeptide by Western blotting (Fig. 1B). Whole-cell extracts were prepared from the ponasterone A-induced cell lines and were used in the double-affinity chromatography procedure, which consisted of successive steps with IgG-Sepharose and calmodulin beads (71). The eluates were frozen and analyzed by different assays.
FIG. 1.
TAP of human transcription factors. (A) Overview of the TAP procedure. (B) Data from a pilot experiment comparing the expression levels of a TAP-tagged polypeptide and its endogenous counterpart upon induction with ponasterone A. TFIIB-TAP expression in EcR-293 cells after induction with different concentrations of ponasterone A (0, 1, and 3 μM) for 24 h was compared to that of endogenous TFIIB by Western blotting with an antibody raised against TFIIB.
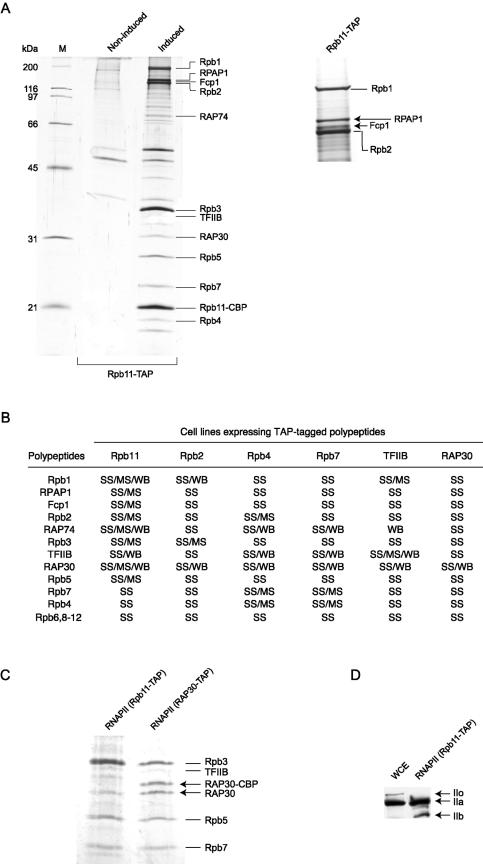
We first expressed the Rpb11 subunit of RNAPII in EcR-293 cells and used the lysate of ponasterone A-induced cells for double-affinity purification. The eluate was subjected to SDS-PAGE and the bands were identified by mass spectrometry. A total of 17 polypeptides that are not present in mock-induced cells and that represent major components of the eluate were identified (Fig. 2A). The other major bands that can be visualized on the gel have all been identified and correspond to proteins that bind nonspecifically to our affinity columns (data not shown). The complex contained the 12 subunits of RNAPII, TFIIB, both subunits of TFIIF (RAP30 and RAP74), Fcp1, the TFIIF-associated CTD phosphatase, and a novel human polypeptide that we named RPAP1.
FIG.2.
Purification of TAP-tagged human RNAPII complex. (A) Whole-cell extracts prepared from induced or noninduced EcR-293 cells programmed to express TAP-tagged Rpb11 (Rpb11-TAP) were purified by the TAP procedure, and the eluates were analyzed by SDS-PAGE. Gel slices containing the most abundant polypeptides were excised, digested with trypsin, and analyzed by peptide mass fingerprinting using MALDI-TOF analysis. The positions of core RNAPII subunits, including Rpb11 carrying the residual CBP domain (Rpb11-CBP), TFIIB, RAP74, RAP30, Fcp1, and RPAP1, a human gene product (DKFZP727M111 protein) of unknown function, are indicated. (B) TAP tagging of Rpb2, Rpb4, Rpb7, Rpb11, TFIIB, and RAP30 allowed for purification of the same 17-polypeptide RNAPII complex. The components of the complex were identified according to their molecular weights in SDS gels stained with silver (SS), their peptide mass fingerprints by MALDI-TOF analysis (MS), and their immunoreactivities in Western blots (WB). (C) SDS gel showing that both tagged (RAP30-CBP) and nontagged RAP30 are present in the RNAPII complex purified with TAP-tagged RAP30. Only nontagged RAP30 is found in the complex purified with TAP-tagged Rpb11. (D) The TAP-tagged RNAPII complex contains a hypophosphorylated CTD. The N-20 antibody, which specifically recognizes the N terminus of Rpb1, was used to detect both the hypophosphorylated (IIa) and hyperphosphorylated (IIo) forms of RNAPII in the whole-cell extract (WCE) and the Rpb11-TAP eluate. A truncated form of Rpb1 lacking the CTD (IIb) was also detected.
To validate the existence of a unique 17-subunit complex containing RNAPII, we tagged other subunits of the complex, including Rpb2, Rpb4, Rpb7, TFIIB, and RAP30, expressed them in stably transfected EcR-293 cells, and used them as baits in the TAP procedure. The eluates were analyzed by silver staining of SDS-PAGE gels, Western blotting, and/or mass spectrometry (Fig. 2B). The results indicated that all 17 polypeptides are bona fide components of the human RNAPII complex. Notably, the eluate prepared with TAP-tagged RAP30 contained both tagged RAP30 with its CBP residual domain and nontagged RAP30 (Fig. 2C). The presence of RAP30 lacking the CBP domain was confirmed by Western blotting (data not shown). These results indicate that two molecules of RAP30 are present within the complex, suggesting that TFIIF is associated with RNAPII as an α2β2 heterotetramer (see Discussion). Finally, our Western blot analysis with an antibody specifically directed against the N-terminal part of Rpb1 (N-20) revealed that the tagged RNAPII complex contains the hypophosphorylated form of RNAPII (IIA form) (Fig. 2D).
RPAP1 is structurally and functionally related to human RNAPII.
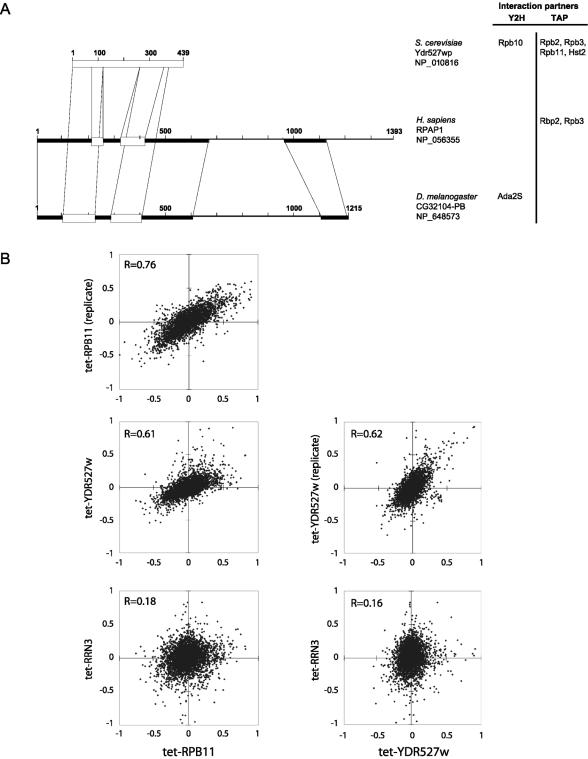
RPAP1 is a previously identified human gene product (DKFZP727M111 protein) of unknown function. BLAST sequence searches against publicly available databases revealed highly significant alignments over the full length of the RPAP1 sequence with proteins of unknown function from Mus musculus (accession no. BAC65787), Rattus norvegicus (accession no. XP_230480), and to a lesser extent, Drosophila melanogaster (accession no. NP_648573 and CG32104-PB). In addition, these proteins show significant similarities to the Saccharomyces cerevisiae Ydr527wp protein (accession no. NP_010816), which is required for cell viability and was shown to bind the Rpb10 subunit of RNAPII in a two-hybrid screen (28, 43). The positions of the different alignments are shown in Fig. 3A. Notably, no particular motif or domain could be predicted with high confidence on the RPAP1 sequence. However, in view of the participation of RPAP1 in the RNAPII complex, an IMPALA analysis against the BLOCK database (74) revealed a very weak similarity (E = 0.23) between the bromodomain-containing protein 4 (Brd4) and the RPAP1 region spanning amino acids 196 to 312 (see below). We also performed a sequence-structure comparison analysis by using the structure-guided alignment program FUGUE against the HOMSTRAD database (56). The result suggests that RPAP1 belongs to the ARM repeat protein superfamily (12). Indeed, the RPAP1 region spanning amino acids 508 to 960 shows an alignment with ARM with a Z score of 4.08 (corresponding to 95% confidence). Interestingly, both Ydr527wp and the RPAP1 D. melanogaster homologue were also predicted, as reported in the Saccharomyces Genome and FlyBase databases, respectively, to belong to the ARM repeat superfamily (http://genome-www4.stanford.edu/cgi-bin/SGD/locus.pl?locus=YDR527W and http://flybase.bio.indiana.edu/.bin/fbidq.html?FBgn0052104).
FIG. 3.
(A) Regions of homology between Homo sapiens RPAP1, D. melanogaster CG32104-PB, and S. cerevisiae Ydr527wp/RBA50, as determined by BLAST analyses. Boxes represent regions with significant alignments. Black boxes, E values ranging from 8 × 10−37 to 1 × 10−10; open boxes, E values ranging from 4 × 10−5 to 0.1. The M. musculus and R. norvegicus orthologues, with 80% identity with the H. sapiens RPAP1 over the full length, were not presented. Reported protein-protein interactions of RPAP1 (this article) and Ydr527wp/RBA50 (33) with transcription factors are indicated on the right. (B) Correlations among mRNA abundance profiles for Tet-promoter mutants of RPB11, YDR527w, and RRN3, measured by using oligonucleotide microarrays. Each mutant was compared to an isogenic control without the tetO7 promoter; log(ratios) are plotted. Tet-RPB11 and Tet-YDR527w were assayed twice, with the replicate cultures being grown, extracted, and assayed on different dates. The correlation of Tet-RPB11 and Tet-YDR527 to a Tet-promoter mutant of RRN3, a gene involved in polymerase I transcription, is shown for contrast.
We created a Tet-promoter allele of the yeast YDR527w gene, turned off gene expression by adding doxycycline to the growth medium for 24 h, and compared gene expression in this mutant to expression patterns from 121 other Tet-promoter mutants affecting genes implicated in diverse biological processes (S. Mnaimneh, A. P. Davierwala, J. Haynes, J. Moffat, W. T. Peng, W. Zhang, X. Yang, J. Pootoolal, G. Chua, A. Lopez, M. Trochesset, D. Morse, N. J. Krogan, S. L. Hiley, Z. Li, Q. Morris, J. Grigull, N. Mitsakakis, C. J. Roberts, J. F. Greenblatt, C. Boone, C. A. Kaiser, B. J. Andrews, and T. R. Hughes, submitted for publication). Among all of these, the effect of the tet-YDR527w allele correlated most strongly with that of tet-RPB11 (Fig. 3B). A clustering analysis did not reveal any specific group of functionally related transcripts that were up- or down-regulated uniquely in these two mutants, supporting the idea that the expression pattern observed is a consequence of an overall perturbation in transcription rather than a disruption of any individual pathway.
To further define the interaction between RPAP1 and RNAPII, we cloned the human RPAP1 cDNA and derived a human cell line that can be induced to express TAP-tagged RPAP1. An analysis of the TAP-tagged RPAP1 eluate revealed the presence of two RNAPII subunits, Rpb2 and Rpb3 (Fig. 3A, right panel). These results further demonstrate the physical interaction between RPAP1 and RNAPII. Recent TAP-tagging experiments in yeast indicated that Ydr527wp associates with Rpb2, Rbp3, and Rpb11 (33). Because Ydr527wp and RPAP1 are both physically and functionally related, the 50-kDa yeast protein encoded by the YDR527W gene was named RBA50 (the name was registered in the Saccharomyces Genome Database).
The TAP-tagged RNAPII complex can be recruited to promoters both in vitro and in vivo and can bind to acetylated histone H3.
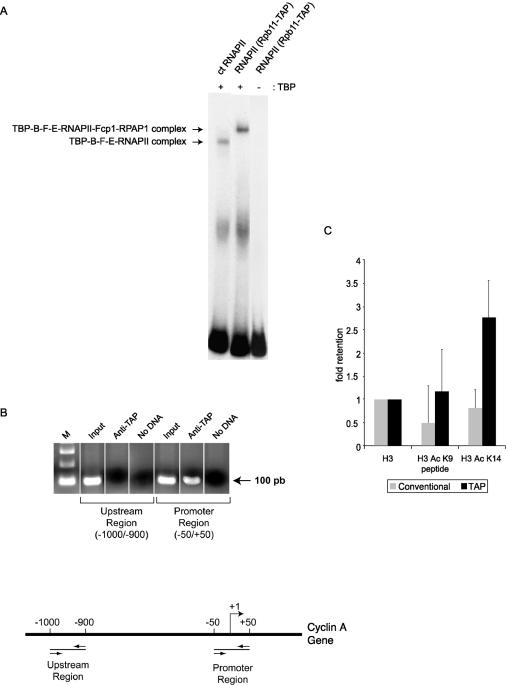
In order to determine whether the TAP-tagged RNAPII complex is functional, we first verified its ability to enter a preinitiation complex in vitro. The electrophoretic mobility shift assay (EMSA) results in Fig. 4A show that the human RNAPII complex can form a preinitiation complex in the presence of TBP, TFIIB, TFIIF, and TFIIE on a radiolabeled probe carrying the AdML promoter. Compared to an equivalent preinitiation complex assembled with highly purified calf thymus RNAPII, the TAP-tagged RNAPII complex had a slightly lower mobility. Because our calf thymus RNAPII contains neither Fcp1 nor RPAP1 (data not shown), our results indicate that the preinitiation complex assembled with the TAP-tagged human RNAPII complex includes at least one of these two polypeptides (Fcp1 and/or RPAP1).
FIG. 4.
TAP-tagged human RNAPII assembles on promoter DNA both in vitro and in vivo and binds to acetylated histones. (A) EMSA performed with 150 ng of purified calf thymus (ct) RNAPII or TAP-tagged human RNAPII in the presence of TBP, TFIIB, TFIIF, and TFIIE. A control reaction assembled in the absence of TBP was included. A radiolabeled DNA fragment containing the AdML promoter was used as a probe. (B) ChIP experiments showing that TAP-tagged RNAPII is specifically recruited to the cyclin A promoter in vivo. PCR amplification using sets of primers specific to chromosomal regions either encompassing (−50 to +50) or located upstream of (−1000 to −900) the transcription start site was used on DNA fragments enriched with IgG beads (anti-TAP). The input lanes correspond to DNA that was not subjected to immunoprecipitation. A PCR control that lacks DNA (no DNA) was included in each case. (C) Peptide binding experiment showing that TAP-tagged RNAPII is retained on acetylated histones. The results of three independent experiments were analyzed with ImageQuant software, corrected for the background, and shown relative to unmodified H3. Conventionally purified RNAPII was used as a negative control. The TAP-tagged RNAPII-bound histone tails acetylated at lysine 14 more efficiently than at lysine 9.
ChIP experiments were used to determine whether the tagged RNAPII complex can also be recruited to promoters in vivo. As shown in Fig. 4B, DNA fragments containing the CCNA1 (cyclin A) promoter (positions −50 to +50) were enriched more efficiently than DNA fragments containing an upstream region (positions −1000 to −900) of the same gene when IgG-Sepharose beads were used to bind the protein A component of the TAP tag. Our results demonstrate that TAP-tagged RNAPII is specifically recruited to the promoter of actively transcribed genes in vivo.
As mentioned above, BLAST searches against publicly available databases revealed weak similarities between the amino acid sequence of RPAP1 and a variety of proteins and domains, including Brd4. Because Brd4 was recently shown to specifically interact with histones H3 and H4 through acetylated lysine residues (19), we tested the ability of the complex to bind to acetylated histones. As shown in Fig. 4C, the TAP-tagged RNAPII complex specifically bound to acetylated histone H3 in vitro, while conventionally purified RNAPII was not retained on acetylated histone H3. TAP-tagged RNAPII bound histone tails acetylated at lysine 14 more efficiently than those acetylated at lysine 9. This result suggests that RPAP1 may recruit RNAPII to chromatin through its interaction with acetylated histones.
The TAP-tagged RNAPII complex supports accurately initiated transcription in vitro.
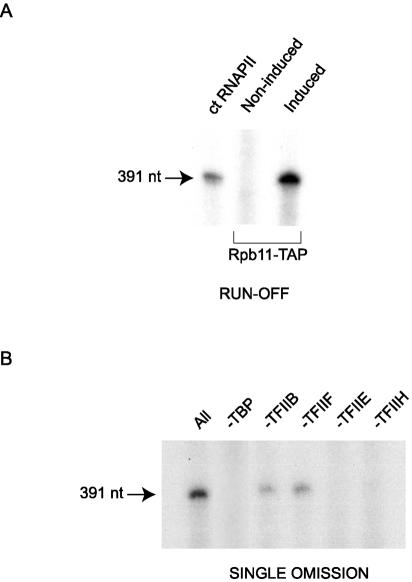
TAP-tagged RNAPII was tested for the ability to initiate transcription in vitro by use of a transcription system reconstituted with purified general initiation factors. Purified calf thymus RNAPII or purified TAP-tagged human RNAPII was incubated with a DNA template containing the AdML promoter in the presence of purified TBP, TFIIB, TFIIF, TFIIE, and TFIIH. The formation of a 391-nt transcript indicated that the tagged RNAPII complex is competent for promoter-selective transcription in vitro (Fig. 5A).
FIG. 5.
TAP-tagged human RNAPII complex can initiate transcription in vitro. (A) In vitro transcription reactions were reconstituted by using 165 ng of purified calf thymus (ct) RNAPII or TAP-tagged human RNAPII in the presence of TBP, TFIIB, TFIIF, TFIIE, and TFIIH. A control reaction performed with an eluate from noninduced cells was included. The linearized DNA template carries the AdML promoter and directs the synthesis of a 391-nt transcript. (B) Single omission assay in which each general transcription factor was omitted from the reconstituted system (all) described for panel A.
When the general initiation factors were individually omitted from the transcription reaction (Fig. 5B), transcription was abolished except in the case of recombinant TFIIB or TFIIF. These data fully support our conclusion that TFIIB and TFIIF are components of the tagged RNAPII complex and establish that they are functionally active within this complex.
Purification and functional analysis of a human RNAPII variant with a site-directed mutation in fork loop 1.
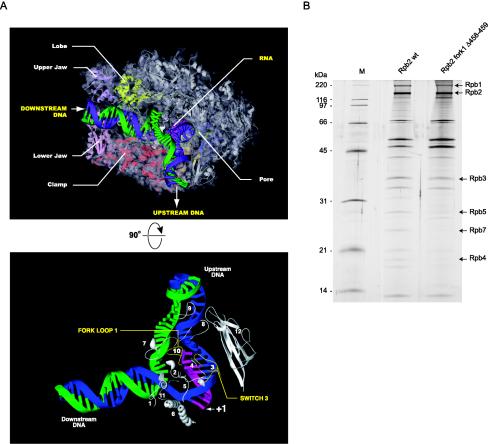
As part of an effort to understand the fine molecular details of the transcription reaction, we used the TAP procedure to generate and isolate RNAPII variants with site-directed mutations in structural elements predicted to have roles in the transcription reaction according to the crystal structure of the yeast enzyme. Because the Rpb2 element known as fork loop 1 (Fig. 6A) is highly conserved from yeast to humans, absent from bacterial RNAP, and proposed to be involved in maintaining the transcription bubble (17, 30) or in RNA-DNA strand separation (85), we first chose to modify this element. In yeast RNAPII, fork loop 1 amino acids Lys471 and Arg476 appear to contact the RNA phosphates at positions −5, −6, and −7 in the hybrid region. Deletion of the whole 13-amino-acid loop totally abolished the ability of Rpb2 to assemble with the other subunits of RNAPII (data not shown), probably because the deletion destabilized Rpb2, which then became misfolded. A smaller deletion of two amino acids located near the center of the loop (Δ458-459), which corresponded to amino acids Lys471 and Ala472 in the yeast enzyme, was successfully used to purify the 17-subunit RNAPII complex (Fig. 6B). This mutant was named Rpb2 fork1 Δ458-459. The silver-stained SDS gel in Fig. 6B shows that the relative abundance of some components of the tagged RNAPII complex varied between the wild type and the mutant, indicating that the mutation may have slightly affected complex assembly. By quantifying the relative intensity of each subunit, we established that the most important difference concerned Rpb1, which was at the very most fivefold less abundant in the mutant than in the wild-type complex.
FIG. 6.
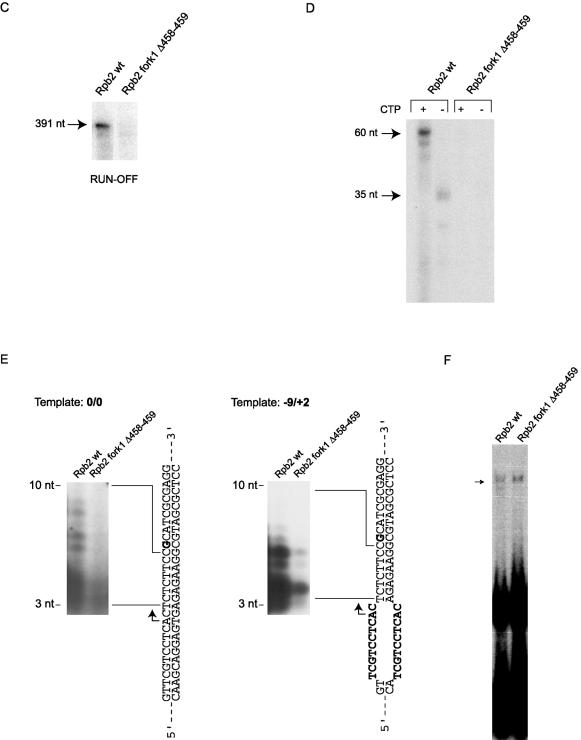
Purification and functional analysis of TAP-tagged human RNAPII with a mutation in fork loop 1. (A) (Top) Model showing elongating yeast RNAPII (30). The template DNA strand (blue) from −3 to +10 and the 9-nt RNA (pink) were placed according to the crystal structure. The remainder of the template strand and the coding strand (green) were modeled in Cinema 4D, and their exact positions are speculative. (Bottom) Simplified view of yeast RNAPII catalytic center. Different domains of Rpb1 and Rpb2 located near the DNA-RNA hybrid are shown. Numbers: 1 to 5, switches; 6, bridge helix; 7, rudder; 8, lid; 9, zipper; 10, fork loop 1; 11, fork loop 2; 12, wall. The parts of the lid (amino acids 250 to 258) and the rudder (amino acids 315 to 320) that are absent in the PDB files corresponding to elongating RNAPII were reconstructed based on the crystal structure of free yeast RNAPII (18). The missing parts of fork loop 1 (amino acids 468 to 476) and fork loop 2 (amino acids 503 to 508) were modeled in Cinema 4D, and their exact positions are speculative. (B) Silver-stained SDS gel of wild type TAP-tagged RNAPII (Rpb2 wt) and a mutant of human RNAPII carrying a two-amino-acid deletion in the Rpb2 fork loop 1 (Rpb2 fork1 Δ458-459) domain. The positions of some RNAPII subunits and molecular size markers are indicated. (C) In vitro transcription reactions (runoff) contained either 24 ng of wild-type RNAPII (Rpb2 wt) or 120 ng of Rpb2 fork1 Δ458-459 in the presence of TBP, TFIIB, TFIIF, TFIIE, and TFIIH. (D) Elongation assays were performed with a C-tailed template carrying a 15-C extension in the absence of general transcription factors. The coding strand lacks CMP except in the +25-to-+27 region. The positions of transcripts produced by RNAPII in the presence (+CTP; 60 nt) or absence (−CTP; 35 nt) of CTP are indicated. The reactions contained 88 ng of Rpb2 wt and 440 ng of Rpb2 fork1 Δ458-459. (E) Abortive initiation assays were performed with 100 ng of Rpb2 wt and 500 ng of Rpb2 fork1 Δ458-459 in the presence of TBP, TFIIB, TFIIF, and TFIIE on closed (0/0) or premelted (−9/+2) templates in the absence of GTP. The templates are schematized and the 3- to 10-nt abortive transcripts are indicated. (F) EMSAs were performed with the AdML promoter in the presence of TBP, TFIIB, TFIIF, and TFIIE. Two hundred nanograms of Rpb2 wt and 500 ng of Rpb2 fork 1 Δ58-459 were used in the reactions.
In order to elucidate the function of fork loop 1 in transcription, we compared Rpb2 fork1 Δ458-459 with wild-type RNAPII in several different assays. The assays shown in Fig. 6C to E used fivefold larger amounts of the mutant complex than of the wild type to ensure that the amount of the mutant complex was not underestimated (see above). As shown in Fig. 6C, Rpb2 fork1 Δ458-459 was defective in runoff transcription assays when we used the AdML promoter in the presence of all of the general initiation factors.
In order to delineate with more precision the molecular defect that impairs the transcriptional activity of Rpb2 fork1 Δ458-459 in vitro, we tested its activity in a promoter-independent assay. As described previously (29, 79), RNAPII can transcribe from a template carrying a 3′ C extension (C-tailed template) in the absence of the general initiation factors. The template used for Fig. 6D carried a 15-C extension on the 3′ strand. The transcribed strand was devoid of dCMP except at positions +25 to +27 (+1 corresponds to the single-stranded-double-stranded junction). In the absence of CTP, wild-type RNAPII produced a transcript of 35 nt. In the presence of CTP, the major transcript was 60 nt in length, indicating that the polymerase starts transcription 10 nt upstream of the double-stranded junction. In contrast to the RNAPII complex carrying wild-type Rpb2, Rpb2 fork1 Δ458-459 was inactive in the C-tailed template elongation assay.
Figure 6E shows that Rpb2 fork1 Δ458-459 was also defective in an abortive initiation assay carried out with a short double-stranded oligonucleotide carrying the AdML promoter in the presence of the general initiation factors, but in the absence of GTP. A G nucleotide strategically located at +11 in the template blocked transcription at position +10 (Fig. 6E). When we used a fully double-stranded template (0/0), Rpb2 fork1 Δ458-459 did not produce abortive transcripts. We rationalized that if Rpb2 fork1 Δ458-459 was defective in promoter melting, a premelted template would counteract the defect in the mutant. The premelted template (−9/+2), which carried an unpaired DNA region from −9 to +2 that mimicked the fully melted promoter, supported the synthesis of abortive transcripts, although with a poor efficiency compared to that of the wild type. This finding indicated that a mutation in fork loop 1 does not simply affect promoter melting and its maintenance, but rather affects the interaction of RNAPII with the melted promoter region.
We finally assessed the ability of Rpb2 fork1 Δ458-459 to assemble into a preinitiation complex on promoter-containing DNA. EMSA experiments showed that Rpb2 fork1 Δ458-459 can enter a preinitiation complex with the general initiation factors as efficiently as wild-type RNAPII (Fig. 6F).
Human RNAPII variant with a site-directed mutation in switch 3.
The structure named switch 3 is the only loop of Rpb2 located near the DNA-RNA hybrid that is ordered in the crystal structure of elongating yeast RNAPII (30). Switch 3 makes direct contact with the template DNA near positions −5 (arginine 1122) and −2 (arginine 1129) (Fig. 6A). Since R1129 is juxtaposed with one of the Rpb2 domains that mediate the interaction with Rpb1, we reasoned that disruption of this amino acid would likely affect the assembly of RNAPII and decided to turn our attention to R1122. We produced a mutant in which amino acids R1122, S1123, and R1124 were replaced with alanine residues. In human Rpb2, these amino acids correspond to R1078, S1079, and R1080. This mutant was named Rpb2 sw3-1078.
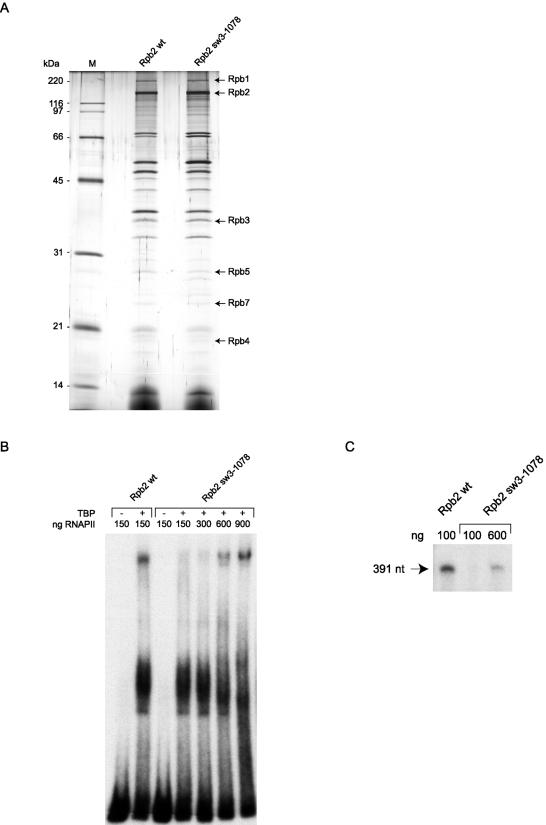
Figure 7A shows that TAP-tagged Rpb2 with the sw3-1078 mutation is as efficient as the wild type at forming the 17-subunit RNAPII complex. However, EMSA experiments (Fig. 7B) indicated that purified Rpb2 sw3-1078 is not efficient at entering a preinitiation complex on promoter DNA in the presence of the general initiation factors. Increasing the concentration of Rpb2 sw3-1078 by a factor of 6 compared to the wild type was necessary in order to achieve efficient formation of a preinitiation complex. As expected on the basis of the EMSA results, Rpb2 sw3-1078 did not support transcription in vitro (Fig. 7C). Increasing the concentration of Rpb2 sw3-1078 by a factor of 6 compared to the wild type led to the production of runoff transcripts, but the transcription efficiency remained slightly lower than that of the wild type, indicating that the mutation in switch 3 not only affects the ability of Rpb2 sw3-1078 to assemble into a preinitiation complex, but also, to a lesser extent, affects its capacity to support transcription.
FIG. 7.
Purification and functional analysis of TAP-tagged human RNAPII with a mutation in switch 3. (A) Silver-stained SDS gel of wild-type TAP-tagged RNAPII (Rpb2 wt) and a mutant of human RNAPII carrying a triple alanine substitution in the Rpb2 switch 3 (Rpb2 sw3-1078) domain. (B) EMSAs were performed with the AdML promoter with TFIIB, TFIIF, and TFIIE in either the presence or the absence of TBP. The amounts used for the wild type and the mutant are indicated. (C) In vitro transcription reactions (runoff) contained different amounts of Rpb2 wt and Rpb2 sw3-1078 in the presence of TBP, TFIIB, TFIIF, TFIIE, and TFIIH.
Mutants Rpb2 fork1 Δ458-459 and Rpb2 sw3-1078 are defective in vivo.
To assess the defects of Rpb2 fork1 Δ458-459 and Rpb2 sw3-1078 in vivo, we used ChIP experiments to evaluate their association with both the promoter and the transcription unit of two genes that are actively transcribed in human cells, the guanine nucleotide binding protein beta polypeptide 2-like 1 (GNB2L1) gene and the ferritin light polypeptide (FTL) gene. As shown in Fig. 8, Rpb2 fork1 Δ458-459 is recruited to both promoters with an efficiency similar to that of a wild-type TAP-tagged RNAPII complex, but it is not found in association with the transcription units. In contrast, Rpb2 sw3-1078 is not enriched in the promoter regions or the transcribed regions. The mutant Rpb2 fork1 Δ453-465, which carries a complete deletion of the 13-amino-acid loop and does not assemble into a bona fide 17-subunit RNAPII complex, was used as a negative control that was enriched at neither the promoter nor the transcription unit of either the GNB2L1 or FTL gene. These in vivo results fully support our in vitro studies indicating that fork loop 1 is essential for transcription but is not involved in preinitiation complex assembly, whereas switch 3 is required for efficient preinitiation complex formation.
DISCUSSION
Simple, efficient purification of human RNAPII under native conditions.
Classical approaches for the purification of mammalian RNAPII and its general transcription factors have typically used many different chromatography steps, including ion-exchange chromatography. Eluting these columns with buffers containing high salt concentrations most certainly affected the polypeptide composition of the purified complexes. For example, regulatory subunits, which are often less tightly associated with the enzyme they regulate, may have been lost during the purification procedure. Seraphin and colleagues (71) developed the TAP procedure to allow for the purification of protein complexes under native conditions. Two affinity chromatography steps are used. Elution from an IgG column is performed by a proteolytic treatment with the tobacco etch virus protease while elution from a calmodulin column uses EGTA as a chelating agent. For this paper, we adapted the TAP procedure for the purification of protein complexes from human cells, using an ecdysone-inducible expression system that allows for the expression of the TAP-tagged polypeptides at near-physiological levels. The expression levels were modulated by adjusting the concentration of the inducer, ponasterone A (Fig. 1B). By preventing overexpression of the tagged polypeptides, we limited the formation of nonspecific interactions.
We report here the use of the TAP procedure with human cells for the easy and efficient purification of a complex containing the 12 subunits of RNAPII, both subunits of TFIIF, TFIIB, Fcp1, and RPAP1, a novel polypeptide of unknown function. This 17-subunit complex was purified through the alternate TAP tagging of six of its subunits, demonstrating that all of the components are part of a unique complex. Because (i) the TAP-tagged polypeptides are expressed at near-physiological levels in stably transfected human cells, (ii) protein extraction is performed under gentle conditions, and (iii) the affinity purification steps are achieved under native conditions, our results indicate that this TAP-tagged RNAPII complex is the most abundant soluble, stable form of RNAPII in human cells. If a mediator complex (70) is associated with RNAPII in human cell extracts, either its concentration is much lower than that of RNAPII or its association with RNAPII is too weak to survive double affinity purification.
The presence of the TAP tag at the C termini of subunits of the RNAPII-containing complex does not alter its activity. The TAP-tagged RNAPII complex is fully active in vitro, as it can form a preinitiation complex with the general initiation factors on the AdML promoter, accurately initiate transcription, and elongate the transcripts (Fig. 4 and 5). In addition, both the TFIIB and TFIIF components of the complex are functionally active in transcription reactions. Notably, TAP-tagged RNAPII can be recruited to transcriptionally active promoters in vivo, an indication that the purified human RNAPII complex is biologically active.
Five different regulatory subunits are stably associated with soluble RNAPII in human cells.
The soluble form of RNAPII is expected to be the form of the enzyme that is recruited to promoters upon activation signals. In support of this hypothesis, the CTD of Rpb1 in the TAP-tagged RNAPII complex is in the hypophosphorylated state, the form of the enzyme that is recruited to promoter DNA. A possible function of Fcp1 in this complex could be to maintain the CTD in this unphosphorylated state, as previously proposed (47). Our EMSAs suggested that Fcp1 can enter the preinitiation complex with the RNAPII complex. The entry of Fcp1 with RNAPII at the promoter is supported by the finding by Buratowski and colleagues that the CTD phosphatase colocalizes with RNAPII at active promoters in yeast (10). Therefore, the role of Fcp1 in maintaining RNAPII in the hypophosphorylated state may be required until RNAPII becomes processive and clears the promoter.
Both TFIIB and TFIIF have previously been shown to regulate the recruitment of RNAPII to promoter DNA (15, 54, 80). TFIIB binds to TBP and forms a TBP-TFIIB-promoter complex (4, 59). TFIIB binds directly to a DNA motif named the TFIIB recognition element, a core element found in several RNAPII promoters (51). TFIIF binds to both RNAPII and TFIIB and is also involved in RNAPII recruitment to core promoters (15, 54, 80). In addition, TFIIF was shown to regulate the contacts of RNAPII with promoter DNA before transcription initiation (20, 25, 72). TFIIF plays a central role in promoter bending and wrapping against the mobile clamp of RNAPII, a process that is essential for transcription initiation (24). The function of TFIIF in promoter wrapping and transcriptional initiation requires a homomeric interaction region of RAP74, called HIR-1, that maintains TFIIF as a α2β2 heterotetramer (72). Our results showing that two molecules of RAP30 are present within the tagged RNAPII complex support the notion that TFIIF works as a α2β2 heterotetramer in vivo (13, 23).
Notably, both TFIIF and TFIIB can also bind to Fcp1 in a competitive manner and with opposite effects, with TFIIF stimulating and TFIIB inhibiting the phosphatase activity (9). Fcp1 may not efficiently dephosphorylate RNAPII at Ser2 of the CTD in the region downstream of the promoter (10) because TFIIF appears not to be present in this complex (50, 68). Clearly, TFIIB, TFIIF, and Fcp1 participate in an important network of regulatory interactions during RNAPII transcription.
The TAP-tagged RNAPII complex also contains at least one polypeptide that can specifically bind to acetylated histones in vitro. One putative candidate is RPAP1. Whether or not RPAP1 contains a bromodomain remains unclear because the definitive identification of a bromodomain requires structural data. An essential yeast polypeptide with regions of homology with RPAP1 was previously shown to interact with the Rpb10 subunit of RNAPII (28, 43) and also affected global gene expression in the same way as Rpb11 in our Tet-promoter mutant experiments (Fig. 3). These findings support the notion that the association of RPAP1 with RNAPII is physiologically relevant, although its exact role remains to be elucidated.
Structure-guided alignments indicated that RPAP1 could belong to the ARM repeat protein superfamily (12). ARM repeat proteins contain tandem copies of a degenerate 40-amino-acid sequence motif that forms a conserved three-dimensional structure. A single ARM repeat consists of three α helices. Such repeats are found in a variety of proteins, including the mammalian armadillo homologs β-catenin and importin α (40). β-catenin is mainly detected in adherens junctions where it links the membrane-anchored cadherin to actin filament. Cytoplasmic β-catenin is degraded by the ubiquitin-proteasome system. However, when Wnt signaling is activated, cytoplasmic β-catenin is stabilized. The increased level of β-catenin leads to its nuclear translocation, where it affects transcriptional activation. Although it was reported to be a cytoplasmic protein (41), Ydr527wp/RBA50 is found in complex with three RNAPII subunits and appears to affect transcription. Similar to β-catenin, Ydr527wp/RBA50 may have a dual cellular location and may be translocated to the nucleus.
Proteins that were previously shown to interact with RNAPII, such as the mediator and various elongation factors, were not found to be major components of our TAP-tagged RNAPII eluate. This is likely because these RNAPII-interacting proteins are associated with RNAPII in the insoluble fraction and, consequently, are mainly lost during the purification procedure. However, these complexes may represent minor forms of RNAPII complexes in our TAP-tagged eluates.
Structure-function analysis of the human RNAPII active center.
The crystal structure of eukaryotic RNAPII was used to predict the function of many structural elements found in the active center. For example, some loops and helices that either directly contact or closely approach the DNA-RNA duplex in the active site of the elongating enzyme were suggested to regulate the formation and maintenance of both the transcription bubble and the RNA-DNA hybrid. In order to begin assessing the functions of these structural elements, we expressed TAP-tagged Rpb2 mutants carrying site-directed mutations and purified the mutant forms of RNAPII. Our data indicate that the assembly and/or stability of the RNAPII complex is affected by mutations in the active site. For example, the full deletion of fork loop 1 led to defects in the assembly of the 17-subunit RNAPII complex. However, a shorter mutant with a deletion of only two amino acids from Rpb2 was found to assemble correctly in solution in vivo (Fig. 6) and allowed us to analyze the function of this element during the transcription reaction. The Rpb2 fork1 Δ458-459 mutant was found to enter a preinitiation complex with the general initiation factors with an efficiency similar to that of the wild-type complex, but it was incapable of transcription initiation and elongation in vitro. When we used a premelted template in which the transcription bubble was fully open between positions −9 and +2 in our initiation assay, Rpb2 fork1 Δ458-459 was able to initiate transcript formation, but not very efficiently. These findings indicate that there may be a defect at the level of the interaction between fork loop 1 and the melted DNA at very early stages of the transcription reaction. For example, the two-amino-acid-shorter fork loop 1 in the RNAPII mutant may be unable to contact one of the DNA strands in the transcription bubble, leading to incorrect positioning of the DNA template in the active site and impairing the ability of the enzyme to initiate transcription efficiently. Since this mutant supported transcription initiation much less efficiently even when a fully open template was used, our results do not support a direct role for fork loop 1 in promoter melting.
Given the recent crystal structure of elongating yeast RNAPII showing an interaction between Lys 471 (Lys 458 in human RNAPII) and the RNA around positions −5 to −7 (85), an alternative possibility to explain the defect of Rpb2 fork 1 Δ458-459 is that during transcription initiation, the mobile fork loop 1 makes crucial contacts with the first arriving ribonucleotides, making it essential for phosphodiester bond formation. According to this speculative view, fork loop 1 would keep contact with the RNA as transcription progresses and be displaced until it reaches the position seen in the crystal structure, where it would participate in RNA-DNA strand separation.
The five switches located at the bottom of the cleft of RNAPII were first proposed to be involved in coupling the closure of the mobile clamp to the presence of a DNA-RNA hybrid (18, 30). This hypothesis was based on the fact that most of the switches are disordered in the crystal structure of the 10-subunit RNAPII complex in solution (e.g., lacking the Rpb4-Rpb7 dimer), whereas they are ordered in the elongating RNAPII structure. The ordering of the switches coincided with the movement of the clamp, which was in an open state in the structure of the free enzyme to permit the entry of DNA into the cleft and in a closed state in the structure of the RNAPII-DNA-RNA ternary complex to ensure the stability of the elongating complex. Since some switches, including switch 3, directly contact the DNA in the DNA-RNA hybrid region, they were proposed to be involved in sensing the presence of nascent RNA and coupling it to the closure of the clamp on the DNA. However, the recent crystal structures of the 12-subunit RNAPII complex (2, 7) revealed that the clamp is in a closed state in the presence of the Rpb4-Rpb7 dimer. These results led the authors to propose that the DNA is first loaded onto the clamp far from the active site in a way that is similar to that of the bacterial RNA polymerase holoenzyme-promoter DNA complex (57) and that it reaches the cleft only after DNA melting has occurred. In this situation, switch 3 could only contact DNA after the opening of the promoter and entry of the DNA into the cleft.
Our results indicate that switch 3 is important for the assembly of RNAPII into a preinitiation complex on promoter DNA. Mutating amino acids 1078 to 1080 clearly reduced the ability of human RNAPII to form a preinitiation complex in the presence of the general transcription factors both in vitro and in vivo. In vitro, sixfold larger amounts of Rpb2 sw3-1078 were necessary to form a complex in gel shift experiments. Our results suggest that switch 3 is necessary for the accurate interaction of the clamp with the promoter DNA before opening of the transcription bubble and that this is the primary defect in our switch 3 mutant. Because the structure of the complete polymerase shows that a duplex DNA cannot bind switch 3 before melting, it is likely that the effect of the switch 3 mutation is either allosteric or mediated by interactions with a general initiation factor such as TFIIB or TFIIF. The importance of the interaction of switch 3 with the template DNA at positions −2 and −5 after promoter melting might be reflected by the fact that the transcription reactions carried out in the presence of sixfold larger amounts of the sw3-1078 mutant did not yield as many transcripts as did the wild type. This may also suggest that the preinitiation complexes formed with the sw3-1078 mutant are not fully functional, presumably because the interactions of switch 3 with the DNA template in the DNA-RNA hybrid region are impaired.
Acknowledgments
We are grateful to the members of our laboratory for helpful discussions. We thank Diane Bourque for artwork and Julie Edwards for critical reading of the manuscript. We also thank Takahiro Nagase from the Kazusa DNA Research Institute for kindly providing the cDNA encoding RPAP1.
This work was supported by grants from the Canadian Institutes for Health Research (to B.C.), Genome Canada (to B.C., J.G., and T.R.H.), Genome Québec (to B.C.), the Ontario Genomics Institute (to J.G. and T.R.H.), and the Natural Sciences and Engineering Research Council of Canada (to T.R.H.). C.J. holds a studentship from the Canadian Institutes for Health Research and M.F.L. is supported by the Natural Sciences and Engineering Research Council of Canada and the Fonds Québécois de la Recherche sur la Nature et les Technologies. A.P.D. is supported by a C. H. Best Postdoctoral Fellowship. B.C. is a senior scholar from the Fonds de la Recherche en Santé du Québec.
REFERENCES
- 1.Acker, J., M. Wintzerith, M. Vigneron, and C. Kedinger. 1992. Primary structure of the second largest subunit of human RNA polymerase II (or B). J. Mol. Biol. 226:1295-1299. [DOI] [PubMed] [Google Scholar]
- 2.Armache, K. J., H. Kettenberger, and P. Cramer. 2003. Architecture of initiation-competent 12-subunit RNA polymerase II. Proc. Natl. Acad. Sci. USA 100:6964-6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Awrey, D. E., R. G. Weilbaecher, S. A. Hemming, S. M. Orlicky, C. M. Kane, and A. M. Edwards. 1997. Transcription elongation through DNA arrest sites. A multistep process involving both RNA polymerase II subunit RPB9 and TFIIS. J. Biol. Chem. 272:14747-14754. [DOI] [PubMed] [Google Scholar]
- 4.Bagby, S., S. Kim, E. Maldonado, K. I. Tong, D. Reinberg, and M. Ikura. 1995. Solution structure of the C-terminal core domain of human TFIIB: similarity to cyclin A and interaction with TATA-binding protein. Cell 82:857-867. [DOI] [PubMed] [Google Scholar]
- 5.Bryant, G. O., and M. Ptashne. 2003. Independent recruitment in vivo by Gal4 of two complexes required for transcription. Mol. Cell 11:1301-1309. [DOI] [PubMed] [Google Scholar]
- 6.Burton, Z. F., L. G. Ortolan, and J. Greenblatt. 1986. Proteins that bind to RNA polymerase II are required for accurate initiation of transcription at the adenovirus 2 major late promoter. EMBO J. 5:2923-2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bushnell, D. A., and R. D. Kornberg. 2003. Complete, 12-subunit RNA polymerase II at 4.1-A resolution: implications for the initiation of transcription. Proc. Natl. Acad. Sci. USA 100:6969-6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell, E. A., O. Muzzin, M. Chlenov, J. L. Sun, C. A. Olson, O. Weinman, M. L. Trester-Zedlitz, and S. A. Darst. 2002. Structure of the bacterial RNA polymerase promoter specificity sigma subunit. Mol. Cell 9:527-539. [DOI] [PubMed] [Google Scholar]
- 9.Chambers, R. S., B. Q. Wang, Z. F. Burton, and M. E. Dahmus. 1995. The activity of COOH-terminal domain phosphatase is regulated by a docking site on RNA polymerase II and by the general transcription factors IIF and IIB. J. Biol. Chem. 270:14962-14969. [DOI] [PubMed] [Google Scholar]
- 10.Cho, E. J., M. S. Kobor, M. Kim, J. Greenblatt, and S. Buratowski. 2001. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev. 15:3319-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choder, M., and R. A. Young. 1993. A portion of RNA polymerase II molecules has a component essential for stress responses and stress survival. Mol. Cell. Biol. 13:6984-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coates, J. C. 2003. Armadillo repeat proteins: beyond the animal kingdom. Trends Cell Biol. 13:463-471. [DOI] [PubMed] [Google Scholar]
- 13.Conaway, J. W., and R. C. Conaway. 1989. A multisubunit transcription factor essential for accurate initiation by RNA polymerase II. J. Biol. Chem. 264:2357-2362. [PubMed] [Google Scholar]
- 14.Conaway, J. W., A. Shilatifard, A. Dvir, and R. C. Conaway. 2000. Control of elongation by RNA polymerase II. Trends Biochem. Sci. 25:375-380. [DOI] [PubMed] [Google Scholar]
- 15.Conaway, R. C., K. P. Garrett, J. P. Hanley, and J. W. Conaway. 1991. Mechanism of promoter selection by RNA polymerase II: mammalian transcription factors alpha and beta gamma promote entry of polymerase into the preinitiation complex. Proc. Natl. Acad. Sci. USA 88:6205-6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coulombe, B., and Z. F. Burton. 1999. DNA bending and wrapping around RNA polymerase: a “revolutionary” model describing transcriptional mechanisms. Microbiol. Mol. Biol. Rev. 63:457-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cramer, P., D. A. Bushnell, J. Fu, A. L. Gnatt, B. Maier-Davis, N. E. Thompson, R. R. Burgess, A. M. Edwards, P. R. David, and R. D. Kornberg. 2000. Architecture of RNA polymerase II and implications for the transcription mechanism. Science 288:640-649. [DOI] [PubMed] [Google Scholar]
- 18.Cramer, P., D. A. Bushnell, and R. D. Kornberg. 2001. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science 292:1863-1876. [DOI] [PubMed] [Google Scholar]
- 19.Dey, A., F. Chitsaz, A. Abbasi, T. Misteli, and K. Ozato. 2003. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc. Natl. Acad. Sci. USA 100:8758-8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Douziech, M., F. Coin, J. M. Chipoulet, Y. Arai, Y. Ohkuma, J. M. Egly, and B. Coulombe. 2000. Mechanism of promoter melting by the xeroderma pigmentosum complementation group B helicase of transcription factor IIH revealed by protein-DNA photo-cross-linking. Mol. Cell. Biol. 20:8168-8177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feaver, W. J., O. Gileadi, Y. Li, and R. D. Kornberg. 1991. CTD kinase associated with yeast RNA polymerase II initiation factor b. Cell 67:1223-1230. [DOI] [PubMed] [Google Scholar]
- 22.Finkelstein, A., C. F. Kostrub, J. Li, D. P. Chavez, B. Q. Wang, S. M. Fang, J. Greenblatt, and Z. F. Burton. 1992. A cDNA encoding RAP74, a general initiation factor for transcription by RNA polymerase II. Nature 355:464-467. [DOI] [PubMed] [Google Scholar]
- 23.Flores, O., I. Ha, and D. Reinberg. 1990. Factors involved in specific transcription by mammalian RNA polymerase II. Purification and subunit composition of transcription factor IIF. J. Biol. Chem. 265:5629-5634. [PubMed] [Google Scholar]
- 24.Forget, D., M. F. Langelier, C. Therien, V. Trinh, and B. Coulombe. 2004. Photo-cross-linking of a purified preinitiation complex reveals central roles for the RNA polymerase II mobile clamp and TFIIE in initiation mechanisms. Mol. Cell. Biol. 24:1122-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forget, D., F. Robert, G. Grondin, Z. F. Burton, J. Greenblatt, and B. Coulombe. 1997. RAP74 induces promoter contacts by RNA polymerase II upstream and downstream of a DNA bend centered on the TATA box. Proc. Natl. Acad. Sci. USA 94:7150-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu, J., A. L. Gnatt, D. A. Bushnell, G. J. Jensen, N. E. Thompson, R. R. Burgess, P. R. David, and R. D. Kornberg. 1999. Yeast RNA polymerase II at 5 A resolution. Cell 98:799-810. [DOI] [PubMed] [Google Scholar]
- 27.Gerard, M., L. Fischer, V. Moncollin, J. M. Chipoulet, P. Chambon, and J. M. Egly. 1991. Purification and interaction properties of the human RNA polymerase B(II) general transcription factor BTF2. J. Biol. Chem. 266:20940-20945. [PubMed] [Google Scholar]
- 28.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. P. Arkin, A. Astromoff, M. El Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-391. [DOI] [PubMed] [Google Scholar]
- 29.Gnatt, A., J. Fu, and R. D. Kornberg. 1997. Formation and crystallization of yeast RNA polymerase II elongation complexes. J. Biol. Chem. 272:30799-30805. [DOI] [PubMed] [Google Scholar]
- 30.Gnatt, A. L., P. Cramer, J. Fu, D. A. Bushnell, and R. D. Kornberg. 2001. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science 292:1876-1882. [DOI] [PubMed] [Google Scholar]
- 31.Ha, I., W. S. Lane, and D. Reinberg. 1991. Cloning of a human gene encoding the general transcription initiation factor IIB. Nature 352:689-695. [DOI] [PubMed] [Google Scholar]
- 32.Hampsey, M. 1998. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol. Mol. Biol. Rev. 62:465-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hazbun, T. R., L. Malmstrom, S. Anderson, B. J. Graczyk, B. Fox, M. Riffle, B. A. Sundin, J. D. Aranda, W. H. McDonald, C. H. Chiu, B. E. Snydsman, P. Bradley, E. G. Muller, S. Fields, D. Baker, J. R. Yates III, and T. N. Davis. 2003. Assigning function to yeast proteins by integration of technologies. Mol. Cell 12:1353-1365. [DOI] [PubMed] [Google Scholar]
- 34.Hegde, P., R. Qi, K. Abernathy, C. Gay, S. Dharap, R. Gaspard, J. E. Hughes, E. Snesrud, N. Lee, and J. Quackenbush. 2000. A concise guide to cDNA microarray analysis. BioTechniques 29:548-556. [DOI] [PubMed] [Google Scholar]
- 35.Ho, C. K., and S. Shuman. 1999. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol. Cell 3:405-411. [DOI] [PubMed] [Google Scholar]
- 36.Hodo, H. G., III, and S. P. Blatti. 1977. Purification using polyethylenimine precipitation and low molecular weight subunit analyses of calf thymus and wheat germ DNA-dependent RNA polymerase II. Biochemistry 16:2334-2343. [DOI] [PubMed] [Google Scholar]
- 37.Holstege, F. C., U. Fiedler, and H. T. Timmers. 1997. Three transitions in the RNA polymerase II transcription complex during initiation. EMBO J. 16:7468-7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holstege, F. C., D. Tantin, M. Carey, P. C. van der Vliet, and H. T. Timmers. 1995. The requirement for the basal transcription factor IIE is determined by the helical stability of promoter DNA. EMBO J. 14:810-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holstege, F. C., P. C. van der Vliet, and H. T. Timmers. 1996. Opening of an RNA polymerase II promoter occurs in two distinct steps and requires the basal transcription factors IIE and IIH. EMBO J. 15:1666-1677. [PMC free article] [PubMed] [Google Scholar]
- 40.Huber, A. H., W. J. Nelson, and W. I. Weis. 1997. Three-dimensional structure of the armadillo repeat region of beta-catenin. Cell 90:871-882. [DOI] [PubMed] [Google Scholar]
- 41.Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson, J. S. Weissman, and E. K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature 425:686-691. [DOI] [PubMed] [Google Scholar]
- 42.Ingles, C. J., M. Shales, W. D. Cress, S. J. Triezenberg, and J. Greenblatt. 1991. Reduced binding of TFIID to transcriptionally compromised mutants of VP16. Nature 351:588-590. [DOI] [PubMed] [Google Scholar]
- 43.Ito, T., T. Chiba, R. Ozawa, M. Yoshida, M. Hattori, and Y. Sakaki. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98:4569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang, Y., S. J. Triezenberg, and J. D. Gralla. 1994. Defective transcriptional activation by diverse VP16 mutants associated with a common inability to form open promoter complexes. J. Biol. Chem. 269:5505-5508. [PubMed] [Google Scholar]
- 45.Kim, J. L., D. B. Nikolov, and S. K. Burley. 1993. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature 365:520-527. [DOI] [PubMed] [Google Scholar]
- 46.Kim, Y., J. H. Geiger, S. Hahn, and P. B. Sigler. 1993. Crystal structure of a yeast TBP/TATA-box complex. Nature 365:512-520. [DOI] [PubMed] [Google Scholar]
- 47.Kimura, M., H. Suzuki, and A. Ishihama. 2002. Formation of a carboxy-terminal domain phosphatase (Fcp1)/TFIIF/RNA polymerase II (Pol II) complex in Schizosaccharomyces pombe involves direct interaction between Fcp1 and the Rpb4 subunit of Pol II. Mol. Cell. Biol. 22:1577-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kobor, M. S., J. Archambault, W. Lester, F. C. Holstege, O. Gileadi, D. B. Jansma, E. G. Jennings, F. Kouyoumdjian, A. R. Davidson, R. A. Young, and J. Greenblatt. 1999. An unusual eukaryotic protein phosphatase required for transcription by RNA polymerase II and CTD dephosphorylation in S. cerevisiae. Mol. Cell 4:55-62. [DOI] [PubMed] [Google Scholar]
- 49.Kobor, M. S., L. D. Simon, J. Omichinski, G. Zhong, J. Archambault, and J. Greenblatt. 2000. A motif shared by TFIIF and TFIIB mediates their interaction with the RNA polymerase II carboxy-terminal domain phosphatase Fcp1p in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:7438-7449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krogan, N. J., M. Kim, S. H. Ahn, G. Zhong, M. S. Kobor, G. Cagney, A. Emili, A. Shilatifard, S. Buratowski, and J. F. Greenblatt. 2002. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol. Cell. Biol. 22:6979-6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lagrange, T., A. N. Kapanidis, H. Tang, D. Reinberg, and R. H. Ebright. 1998. New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB. Genes Dev. 12:34-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Langelier, M. F., D. Forget, A. Rojas, Y. Porlier, Z. F. Burton, and B. Coulombe. 2001. Structural and functional interactions of transcription factor (TF) IIA with TFIIE and TFIIF in transcription initiation by RNA polymerase II. J. Biol. Chem. 276:38652-38657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu, H., L. Zawel, L. Fisher, J. M. Egly, and D. Reinberg. 1992. Human general transcription factor IIH phosphorylates the C-terminal domain of RNA polymerase II. Nature 358:641-645. [DOI] [PubMed] [Google Scholar]
- 54.Maldonado, E., I. Ha, P. Cortes, L. Weis, and D. Reinberg. 1990. Factors involved in specific transcription by mammalian RNA polymerase II: role of transcription factors IIA, IID, and IIB during formation of a transcription-competent complex. Mol. Cell. Biol. 10:6335-6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marton, M. J., J. L. DeRisi, H. A. Bennett, V. R. Iyer, M. R. Meyer, C. J. Roberts, R. Stoughton, J. Burchard, D. Slade, H. Dai, D. E. Bassett, Jr., L. H. Hartwell, P. O. Brown, and S. H. Friend. 1998. Drug target validation and identification of secondary drug target effects using DNA microarrays. Nat. Med. 4:1293-1301. [DOI] [PubMed] [Google Scholar]
- 56.Mizuguchi, K., C. M. Deane, T. L. Blundell, and J. P. Overington. 1998. HOMSTRAD: a database of protein structure alignments for homologous families. Protein Sci. 7:2469-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murakami, K. S., S. Masuda, E. A. Campbell, O. Muzzin, and S. A. Darst. 2002. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science 296:1285-1290. [DOI] [PubMed] [Google Scholar]
- 58.Murakami, K. S., S. Masuda, and S. A. Darst. 2002. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 A resolution. Science 296:1280-1284. [DOI] [PubMed] [Google Scholar]
- 59.Nikolov, D. B., H. Chen, E. D. Halay, A. A. Usheva, K. Hisatake, D. K. Lee, R. G. Roeder, and S. K. Burley. 1995. Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature 377:119-128. [DOI] [PubMed] [Google Scholar]
- 60.Nissen, R. M., and K. R. Yamamoto. 2000. The glucocorticoid receptor inhibits NFkappaB by interfering with serine-2 phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 14:2314-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.No, D., T. P. Yao, and R. M. Evans. 1996. Ecdysone-inducible gene expression in mammalian cells and transgenic mice. Proc. Natl. Acad. Sci. USA 93:3346-3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ohkuma, Y., S. Hashimoto, C. K. Wang, M. Horikoshi, and R. G. Roeder. 1995. Analysis of the role of TFIIE in basal transcription and TFIIH-mediated carboxy-terminal domain phosphorylation through structure-function studies of TFIIE-alpha. Mol. Cell. Biol. 15:4856-4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ohkuma, Y., and R. G. Roeder. 1994. Regulation of TFIIH ATPase and kinase activities by TFIIE during active initiation complex formation. Nature 368:160-163. [DOI] [PubMed] [Google Scholar]
- 64.Ohkuma, Y., H. Sumimoto, A. Hoffmann, S. Shimasaki, M. Horikoshi, and R. G. Roeder. 1991. Structural motifs and potential sigma homologies in the large subunit of human general transcription factor TFIIE. Nature 354:398-401. [DOI] [PubMed] [Google Scholar]
- 65.Orphanides, G., T. Lagrange, and D. Reinberg. 1996. The general transcription factors of RNA polymerase II. Genes Dev. 10:2657-2683. [DOI] [PubMed] [Google Scholar]
- 66.Pan, G., and J. Greenblatt. 1994. Initiation of transcription by RNA polymerase II is limited by melting of the promoter DNA in the region immediately upstream of the initiation site. J. Biol. Chem. 269:30101-30104. [PubMed] [Google Scholar]
- 67.Peterson, M. G., J. Inostroza, M. E. Maxon, O. Flores, A. Admon, D. Reinberg, and R. Tjian. 1991. Structure and functional properties of human general transcription factor IIE. Nature 354:369-373. [DOI] [PubMed] [Google Scholar]
- 68.Pokholok, D. K., N. M. Hannett, and R. A. Young. 2002. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell 9:799-809. [DOI] [PubMed] [Google Scholar]
- 69.Puig, O., F. Caspary, G. Rigaut, B. Rutz, E. Bouveret, E. Bragado-Nilsson, M. Wilm, and B. Seraphin. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24:218-229. [DOI] [PubMed] [Google Scholar]
- 70.Rachez, C., and L. P. Freedman. 2001. Mediator complexes and transcription. Curr. Opin. Cell Biol. 13:274-280. [DOI] [PubMed] [Google Scholar]
- 71.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 72.Robert, F., M. Douziech, D. Forget, J. M. Egly, J. Greenblatt, Z. F. Burton, and B. Coulombe. 1998. Wrapping of promoter DNA around the RNA polymerase II initiation complex induced by TFIIF. Mol. Cell 2:341-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rodriguez, C. R., E. J. Cho, M. C. Keogh, C. L. Moore, A. L. Greenleaf, and S. Buratowski. 2000. Kin28, the TFIIH-associated carboxy-terminal domain kinase, facilitates the recruitment of mRNA processing machinery to RNA polymerase II. Mol. Cell. Biol. 20:104-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schaffer, A. A., Y. I. Wolf, C. P. Ponting, E. V. Koonin, L. Aravind, and S. F. Altschul. 1999. IMPALA: matching a protein sequence against a collection of PSI-BLAST-constructed position-specific score matrices. Bioinformatics 15:1000-1011. [DOI] [PubMed] [Google Scholar]
- 75.Serizawa, H., R. C. Conaway, and J. W. Conaway. 1992. A carboxyl-terminal-domain kinase associated with RNA polymerase II transcription factor delta from rat liver. Proc. Natl. Acad. Sci. USA 89:7476-7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Serizawa, H., T. P. Makela, J. W. Conaway, R. C. Conaway, R. A. Weinberg, and R. A. Young. 1995. Association of Cdk-activating kinase subunits with transcription factor TFIIH. Nature 374:280-282. [DOI] [PubMed] [Google Scholar]
- 77.Shiekhattar, R., F. Mermelstein, R. P. Fisher, R. Drapkin, B. Dynlacht, H. C. Wessling, D. O. Morgan, and D. Reinberg. 1995. Cdk-activating kinase complex is a component of human transcription factor TFIIH. Nature 374:283-287. [DOI] [PubMed] [Google Scholar]
- 78.Skaar, D. A., and A. L. Greenleaf. 2002. The RNA polymerase II CTD kinase CTDK-I affects pre-mRNA 3′ cleavage/polyadenylation through the processing component Pti1p. Mol. Cell 10:1429-1439. [DOI] [PubMed] [Google Scholar]
- 79.Sluder, A. E., D. H. Price, and A. L. Greenleaf. 1988. Elongation by Drosophila RNA polymerase II. Transcription of 3′-extended DNA templates. J. Biol. Chem. 263:9917-9925. [PubMed] [Google Scholar]
- 80.Sopta, M., R. W. Carthew, and J. Greenblatt. 1985. Isolation of three proteins that bind to mammalian RNA polymerase II. J. Biol. Chem. 260:10353-10360. [PubMed] [Google Scholar]
- 81.Sumimoto, H., Y. Ohkuma, E. Sinn, H. Kato, S. Shimasaki, M. Horikoshi, and R. G. Roeder. 1991. Conserved sequence motifs in the small subunit of human general transcription factor TFIIE. Nature 354:401-404. [DOI] [PubMed] [Google Scholar]
- 82.Takahashi, Y., J. B. Rayman, and B. D. Dynlacht. 2000. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 14:804-816. [PMC free article] [PubMed] [Google Scholar]
- 83.Tirode, F., D. Busso, F. Coin, and J. M. Egly. 1999. Reconstitution of the transcription factor TFIIH: assignment of functions for the three enzymatic subunits, XPB, XPD, and cdk7. Mol. Cell 3:87-95. [DOI] [PubMed] [Google Scholar]
- 84.Vassylyev, D. G., S. Sekine, O. Laptenko, J. Lee, M. N. Vassylyeva, S. Borukhov, and S. Yokoyama. 2002. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature 417:712-719. [DOI] [PubMed] [Google Scholar]
- 85.Westover, K. D., D. A. Bushnell, and R. D. Kornberg. 2004. Structural basis of transcription: separation of RNA from DNA by RNA polymerase II. Science 303:1014-1016. [DOI] [PubMed] [Google Scholar]
- 86.Wu, L. F., T. R. Hughes, A. P. Davierwala, M. D. Robinson, R. Stoughton, and S. J. Altschul. 2002. Large-scale prediction of Saccharomyces cerevisiae gene function using overlapping transcriptional clusters. Nat. Genet. 31:255-265. [DOI] [PubMed] [Google Scholar]
- 87.Yang, Y. H., and T. Speed. 2002. Design issues for cDNA microarray experiments. Nat. Rev. Genet. 3:579-588. [DOI] [PubMed] [Google Scholar]
- 88.Young, R. A. 1991. RNA polymerase II. Annu. Rev. Biochem. 60:689-715. [DOI] [PubMed] [Google Scholar]
- 89.Zhang, G., E. A. Campbell, L. Minakhin, C. Richter, K. Severinov, and S. A. Darst. 1999. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 A resolution. Cell 98:811-824. [DOI] [PubMed] [Google Scholar]