Summary
Background
West Nile virus (WNV) can be transmitted by transfusion through infected blood components. In Germany, a 28-day deferral for blood donors of therapeutic blood components who had spent at least 2 days in WNV-endemic areas from June 1 to November 30, 2014 was enforced. Otherwise, screening of blood donors for WNV RNA or the application of pathogen reduction techniques are appropriate alternatives.
Methods
In the present study, we evaluated NAT screening for the detection of WNV in blood components. A total of 58 minipools consisting of 357 individual blood donors were screened for the presence of WNV RNA employing an automated high-volume extraction method using the RealStar WNV RT-PCR Kit. Additionally, different WNV reference reagents were quantified to prove the status quo of standardization. Four different WNV real-time NAT kits were compared using samples of an external quality assessment panel.
Results
The 95% lower detection limit of the WNV MP-NAT was determined to 30.2 copies/ml (95% CI 24.2-45.4 copies/ml). No WNV RNA-positive minipool was detected. Quantification of WNV reference reagents revealed shortcomings in standardization. Comparison of several WNV NAT assays showed considerable differences in assay sensitivities and particularly a missing detection of WNV lineage 2. Implementation of seasonal WNV MP-NAT screening was demonstrated.
Conclusion
Actually, WNV infections in Germany are rare events introduced by returning travelers, but surveillance of these emerging infections is important for safety in blood supply. The validation study pointed out the need for standardization of WNV NAT because of current lack of an international standard for WNV RNA.
Keywords: Blood donation testing, Blood safety, NAT screening, Quality control
Introduction
West Nile Virus (WNV) is a neurotropic human pathogen that is the causative agent of West Nile fever (WNF) and encephalitis [1]. This arbovirus, a member of the Flaviviridae family, was introduced into North America in 1999. In humans, most of WNV infections remain asymptomatic, but approximately 20% of the individuals develop WNF. In a small percentage (<1%) of those infected, severe outcome with neurological symptoms occurs [2]. In contrast, in the USA West Nile neuro-invasive disease (WNND) is described in 61% of reported WNV disease cases in the year 2014 [3]. This led to a reassessment of previous WNV outbreaks in Europe and an intensification of the epidemiological surveillance [2]. An increasing number of WNV infections in humans in the Mediterranean Basin (Israel, Italy, Greece, and North Africa), Romania, Hungary, Austria, Russia, and several Central Asian countries has been reported in recent years (www.ECDC.europa.eu; [4,5]). The infections are caused by WNV lineage 1 (WNV-L1) and lineage 2 (WNV-L2). The European Centre for Disease Prevention and Control (ECDC) has published weekly updates on the spatial distribution of WNF cases in the European Union (EU) and neighboring countries on its website and identifies individual European regions. An intense debate on the current hazard potential and the introduction of preventive measures to minimize the risk of WNV transmission through blood components was started in the committee of the European Commission due to the surveillance data reported [6]. Deferral from blood donation after returning from travels to North America or other (tropical) endemic regions has already been implemented in Germany since 2003 [7,8].
Taking into account the epidemiological situation in Europe and the Mediterranean Basin, the Paul-Ehrlich Institute (PEI) decided that in the period from June 1 to November 30, 2014 blood donations for cellular blood components, therapeutic individual plasma, and stem cell preparations for hematopoietic reconstitution must be excluded, from donors for at least 28 days after their return from regions or countries with ongoing transmission of WNV to humans, including the North American continent and Mexico [9]. Blood donors tested negative for the WNV genome or pathogen-inactivated blood components are not affected by this regulatory requirement. A minimum analytical sensitivity of 250 copies/ml WNV RNA for the single donation for the WNV nucleic acid amplification tests (NAT) was required by the PEI. Due to the lack of an international WNV NAT standard, the reference preparation for quantification was the Health Canada WNV Reference Reagent [10] or secondary references calibrated to that. The NAT assay should detect WNV-L1 and WNV-L2 with an equal sensitivity.
In the present study, we evaluated a routine in-process screening procedure for the detection of WNV RNA in order to increase blood safety.
Material and Methods
Blood Donors
A total of 357 allogeneic blood donations (geographic origin: North Rhine-Westphalia, Lower Saxony, Hesse) were screened for WNV RNA by the Uni.Blutspendedienst OWL between June and December 2014. Thereof, we tested 55.5% male (age 35.5 ± 12.6 years) and 44.5 %, female donors (age 31.9 ± 13.0 years). Donors were asked whether they had traveled to WNV-endemic regions or had general symptoms of an infection/disease. In order to keep the number of donor deferrals small, we differentiated according to travel history regarding WNV-endemic areas based on the mandatory list of countries and territories for the donor resets published monthly by the PEI. The regulation of the PEI includes the definition of WNV-endemic areas differentiated by German riparian states and non-riparian states. The latter can also be differentiated in the responsibility of the pharmaceutical manufacturer in WNV-endemic regions defining hotspots instead of considering the whole country as WNV-endemic. The Uni.Blutspendedienst OWL routinely tests WNV samples in pools of eight. Nucleic acids are extracted from the pools using an automated, robotic system, which is a combination of a Tecan Evo 200 (Tecan Germany, Crailsheim, Germany) and a Chemagen separation module MSM1 for viral nucleic extraction. Routine viral NAT screening for HCV, HIV-1, HBV, HAV, and parvovirus B19 was performed with minipools (MPs) of 96 donations using the Chemagen nucleic extraction protocol for 4.8 ml plasma. In order to meet the analytical sensitivity for WNV NAT, we performed additional pooling on the automated device by pipetting MPs of 8 donations of 300 µl EDTA plasma per single donor each. Pools with fewer than 8 samples were filled up to 2.4 ml with negative human plasma. Donors of reactive pools were tested singularly for identification of the reactive individual donor. All donors underwent a predonation medical examination which negated current diseases or any known risk factors for viral infection.
Negative and Positive Samples
The negative samples were prepared using a citrate plasma pool made up of 5 donations that had tested negative for HCV, HBV, HIV, and WNV by serological and NAT tests.
Firstly, the PEI reference preparation for WNV RNA (Paul-Ehrlich Institute, Langen, Germany) was used containing 140,000 copies/ml (PEI_WNVL1_9002/14; lineage 1, New York isolate from 1999) for calibration of the NATrol™ WNV control target NATWNV-0005 (50,000 copies/ml, lineage 1; ZeptoMetrix Corporation, Buffalo, NY, USA). The determination of lower detection limit was performed with the calibrated NATWNV-0005. In addition, four WNV reference preparations were obtained from the National Centre for Immunobiologicals Research and Evaluation (CRIVIB), ISS, Rome, Italy, used for determination of analytical specificity and sensitivity:
-
–
ISS_WNVL1-0213 (WNV-L1, Ref Prep batch 0213; 1,500 copies/ml traceable to the Health Canada Reference Preparation (HC-SC WNV Nat Ref 001/03))
-
–
ISS_WNVL1-0410 (WNV-L1, Ref Prep batch 0410, 150,000 copies/ml)
-
–
ISS_WNVL2-0411 (WNV-L2, Ref Prep batch 0411, 17,000 copies/ml)
-
–
ISS_WNVL2-0513 (WNV-L2, Ref Prep batch 0513, about 12,600,000 copies/ml).
Nucleic Acid Extraction
An amount of 2.4 ml of pooled plasma (8 donations) was extracted with the chemagic viral DNA/RNA reagent kit (Viral 2k; PerkinElmer Chemagen Technologie GmbH, Baesweiler, Germany) combined with the automated chemagic magnetic separation module MSMI (PerkinElmer Chemagen Technologie GmbH). Briefly, 2.4 ml of pooled plasma was mixed with 2.4 ml lysis buffer, 20 µl protease, 7 µl polyA, and 10 µl WNV internal control (Real-Star WNV RT-PCR Kit 1.0, Altona Diagnostics, Hamburg, Germany). Samples were incubated at 55 °C for 10 min. Subsequently, lysates were mixed with 7.5 ml binding buffer containing 100 µl magnetic beads. The MSMI module performed the nucleic acid extraction process automatically, including binding, washing twice and elution in a final volume of 100 µl elution buffer. Total RNA from individual donations was extracted from 600 µl plasma and filled up to 2.4 ml with negative human plasma using the same protocol.
Real-Time PCR
The amplification step for WNV screening of blood donations, using Real-Star WNV RT-PCR Kit 1.0 (Altona Diagnostics), was performed on the Rotor-Gene 3000 system (Corbett Life Sciences, Sydney, Australia), according to the manufacturer's instructions. RT-PCR reactions were carried out in 0.2 ml tubes containing 25 µl reaction mix and 25 µl nucleic acid extract.
Moreover, we compared the analytical performance of four WNV RT-PCR assays that could be used on open device platforms for nucleic acid extraction and RNA amplification/detection.
The WNV PCR Kit (GFE Blut mbH, Frankfurt/M., Germany) was used on the LightCycler 480-II real-time PCR system (Roche Diagnostics, Mannheim, Germany). An amount of 30 µl of RNA extract was amplified in a final volume of 75 µl PCR reactions mix according to the manufacturer's instructions.
The artus WNV LC RT-PCR Kit (Qiagen, Hilden, Germany) was used on the LightCycler 2.0 real-timePCR system (Roche Diagnostics, Mannheim, Germany). An amount of 20 µl PCR reactions consisted of 15 µl reaction mix and 5 µl RNA extract. Amplification using the artus WNV LC RT-PCR Kit was performed according to the manufacturer's instructions.
The LightMix WNV RT-PCR Kit (TIB Molbiol, Berlin, Germany) was performed on the LightCycler 480-II real-time PCR system. The reaction mix consisted of 15 µl reaction mix containing 10 µl 2× Superscript III One-Step RT-PCR reaction buffer including 6 mmol/l MgSO4, 2 µl reagent mix, 2 µl IC mix, 1 µl Superscript III Platinum Taq polymerase Mix (Life Technologies GmbH, Darmstadt, Germany) and 5 µl RNA extract.
The Genesig Quantification WNV Kit (PrimerDesign Ltd., Southampton, UK) was used on the LightCycler 480-II real-time PCR system. The reaction mix consisted of 15 µl reaction mix containing 10 µl 2× oasig OneStep qRT-PCR mastermix (PrimerDesign), 1 µl WNV primer/probe mix, 1 µl IC primer/probe mix, 0.5 µl IC RNA, 2.5 µl H2O and 5 µl RNA extract.
Analytical Sensitivity and Comparison of Different Amplification Methods
The analytical sensitivity expressed as 95% limit of detection (LOD), and the precision of the RealStar WNV RT-PCR 1.0 assay in combination with the 2.4 ml nucleic acid extraction protocol was determined using a twofold dilution series of plasma inoculated with the NATrol™ WNV control target NATWNV-005 in six dilution steps and 24 replicates. First of all, the NATrol NATWNV-005 was calibrated to the Health Canada Reference Reagent (HC-SC WNV Nat Ref 001/03), as described in the Results section. The 95% LOD was calculated by the log-converted calculation mode of probit analysis using the SPSS software (IBM SPSS Statistics, version 21; IBM GmbH Ehningen, Germany).
External Quality Assessment
The Uni.Blutspendedienst OWL participated in the External Quality Assessment (EQA) 2014 scheme, organized by the Italian National Blood Centre (NBC) and the CRIVIB. The scheme, addressed to the Italian Blood Transfusion Centers Laboratories that carry out NAT testing of plasma for the detection of WNV RNA, was also open to foreign laboratories. The EQA scheme was carried out in two phases with distribution of two EQA panels of 10 blinded samples each. The first EQA panel (distributed in July 2014) consisted of 4 samples of WNV-L1 and 4 samples of WNV-L2, containing 600, 300, 150 or 75 copies/ml; an additional two samples negative for WNV were included in the panel. The second EQA panel (distributed in August 2014) consisted of the same samples in a different coding.
Results
Test Design
The aim of this study was the implementation of a routine WNV RNA screening according to the setup applied for our routine viral nucleic acid screening (HIV-1, HBV, HCV, HAV, PVB19) using the RealStar WNV RT-PCR Kit 1.0. The combination of the nucleic acid extraction method and real-time NAT was evaluated according to the requirements for validation and routine use of NAT for the detection of viral nucleic acids in blood donors defined by the PEI according to Committee for Proprietary Medicinal Products (CPMP) guidelines [11,12].
Standardization of WNV NAT
Due to lack of international NAT standards for WNV RNA, the calibration in copies/ml against the Health Canada standard (HCS) of positive controls used for validation of WNV NAT is recommended. In fact, HCS was previously used for validation studies of the two CE-marked commercial kits for WNV screening in blood: the cobas TaqScreen WNV test (Roche Diagnostics, Pleasanton, CA, USA) and the Procleix WNV assay (Grifols Diagnostic Solutions Inc., Emeryville, CA, USA) [10,13]. The PEI advised German Blood Centers to determine the 95% LOD of the WNV NAT assays for blood donor screening in Germany using a reference material with a WNV RNA content traceable to HCS. Because the HCS was no longer available, we were forced to use the NATtrol WNV control target NATWNV-005 (Zeptometrix). In a second investigation, in order to verify the WNV RNA content of the NATtrol WNV control target, we quantified it against two secondary reference preparations calibrated against the HCS from PEI and ISS. Serial dilutions of the reference preparations from PEI and ISS and NATtrol WNV control target were used for nucleic acid extractions and amplification using the RealStar WNV RT-PCR Kit 1.0. The PEI or ISS WNV RNA references were used for quantification of the NATtrol WNV control target that showed aWNV RNA content lower than that described by the manufacturer. Two batches of NATtrol WNV control target were calibrated and detected an over-quantification of factor 1.4 or 4.1 (table 1).
Table 1.
Quantification of the WNV RNA reagent against different reference preparations
| Name | NATtrol NATWNV-005 (Zeptometrix) |
||||
|---|---|---|---|---|---|
| Batch | Lot no. #309897 |
Lot no. #312649 |
|||
| manufacturer's specifications | ISS_WNVL10213a | manufacturer's specifications | PEI_WNVL19002/14b | ISS_WNVL10213a | |
| Result of quantification, copies/ml | 1,000 | 719 | 1,000 | 250 | 245 |
| Conclusion | over-quantification referred to HCS: factor 1.4 | over-quantification referred to HCS: factor 4.1 | |||
ISS_WNVL1-0213 (National Center for Immunobiologicals Research and Evaluation, ISS, Rome, Italy).
PEI reference preparation for WNV RNA (Paul-Ehrlich Institute, Langen, Germany).
Analytical Sensitivity
A twofold dilution series of HCS-calibrated NATtrol WNV control target NATWNV-005 was used to evaluate the 95% LOD of the NAT assay applied for WNV screening in blood donors, because other WNV reference reagents (PEI, ISS: see ‘Material and Methods’) were available at a later date after validation. The 95% LOD, as determined by probit analysis, was estimated to be 30.2 copies/ml (95% CI 24.2-45.4 copies/ ml). The corresponding concentration for achieving a 50% probability was 12.8 copies/ml (95% CI 9.2-16.5 copies/ml) (table 2).
Table 2.
WNV NAT RealStar WNV RT-PCR Kit in combination with chemagic viral 2K (from 2.4 ml plasma) extraction protocol
| WNV-RNA concentration, copies/mla | Reactive samples/number tested | Percent positive |
|---|---|---|
| 179 | 24/24 | 100 |
| 89.5b | 24/24 | 100 |
| 44.8 | 24/24 | 100 |
| 22.4 | 19/24 | 75 |
| 11.2 | 12/24 | 50 |
| 5.6 | 5/24 | 21 |
| WNV-RNA concentration, copies/ml | ||
| 95 % detection limit (95% CI) | 30.2 (24.2-45.4) | |
| Intra-assay WNVb, mean ±SD / CV | 33.20 ± 0.42 / 1.27 | |
| Interassay WNVb, mean ±SD / CV | 32.62 ± 0.86 / 2.63 | |
NATtrol(tm) WNV (NATWNV-0005, lot number 309897); calibrated to Health Canada WNV Reference preparation.
Intra- and interassay variabilities were calculated exemplarily for the concentration of 89.5 copies/ml.
The NAT assay should equally detect WNV-L1 and WNV-L2. Therefore, quantified WNV-positive reference preparations for WNV-L1 (PEI_WNVL1_9002/14, ISS_WNVL1-0213, ISS_WNVL1- 0410), andWNV-L2 (ISS_WNVL2-0411, ISS_WNVL2-0513) were adjusted to 250 copies/ml and used in WNV NAT. This viral titer represents the minimum analytical sensitivity for the single donation for the WNV NAT. All samples were detected to the same extent (data not shown).
Assay Reproducibility
The reproducibility of the NAT assay was demonstrated by analyzing the intra- and inter-assay precision for the crossing threshold (CT) values. The intra-assay precision was calculated from 8 replicates with the WNV target containing 89 copies/ml, with a CT mean value of 33.20 (SD 0.42, CV 1.27%) (table 2). The inter-assay variability was determined from three independent PCR runs with 8 replicates per run, with CT mean values of 32.62 (DS 0.86, CV 2.63%) for the WNV target (89 copies/ml).
Screening of Blood Donors
The NAT method has been in use as a routine setting since June 2014 for WNV RNA blood donor screening, according to the PEI decision [9]. Donors were asked whether they had traveled to WNV-endemic areas (published monthly by the PEI according to the ECDC publications) during the transmission season (table 3). A total of 357 donations in 58 MPs during the first 7 months of routine WNV NAT screening (June to December, 2014) were tested without any reactive result. None of the donors did report WNF symptoms retrospectively. It should be noted that the 357 donations correspond to 0.9% of all whole blood donations which were taken by our blood transfusion service during this period.
Table 3.
Cumulative number of blood donors returning from WNV-endemic countries tested in this study compared to WNF cases reported by the ECDC for the years 2013 and 2014
| WNV endemic country | Cumulative number of WNV NAT screenings by Uni. BSPD OWL 2014 |
Cumulative number of reported WNF cases by country |
||||
|---|---|---|---|---|---|---|
| ECDC 12/2013b |
ECDC 12/20140c |
|||||
| no. | % | no. | % | no. | % | |
| Italy | 103 | 28.8 | 69 | 8.8 | 24 | 11.4 |
| Croatia | 42 | 11.8 | 16 | 2.0 | - | - |
| Greece | 40 | 11.2 | 86 | 10.9 | 15 | 7.1 |
| Hungary | 26 | 7.3 | 31 | 3.9 | 11 | 5.2 |
| Bulgaria | 19 | 5.3 | - | - | - | - |
| Russian Federation | 11 | 3.1 | 177 | 22.5 | 29 | 13.8 |
| Tunisia | 10 | 2.8 | 6 | <1.0 | - | - |
| Austria | 7 | 2.0 | - | - | 1 | <1.0 |
| Romania | 4 | 1.1 | 24 | 3.1 | 23 | 11.0 |
| Serbia | 3 | <1.0 | 302 | 38.5 | 76 | 36.2 |
| Bosnia and Herzegovina | 3 | <1.0 | 3 | <1.0 | 13 | 6.2 |
| Israel | 2 | <1.0 | 63 | 8.0 | 17 | 8.1 |
| Czech Republic | 2 | <1.0 | 1 | <1.0 | - | - |
| Other | 8a | <1.0 | 8 | 1.0 | 1 | <1.0 |
| Not reported | 27 | 7.6 | - | - | - | - |
| USA | 41 | 11.5 | n.r. | n.r. | n.r. | n.r. |
| Canada | 8 | 2.2 | n.r. | n.r. | n.r. | n.r. |
| Total | 357 | 100 | 785 | 100 | 210 | 100 |
n.r. = Not reported by the ECDC.
n = 1: Ukraine, Cuba, Morocco, Egypt, United Arab Emirates, Slovenia, Moldova, Jordan.
Cumulative number of WNF cases, by affected area in 2013, as of 6/6/2014 (ECDC).
Cumulative number of WNF cases, by affected area, as of 20/11/2014 (ECDC).
Amplification failures were monitored in each single-pool RNA extract by in-tube amplification of a control RNA sequence, detected with a heterologous DNA probe. There were no invalid results (failed amplification of internal control RNA). The low level run control (threefold 95% LOD corresponding to 90 copies/ml) was positive in 100% of WNV RNA tests, as shown in figure 1. Two different batches of WNV run control were tested with a CT mean value of 31.0 (SD 0.93 CV 3.0%) and 31.0 (SD 1.06, CV 3.4%), respectively (fig. 1).
Fig. 1.
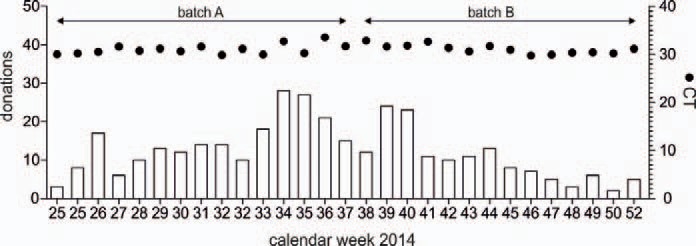
Number of WNV NAT screenings in the season 2014. WNV NAT screening in the Uni.Blutspendedienst OWL was performed weekly from calendar week 25 (June 1) to calendar week 52 of 2014 (December 31). The CT values (•) of the low level run control (90 copies/ml), tested in two different batches, are displayed.
EQA for WNV RNA
Participation in EQA programs is recognized as an important factor for quality assurance, as laid down in Directive 2005/62/EC80. Testing laboratories should verify the quality of their techniques through ongoing participation in the EQA program. A regular EQA scheme for WNV detection by NAT has been organized by the Italian NBC and CRIVIB annually in Italy since 2010 [14,15,16]. The 2014 EQA scheme for WNV was run in the summer-autumn period in two phases. The Uni.Blutspendedienst OWL laboratory participated in this scheme using the NAT method described in this study in addition to other commercially available WNV NAT kits (table 4).
Table 4.
Results of EQA panel 1 and 2 (2014)
| Copies/ml (1:8 dilution in MP) | Lineagea | RealStar WNV RT-PCR Kit (ALTONA DIAGNOSTICS) Rotorgene Q |
WNV RT-PCR Kit (GFE Blut) LightCycler 480-II |
Genesig Quantification WNV Kit (Primerdesign) LightCycler 480-II |
LightMix WNV RT-PCR Kit (TIB Molbiol) LightCycler 480-II |
artus WNV LC RT-PCR Kit (Qiagen) LightCycler 2.0 |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MP8 (0.3 ml) | ID (1.4 ml) | MP8 (0.3 ml) | ID (1.4 ml) | MP8 (0.3 ml) | ID (1.4 ml) | MP8 (0.3 ml) | ID (1.4 ml) | MP8 (0.3 ml) | ID (1.4 ml) | ||
| EQA Panel 1/2014 | |||||||||||
| 600 (75) | 1 | 31.98 | 30.67 | 31.31 | 29.02 | >50 | 33.99 | 36.19 | 33.80 | 37.15 | 36.27 |
| 300 (38) | 1 | 36.19 | 31.57 | 32.56 | 30.52 | >50 | 35.13 | >50 | 35.03 | >45 | 37.97 |
| 150 (18) | 1 | 35.98 | 32.04 | 33.52 | 31.12 | >50 | 34.95 | >50 | 35.32 | >45 | 37.21 |
| 75 (10) | 1 | 36.71 | 31.53 | 34.24 | 31.24 | >50 | > 50 | >50 | 35.47 | >45 | 37.92 |
| 600 (75) | 2 | 30.76 | 28.08 | 31.10 | 28.61 | 34.48 | 33.78 | >50 | >50 | 39.25 | 34.28 |
| 300 (38) | 2 | 32.54 | 28.91 | 32.00 | 29.33 | 34.93 | 33.95 | >50 | >50 | 37.26 | 36.11 |
| 150 (18) | 2 | 33.65 | 30.08 | 33.64 | 30.30 | >50 | 33.62 | >50 | >50 | >45 | 36.78 |
| 75 (10) | 2 | 33.83 | 30.67 | 33.55 | 31.55 | >50 | >50 | >50 | >50 | >45 | 37.52 |
| Negative | - | >45 | >45 | >45 | >45 | >50 | >50 | >50 | >50 | >45 | >45 |
| Negative | - | >45 | >45 | >45 | >45 | >50 | >50 | >50 | >50 | >45 | >45 |
| EQA Panel 2/2014 | |||||||||||
| 600 (75) | 1 | 31.58 | 29.84 | 30.76 | 29.61 | 34.35 | 33.85 | >50 | 34.86 | 38.25 | 35.08 |
| 300 (38) | 1 | 32.67 | 30.74 | 31.82 | 29.76 | >50 | 33.62 | >50 | 36.33 | >50 | 39.29 |
| 150 (18) | 1 | 32.94 | 31.37 | 31.91 | 30.86 | >50 | >50 | >50 | 36.13 | >50 | 36.87 |
| 75 (10) | 1 | 34.53 | 32.45 | 33.28 | 31.48 | >50 | >50 | >50 | >50 | >50 | 38.07 |
| 600 (75) | 2 | 30.47 | 27.78 | 29.37 | 27.60 | 34.09 | 33.25 | >50 | >50 | 35.40 | 34.02 |
| 300 (38) | 2 | 31.84 | 28.27 | 30.86 | 28.34 | 34.54 | 33.69 | >50 | >50 | 36.09 | 34.78 |
| 150 (18) | 2 | 31.52 | 29.79 | 31.32 | 29.79 | >50 | 33.51 | >50 | >50 | >50 | 35.56 |
| 75 (10) | 2 | 33.07 | 30.90 | 32.21 | 30.82 | >50 | 34.01 | >50 | >50 | >50 | 37.76 |
| Negative | - | >45 | >45 | >45 | >45 | >50 | >50 | >50 | >50 | >50 | >50 |
| Negative | - | >45 | >45 | >45 | >45 | >50 | >50 | >50 | >50 | >50 | >50 |
Italics = false-negative results.
WNV-L1 Ref Prep batch 0410; WNV-L2 Ref Prep batch 0513.
In order to compare the applicability of the different PCR methods for WNV blood donors in screening of MP8, subsequent plasma samples of WNV EQA samples were diluted with negative human plasma to simulate master pools of 8 donations mimicking a routine pool screening procedure. Simulation of master pools was set up by combining 300 µl of EQA sample with 2.1 ml negative human plasma to achieve a level of 2.4 ml. The individual testing was performed with a 1.4 ml EQA sample filled-up with 1 ml negative human plasma. Nucleic acid was extracted from all samples with the 2.4 ml protocol on the Chemagen MSMI platform.
Thanks to the higher sensitivity of the RealStar WNV RT-PCR Kit 1.0 and the WNV PCR Kit (GFE Blut mbH), these two NAT assays were able to correctly detect all 8 WNV RNA-positive samples at different concentrations. Remarkably, both the individual (1.4 ml plasma input per extraction) and pool testing (0.3 ml plasma input per extraction) showed a correct result. WNV-L1 and WNV-L2 were equally detected.
On the contrary, the artus WNV LC RT-PCR Kit (Qiagen) detected all WNV RNA-positive samples only in the individual testing, and samples with 300-600 copies/ml in MP8 testing. The other two WNV RT-PCR assays, Genesig Quantification WNV Kit (PrimerDesign Ltd) and LightMix WNV RT-PCR Kit (TIB Molbiol), showed a poor sensitivity in line with the 95% LOD stated by the respective manufacturers. Additionally, the LightMix WNV RT-PCR Kit could not detect WNV-L2 in any sample.
Discussion
In this study, we demonstrate the technical and analytical feasibility of WNV blood donor screening on a routine basis. Our validation study shows that the 95% LOD of the WNV NAT was 30.2 copies/ml. Even if the 95% LOD was evaluated only for WNV-L1, we can conclude from results reported in the EQA study and from spiking experiments with WNV-L2 reference samples conducted in our laboratory (data not shown) that WNV-L2 is detected as well as WNV-L1. The setting of WNV NAT is compatible to the virus NAT screening used in our blood transfusion service. Therefore, our WNV NAT is also suitable for blood donor screening and comparable to currently commercially available WNV NAT screening methods (table 5), in spite of a lower automation grade and throughput.
Table 5.
Overview of currently commercially available WNV NAT screening methods
| Kit Name | RealStar WNV RT-PCR Kit 1.0 | Cobas WNV Test | Procleix WNV Assay | WNV PCR Kit | |
| Manufacturer | Altona Diagnostics | Roche Diagnostics | Grifols | GFE Blut | |
| Automation | no | full automation cobas® | full automation Procleix® | full automation Procleix® | autoX |
| 6800/8800 Systems | Panther® System | Tigris® System | |||
| FDA/CE-IVD | no/yes | no/yes | no/yes | yes/yes | no/yes |
| Sample preparation / nucleic acid extraction | |||||
| Virus enrichment pre-extraction | no | no | no | pool plasma (8 × 1 ml) centrifugation 60 min,55,000 × g | |
| Maximal MP sizea | 8 | 19 | 21 | 28 | 8 |
| Nucleic acid extraction procedure | chemagic viral RNA/DNA Kit on MSM-I (2.4 ml protocol) | magnetic glass particles for fully automated NA-extraction | target-specific extraction - magnetic microparticles capture viral nucleic acids with viral-specific probes (fully automated) | autoX extraction kit | |
| Processed sample volume, plasma ml | 2.4 | 0.85 | 0.5 | 1.0 | |
| Elution volume, µl | 100 | 50 | N/A, single tube format | 80 | |
| NAT/detection | |||||
| Principle of NAT detection | RT-PCR, TaqMan probes | RT-PCR, TaqMan probes | transcription-mediated an | plification | RT-PCR, TaqMan probes |
| NAT instrument | Rotorgene Q | cobas® 6800/8800 Systems | Procleix® Panther® System | Procleix® Tigris® System | Roche LightCycler 480 |
| Target (gene region) | none 3'UTR | 3'UTR | NN | 3'UTR | |
| Eluat/PCR volume, µl/µl | 25/50 | 25/50 | 100 % of the sample is processed and used in the amplification reaction | 30/75 | |
| Test specifications | |||||
| Analytical sensitivity | 30.2 (HCS) | 12,9b (lineage 1, HCS) | 11.9 copies/ml (HCS) | 8.9 copies/ml (HCS) | 15.4b, c (22.2 pool) |
| (95 % LOD), copies/ml based on standard 1) | (241.6 pool) | (245.1 pool)6.2b (lineage 2) | |||
| Specificity (WNV lineage) | quantitative detection of WNV lineage 1 and 2 | 100% WNV lineage 1 and 2 | 100% (N = 3,933) | 99.91% (N = 60,388) | qualitative detection of WNV lineage 1(abc) and 2 |
| Accomplishment | |||||
| Hands-on-time, min | 15 | 15 | 25 | ||
| Time-to-result, h | 4 | 3 | 3.5 | 3.5 | 5 |
| Throughput | 80 results (10 pools of 8 samples) in 4 h | 1,786 results (94 pools of 19 samples) in 3 h 384 results in 8 h shift - cobas® 6,800 | 5,775 results (275 pools of 21 samples) in 8 h 10,500 results (500 pools of 21 samples) in 12 h | 13,776 results (492 pools of 28 samples) in 9 hours and 27,776 results (992 pools of 28 samples) in 14 h | 1,056 in 9 h |
| Remarks | intended use includes cadaveric (non-heart beating) donors | ||||
With regard to PEI sensitivity requirement of 250 copies/ml.
Based on HCS: Health Canada WNV Reference Standard.
Based on WNV-Lineage 2-Isolat B956 (GenBank AY532665) Standard des Istituto Superiore di Sanita, Rome.
WNV-L1 has been identified in the majority of outbreaks in animals and humans in Europe [5]. In recent years a relevant spread of WNV-L2 across Europe and the Mediterranean Basin was observed, with zones where the circulation of the two lineages coexists. The WNV NAT assays used for blood screening detected positive donations of both lineages (table 5) and demonstrate good performance in routine testing. Even if the Procleix WNV assay showed a little different performance for both lineages due to possible mismatches in primers or probes [17], the 95% LOD is about 20 copies/ml for WNV-L2 according to the manufacturer's instruction, which is quite sufficient for routine WNV NAT screening. In most settings this assay is used in ID testing. Other WNV NAT assays, for example, the LightMix WNV RT-PCR Kit (TIB Molbiol) tested in our study, did not detect any WNV-L2 strain. Problems in the detection of WNV-L2 were noticed for the assays developed in-house in another EQA with both WNV lineages in 2010 [18]. In this study, 16 out of 30 laboratories failed to detect WNV-L2; therefore, improvement of some PCR assays is essential. Since WNV-L2 was recently introduced into Europe, the detection of both WNV lineages by NAT has been mandatory for diagnosis [19].
The analytical sensitivity (95% LOD) of WNV NAT assays ranges from 8.9 to 32 copies/ml. It has to be pointed out that the lack of an international NAT standard is the main problem. The determination of 95% LOD was performed using the HCS [20] that is no longer available. Therefore, secondary reference preparations or commercially available quantified controls both calibrated against HCS should be used. The NATrol NATWNV control (Zeptometrix), for example, is often used for this purpose. In our hands, the quantification against ISS or the novel PEI WNV-RNA reference preparations, both calibrated to HCS, proved an over-quantification of NATrol NAT WNV control with a factor up to 4.1. This, again, stresses the importance of establishing an international standard for WNV NAT.
The disadvantage of currently commercially available WNV NAT assays is their requirement of special screening platforms being fully integrated and automated and not as flexible as open NAT platforms. In our setting, we use the fully automated nucleic acid extraction method chemagic Viral DNA/RNA Kit that allows the processing of large plasma volumes. Due to small pools of 8 donors and restricted sample volume, we chose the 2.4 ml protocol. However, this method has the potential to isolate viral nucleic acids from MPs of up to 10 ml of plasma, which should potentially result in a higher sensitivity in combination with large pool sizes, as described for other viruses [21].
The pool size of 8 samples was selected to meet the mandated detection limit of 250 copies/ml and enable a cost-effective blood screening strategy.
In contrast to observations in WNV-endemic countries, such as the USA or Italy, WNV in Germany was not detected during ongoing surveillance programs in mosquitoes, birds, and blood donors. The seroprevalence (anti-WNV IgG) and WNV RNA incidence among 14,437 and 9,976 blood donors from Germany was analyzed in 2004 [22]. There was a high rate of initially anti-WNV reactives (5.9%), but only a few cases (0.03%) were confirmed as anti-WNV-positive by neutralization assay. No WNV RNA-positive blood donor was identified in this study. Following the first confirmed imported WNV infection in 2011, the number of imported WNV infections to Germany increased in 2012 [23]. Two cases of WNND and two cases of WNF were reported. The WNND cases were imported from Montenegro and Greece, including the first fatal case for Germany. The WNF cases were imported from Tunisia and Egypt. At present, there is no evidence for indigenous WNV infections in birds [24,25] or horses [26] in Germany. However, the presence of low-level viremic blood donors during epidemic periods and the impact of MP dilutions on sensitivity suggested the need for further improvements in sensitivity as well as a role for targeted ID NAT in epidemic regions [27].
In the USA, an adopted screening strategy is favored that changes from a MP to individual testing when the incidence of WNF increases [27,28]. In order to increase detection of low-level viremic units during epidemic periods, the reduction of the pool size was recommended whenever two MP-NAT reactive and a weekly rate of 1:1,000 occur [28]. Alternatively, initiating ID NAT in any region after the identification of four RNA-positive donations and consequent calculation of a detection frequency of 1:1,000 based on the date of collection of the first reactive donation was suggested [29]. In Italy, a national veterinary surveillance plan for WNV has been in place since 2002, following the first outbreak of WNV infection in horses reported in 1998 [16]. This plan included the monitoring of WNV circulation based on wild bird mortality as well as on entomological and sentinel animal surveillance, an on checking for WNV seroconversion in horses. The first 3 human cases of WNND were reported in 2008, and that led to the introduction of WNV NAT to test all blood, peripheral, bone marrow and cord blood stem cell donations collected in WNV-epidemic areas, and a nationwide 28-day deferral for blood donors who had spent at least 1 night in these areas. The safety measures for transfusion transmission of WNV (TT-WNV) were adapted from 2008 to 2013. Finally, in 2013, the Italian NBC implemented seasonal measures to prevent the WNV transmission through blood, including NAT in the period from July 15 to November 30. These preventive measures appear to have effectively improved the safety of the blood supply as no cases of TT-WNV have been reported so far [16]. In the 2014 season, first experience with the WNV MP-NAT screening has been made, demonstrating the feasibility in routine setting. Due to the small number of tested donations, only limited statements regarding the infectious risk of staying in WNV-endemic areas can be currently made for German blood donors. Possible future strategies could be a combination of phases of donor deferral and NAT testing to take into account the cost side. In future, a provision of donations after the peak tourist activity in October could be a possible variant in the WNV strategy. On the other hand, the satisfaction of donors also plays an important role. Donor deferral during holiday season, a season known to be problematic for the blood supply anyway (e.g. rare blood groups), might result in an undesired absence of donors for the next season due to previous demotivating experiences.
In conclusion, we demonstrated short-term and uncomplicated implementation of seasonal WNV NAT in routine blood donor screening in Germany. The NAT method was satisfactorily validated according to the PEI requirements using MPs of 8 donations. The requirement of an equal detection of both WNV lineages was proven by regular participation in EQA programs. Actually, the major problem is the lack of an international WNV NAT standard that will allow standardization of WNV NAT and reasonable comparability of NAT assays. Overall, the implementation of seasonal measures to prevent TT-WNV in Germany, including NAT, pathogen reduction techniques or blood donor deferral, is to date a special precaution; but in the future, TT-WNV could be more important because of environmental factors such as climate change and the increasing mobility of people. Therefore, continuous surveillance is important for the safety of the blood supply.
Disclosure Statement
The authors declared no conflict of interest.
Acknowledgements
The authors thank Bärbel Kammel, Birgit Drawe and staff of the Nucleic Acid Testing and Infectious Disease Screening Laboratories of Uni.Blutspendedienst OWL for their technical assistance and for donor testing results. We thank Philip Saunders for his linguistic advice.
References
- 1.Colpitts TM, Conway MJ, Montgomery RR, Fikrig E. West Nile virus: biology, transmission, and human infection. Clin Microbiol Rev. 2012;25:635–648. doi: 10.1128/CMR.00045-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pauli G, Bauerfeind U, Blumel J, Burger R, Drosten C, Groner A, Gurtler L, Heiden M, Hildebrandt M, Jansen B, Montag-Lessing T, Offergeld R, Seitz R, Schlenkrich U, Schottstedt V, Strobel J, Willkommen H. West Nile virus. Transfus Med Hemother. 2013;40:265–284. doi: 10.1159/000353698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention West Nile virus disease cases and deaths reported to CDC by year and clinical presentation, 1999-2014 www.cdc.gov/westnile/resources/pdfs/data/1-wnv-disease-cases-by-year_1999-2014_06042015.pdf, (last accessed September 11, 2015).
- 4.European Centre for Disease Prevention and Control Annual epidemiological report 2014 - emerging and vector-borne diseases http://ecdc.europa.eu/en/publications/_layouts/forms/Publication_DispForm.aspx? List=4f55ad51-4aed-4d32-b960-af70113dbb90&ID= 1196 (last accessed September 11, 2015).
- 5.European Centre for Disease Prevention and Control Epidemiological update: End of West Nile virus transmission season in Europe http://ecdc.europa.eu/en/press/news/_layouts/forms/News_DispForm.aspx? List=8db7286c-fe2d-476c-9133-18ff4cb1b568&ID= 1123 (last accessed September 11, 2015).
- 6.Working Group on Blood Safety and WNV West Nile virus and blood safety- introduction to a preparedness plan in Europe http://ec.europa.eu/health/blood_tissues_ organs/docs/wnv_preparedness_plan_2012.pdf (last accessed September 11, 2015).
- 7.Paul-Ehrlich Institut Abwehr von Arzneimittelrisiken - Anordnung des Ausschlusses von Blutspendern zur Verhinderung einer möglichen Übertragung des West-Nil-Virus durch zelluläre Blutprodukte oder gefrorenes Frischplasma Bundesanzeiger. 2003;180:21. [Google Scholar]
- 8.Paul-Ehrlich Institut Änderung der Anordnung vom 2. September 2003 über den Ausschluss von Blutspendern zur Verhinderung einer möglichen Übertragung des West-Nil-Virus durch zelluläre Blutprodukte oder gefrorenes Frischplasma. Bundesanzeiger. 2004;118:13. [Google Scholar]
- 9.Paul-Ehrlich Institut Bekanntmachung über die Zulassung von Arzneimitteln - Abwehr von Arzneimittelrisiken Stufe II - Änderung des Bescheides über die Anordnung des Ausschlusses von Blutspendern zur Verhinderung einer möglichen Übertragung des West-Nil-Virus durch nicht pathogen-inaktivierte Blutkomponenten. Bundesanzeiger 07.03.2014 B6.
- 10.Pai A, Kleinman S, Malhotra K, Lee-Haynes L, Pietrelli L, Saldanha J. Performance characteristics of the Food and Drug Administration-licensed Roche cobas TaqScreen West Nile virus assay. Transfusion. 2008;48:2184–2189. doi: 10.1111/j.1537-2995.2008.01861.x. [DOI] [PubMed] [Google Scholar]
- 11.European Medicines Agency (EMA) Note for guidance on validation of analytical procedures: Methodology (CPMP/ICH/281/95). EMA, 1995.
- 12.European Medicines Agency (EMA) Note for guidance on validation of analytical methods: Definitions and terminology (CPMP/ICH/ 381/95) EMA, 1995.
- 13.Linnen JM, Deras ML, Cline J, Wu W, Broulik AS, Cory RE, Knight JL, Cass MM, Collins CS, Giachetti C. Performance evaluation of the Procleix West Nile virus assay on semi-automated and automated systems. J Med Virol. 2007;79:1422–1430. doi: 10.1002/jmv.20930. [DOI] [PubMed] [Google Scholar]
- 14.Pisani G, Pupella S, Cristiano K, Marino F, Simeoni M, Luciani F, Scuderi G, Sambri V, Rossini G, Gaibani P, Pierro A, Wirz M, Grazzini G. Detection of West Nile virus RNA (lineages 1 and 2) in an external quality assessment programme for laboratories screening blood and blood components for West Nile virus by nucleic acid amplification testing. Blood Transfus. 2012;10:515–520. doi: 10.2450/2012.0036-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pisani G, Pupella S, Marino F, Gaggioli A, Sambri V, Rossini G, Wirz M, Grazzini G. Interlaboratory study to evaluate the performance of laboratories involved in West Nile virus RNA screening of blood and blood components by nucleic acid amplification testing in Italy. Blood Transfus. 2011;9:425–429. doi: 10.2450/2011.0025-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pupella S, Pisani G, Cristiano K, Catalano L, Grazzini G. West Nile virus in the transfusion setting with a special focus on Italian preventive measures adopted in 2008-2012 and their impact on blood safety. Blood Transfus. 2013;11:563–574. doi: 10.2450/2013.0077-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pisani G, Cristiano K, Pupella S, Grazzini G. Could the different performance of the Procleix West Nile virus assay towards WNV L1 and L2 represent an issue in the blood transfusion setting? Blood Transfus. 2014;12:619–620. doi: 10.2450/2014.0060-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Linke S, Mackay WG, Scott C, Wallace P, Niedrig M. Second external quality assessment of the molecular diagnostic of West Nile virus: are there improvements towards the detection of WNV? J Clin Virol. 2011;52:257–260. doi: 10.1016/j.jcv.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 19.Pupella S, Pisani G, Cristiano K, Catalano L, Grazzini G. Update on West Nile virus in Italy. Blood Transfus. 2014;12:626–627. doi: 10.2450/2014.0110-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saldanha J, Shead S, Heath A, Drebot M. Collaborative study to evaluate a working reagent for West Nile virus RNA detection by nucleic acid testing. Transfusion. 2005;45:97–102. doi: 10.1111/j.1537-2995.2005.04151.x. [DOI] [PubMed] [Google Scholar]
- 21.Hourfar MK, Schmidt M, Seifried E, Roth WK. Evaluation of an automated high-volume extraction method for viral nucleic acids in comparison to a manual procedure with preceding enrichment. Vox Sang. 2005;89:71–76. doi: 10.1111/j.1423-0410.2005.00649.x. [DOI] [PubMed] [Google Scholar]
- 22.Pfleiderer C, Blumel J, Schmidt M, Roth WK, Houfar MK, Eckert J, Chudy M, Menichetti E, Lechner S, Nubling CM. West Nile virus and blood product safety in Germany. J Med Virol. 2008;80:557–563. doi: 10.1002/jmv.21110. [DOI] [PubMed] [Google Scholar]
- 23.Gabriel M, Emmerich P, Frank C, Fiedler M, Rashidi-Alavijeh J, Jochum C, Gunther S, Auerhammer K, Rupprecht HJ, Blank RT, Sacher N, Pertzborn L, Stark K, Schrauzer T, Schmidt-Chanasit J. Increase in West Nile virus infections imported to Germany in 2012. J Clin Virol. 2013;58:587–589. doi: 10.1016/j.jcv.2013.08.027. [DOI] [PubMed] [Google Scholar]
- 24.Ziegler U, Seidowski D, Globig A, Fereidouni SR, Ulrich RG, Groschup MH. Sentinel birds in wild-bird resting sites as potential indicators for West Nile virus infections in Germany. Arch Virol. 2010;155:965–969. doi: 10.1007/s00705-010-0618-z. [DOI] [PubMed] [Google Scholar]
- 25.Timmermann U, Becker N. Mosquito-borne west nile virus (WNV) surveillance in the upper rhine valley, Germany. J Vector Ecol. 2010;35:140–143. doi: 10.1111/j.1948-7134.2010.00039.x. [DOI] [PubMed] [Google Scholar]
- 26.Ziegler U, Angenvoort J, Klaus C, Nagel-Kohl U, Sauerwald C, Thalheim S, Horner S, Braun B, Kenklies S, Tyczka J, Keller M, Groschup MH. Use of competition ELISA for monitoring of West Nile virus infections in horses in Germany. Int J Environ Res Public Health. 2013;10:3112–3120. doi: 10.3390/ijerph10083112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ziermann R, Sanchez-Guerrero SA. Procleix West Nile virus assay based on transcription-mediated amplification. Expert Rev Mol Diagn. 2008;8:239–245. doi: 10.1586/14737159.8.3.239. [DOI] [PubMed] [Google Scholar]
- 28.Custer B, Tomasulo PA, Murphy EL, Caglioti S, Harpool D, McEvoy P, Busch MP. Triggers for switching from minipool testing by nucleic acid technology to individual-donation nucleic acid testing for West Nile virus: analysis of 2003 data to inform 2004 decision making. Transfusion. 2004;44:1547–1554. doi: 10.1111/j.0041-1132.2004.04227.x. [DOI] [PubMed] [Google Scholar]
- 29.Stramer SL, Fang CT, Foster GA, Wagner AG, Brodsky JP, Dodd RY. West Nile virus among blood donors in the United States, 2003 and 2004. N Engl J Med. 2005;353:451–459. doi: 10.1056/NEJMoa044333. [DOI] [PubMed] [Google Scholar]


