Abstract
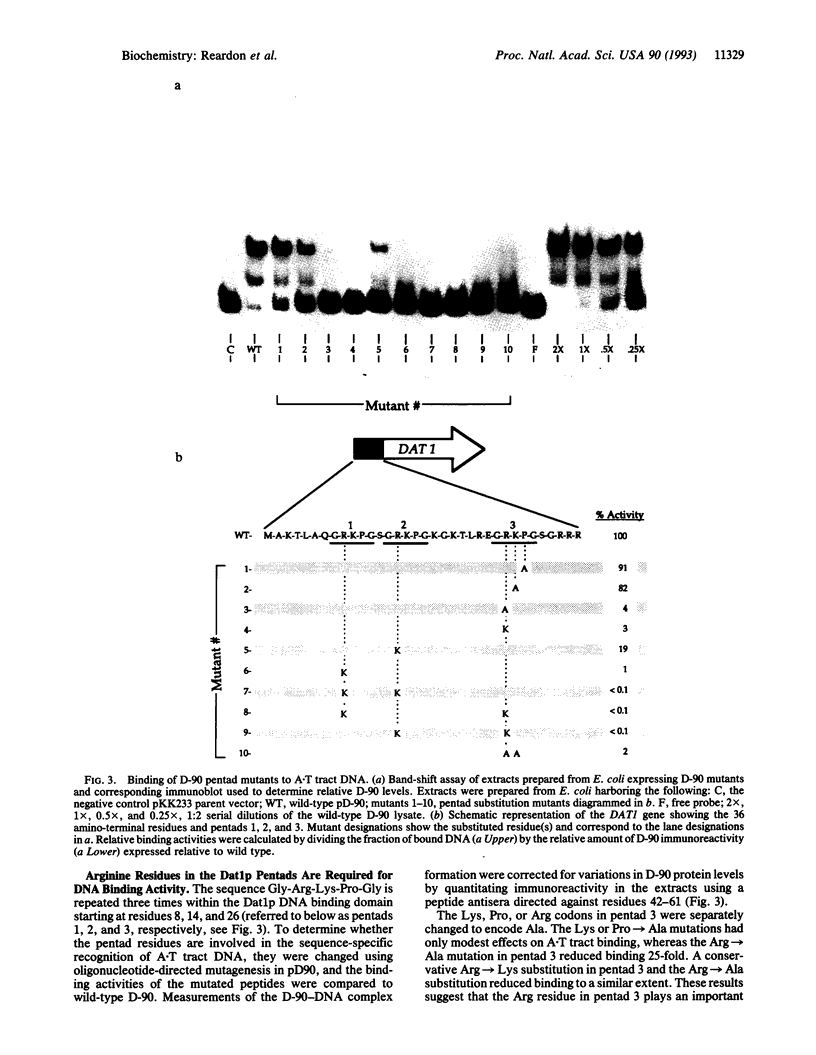
The DAT1 gene of Saccharomyces cerevisiae encodes a DNA binding protein that specifically interacts with nonalternating oligo(A).oligo(T) tracts (A.T tracts). Deletion analysis of DAT1 coding information showed that the amino-terminal 36 residues are sufficient for specific DNA binding activity. Furthermore, a 35-residue synthetic peptide corresponding to amino acids 2-36 bound to A.T tracts with an equilibrium dissociation constant of 4 x 10(-10) M. Within this region the pentad Gly-Arg-Lys-Pro-Gly is repeated three times. Mutational analysis revealed that the Arg side chains are required for high-affinity binding, whereas the other pentad side chains are dispensable. Chemical interference experiments showed that the DAT1 protein interacts with the minor groove of the double helix. The data suggest that the pentad arginines interact in a cooperative manner with a repeated minor groove feature of A.T tract DNA to achieve high-affinity recognition. Amino acid similarities with other DNA binding proteins suggest that the DAT1 protein pentad represents a specialized example of a widespread motif used by proteins to recognize A.T base pairs.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aggarwal A. K., Rodgers D. W., Drottar M., Ptashne M., Harrison S. C. Recognition of a DNA operator by the repressor of phage 434: a view at high resolution. Science. 1988 Nov 11;242(4880):899–907. doi: 10.1126/science.3187531. [DOI] [PubMed] [Google Scholar]
- Ashley C. T., Pendleton C. G., Jennings W. W., Saxena A., Glover C. V. Isolation and sequencing of cDNA clones encoding Drosophila chromosomal protein D1. A repeating motif in proteins which recognize at DNA. J Biol Chem. 1989 May 15;264(14):8394–8401. [PubMed] [Google Scholar]
- Ballard M. J., Tyndall W. A., Shingle J. M., Hall D. J., Winter E. Tyrosine phosphorylation of a yeast 40 kDa protein occurs in response to mating pheromone. EMBO J. 1991 Dec;10(12):3753–3758. doi: 10.1002/j.1460-2075.1991.tb04944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Tabor S., Struhl K. Distinguishing between mechanisms of eukaryotic transcriptional activation with bacteriophage T7 RNA polymerase. Cell. 1987 Sep 25;50(7):1047–1055. doi: 10.1016/0092-8674(87)90171-1. [DOI] [PubMed] [Google Scholar]
- Churchill M. E., Travers A. A. Protein motifs that recognize structural features of DNA. Trends Biochem Sci. 1991 Mar;16(3):92–97. doi: 10.1016/0968-0004(91)90040-3. [DOI] [PubMed] [Google Scholar]
- Coll M., Frederick C. A., Wang A. H., Rich A. A bifurcated hydrogen-bonded conformation in the d(A.T) base pairs of the DNA dodecamer d(CGCAAATTTGCG) and its complex with distamycin. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8385–8389. doi: 10.1073/pnas.84.23.8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGabriele A. D., Sanderson M. R., Steitz T. A. Crystal lattice packing is important in determining the bend of a DNA dodecamer containing an adenine tract. Proc Natl Acad Sci U S A. 1989 Mar;86(6):1816–1820. doi: 10.1073/pnas.86.6.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K. R., Lehn D. A., Elton T. S., Barr P. J., Reeves R. Complete murine cDNA sequence, genomic structure, and tissue expression of the high mobility group protein HMG-I(Y). J Biol Chem. 1988 Dec 5;263(34):18338–18342. [PubMed] [Google Scholar]
- Karlin S., Blaisdell B. E., Sapolsky R. J., Cardon L., Burge C. Assessments of DNA inhomogeneities in yeast chromosome III. Nucleic Acids Res. 1993 Feb 11;21(3):703–711. doi: 10.1093/nar/21.3.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissinger C. R., Liu B. S., Martin-Blanco E., Kornberg T. B., Pabo C. O. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 A resolution: a framework for understanding homeodomain-DNA interactions. Cell. 1990 Nov 2;63(3):579–590. doi: 10.1016/0092-8674(90)90453-l. [DOI] [PubMed] [Google Scholar]
- Laughon A. DNA binding specificity of homeodomains. Biochemistry. 1991 Dec 3;30(48):11357–11367. doi: 10.1021/bi00112a001. [DOI] [PubMed] [Google Scholar]
- Levinger L., Varshavsky A. Protein D1 preferentially binds A + T-rich DNA in vitro and is a component of Drosophila melanogaster nucleosomes containing A + T-rich satellite DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7152–7156. doi: 10.1073/pnas.79.23.7152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lue N. F., Buchman A. R., Kornberg R. D. Activation of yeast RNA polymerase II transcription by a thymidine-rich upstream element in vitro. Proc Natl Acad Sci U S A. 1989 Jan;86(2):486–490. doi: 10.1073/pnas.86.2.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson H. C., Finch J. T., Luisi B. F., Klug A. The structure of an oligo(dA).oligo(dT) tract and its biological implications. Nature. 1987 Nov 19;330(6145):221–226. doi: 10.1038/330221a0. [DOI] [PubMed] [Google Scholar]
- Ner S. S. HMGs everywhere. Curr Biol. 1992 Apr;2(4):208–210. doi: 10.1016/0960-9822(92)90541-h. [DOI] [PubMed] [Google Scholar]
- Otting G., Qian Y. Q., Billeter M., Müller M., Affolter M., Gehring W. J., Wüthrich K. Protein--DNA contacts in the structure of a homeodomain--DNA complex determined by nuclear magnetic resonance spectroscopy in solution. EMBO J. 1990 Oct;9(10):3085–3092. doi: 10.1002/j.1460-2075.1990.tb07505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves R., Nissen M. S. The A.T-DNA-binding domain of mammalian high mobility group I chromosomal proteins. A novel peptide motif for recognizing DNA structure. J Biol Chem. 1990 May 25;265(15):8573–8582. [PubMed] [Google Scholar]
- Seeman N. C., Rosenberg J. M., Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc Natl Acad Sci U S A. 1976 Mar;73(3):804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluka J. P., Horvath S. J., Glasgow A. C., Simon M. I., Dervan P. B. Importance of minor-groove contacts for recognition of DNA by the binding domain of Hin recombinase. Biochemistry. 1990 Jul 17;29(28):6551–6561. doi: 10.1021/bi00480a002. [DOI] [PubMed] [Google Scholar]
- Solomon M. J., Strauss F., Varshavsky A. A mammalian high mobility group protein recognizes any stretch of six A.T base pairs in duplex DNA. Proc Natl Acad Sci U S A. 1986 Mar;83(5):1276–1280. doi: 10.1073/pnas.83.5.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Naturally occurring poly(dA-dT) sequences are upstream promoter elements for constitutive transcription in yeast. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8419–8423. doi: 10.1073/pnas.82.24.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm R., Baumruker T., Franza B. R., Jr, Herr W. A 100-kD HeLa cell octamer binding protein (OBP100) interacts differently with two separate octamer-related sequences within the SV40 enhancer. Genes Dev. 1987 Dec;1(10):1147–1160. doi: 10.1101/gad.1.10.1147. [DOI] [PubMed] [Google Scholar]
- Weir H. M., Kraulis P. J., Hill C. S., Raine A. R., Laue E. D., Thomas J. O. Structure of the HMG box motif in the B-domain of HMG1. EMBO J. 1993 Apr;12(4):1311–1319. doi: 10.1002/j.1460-2075.1993.tb05776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter E., Varshavsky A. A DNA binding protein that recognizes oligo(dA).oligo(dT) tracts. EMBO J. 1989 Jun;8(6):1867–1877. doi: 10.1002/j.1460-2075.1989.tb03583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]