Abstract
Background:
There are more than 300 genetic loci that have been found to be related to hereditary hearing impairment (HHI), including 92 causative genes for nonsyndromic hearing loss, among which 34 genes are related to autosomal dominant nonsyndromic HHI (ADNSHHI). Traditional linkage analysis and candidate gene sequencing are not effective at detecting the ADNSHHI, especially for the unconditional families that may have more than one pathogenic cause. This study identified two disease-causing genes TJP2 and GJB2 in a Chinese family with unconditional ADNSHHI.
Methods:
To decipher the genetic code of a Chinese family (family 686) with ADNSHHI, different gene screening techniques have been performed, including linkage analysis, candidate genes screening, high-throughput sequencing and Sanger sequencing. These techniques were done on samples obtained from this family over a period of 10 years.
Results:
We identified a pathogenic missense mutation, c. 2081G>A (p.G694E), in TJP2, a gene that plays a crucial role in apoptosis and age-related hearing loss (ARHL). The mutation was co-segregated in this pedigree in all, but not in the two patients who presented with different phenotypes from the other affected family members. In one of the two patients, we confirmed that the compound heterozygosity for p. Y136* and p.G45E in the GJB2 gene may account for the phenotype shown in this patient.
Conclusions:
We identified the co-occurrence of two genetic causes in family 686. The possible disease-causing missense mutation of TJP2 in family 686 presents an opportunity for further investigation into ARHL. It is necessary to combine various genes screening methods, especially for some unconventional cases.
Keywords: Autosomal Dominant Nonsyndromic Hereditary Hearing Impairment, GJB2, TJP2
INTRODUCTION
Hearing impairment is the most common sensory deficit in the world. Hearing loss is caused by genetic factors in more than 60% of the patients. Until date, more than 300 genetic loci have been found to be related with nonsyndromic hearing loss. Totally 92 of these disease-causing genes have been identified (http://hereditaryhearingloss.org). The strategy used to identify causes of hereditary hearing impairment (HHI) is continuously improving. Traditional linkage analysis and candidate gene sequencing have been proven to be useful in identifying the genes that are responsible for autosomal dominant nonsyndromic HHI (ADNSHHI). However, this method is not effective to detect all of the cases of ADNSHHI. Mapping the genes that are responsible for deafness is usually followed by finding the causative gene within the mapped locus. Recently, next generation high-throughput sequencing (NGS) has been verified to be a valuable molecular diagnostic tool for HHI patients.[1]
TJP2, also known as ZO2, encodes the zonula occludens 2, which belongs to the family of the membrane-associated guanylate kinase homolog with three PDZ domains, one SH3 module, and a GuK domain.[2] It is predominantly expressed at the apical region between the hair cells and supporting cells in the organ of Corti. Its expression decreases with age from embryonic development to adulthood, aiding the maintenance of the barrier between the endolymph and perilymph. TJP2 overexpression is associated with the induction of apoptosis, which was the reason of ADNSHHI in a family of Jewish ancestry.[3] A missense mutation or a more complex gain-of-function mutation of TJP2 may be a viable and identifiable cause of ADNSHHI in human patients.[4] However, complete loss of TJP2 leads to embryonic lethality and hence, TJP2 knockout mice are not viable.[5]
Our research group has identified a Chinese genealogy family (family 686) with ADNSHHI in five generations. Through a series study conducted over the last 10 years, we deciphered the genetic code of an unconditional Chinese family with ADNSHHI, while simultaneously demonstrating the numerous strategies for understanding the pathogenesis of HHI.
METHODS
This study was approved by the Committee of Medical Ethics of Chinese People's Liberation Army (PLA) General Hospital. We obtained written informed consent from all the participants in this study. Written informed consents were obtained from the next of kin on the behalf of the minors/children participants involved in this study.
Family recruitment and clinical evaluations
A five-generation pedigree with 99 members showing ADNSHHI was ascertained from the Department of Otolaryngology, Head and Neck Surgery, at the Institute of Otolaryngology of PLA, Chinese PLA General Hospital 10 years ago [Figure 1a]. Twenty-one members of the family chosen participated in our study. Personal and familial medical history, including hearing loss, tinnitus, vestibular symptoms, use of aminoglycosides, and other clinical abnormalities was taken by a team of experienced doctors and audiologists, Audiometric evaluations included pure tone audiometry, auditory brainstem responses, and distortion product otoacoustic emissions. High-resolution computed tomography (HRCT) was also performed on some subjects to verify whether the family members had other complications other than hearing disorders.
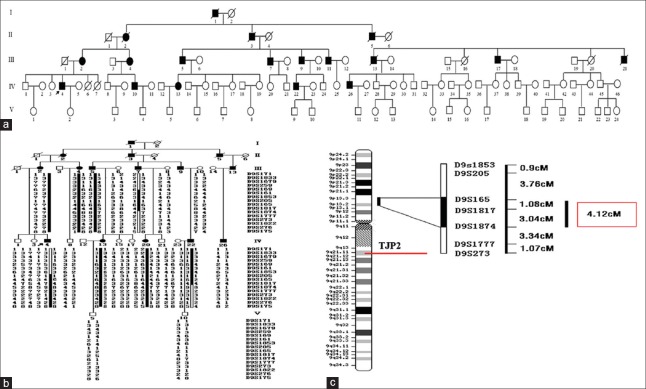
Figure 1.
Pedigrees and haplotype analysis of family 686. (a) Pedigrees of family 686. Filled symbols for males (squares) and female (circles) represent affected individuals, and empty represent unaffected individuals. (b) Single nucleotide polymorphism haplotype analysis in family 686. A total of four recombination events were observed in Family 686, including two recombination events in III7 between D9S259 and D9S169, D9S1874 and D9S1777 respectively, one recombination event in IV4 in which the exchange was between D9S169 and D9S1853, and one significant recombination in V5 in which the exchange occurred at D9S205. V5 carried the disease-causing allele from her affected mother at D9S205. D9S205 was excluded because V5 had normal hearing. (c) Construction of the genetics map. Figure shows the cytogenetic chromosome bands and regional distribution from the 9p13.2 to the 9p13.3 region. Black mark column on the right of the chromosomal region is the positioning region of 686 lines, locating between 9p13.2 and 9p13.3. Distance between markers is the genetic distance. The red box shows the distance positioning segment.
Linkage analysis and candidate gene screening
We performed a genome-wide linkage scan, fine-mapping, and haplotype analysis on family 686. Genomic DNA from 21 participants have been used for genetic analysis (family members III2, III4, III5, III6, III7, III8, III10, IV1, IV3, IV4, IV8, IV13, IV15, IV17, IV20, IV21, IV22, IV23, IV26, V5, and V10). The polymorphic loci single nucleotide polymorphism (SNP) microsatellite markers came from the PRISM® Linkage Mapping Sets-MD10 Version 2.0 kit (ABI, USA). A genome-wide screening was carried out using 386 microsatellite markers, distributed with an average spacing of 10 centimorgan on 22 chromosomes, excluding X and Y (ABI Prism Linkage Mapping Set 2, Applied Biosystems, Foster City, CA, USA). Multiplex polymerase chain reaction (PCR) was performed following the manufacturer's recommendations. We followed the procedures using a PE9600 thermocycler made by Applied Biosystems®. PCR products were loaded onto a 6% denaturing polyacrylamide gel (7M urea) and visualized on an ABI Prism® 3700 sequencer. Alleles were analyzed with ABI GeneMapper software, version 3.0 (http://depts.washington.edu/bsc/genetics/GeneMapper.html).
Further screening within the linked region on chromosome 9 was performed with markers from the Marshfield 9 map (http://research.Marshfieldclinic.org/genetics) for fine mapping. Linkage analysis between the disease locus and the markers was performed using the MLINK program of the LINKAGE version 5.1 software package (http://linkage.rockefeller.edu/pub/). The two-point Log odds (LOD) score was calculated under a 90% penetrance autosomal dominant mode of inheritance, considering the genetic heterogeneity of hereditary hearing loss, and setting the disease allele frequency of 0.0001. Haplotype analysis was constructed using Cyrillic software, version 2.1 (Cyrillic Software, Wallingford, UK).
In accordance with the linkage analysis results, the AQP3 gene was chosen as the candidate gene and Sanger sequencing was first performed in one affected family member and one unaffected member. Design primers were used to amplify the six exons of AQP3 [Supplementary Table 1]. According to standard conditions, we completed all the PCRs. Bi-directional sequencing was carried out using both the forward and reverse primers and was performed using the ABI PRISM Big Dye DNA-sequencer (Applied Biosystems, USA).
Supplementary Table 1.
Primer sequences of AQP3
| Exon | Product length (bp) | TM (°C) | Primer sequences (5’–3’) | |
|---|---|---|---|---|
| Exon 1 | 513 | 70–65 | F | AGGCCACCCGTCCCTCAAAGCTCCT |
| R | GCGGTTAAGCGTGGGGGTCACAGCT | |||
| Exon 2 | 300 | 57 | F | GCATGTTGCTGGCTTCAC |
| R | CTGTGACCTGCCCTTAGGAA | |||
| Exon 3 | 300 | 57 | F | CTCTCTGCACCCCTTCTCAG |
| R | TGCCCAACTTGTTTCTTTCC | |||
| Exon 4 | 500 | 59 | F | CCTCTGCCTGCTGCAATAC |
| R | CTTGCCACCATGTTCTGATG | |||
| Exon 5 and 6 | 600 | 59 | F | AGCACCATTGCTTTCAGGAG |
| R | CCCTTGGACAGTCAGTGGAT | |||
Targeted genes capture and high-throughput sequencing
A customized capture array (NimbleGen, Roche) was performed on the proband, IV26 who had a different phenotype from other affected family members, was then added to sequence. The array can capture exons, splicing sites and immediate flanking intron sequences of 307 human kayo genes responsible for either human or mouse deafness and all the mitochondrial genes [Supplementary Table 2]. The targeted region captured by array was constructed a library and then sequenced on Illumina HiSeq2000 to generate paired end reads (90 bps at each end). Raw image files were processed by Illumina Pipeline (version 1.3.4) for base-calling with default parameters. Reads were aligned to NCBI37/hg19 reference human genome using BWA (http://bio-bwa.sourceforge.net/index.shtml). SNPs and InDels (inserts and deletions) were detected using GATK (http://www.broadinstitute.org/gatk/index.php).[6]
Supplementary Table 2.
Targeted captured genes list
| Gene name |
|---|
| ABR |
| ACAN |
| ACTG1 |
| AIFM1 |
| AKAP12 |
| ALDH1A2 |
| ALMS1 |
| AP3D1 |
| APAF1 |
| APOA1 |
| AQP4 |
| ATF2 |
| ATOH1 |
| ATP2B2 |
| ATP8B1 |
| AXIN1 |
| BARHL1 |
| BBS1 |
| BBS4 |
| BCR |
| BDNF |
| BMP4 |
| BSN |
| BSND |
| C17orf48 |
| C1orf125 |
| CACNA1D |
| CACNB2 |
| CACNG2 |
| CASP3 |
| CCDC50 |
| CD36 |
| CDH23 |
| CDKN1B |
| CDKN2D |
| CEACAM16 |
| CELSR1 |
| CHD7 |
| chrM |
| CHRNA9 |
| CKB |
| CLDN11 |
| CLDN14 |
| CLDN9 |
| CLIC5 |
| CLRN1 |
| COCH |
| COL11A1 |
| COL11A2 |
| COL2A1 |
| COL4A3 |
| COL4A4 |
| COL4A5 |
| COL9A1 |
| COL9A2 |
| CPLX1 |
| CRYM |
| DBH |
| DDR1 |
| DFNA5 |
| DFNB31 |
| DFNB59 |
| DIABLO |
| DIAPH 1 |
| DIAPH 3 |
| DIO2 |
| DIO3 |
| DLX2 |
| DLX5 |
| DMD |
| DNAH7 |
| DNAH9 |
| DPYS |
| DSPP |
| DVL1 |
| DVL2 |
| DVL3 |
| EDN3 |
| EDNRB |
| EGFLAM |
| EPHB1 |
| EPHB2 |
| EPHB3 |
| ERBB4 |
| ESPN |
| ESR2 |
| ESRRB |
| EYA1 |
| EYA4 |
| FABP4 |
| FAS |
| FBXO2 |
| FGF20 |
| FGF3 |
| FGFR1 |
| FGFR2 |
| FGFR3 |
| FIGN |
| FOXG1 |
| FOXI1 |
| FXN |
| FZD3 |
| FZD6 |
| GAS7 |
| GATA3 |
| GBX2 |
| GFI1 |
| GIPC3 |
| GJA1 |
| GJB1 |
| GJB2 |
| GJB3 |
| GJB6 |
| GLI3 |
| GOT1L1 |
| GPR98 |
| GPSM2 |
| GPX1 |
| GRHL2 |
| GRID1 |
| GRXCR1 |
| GUSB |
| HAL |
| HES1 |
| HES5 |
| HGF |
| HMX2 |
| HMX3 |
| HOXA1 |
| HOXA2 |
| HOXB1 |
| HS6ST2 |
| IFT88 |
| IGF1 |
| ILDR1 |
| ITGA8 |
| JAG1 |
| JAG2 |
| KCNE1 |
| KCNJ10 |
| KCNMA1 |
| KCNQ1 |
| KCNQ4 |
| KIT |
| KITLG |
| LAMA2 |
| LARGE |
| LFNG |
| LHFPL5 |
| LMO4 |
| LMX1A |
| LOXHD1 |
| LRIG3 |
| LRP2 |
| LRTOMT |
| MAFB |
| MAP1A |
| MARVELD2 |
| MCOLN3 |
| MGAT4B |
| MIR182 |
| MIR183 |
| MIR96 |
| MITF |
| MKKS |
| MON2 |
| MOS |
| MPV17 |
| MPZ |
| MSRB3 |
| MSX2 |
| MTAP |
| MUC4 |
| MUC6 |
| MUTED |
| MYH1 |
| MYH13 |
| MYH14 |
| MYH2 |
| MYH3 |
| MYH4 |
| MYH8 |
| MYH9 |
| MYO15A |
| MYO1A |
| MYO3A |
| MYO6 |
| MYO7A |
| NAV2 |
| NAV3 |
| NDP |
| NDRG1 |
| NEFL |
| NEU1 |
| NEURL |
| NEUROD1 |
| NEUROG1 |
| NF1 |
| NOTCH1 |
| NOX3 |
| NOXO1 |
| NR4A3 |
| NTF3 |
| NTN1 |
| NTRK2 |
| NTRK3 |
| OC90 |
| OPA1 |
| OR2T4 |
| OTOA |
| OTOF |
| OTOG |
| OTOP1 |
| OTX1 |
| OTX2 |
| PAX2 |
| PAX3 |
| PCDH15 |
| PDE8B |
| PDSS1 |
| PDZD7 |
| PHEX |
| PLDN |
| PMP22 |
| PNOC |
| POU1F1 |
| POU3F4 |
| POU4F3 |
| PROP1 |
| PRPS1 |
| PRRX1 |
| PRRX2 |
| PTK7 |
| PTPRQ |
| RARA |
| RARB |
| RARG |
| RASA1 |
| RDX |
| S1PR2 |
| SCARB2 |
| SCO1 |
| SCRIB |
| SEMA3E |
| SERAC1 |
| SERPINB6 |
| SFTPC |
| SIX1 |
| SIX5 |
| SLC12A2 |
| SLC12A6 |
| SLC12A7 |
| SLC17A8 |
| SLC19A2 |
| SLC1A3 |
| SLC26A4 |
| SLC26A5 |
| SLC30A4 |
| SLC4A11 |
| SLC4A7 |
| SLC9A1 |
| SLCO2B1 |
| SMAD4 |
| SMPX |
| SMS |
| SNAI2 |
| SOBP |
| SOD1 |
| SORBS1 |
| SOX10 |
| SOX2 |
| SOX9 |
| SPRY2 |
| ST3GAL5 |
| STRC |
| TAF10 |
| TBX1 |
| TBX10 |
| TCOF1 |
| TECTA |
| TGFA |
| TGFB2 |
| THRA |
| THRB |
| TIMM8A |
| TJP2 |
| TMC1 |
| TMEM126A |
| TMEM220 |
| TMIE |
| TMPRSS13 |
| TMPRSS3 |
| TNC |
| TNFRSF11B |
| TPRN |
| TRIOBP |
| TRPV4 |
| TSHR |
| TUB |
| TYRP1 |
| UCN |
| USH1C |
| USH1G |
| USH2A |
| USP15 |
| VANGL2 |
| WFS1 |
| YME1L1 |
ABR: Auditory brainstem responses.
Sanger sequencing
After filtering against common SNPs reported by public databases, Sanger sequencing was used in all available members from family 686 to determine whether the potential mutation c. 2081G>A in TJP2 co-segregated with the disease phenotype in these individuals or not. Direct PCR products were sequenced using Bigdye terminator version 3.1 cycle sequencing kits (Applied Biosystems. Foster City, CA) and analyzed using ABI 3700XL Genetic Analyzer.
A total of 100 DNA samples from a panel of unaffected individuals from a Chinese background made up the control genomic DNA samples. BLAST was applied to compare the alignment of the TJP2 protein between different species.
RESULTS
Clinical description
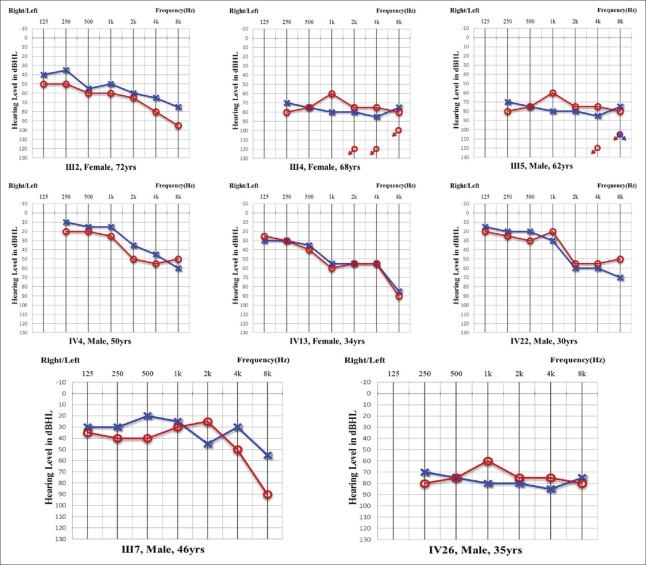
A total of 21 members in the family 686, composed of 8 clinically affected and 13 unaffected individuals, were recruited in this study. Age of onset in the family ranged from 21 to 68 years old. Affected members showed a postlingual, symmetrical, and bilateral nonsyndromic sensorineural hearing loss. Most of the patients had relatively consistent audiograms, except for III7 and IV26, initially presenting at 2 kHz, 4 kHz, and 8 kHz. The hearing loss of these three frequencies declined rapidly in the first decade, with the thresholds worse than 40–50 dB HL. The audiograms were the downslope type. The hearing impairments then implicated the 1 kHz and the 0.25 kHz frequencies, and finally 0.5 kHz. The subsequent progression of hearing loss to a severe level at all frequencies at a later age was gradual in most cases. All patients had associated lofty-tone tinnitus, but no vestibular symptoms or signs were reported [Table 1 and Figure 2]. HRCT results of the temporal bone in some of the patients showed normal middle ears structure, including a normal vestibular aqueduct and internal auditory canal. None of the affected members had a history of exposure to aminoglycosides, noise, or other causes that may account for the hearing impairment.
Table 1.
Summary of clinical data for hearing impaired members in family 686
| Subject | Gender | Age of test (years) | Age of onset (years) | PTA (dB HL)* | Hearing impairment† | Audiogram | Tinnitus |
|---|---|---|---|---|---|---|---|
| III: 2 | Female | 72 | 68 | Left: 57.50 | Moderate | Downslope | Left: Durative |
| Right: 66.25 | Moderate | Downslope | Right: Durative | ||||
| III: 4 | Female | 68 | 48 | Left: 72.50 | Severe | Downslope | Left: Durative |
| Right: 93.75 | Severe | Downslope | Right: Durative | ||||
| III: 5 | Male | 62 | 55 | Left: 90.00 | Severe | Downslope | Left: Durative |
| Right: 90.00 | Severe | Downslope | Right: Durative | ||||
| III: 7 | Male | 46 | 42 | Left: 30.00 | Mild | Irregular | Left: Occasional |
| Right: 36.50 | Mild | Irregular | Right: Occasional | ||||
| IV: 4 | Male | 50 | 45 | Left: 45.00 | Moderate | Downslope | Left: Durative |
| Right: 42.50 | Moderate | Downslope | Right: Durative | ||||
| IV: 13 | Female | 34 | 31 | Left: 50.00 | Moderate | Downslope | Left: Occasional |
| Right: 52.50 | Moderate | Downslope | Right: Occasional | ||||
| IV: 22 | Male | 30 | 21 | Left: 45.00 | Moderate | Downslope | Left: Occasional |
| Right: 40.00 | Moderate | Downslope | Right: Occasional | ||||
| IV: 26 | Male | 35 | 32 | Left: 78.75 | Severe | Flat | Left: Durative |
| Right: 71.25 | Severe | Flat | Right: Durative |
*PTA: Pure-tone air-conduction averages (0.5, 1, 2 and 4 kHz) for the better-hearing ear of affected subjects in family 727; †Diagnosed at the time of test. The severity of hearing impairment was defined as mild (26–40 dB HL), moderate (41–55 dB HL), moderately severe (56–70 dB HL), severe (71–90 dB HL) and profound (>90 dB HL). HL: Hearing level.
Figure 2.
Audiograms of both ears from affected subjects in family 686. Symbols “o” and “x” denote air conduction pure-tone thresholds at different frequencies in the right and left ear, respectively. dB: decibels; Hz: Hertz.
Linkage analysis and candidate gene sequencing
The genome-wide linkage study located the hearing impairment gene on the short arm of chromosome 9 (9p13.2-p13.3) and excluded the remainder of the genome. The maximum two-point LOD score of 3.25 (θ = 0) was observed at marker D9S1817, between D9S171 and D9S175. Fine mapping of the gene with dense microsatellite markers from the initially targeted area refined the position of the section to D9S165 and D9S1874, between which the genetic distance is 4.12 centimorgan (cM). This candidate region showed no overlap with the critical intervals identified in the previously studied families [Table 2].
Table 2.
Two-point LOD scores between 9q microsatellite markers for family 686
| Markers | LOD score ATθ | Zmax | θmax | |||||
|---|---|---|---|---|---|---|---|---|
| 0.0 | 0.1 | 0.2 | 0.3 | 0.4 | 0.5 | |||
| D9s259 | −1.74 | 1.23 | 0.96 | 0.59 | 0.25 | 0.00 | 1.25 | 0.0790 |
| D9s169 | 0.31 | 2.46 | 1.95 | 1.28 | 0.57 | 0.00 | 2.55 | 0.0570 |
| D9s161 | 1.21 | 0.88 | 0.58 | 0.33 | 0.13 | 0.00 | 1.21 | 0.0000 |
| D9s1853 | −5.82 | −0.59 | −0.06 | 0.09 | 0.09 | 0.00 | 0.10 | 0.3480 |
| D9s205 | 0.06 | 0.09 | 0.04 | −0.01 | −0.03 | 0.00 | 0.09 | 0.0670 |
| D9s165 | −2.10 | 0.60 | 0.58 | 0.40 | 0.19 | 0.00 | 0.63 | 0.1370 |
| D9s1817 | 3.25 | 2.58 | 1.89 | 1.19 | 0.53 | 0.00 | 3.25 | 0.0000 |
| D9s1874 | −5.82 | 0.43 | 0.84 | 0.76 | 0.43 | 0.00 | 0.85 | 0.2220 |
| D9s1777 | −2.17 | 0.69 | 0.55 | 0.35 | 0.16 | 0.00 | 0.69 | 0.0940 |
| D9s273 | 0.80 | 0.67 | 0.45 | 0.24 | 0.09 | 0.00 | 0.80 | 0.0000 |
| D9s1822 | −5.23 | 0.56 | 0.56 | 0.34 | 0.12 | 0.00 | 0.61 | 0.1440 |
LOD: Log odds score, the LOD scores were computed under an autosomal dominant mode of inheritance.
A total of four recombination events were observed in family 686. The gene was located between the D9S165 and D9S1874 interval, which is about 4.12 cM (the genetic distance was calculated from http://www.ensemble.org/per1) [Figure 1b and 1c].
Twenty-one family members were screened for AQP3, including 8 affected members and 13 unaffected members. Two polymorphisms in AQP3 were found: 390C>T/390C>T and 394G>A/WT. All the members in this family have 390C>T/390C >T and only four members have 394G>A/WT, including three patients and one unaffected member, who was 32 years old. The latter mutation can change the 132nd amino acid from Asp to Asn, possibly leading to changes in both the functionality and spatial structure of the resulting protein. It is reasonable to assume that the unaffected member with this functional polymorphism might show the disease at an older age. However, not all of the patients in this family had this variant.
Targeted genes capture and high-throughput sequencing
Approximately, 1.44 Mbp of the exons and adjacent intronic regions of the 307 genes were captured and sequenced. The average sequencing depth for the target region was approximately 468X, and the average coverage for the targeted region with >20X was 97.51%, satisfying the requirements for identifying SNPs and InDels. For the proband of family 686, through function and frequency filtration, the missense variant c. 2081G>A (p.G694E) in exon14 of TJP2 (NM_201629.3) was identified and no other possibly pathogenic variants were found. In IV26, who was an affected member without TJP2 c. 2081G>A [Supplementary Figure 1 (1.1MB, tif) ], two previously reported missense mutations, c. 408C>A (p.Y136*) and c. 134G>A (p.G45E) in GJB2, were identified [Supplementary Figure 2 (804.3KB, tif) ].[7]
DNA sequence chromatograms showing the heterozygous missense mutation c.2081G>A of TJP2 gene in affected individuals III2, III4, III5, IV4, IV13, IV22. IV8 was the unaffected control in this family. III7 and IV26 were affected members without the TJP2 c.2081G>A mutation.
DNA sequence chromatograms showing the two heterozygous missense mutations c.134C>A and c.408G>A of GJB2 gene in affected individuals (lower panel) compared with the wild type controls (upper panel).
Mutation detection and analysis
All identified family members were tested for c. 2081G>A. Results showed that all the patients carried the variant except for III7 and IV26, while none of the unaffected members of the family carried the variant [Supplementary Figure 1 (1.1MB, tif) ]. Sanger sequencing confirmed the co-segregation of p.G694E with the disease phenotype in family 686, with the exception of III7 and IV26. The TJP2 mutation occurred at highly conserved amino acids, and the SIFT, Polyphen2, LRT, Mutation Taster, GERP++, and PhyloP programs all predicted it to be deleterious. The amino acid sequence encoded by TJP2 was compared between different species, including mouse, rat, dog, cattle, and human, and it was found that c. 2081G>A (p.G694E) is located in a highly conserved region.
No TJP2 c. 2081G>A variation was identified in 100 control genomic DNA samples from a panel of unaffected individuals [Supplementary Figure 3 (1.9MB, tif) ]. Based on these results and the phenotypes of family 686, we concluded that the mutations in TJP2 and GJB2 are responsible for hearing loss in this family.
A flow chart of the series studies of Family 686.
DISCUSSION
Co-occurrence of disease-causing mutations identified in family 686
To the best of our knowledge, this study is the third report on disease-causing missense mutations in TJP2 in patients with HHI.[8,9] The c. 2081G>A mutation that was predicted to be pathogenic was detected in six patients from the family 686, and was absent in unaffected members. This mutation caused a transition from a guanine to adenine at nucleotide position 2081, causing a glycine to glutamic acid substitution at amino acid position 694 (p.G694E). This variant was initially reported as an SNP.[9] However, the frequency of this mutation in dbSNP and 1k genome database was 0.0000 and 0.0009, respectively, suggesting that this mutation may be disease-causing.
Two patients, III7 and IV26, did not carry the c. 2081G>A mutation. Further analysis of the phenotypes of these two patients revealed that they presented with certain symptoms different from those found in other patients in the family. The hearing curve for patient III7 was different from other affected members in this family in two ways: (1) High-frequency hearing impairment at 2 kHz and 4 kHz was not obvious and thresholds were much better than at 8 kHz, while other patients from family 686 had a more synchronized hearing decline, (2) low-frequency hearing loss occurred earlier than in other family members. Hearing loss caused by TJP2 mutation presented as late-onset and typically involved the high frequencies first. However, III7 had no characteristics typical of hearing loss caused by TJP2 mutation, making it difficult to elucidate its cause. Therefore, hearing impairment of III7 may have been caused by other reasons.
In the case of patient IV26, hearing impairment began at the age of 32 years, and the hearing curve showed bilateral, severe hearing loss across all frequencies at the age of 35 years, progressing rapidly during the first 10 years of onset. The patient did not present with the pattern of high-frequency hearing being affected first, followed by all other frequencies. His hearing threshold was much higher than that of other patients in this family. Further investigation using targeted gene capture and NGS showed that this patient was a compound heterozygote for two mutations in the GJB2 gene, a condition that has been reported to cause HHI. ADNSHHI has complicated genotype-phenotype correlations because of its extreme heterogeneity.[4] The heterogeneity observed in one family was also reported in more than ten other families who experienced hearing loss.[10]
Until date, three missense mutations in TJP2 and a tandem inverted genomic duplication that includes the TJP2 gene have been reported to be related with HHI.[3,8,9] With the exception of III7 and IV26, the phenotype of affected members of family 686 is similar to that of the previously studied Tunisian family of Jewish ancestry, in terms of age of onset and audiological results. In the Jewish family, the ages of onset were in the fourth decade and hearing loss first involved high frequencies, followed by a gradual loss of hearing at mid and low frequencies. TJP2 is also predicted to be a candidate gene for age-related hearing loss (ARHL). Families with TJP2 mutations have patterns of hearing loss similar to those associated with ARHL, in that the high frequencies are affected first. Most cases of ADNSHHI are characterized by postlingual progressive sensorineural hearing loss, with the age of onset mostly being in the second or the 3rd year, such as is the case with KCNQ4,[11] GJB3,[12] and MYH14.[13] Some other cases of ADNSHHI that are caused by genetic mutations do not manifest until the third or 4th year, such as COCH[14] and TJP2,[3] which are thought to be related to ARHL. However, both ARHL and TJP2 mutation-related hearing impairments are most likely associated with apoptosis of hair cells in the inner ear. TJP2 is one of the three apoptosis-associated genes that have been reported in connection with ADNSHHI, in addition to DFNA5 and MSRB3.[3,15,16] Further research into TJP2 transcription and protein function may play a crucial role in understanding the molecular mechanism behind the pathogenesis of ARHL, and in deciphering the possible theory of hair cell death and the connection between gene mutations and progressive hearing loss.
Genetic research strategies for hereditary hearing impairment
Identification of causative genes for medical conditions such as HHI can be very challenging. Traditionally, strategies for identifying genes associated with HHI have included linkage analysis and candidate gene screening, which has led to the mapping of more than 60 loci for ADNSHHI. However, only 34 genes have been identified since 1992 (http://hereditaryhearingloss.org). In this study, we located a gene responsible for hearing impairment on the short arm of chromosome 9 (9p13.2-p13.3) using traditional approaches, but without positive results after sequencing candidate genes in all of the identified family members. Based on our experience, it is necessary to expand the physical scope of the location region moderately to avoid the deviation of the genetic distance calculation, which may be caused by neglecting some relatively small chromosomes exchange. This can lead to the real disease-causing gene being located near the targeted area instead of in the targeted region strictly. It is more difficult to identify the disease-causing gene than to map it onto a specific region on the chromosome, due to limitations in technology, the large size of many genes and the high-cost of Sanger sequencing.
Since 2009, NGS has played a crucial role in studying the genetics of HHI, with more than 10 genes having been recently identified (http://hereditaryhearingloss.org). NGS, also known as high-throughput sequencing or massively parallel sequencing, is quickly replacing many single gene tests for hearing impairment. It is useful for assessing patients whose phenotypes are not clinically distinguishable and should be considered by clinicians if single gene tests yield no diagnosis.[17] Among the NGS technologies, targeted capture panel tests are superior to whole exome sequencing, which looks at 20,000 genes and possible missing parts of the genes, offering more depth of coverage in genes associated with hearing loss.
In conclusion, through a series of studies over a period 10 years, we identified the co-occurrence of disease-causing genes TJP2 and GJB2 in family 686. The mechanism underlying its role in apoptosis and its association with ARHL may be useful in understanding the pathogenesis of ARHL. The combined GJB2 compound heterozygote elucidated the heterogeneity and complexity of HHI. In addition, we have summarized the various strategies to study the pathogenesis of ADNSHHI in this Chinese family. We concluded that none of the techniques are conclusive in every situation, and it is, therefore, necessary to combine the knowledge and skills from clinical medicine, genetics, bioinformatics and IT, while applying lateral thinking and constantly proposing new solutions. Therefore, it is necessary to address difficulties with an open mind and to use a combination of methods.
Supplementary information is linked to the online version of the paper on the Chinese Medical Journal website.
Financial support and sponsorship
This work was supported by grants from the National Key Basic Research Program of China (No. 2014CB943001) and the National Natural Science Foundation of China (No. 81120108009 and 81530032).
Conflicts of interest
There are no conflicts of interest.
Footnotes
Edited by: Yuan-Yuan Ji
REFERENCES
- 1.Brownstein Z, Bhonker Y, Avraham KB. High-throughput sequencing to decipher the genetic heterogeneity of deafness. Genome Biol. 2012;13:245. doi: 10.1186/gb-2012-13-5-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Köhler K, Zahraoui A. Tight junction: A co-ordinator of cell signalling and membrane trafficking. Biol Cell. 2005;97:659–65. doi: 10.1042/BC20040147. [DOI] [PubMed] [Google Scholar]
- 3.Walsh T, Pierce SB, Lenz DR, Brownstein Z, Dagan-Rosenfeld O, Shahin H, et al. Genomic duplication and overexpression of TJP2/ZO-2 leads to altered expression of apoptosis genes in progressive nonsyndromic hearing loss DFNA51. Am J Hum Genet. 2010;87:101–9. doi: 10.1016/j.ajhg.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lenz DR, Avraham KB. Hereditary hearing loss: From human mutation to mechanism. Hear Res. 2011;281:3–10. doi: 10.1016/j.heares.2011.05.021. [DOI] [PubMed] [Google Scholar]
- 5.Xu J, Kausalya PJ, Phua DC, Ali SM, Hossain Z, Hunziker W. Early embryonic lethality of mice lacking ZO-2, but Not ZO-3, reveals critical and nonredundant roles for individual zonula occludens proteins in mammalian development. Mol Cell Biol. 2008;28:1669–78. doi: 10.1128/MCB.00891-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang H, Zhao Y, Yi Y, Gao Y, Liu Q, Wang D, et al. Targeted high-throughput sequencing identifies pathogenic mutations in KCNQ4 in two large Chinese families with autosomal dominant hearing loss. PLoS One. 2014;9:e103133. doi: 10.1371/journal.pone.0103133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuse Y, Doi K, Hasegawa T, Sugii A, Hibino H, Kubo T. Three novel connexin26 gene mutations in autosomal recessive non-syndromic deafness. Neuroreport. 1999;10:1853–7. doi: 10.1097/00001756-199906230-00010. [DOI] [PubMed] [Google Scholar]
- 8.Hilgert N, Alasti F, Dieltjens N, Pawlik B, Wollnik B, Uyguner O, et al. Mutation analysis of TMC1 identifies four new mutations and suggests an additional deafness gene at loci DFNA36 and DFNB7/11. Clin Genet. 2008;74:223–32. doi: 10.1111/j.1399-0004.2008.01053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim MA, Kim YR, Sagong B, Cho HJ, Bae JW, Kim J, et al. Genetic analysis of genes related to tight junction function in the Korean population with non-syndromic hearing loss. PLoS One. 2014;9:e95646. doi: 10.1371/journal.pone.0095646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu Y, Zhou X, Jin Z, Cheng J, Shen W, Ji F, et al. Resolving the genetic heterogeneity of prelingual hearing loss within one family: Performance comparison and application of two targeted next generation sequencing approaches. J Hum Genet. 2014;59:599–607. doi: 10.1038/jhg.2014.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kubisch C, Schroeder BC, Friedrich T, Lütjohann B, El-Amraoui A, Marlin S, et al. KCNQ4, a novel potassium channel expressed in sensory outer hair cells, is mutated in dominant deafness. Cell. 1999;96:437–46. doi: 10.1016/s0092-8674(00)80556-5. [DOI] [PubMed] [Google Scholar]
- 12.Xia JH, Liu CY, Tang BS, Pan Q, Huang L, Dai HP, et al. Mutations in the gene encoding gap junction protein beta-3 associated with autosomal dominant hearing impairment. Nat Genet. 1998;20:370–3. doi: 10.1038/3845. [DOI] [PubMed] [Google Scholar]
- 13.Donaudy F, Snoeckx R, Pfister M, Zenner HP, Blin N, Di Stazio M, et al. Nonmuscle myosin heavy-chain gene MYH14 is expressed in cochlea and mutated in patients affected by autosomal dominant hearing impairment (DFNA4) Am J Hum Genet. 2004;74:770–6. doi: 10.1086/383285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robertson NG, Lu L, Heller S, Merchant SN, Eavey RD, McKenna M, et al. Mutations in a novel cochlear gene cause DFNA9, a human nonsyndromic deafness with vestibular dysfunction. Nat Genet. 1998;20:299–303. doi: 10.1038/3118. [DOI] [PubMed] [Google Scholar]
- 15.Van Laer L, Huizing EH, Verstreken M, van Zuijlen D, Wauters JG, Bossuyt PJ, et al. Nonsyndromic hearing impairment is associated with a mutation in DFNA5. Nat Genet. 1998;20:194–7. doi: 10.1038/2503. [DOI] [PubMed] [Google Scholar]
- 16.Ahmed ZM, Yousaf R, Lee BC, Khan SN, Lee S, Lee K, et al. Functional null mutations of MSRB3 encoding methionine sulfoxide reductase are associated with human deafness DFNB74. Am J Hum Genet. 2011;88:19–29. doi: 10.1016/j.ajhg.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alford RL, Arnos KS, Fox M, Lin JW, Palmer CG, Pandya A, et al. American College of Medical Genetics and Genomics guideline for the clinical evaluation and etiologic diagnosis of hearing loss. Genet Med. 2014;16:347–55. doi: 10.1038/gim.2014.2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DNA sequence chromatograms showing the heterozygous missense mutation c.2081G>A of TJP2 gene in affected individuals III2, III4, III5, IV4, IV13, IV22. IV8 was the unaffected control in this family. III7 and IV26 were affected members without the TJP2 c.2081G>A mutation.
DNA sequence chromatograms showing the two heterozygous missense mutations c.134C>A and c.408G>A of GJB2 gene in affected individuals (lower panel) compared with the wild type controls (upper panel).
A flow chart of the series studies of Family 686.