Abstract
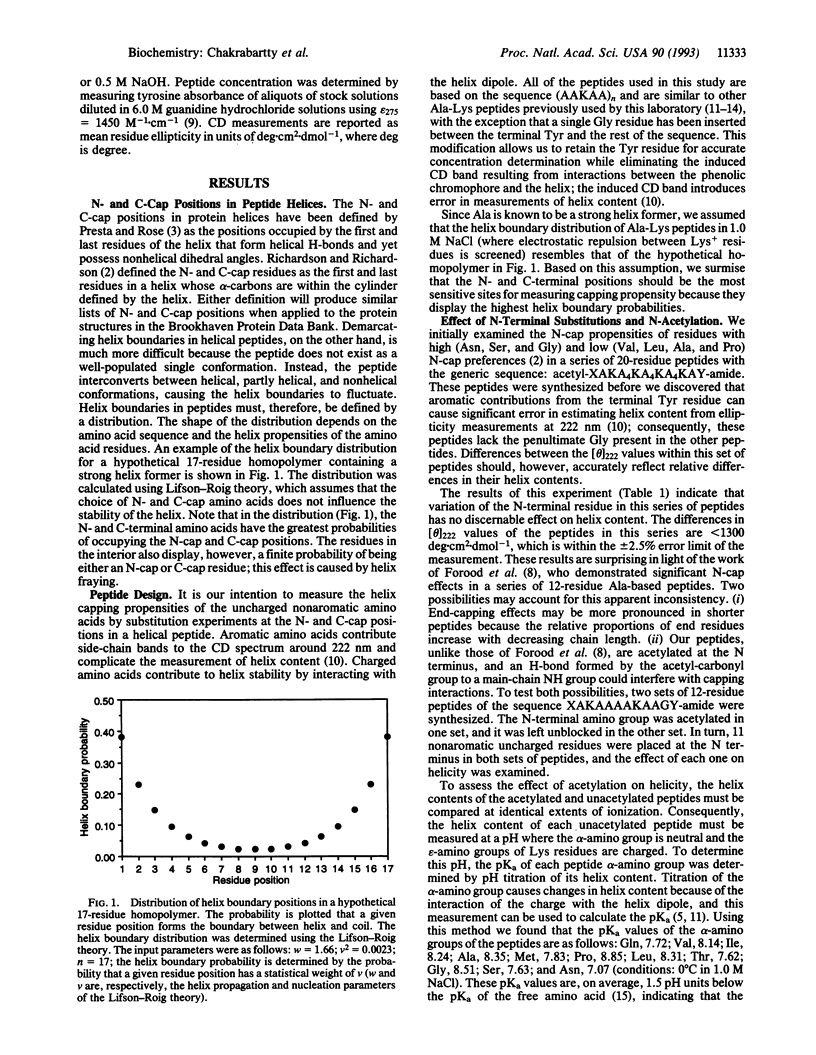
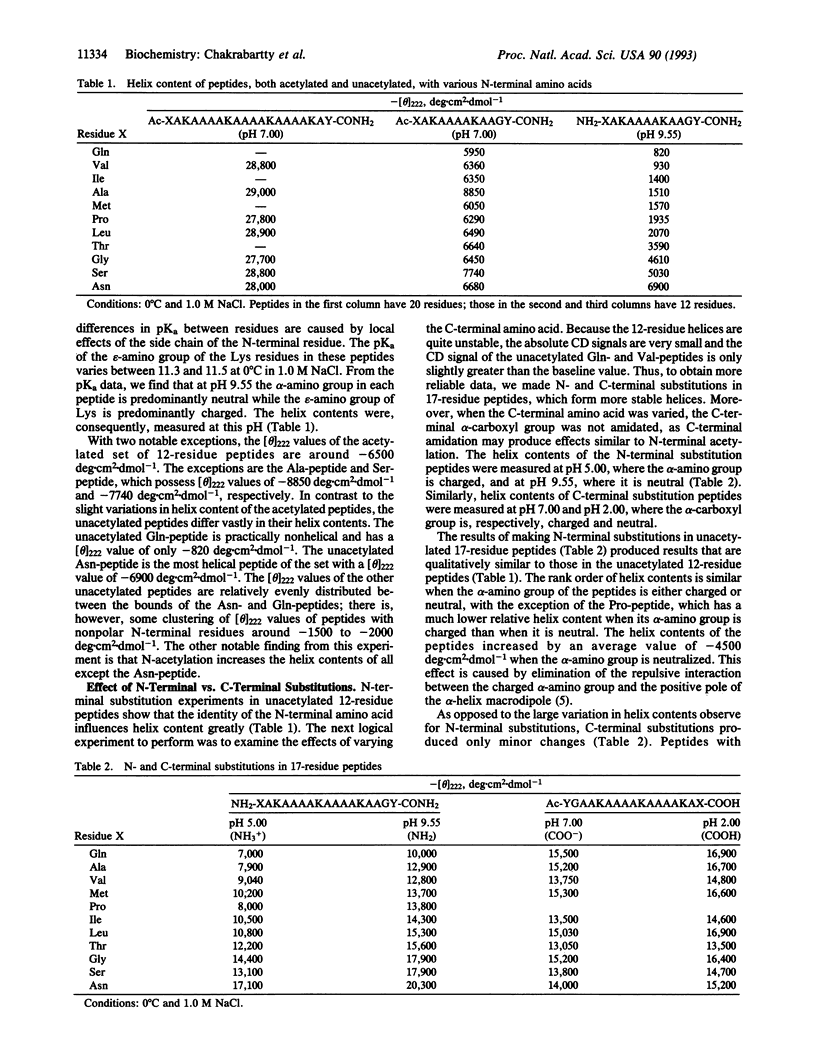
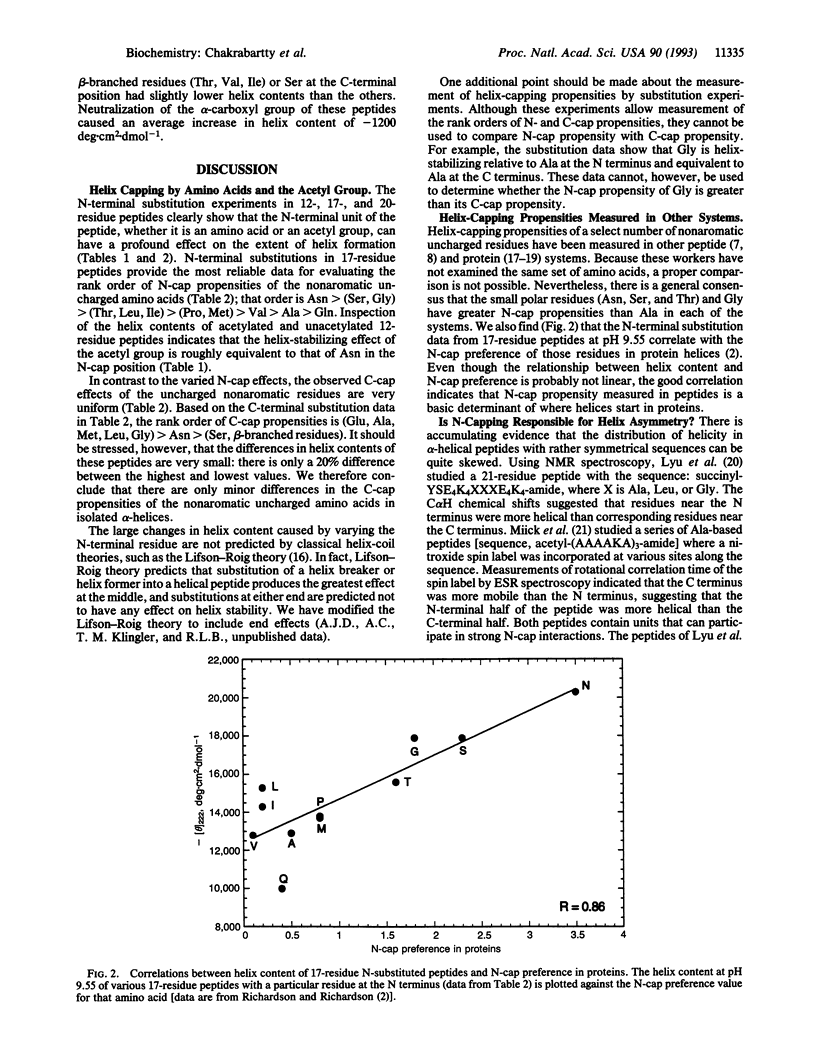
Helix content of peptides with various uncharged nonaromatic amino acids at either the N-terminal or C-terminal position has been determined. The choice of N-terminal amino acid has a major effect on helix stability: asparagine is the best, glycine is very good, and glutamine is the worst helix-stabilizing amino acid at this position. The rank order of helix stabilization parallels the frequencies of these amino acids at the N-terminal boundary (N-cap) position of helices in proteins found by Richardson and Richardson [Richardson, J. S. & Richardson, D. C. (1988) Science 240, 1648-1652], and the N-terminal amino acid in a peptide composed of helix-forming amino acids may be considered as the N-cap residue. The choice of C-terminal amino acid has only a minor effect on helix stability. N-capping interactions may be responsible for the asymmetric distribution of helix content within a given peptide found by various workers. An acetyl group on the N-terminal alpha-amino function cancels the N-cap effect and the acetyl group is equivalent to N-terminal asparagine in an unacetylated peptide. Our results demonstrate a close relationship between the mechanisms of alpha-helix formation in peptides and in proteins.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bell J. A., Becktel W. J., Sauer U., Baase W. A., Matthews B. W. Dissection of helix capping in T4 lysozyme by structural and thermodynamic analysis of six amino acid substitutions at Thr 59. Biochemistry. 1992 Apr 14;31(14):3590–3596. doi: 10.1021/bi00129a006. [DOI] [PubMed] [Google Scholar]
- Blagdon D. E., Goodman M. Letter: Mechanisms of protein and polypeptide helix initiation. Biopolymers. 1975 Jan;14(1):241–245. doi: 10.1002/bip.1975.360140118. [DOI] [PubMed] [Google Scholar]
- Brandts J. F., Kaplan L. J. Derivative sspectroscopy applied to tyrosyl chromophores. Studies on ribonuclease, lima bean inhibitors, insulin, and pancreatic trypsin inhibitor. Biochemistry. 1973 May 8;12(10):2011–2024. doi: 10.1021/bi00734a027. [DOI] [PubMed] [Google Scholar]
- Bruch M. D., Dhingra M. M., Gierasch L. M. Side chain-backbone hydrogen bonding contributes to helix stability in peptides derived from an alpha-helical region of carboxypeptidase A. Proteins. 1991;10(2):130–139. doi: 10.1002/prot.340100206. [DOI] [PubMed] [Google Scholar]
- Chakrabartty A., Kortemme T., Padmanabhan S., Baldwin R. L. Aromatic side-chain contribution to far-ultraviolet circular dichroism of helical peptides and its effect on measurement of helix propensities. Biochemistry. 1993 Jun 1;32(21):5560–5565. doi: 10.1021/bi00072a010. [DOI] [PubMed] [Google Scholar]
- Chakrabartty A., Schellman J. A., Baldwin R. L. Large differences in the helix propensities of alanine and glycine. Nature. 1991 Jun 13;351(6327):586–588. doi: 10.1038/351586a0. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Conformational parameters for amino acids in helical, beta-sheet, and random coil regions calculated from proteins. Biochemistry. 1974 Jan 15;13(2):211–222. doi: 10.1021/bi00699a001. [DOI] [PubMed] [Google Scholar]
- Ebina S., Wüthrich K. Amide proton titration shifts in bull seminal inhibitor IIA by two-dimensional correlated 1H nuclear magnetic resonance (COSY). Manifestation of conformational equilibria involving carboxylate groups. J Mol Biol. 1984 Oct 25;179(2):283–288. doi: 10.1016/0022-2836(84)90469-8. [DOI] [PubMed] [Google Scholar]
- Forood B., Feliciano E. J., Nambiar K. P. Stabilization of alpha-helical structures in short peptides via end capping. Proc Natl Acad Sci U S A. 1993 Feb 1;90(3):838–842. doi: 10.1073/pnas.90.3.838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lattman E. E., Rose G. D. Protein folding--what's the question? Proc Natl Acad Sci U S A. 1993 Jan 15;90(2):439–441. doi: 10.1073/pnas.90.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu P. C., Liff M. I., Marky L. A., Kallenbach N. R. Side chain contributions to the stability of alpha-helical structure in peptides. Science. 1990 Nov 2;250(4981):669–673. doi: 10.1126/science.2237416. [DOI] [PubMed] [Google Scholar]
- Lyu P. C., Wemmer D. E., Zhou H. X., Pinker R. J., Kallenbach N. R. Capping interactions in isolated alpha helices: position-dependent substitution effects and structure of a serine-capped peptide helix. Biochemistry. 1993 Jan 19;32(2):421–425. doi: 10.1021/bi00053a006. [DOI] [PubMed] [Google Scholar]
- Marqusee S., Robbins V. H., Baldwin R. L. Unusually stable helix formation in short alanine-based peptides. Proc Natl Acad Sci U S A. 1989 Jul;86(14):5286–5290. doi: 10.1073/pnas.86.14.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miick S. M., Casteel K. M., Millhauser G. L. Experimental molecular dynamics of an alanine-based helical peptide determined by spin label electron spin resonance. Biochemistry. 1993 Aug 10;32(31):8014–8021. doi: 10.1021/bi00082a024. [DOI] [PubMed] [Google Scholar]
- Padmanabhan S., Marqusee S., Ridgeway T., Laue T. M., Baldwin R. L. Relative helix-forming tendencies of nonpolar amino acids. Nature. 1990 Mar 15;344(6263):268–270. doi: 10.1038/344268a0. [DOI] [PubMed] [Google Scholar]
- Pogorelov V. M., Timkina E. N., Mattes G., Ile R., Miterev GYu, Novikova M. S., Isaev V. G., Kozinetz G. I. The monitoring of acute leukemia patients: Part 1. Basic morphometric characteristics of bone marrow blast cells in acute lymphoblastic leukemia. Leuk Res. 1990;14(9):795–800. doi: 10.1016/0145-2126(90)90073-i. [DOI] [PubMed] [Google Scholar]
- Presta L. G., Rose G. D. Helix signals in proteins. Science. 1988 Jun 17;240(4859):1632–1641. doi: 10.1126/science.2837824. [DOI] [PubMed] [Google Scholar]
- Richardson J. S., Richardson D. C. Amino acid preferences for specific locations at the ends of alpha helices. Science. 1988 Jun 17;240(4859):1648–1652. doi: 10.1126/science.3381086. [DOI] [PubMed] [Google Scholar]
- Richardson J. S., Richardson D. C. Amino acid preferences for specific locations at the ends of alpha helices. Science. 1988 Jun 17;240(4859):1648–1652. doi: 10.1126/science.3381086. [DOI] [PubMed] [Google Scholar]
- Rohl C. A., Scholtz J. M., York E. J., Stewart J. M., Baldwin R. L. Kinetics of amide proton exchange in helical peptides of varying chain lengths. Interpretation by the Lifson-Roig equation. Biochemistry. 1992 Feb 11;31(5):1263–1269. doi: 10.1021/bi00120a001. [DOI] [PubMed] [Google Scholar]
- Serrano L., Fersht A. R. Capping and alpha-helix stability. Nature. 1989 Nov 16;342(6247):296–299. doi: 10.1038/342296a0. [DOI] [PubMed] [Google Scholar]
- Serrano L., Sancho J., Hirshberg M., Fersht A. R. Alpha-helix stability in proteins. I. Empirical correlations concerning substitution of side-chains at the N and C-caps and the replacement of alanine by glycine or serine at solvent-exposed surfaces. J Mol Biol. 1992 Sep 20;227(2):544–559. doi: 10.1016/0022-2836(92)90906-z. [DOI] [PubMed] [Google Scholar]
- Shoemaker K. R., Kim P. S., York E. J., Stewart J. M., Baldwin R. L. Tests of the helix dipole model for stabilization of alpha-helices. Nature. 1987 Apr 9;326(6113):563–567. doi: 10.1038/326563a0. [DOI] [PubMed] [Google Scholar]



