Abstract
The endosomal barrier is a major bottleneck for the effective intracellular delivery of siRNA by nonviral nanocarriers. Here, we report a novel amphotericin B (AmB)-loaded, dual pH-responsive micelleplex platform for siRNA delivery. Micelles were self-assembled from poly(2-(dimethylamino)ethyl methacrylate)-block-poly(2-(diisopropylamino)ethyl methacrylate) (PDMA-b-PDPA) diblock copolymers. At pH 7.4, AmB was loaded into the hydrophobic PDPA core, and siRNA was complexed with a positively charged PDMA shell to form the micelleplexes. After cellular uptake, the PDMA-b-PDPA/siRNA micelleplexes dissociated in early endosomes to release AmB. Live cell imaging studies demonstrated that released AmB significantly increased the ability of siRNA to overcome the endosomal barrier. Transfection studies showed that AmB-loaded micelleplexes resulted in significant increase in luciferase (Luc) knockdown efficiency over the AmB-free control. The enhanced Luc knockdown efficiency was abolished by bafilomycin A1, a vacuolar ATPase inhibitor that inhibits the acidification of the endocytic organelles. These data support the central hypothesis that membrane poration by AmB and increased endosomal swelling and membrane tension by a “proton sponge” polymer provided a synergistic strategy to disrupt endosomes for improved intracellular delivery of siRNA.
Keywords: siRNA delivery, amphotericin B, pH-responsive micelleplexes, endosomal escape, nonviral nanocarrier, polymeric micelles
Graphical Abstract
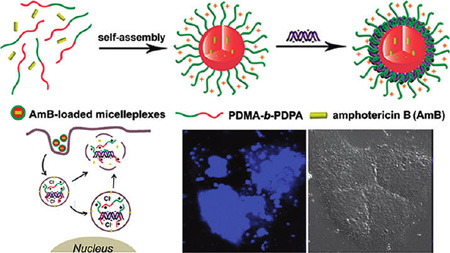
Small interfering RNA (siRNA)-mediated RNA interference (RNAi) has received considerable attention since the discovery of the RNAi mechanism in 1998.1,2 RNAi holds great promise in molecular therapy of intractable and genetic-related human diseases by silencing the target mRNA (mRNA).3–5 For systemic siRNA delivery, naked siRNA is vulnerable to serum and endogenous nuclease degradations. Furthermore, siRNA does not easily cross cellular membranes because of its large size and negatively charged potential. Nonviral carriers have been extensively investigated to improve the siRNA stability, bioavailability, and delivery efficiency to the target tissues or cells.6–8 Endosomal escape is one of the major barriers for nonviral siRNA delivery, since siRNA trapped in endosomes is typically trafficked into lysosomes where siRNA is degraded.5,8 To overcome the endosomal barrier, a variety of stimuli-responsive nonviral vectors have been exploited.9–15Among these, pH-responsive vectors utilizing the acidic intracellular environment via proton buffer effect (e.g., poly(ethyleneimine) (PEI), poly(l-histidine), and poly(β-amino esters))15–17 or reversible PEG shielding were extensively investigated.18–20 Furthermore, amphiphilic peptides (AMP) or virus-derived proteins were covalently conjugated with the polycationic vectors to enhance siRNA endosomal release by forming transmembrane pores or fusing with endosome membranes.11,21,22 Despite these remarkable advances, timely and efficient siRNA endosomal escape remains a considerable challenge.
Recently, we reported a new set of pH-activatable micellar (pHAM) nanoparticles with tunable pH-responsive properties.23 These nanoparticles were produced from a series of diblock copolymers with an ionizable block with controlled hydrophobicity. One such polymer, poly(ethylene glycol)-block-poly(2-(diisopropylamino) ethylmethacrylate) (PEG-b-PDPA), had a pH transition at 6.3.23 The PEG-b-PDPA micelles were specifically activated/dissociated in early endosomes (pH = 5.9–6.2)24 via protonation of the PDPA segment in the first 30 min after cellular uptake and displayed an excellent “proton sponge” effect due to the presence of the diisopropyl-substituted tertiary amines. PEG-b-PDPA nanoparticles are a good candidate for endosome-targeted delivery of hydrophobic small molecules or siRNA. When the nanoparticles were applied for cell culture study, we observed that protonated PEG-b-PDPA unimers were entrapped in late endosome or lysosome vesicles over time, resulting in vesicle enlargement but no visible bursting.23 These data agreed with the theoretical prediction that the “proton sponge” effect alone is not sufficient for endosomal disruption.25
Amphotericin B (AmB), a hydrophobic antifungal drug, is known to transiently increase membrane permeability at sublethal concentrations by forming transmembrane pores.26–28 Bolard et al. reported a cationic lipid AmB derivative with membrane penetration ability for antisense oligonucleotide delivery.29 In this study, we hypothesize that combination of membrane poration by AmB and endosome swelling by polycations can synergistically facilitate the efficient siRNA endosomal escape by disrupting the endosome membrane, thereby improving target gene knockdown efficiencies using specific siRNAs. To test this hypothesis, we established dual pH-responsive poly(2-(dimethylamino)ethyl methacrylate)-block-poly(2-(diisopropylamino)ethyl methacrylate) (PDMA-b-PDPA) micelles for AmB and siRNA co-delivery. The PDPA segment self-assembled into hydrophobic cores for AmB encapsulation at pH 7.4, whereas the PDMA segment formed a positively charged shell that allowed for siRNA complexation (Figure 1). We refer to this micelle-mediated formation of a siRNA complex as a “micelleplex” to differentiate from other compaction forms (e.g., polyplexes where siRNA directly complexes with cationic polymers).30
Figure 1.
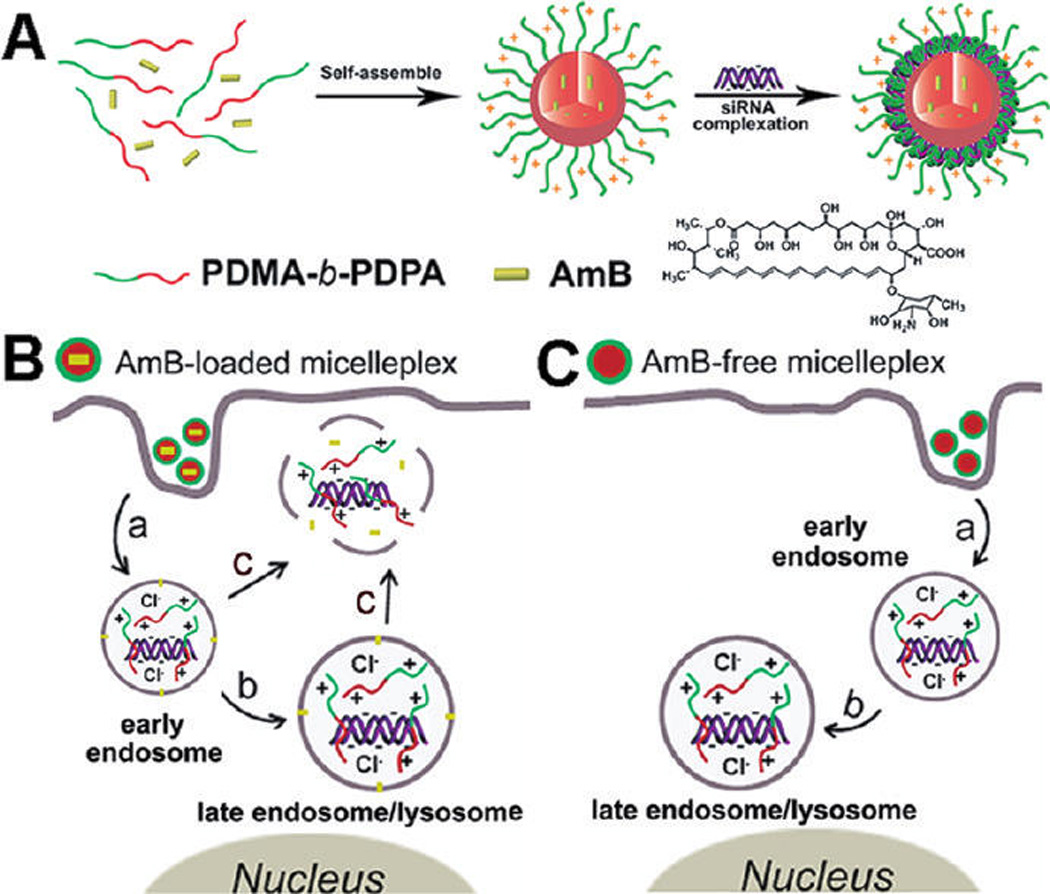
Schematic diagram of AmB-loaded dual pH-responsive micelleplexes for siRNA delivery with enhanced siRNA endosomal escape ability. (A) Production of AmB-loaded PDMA-b-PDPA micelleplexes. AmB was loaded in the hydrophobic PDPA core, and siRNA was complexed with the PDMA corona shell. (B) AmB-facilitated endosome disruption and siRNA cytoplasmic release (a: AmB-loaded micelleplexes dissociated in early endosomes after cell uptake, and AmB molecules are inserted into endosomal membranes; b: protonated PDMA-b-PDPA unimers complexed with siRNA and trafficked from early endosomes into late endosome/lysosomes, causing vesicle swelling; c: AmB-enhanced siRNA release from endosomes into cytoplasm via membrane destabilization). (C) In the case of AmB-free micelleplexes, polymer/siRNA complexes were entrapped in late endosomes or lysosomes without efficient cytoplasmic siRNA release.
RESULTS
Characterization of Dual pH-Responsive PDMA-b-PDPA Copolymer and Micelles
PDMA-b-PDPA copolymer was synthesized by atom transfer radical polymerization (ATRP).31 Several other diblock copolymers (e.g., poly(ethylene glycol)-block-poly(2-(diisopropylamino)ethyl methacrylate) (PEG-b-PDPA), polystyrene-block-poly(2-(dimethylamino)ethyl methacrylate) (PSt-b-PDMA), and poly-(ethylene glycol)-block-poly(2-(dimethylamino)ethyl methacrylate) (PEG-b-PDMA)) were also synthesized by the ATRP method as various controls (Figure 2A for chemical structures, Supporting Information Figure SI 1 and Table SI 1 for polymer syntheses and characterizations).
Figure 2.
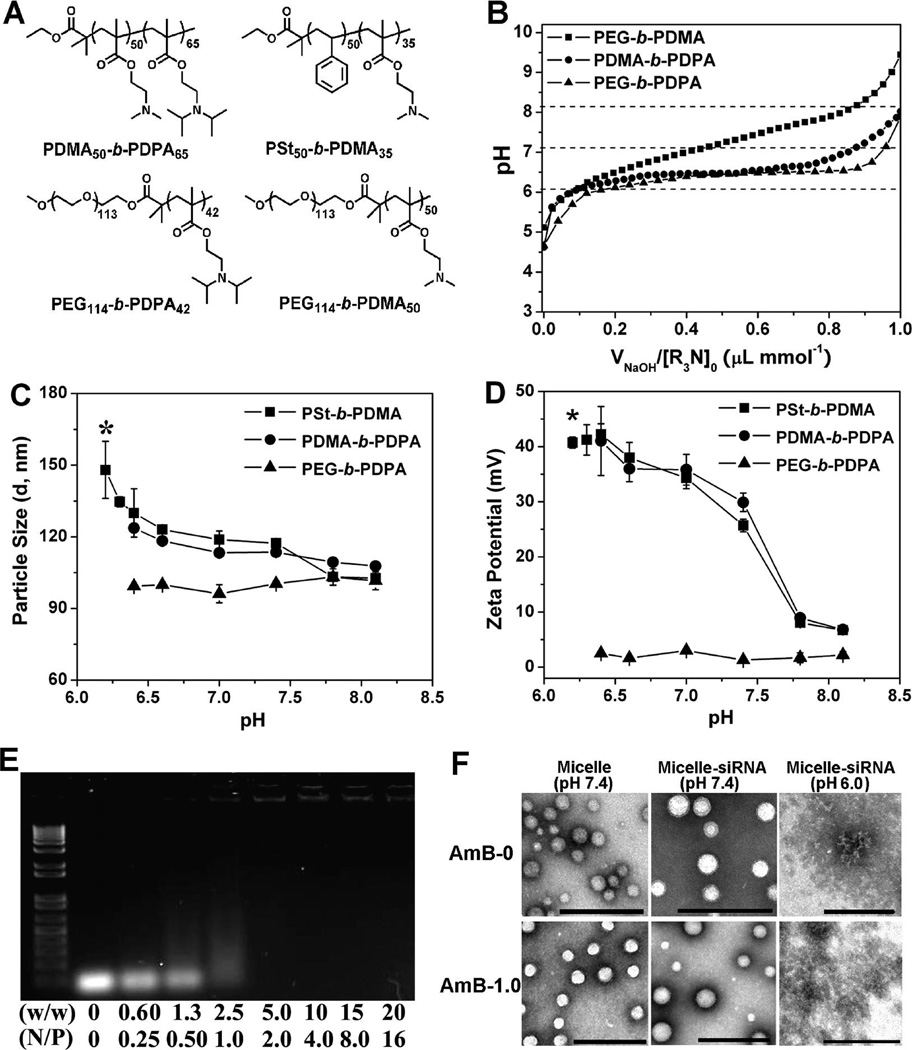
Physical characterization of the dual pH-responsive PDMA-b-PDPA micelles and micelleplexes. (A) Chemical structures of the diblock copolymers used in this study. (B) Titration curves of PDMA-b-PDPA, PEG-b-PDPA, and PEG-b-PDMA copolymers. Change of hydrodynamic diameter (C) and zeta-potential (D) of the PDMA-b-PDPA micelles as a function of buffer pH (*no particles were detected by DLS for PEG-b-PDPA and PDMA-b-PDPA micelles at pH 6.2, indicating their dissociation at acidic pH). The surface charge of PSt-b-PDMA micelles reached a plateau at pH 6.2 owing to the complete protonation of the PDMA segment. (E) Gel shift assay shows the siRNA binding stability by the PDMA50-b-PDPA65 micelleplexes. (F) TEM images of AmB-free or 1.0 wt % AmB-loaded PDMA-b-PDPA micelleplexes at pH 7.4 and 6.0, respectively (scale bars = 200 nm).
pH titration of PDMA-b-PDPA copolymer showed dual pH transitions: one coincided with a PDPA segment (pKa = 6.3) and the other with PDMA (pKa = 7.4) (Figure 2B). PEG-b-PDPA and PEG-b-PDMA copolymers are shown for comparison. As previously demonstrated, hydrophobic PDPA segments led to cooperative deprotonation of PDPA tertiary ammonium groups, resulting in a dramatically sharpened pH transition.23 More specifically, a majority of the deionization (e.g., α = 10% to 90%, where α is defined as the mole fraction of neutral amines: α = [R3N]/([R3N] + [R3NH+])) for PEG-b-PDPA copolymer occurs within 0.5 pH unit, compared to approximately 2 pH units for the hydrophilic PEG-b-PDMA copolymer. PDMA-b-PDPA copolymer showed a composite behavior, where initial titration (α < 0.6) illustrated ultra-pH-sensitive behaviors from the PDPA segment, and later titration (α > 0.6) was similar to that of PDMA. These data suggested a strong buffer capacity of PDMA-b-PDPA copolymers.
To examine the dual pH-responsive properties of the PDMA-b-PDPA micelles, we applied dynamic light scattering (DLS) and zeta-potential analyses to examine their pH-dependent swelling and dissociation properties. As shown in Figure 2C and D, at pH > 6.3, PDMA-b-PDPA micelles were formed, and the micelle size and zeta potential decreased with the increase of pH, primarily due to the deprotonation of the PDMA shells (pKa = 7.4). More specifically, the surface potential decreased from +41 mV at pH 6.4 to +8 mV at pH 8.2. At pH 7.4, the surface charge is around +30 mV, still highly positive for siRNA compaction. In the pH range of 6.4 to 8.2, the inverse correlation of particle size and zeta potential for PDMA-b-PDPA micelles with pH was similar to that from the PSt-b-PDMA micelles. Since the hydrophobic PSt core is pH-insensitive, the decrease of particle size and surface charge of the PSt-b-PDMA micelles should be caused by the deprotonation of the PDMA shell. In contrast, PEG-b-PDPA micelles displayed relatively the same particle size and zeta-potential in this pH range because of the presence of a neutral PEG layer. At pH < 6.3 (i.e., the pKa of the PDPA segment), no PDMA-b-PDPA or PEG-b-PDPA micelles were detected by DLS due to the protonation of the PDPA segment and dissociation of the nanoparticles.
AmB Loading and siRNA Complexation
We controlled the AmB loading density in the PDMA-b-PDPA micelles at 0.5, 1.0, and 2.0 wt % (see Supporting Information Table SI 2). The AmB loading efficiency was over 95%, as determined by UV–vis spectra, suggesting good miscibility between AmB and PDPA block. This in turn contributed to the high stability of the AmB-loaded micelles (e.g., no observable AmB precipitation within 24 h storage at room temperature). The particle size and surface charge of the AmB-encapsulated micelles were investigated by DLS and zeta-potential measurement, respectively (see Table SI 3). The micelles with 2.0 wt % AmB loading were 13 nm larger than the AmB-free ones. This phenomenon agreed well with our previous observation that loading of hydrophobic drug in polymeric micelles led to increased particle size. The surface charge of the AmB-loaded micelles was approximately +32 mV at different AmB loading densities. After siRNA complexation, particle size and zeta-potential were found comparable to that of the AmB-loaded micelles at a weight ratio above 7.5 (Figure SI 2A and B).
The stability of the siRNA-loaded PDMA-b-PDPA micelleplexes was examined by gel electrophoresis assay. At a polymer to siRNA weight ratio of 5.0 or above (“N/P” ratio of 2.0, calculated using the nitrogens from dimethyl amino groups only), the shift of the siRNA band was completely retarded, suggesting high siRNA binding affinity by the PDMA-b-PDPA micelles (Figure 2E).
The PDPA-protonation-induced dissociation of the AmB-loaded micelleplexes was confirmed by TEM examination. At pH 7.4, the AmB-free and AmB-loaded micelles were both found to be spherical. At pH 6.0, only amorphous aggregates of the siRNA/polymer complexes or PDMA-b-PDPA unimers were present, suggesting complete micelle dissociation at endosomal pH (Figure 2F).
Evaluation of the Endosomal Escape Capability of AmB-Free PDMA-b-PDPA Micelleplexes
To investigate the endosomal destabilization capability of the PDMA-b-PDPA micelleplexes in vitro, we introduced a PEG-b-PDPA copolymer where the PDPA segment was covalently labeled with tetramethylrhodamine (TMR) dyes. As demonstrated previously, PEG-b-(PDPA-TMR) micelle assembly at neutral pH resulted in the self-quenching of the TMR fluorescence signal.23 At pH 6.2, the TMR signal of the TMR-labeled PDMA-b-PDPA micelleplex recovered dramatically due to the increased fluorophore distance as a result of micelle dissociation (Figure SI 3). Alexa647-labeled scrambled siRNA (AlexasiRNA-Scr) was complexed to the micelle corona layer, and the intracellular uptake of the resulting micelleplexes A549 human lung cancer cells (with stable luciferase expression, referred as “A549-Luc”) was monitored by confocal laser scanning microscopy (CLSM) examination. As shown in Figure 3A, after 12 h incubation, considerable TMR fluorescent signal was observed owing to the dissociation of the PDMA-b-PDPA micelles. The red dots belonging to TMR colocalized with those of Alexa, indicating the complexation of the protonated PDMA-b-PDPA unimers with siRNA. It should be noted that only a small fraction of the TMR/Alexa fluorescent dots colocalized in the Rab5a-GFP-labeled early endosomes (top panel). Instead, the majority of the TMR/Alexa dots overlapped with the Lamp1-GFP (bottom panel), owing to the trafficking of the micelleplexes from early endosomes into later endosomes or lysosomes. Similar results were obtained at extended incubation time (24 h) because of late endosome/lysosome entrapment of the copolymer/siRNA complexes (Figure SI 4A).
Figure 3.
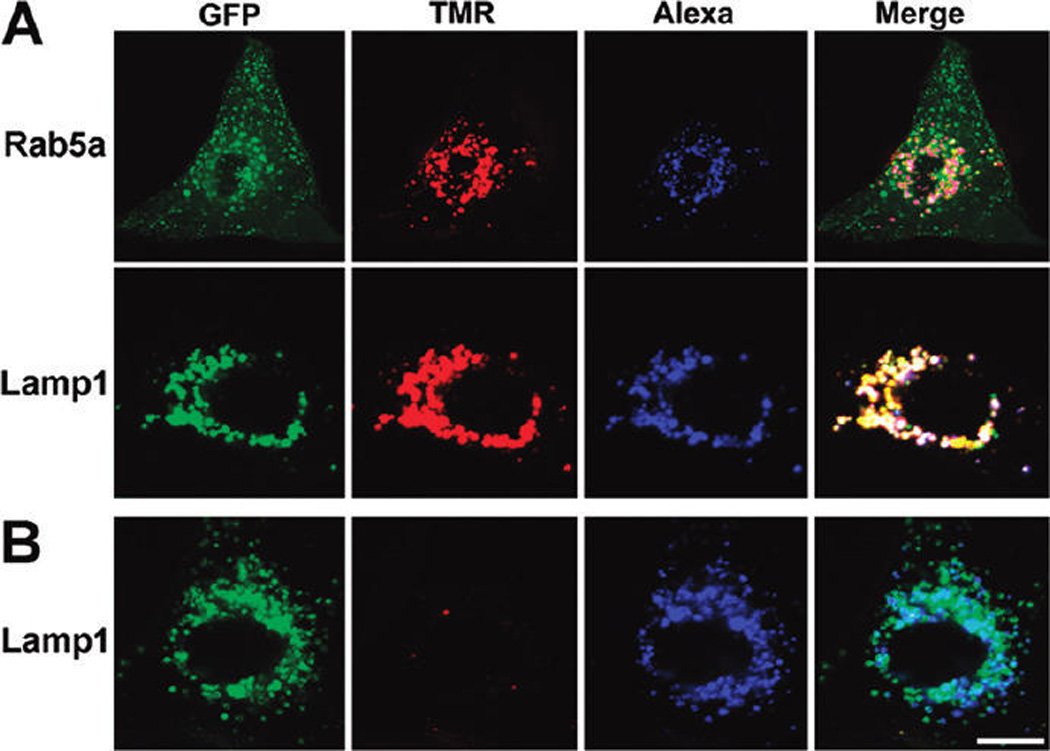
CLSM examination of intracellular dissociation of TMR-labeled PDMA-b-PDPA micelleplexes. (A) Intracellular dissociation of PDMA-b-PDPA micelleplexes in A549-Luc cells containing Rab5a-GFP-labeled early endosomes (top panel) or Lamp1-GFP-labeled late endosomes/lysosomes (middle panel). Dissociation of micelleplexes resulted in the activation of TMR signal (red), which showed colocalization with late endosomes/lysosomes. (B) Baf-A1 inhibited the intracellular dissociation of the PDMA-b-PDPA micelleplexes in A549-Luc cells, as indicated by the lack of TMR fluorescence from the pH-activatable micelles (bottom panel) (scale bar = 10 µm). siRNA molecules were labeled with Alexa dye and are shown as the blue color.
Next, to verify that the micelleplex dissociation was endosome acidification-induced, A549-Luc cells were pretreated with bafilomycin-A1 (Baf-A1), a potent inhibitor for vacuolar-type H+-ATPase (V-ATPase) proton pump. The V-ATPase is responsible for the acidification of the intracellular vesicles (i.e., endosomes or lysosomes).32,33 As shown in Figure 3B, A549-Luc cells displayed “silenced” TMR signals without affecting the emission of Alexa fluorophores after 12 h Baf-A1 treatment. This result is consistent with the inhibition of endosome/lysosome acidification and corresponding lack of dissociation of the PDPA core. Extending the incubation time to 24 h, comparable a TMR fluorescence signal appeared in the Baf-A1-treated cells to that in the untreated cell controls, which was attributed to the reduced intracellular Baf-A1 concentration and recovery of low lysosomal pH (Figure SI 4B). These data were further corroborated by flow cytometry analysis. As shown in the Supporting Information, after 4 h micelleplex incubation, no TMR positive cells were detected in Baf-A1-pretreated cell populations, in significant contrast with the Baf-A1-untreated cells (>50% TMR positive). On the other hand, Baf-A1-treated or untreated cells displayed 63% or 68% Alexa positive percentages, respectively (Figure SI 5), indicating Baf-A1-induced inhibition of vesicle acidification did not interfere with the cellular uptake of micelleplexes.
AmB-Enhanced Endosomal Escape of siRNA
To test our hypothesis that AmB can facilitate the efficient siRNA endosomal escape, we examined the intracellular trafficking of the AmB-loaded micelleplexes by CLSM. To do that, TMR-dextran, a fluidic phase endocytosis marker, was selected to label the intracellular vesicles of A549-Luc cells.34 In contrast to the TMR-conjugated PEG-b-PDPA copolymer, the fluorescence signal of pH-insensitive TMR-dextran can stay ON after endosomal escape into the cytoplasm. Figure 4 shows that the AmB-free micelleplexes had TMR signals (red spots) colocalized well with the Alexa dye (blue spots) due to siRNA trapping in the late endosome or lysosome. When the cells were treated with 0.5 wt % AmB-loaded micelleplexes, the TMR-dextran and Alexa-siRNA diffused throughout the cells, suggesting endosome disruption and release of dextran and siRNA into the cytoplasm. At a high AmB-loading density of 1.0 wt %, a more diffusive siRNA distribution pattern was found (Figure 4 and Figure SI 6). The diffusive pattern of siRNA distribution is consistent with a recent literature report that scrambled siRNA was distributed throughout the cytoplasm and nucleus.35
Figure 4.
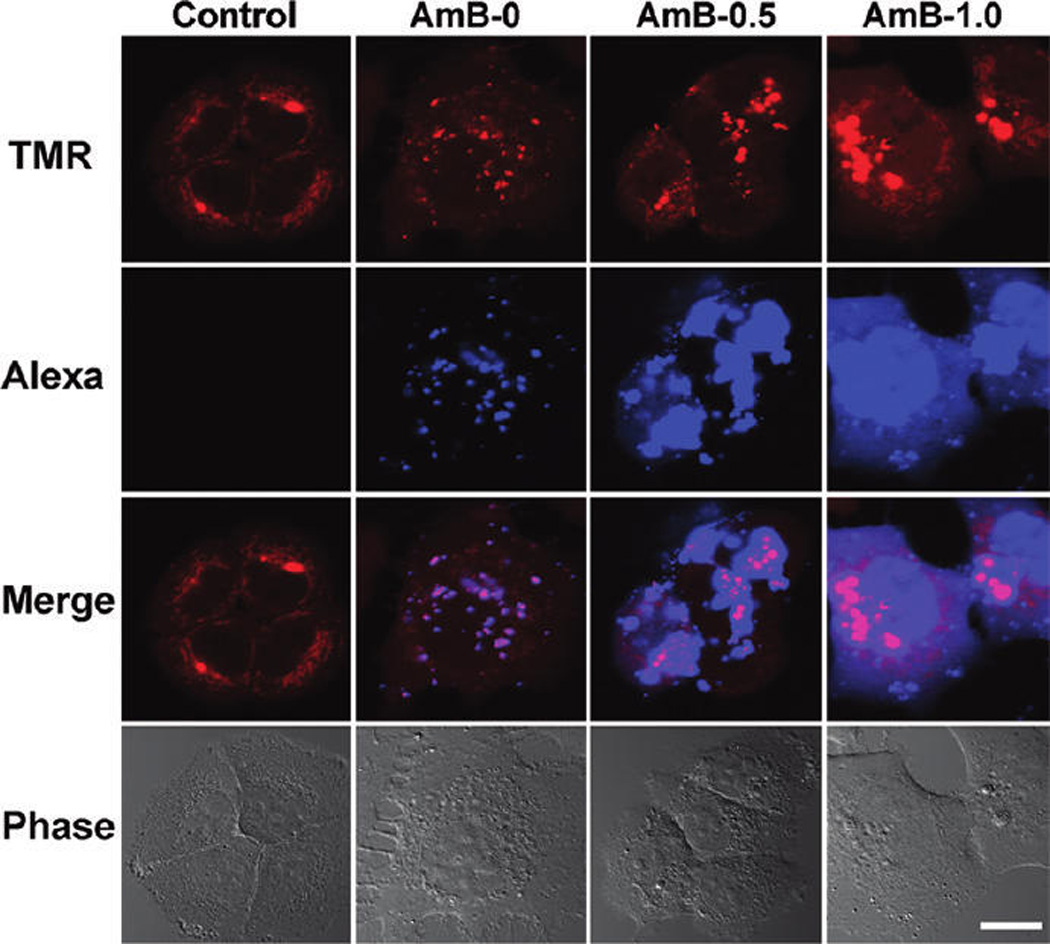
CLSM examination of siRNA endosomal escape in the cells treated with AmB-loaded micelleplexes. The images were taken 12 h after micelleplex incubation. Compared to the AmB-free micelleplexes, AmB-loaded micelleplexes significantly increased the siRNA endosomal escape, as indicated by the diffusive distribution of Alexa-labeled siRNA molecules (control: untreated cells; AmB-0: AmB-free micelleplex-treated cells; AmB-0.5: 0.5 wt % AmB-loaded micelleplexes; AmB-1.0: 1.0 wt % AmB-loaded micelleplexes, scale bar = 20 µm). TMR-dextran polymers were used as an endosomal marker.
AmB-Enhanced Target Gene Knockdown in A549-Luc Cells
Transfection studies were conducted to investigate the correlation between AmB-enhanced siRNA endosomal escape and specific target gene (i.e., luciferase (Luc)) knockdown efficiency. PDMA50-b-PDPA65 copolymer with optimized composition was selected for the preparation of AmB-loaded micelleplexes and siRNA transfection studies. As shown in Figure 5A, 1.0 wt % AmB-loaded micelleplexes (w/w 7.50) silenced >80% Luc protein expression as measured by Luc activity, 1.8-fold higher than the AmB-free control. At an AmB-loading density of 0.5 wt %, the micelleplexes knocked down >90% of Luc protein expression (w/w 10.0), 1.3-fold more efficient than the AmB-free control. The relative cell viability of transfected cells was evaluated by MTT assay, and no statistical difference between the cytotoxicity of the AmB-loaded versus AmB-free micelleplexes was noted (Figure 5B). The AmB-enhanced Luc knockdown was also found in PC-3-Luc prostate and MBA-MD-231-Luc breast cancer cells (data not shown), suggesting that the AmB-enhanced siRNA endosomal escape can serve as a general mechanism to improve siRNA delivery in a variety of cancer cell types.
Figure 5.
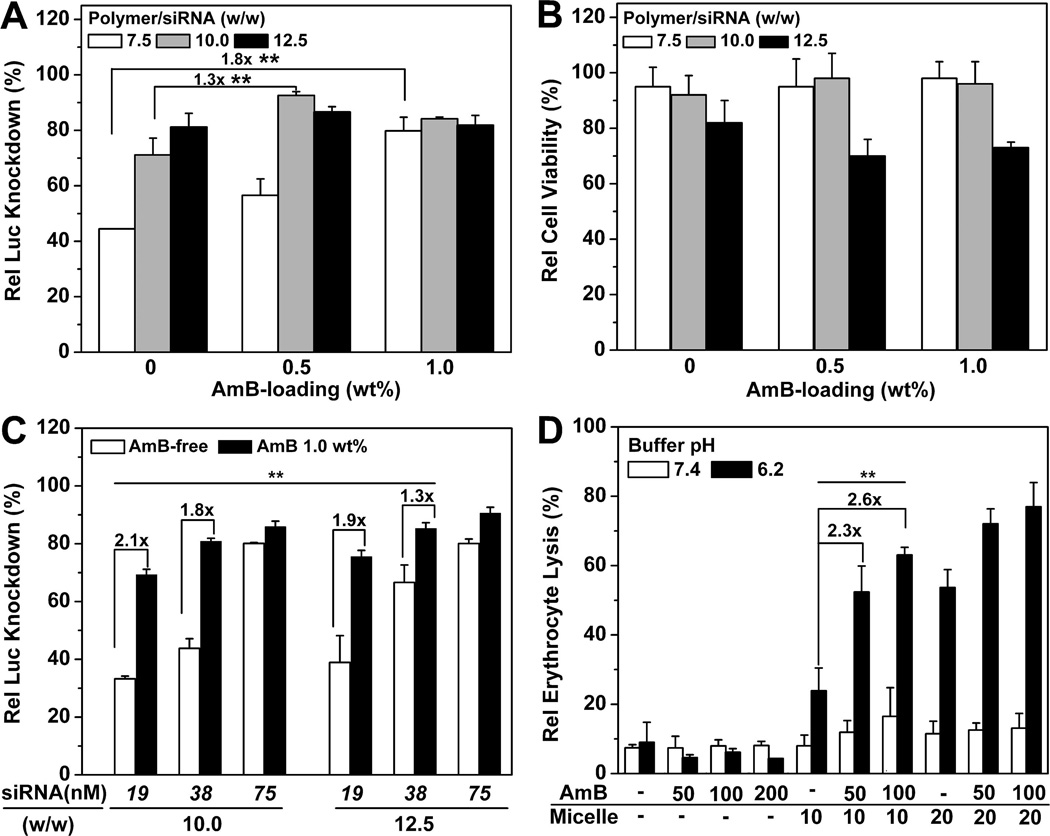
siRNA transfection study of AmB-loaded PDMA-b-PDPA micelleplexes in A549-Luc cells. (A) Influence of AmB loading on Luc knockdown efficiency by the PDMA-b-PDPA micelleplexes (siRNA concentration = 75 nM, or 100 ng per well). The Rel Luc knockdown was determined by normalizing the decreased Luc activity in siRNA-Luc-treated cells over that of the siRNA-Scr-treated control. (B) Relative cell viability of A549-Luc cells transfected by AmB-loaded micelleplexes as determined by the MTT assay. (C) Comparison of AmB-free vs AmB-loaded siRNA/micelleplexes as a function of micelleplex dose and PDMA-b-PDPA/siRNA ratio. Significant increase in Luc knockdown efficiency was observed with AmB loading. (D) Erythrocytic activity of AmB-loaded micelleplexes. At lower pH (6.2), significantly increased hemolytic activities were observed with the combination of AmB and PDMA-b-PDPA copolymer, demonstrating the synergistic effect in membrane destabilization.
To investigate the siRNA dosage effect on Luc knockdown, A549-Luc cells were treated by 1.0 wt % AmB-loaded micelleplexes at the three siRNA concentrations of 19, 38, and 75 nM. As shown in Figure 5C, AmB loading significantly reduced the effective siRNA concentrations for sufficient Luc knockdown. That is, at the same siRNA concentration of 19nM and polymer to siRNA weight ratio of 10, AmB-loaded micelleplexes silenced 70% of Luc protein expression, 2.1-fold more efficient than the AmB-free control. Similar trends were observed at increased siRNA concentrations and polymer to siRNA weight ratios.
To validate the advantage of endosome-targeted AmB and siRNA co-delivery, a mixed solution of free AmB and PDMA-b-PDPA micelleplex was further applied in a transfection study. Free AmB exhibited no positive effect on the Luc knockdown, while elevated cytotoxicity was observed compared to AmB-loaded micelleplexes (Figure 5A and Figure SI 7). This might be caused by free AmB, which induces Na+/K+ ion leakage via formation of transmembrane ion channels in the extracellular membrane.36
AmB Synergizes with PDMA-b-PDPA Unimers in Membrane Disruption
In an attempt to understand the mechanism for AmB-enhanced siRNA endosomal escape, we examined the membrane destabilization capability of the AmB-loaded PDMA-b-PDPA micelles by an erythrocyte lysis assay.37 Figure 5D illustrated that no hemoglobin was released from the red blood cells (RBC) when treated with free AmB at either pH (7.4 and 6.2). This could be explained by the smaller AmB-induced membrane pore size than that of hemoglobin protein.36,38 Hemoglobin was also not released from RBC when treated with AmB-free or AmB-loaded PDMA-b-PDPA micelles at pH 7.4. In contrast, at pH 6.2, 50% or 60% of hemoglobin was dramatically released from RBC treated with 0.5 or 1.0 wt % AmB-loaded micelles, 2.3- and 2.6-fold higher than the cells exposed to AmB-loaded PDMA-b-PDPA micelles at pH 7.4, respectively.
AmB-Enhanced Luc Knockdown Can Be Abolished by Baf-A1
To investigate the influence of endosomal acidification on Luc knockdown, we pretreated the A549-Luc cells with Baf-A1 1 h before micelleplex addition. As shown in Figure 6A, after Baf-A1 treatment, although AmB-loaded micelleplexes showed higher Luc expression knockdown than the AmB-free control, both types of micelleplexes silenced <20% of Luc protein expression (w/w 10), much lower than the Baf-A1-untreated control. This could be explained by Baf-A1-inhibited micelleplex dissociation and, thereby, AmB release from the PDPA core.
Figure 6.
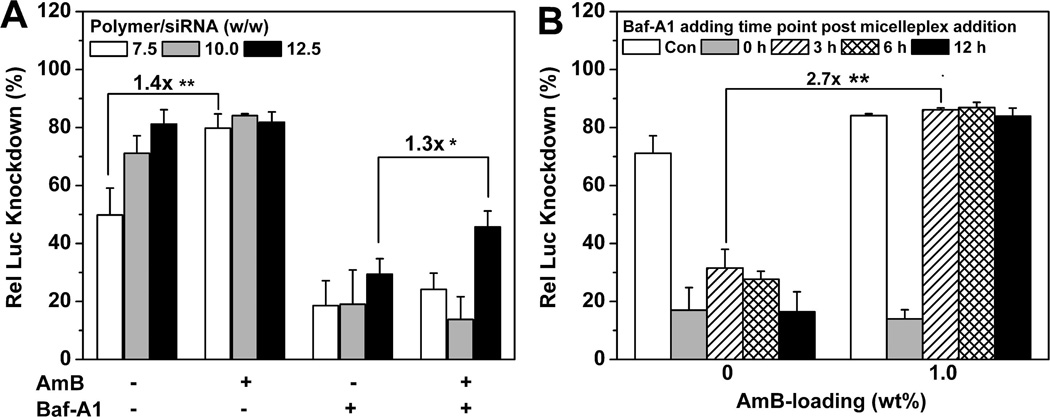
Influence of Baf-A1 treatment on Luc knockdown efficiency by the AmB-loaded PDMA-b-PDPA micelleplexes. (A) Relative Luc knockdown efficiency of AmB-loaded micelleplexes in Baf-A1-pretreated A549-Luc cells. (B) Relative Luc knockdown efficiency vs Baf-A1 addition times. The cells were first treated with micelleplex, and then Baf-A1 was added at different time points (0, 3, 6, or 12 h) after micelleplex addition (1.0 wt % AmB loading, w/w 10, and siRNA concentration of 75 nM were applied) (*p < 0.05, **p < 0.01). Rel Luc knockdown was determined by normalizing the decreased Luc activity in siRNA-Luc-treated cells over that of the siRNA-Scr-treated control.
To further elucidate the role of AmB on Luc knockdown, we delayed the Baf-A1 addition time post micelleplex addition to allow AmB release. When Baf-A1 was added 3 h later after micelleplex addition, AmB-loaded micelleplexes silenced >80% of Luc protein expression, which was 2.7-fold more efficient than AmB-free ones in the same cells (Figure 6B). A similar pattern was found when the cells were treated with Baf-A1 at longer delayed time after micelleplex addition (e.g., 6 or 12 h post micelleplex addition).
DISCUSSION
RNAi is recognized as a promising strategy for treatment of genetic-related human diseases. Here, we established a PDMA-b-PDPA micelleplex platform for siRNA delivery. The PDMA-b-PDPA micelles displayed dual pH-responsive and strong proton buffer properties (pH 6.3–7.4) owing to the stepwise protonation of the PDMA shell and PDPA core. Given the positive charges at neutral pH, PDMA polyplexes or PDMA-modified quantum dots were previously examined for siRNA complexation and delivery.39,40 In contrast, the PDPA segment, which is readily protonated at endosomal pH, serves as a “proton sponge” to destabilize the endosome membrane.41,42 However, neither PEG-b-PDPA micelles nor PDMA-b-PDPA micelleplexes resulted in observable endosome disruptions, as shown in our previous and current studies, despite their selective activation in early endosomes. In this study, we hypothesized that incorporation of AmB, a membrane poration agent, can synergize with the “proton sponge” properties of PDMA-b-PDPA copolymers to overcome the endosomal barrier. As demonstrated in Figure 4 and the Supporting Information (Figure SI 6), AmB-loaded PDMA-b-PDPA micelleplexes displayed much improved ability for siRNA endosomal escape, which was more notable over extended incubation time. AmB-prompted, efficient siRNA cytosol release positively correlated with the Luc knockdown efficiency (Figure 5A).
The Luc knockdown activity of both AmB-loaded and AmB-free micelleplexes was blocked by Baf-A1 pretreatment (Figure 6A). Thus, inhibition of PDMAb-PDPA protonation and micelleplex dissociation prevented siRNA-Luc efficacy. The acidic vesicular pH recovered and PDMA-b-PDPA micelleplexes dissociated 24 h after Baf-A1 incubation (Figure SI 4B); however, this did not lead to silence of Luc protein expression. This was most probably caused by the lysosome degradation of siRNA since all micelleplexes were found to accumulate in late endosomes or lysosomes 12 h postcellular uptake. This information suggests that timely siRNA endosomal escape is crucial for efficient target gene knockdown.43 Cells were treated at different time points after micelleplex addition to allow micelleplex dissociation and AmB release. As predicted, when the cells were treated by Baf-A1 3 h after micelleplex addition, AmB-loaded micelleplexes displayed recovered Luc expression knockdown ability like the Baf-A1-untreated control, presumably due to the AmB-induced, cytoplasmic delivery of siRNA (Figure 6B). In contrast, AmB-free micelleplex-induced Luc protein expression knockdown was blocked by Baf-A1 post-treatment (<20%) regardless of Baf-A1 addition time (i.e., 3, 6, or 12 h), suggesting low efficiency of siRNA intracellular release without AmB incorporation. These data supported the hypothesis that AmB increased/accelerated the siRNA endosomal escape at an early stage of endocytosis.
We proposed an AmB-enhanced siRNA endosomal escape mechanism in Figure 1. Upon endocytosis and micelleplex dissociation, the AmB molecules were released, then inserted into the endosome membrane, which causes dramatic destabilization of the endosome membrane by forming transmembrane pores.26–28 Meanwhile, protonation of PDMA and PDPA tertiary amines led to increased osmotic pressure, and protonated PDMA-b-PDPA unimers attached onto the endosome membrane may further increase membrane tension.44,45 We propose that these three factors synergistically caused disruption of the endosome membranes to release siRNA (Figure 1B). In contrast, AmB-free micelleplexes were entrapped in late endosomes/lysosomes and caused vesicle swelling due to the lack of membrane disruption capacity (Figure 1C).
Compared to conventional siRNA vectors, the AmB-loaded dual pH-responsive PDMA-b-PDPA micelleplex platform possesses several advantages. The PDMA-b-PDPA micelles are composed of a positively charged PDMA shell and a hydrophobic PDPA core at physiological conditions, thereby allowing sequential loading of AmB and siRNA into the micelleplex core and shell, respectively. The PDPA core can be selectively activated in early endosomes, leading to AmB release for endosome membrane destabilization. The nanoparticle-templated PDMA corona can increase the stability of siRNA complexation, as demonstrated by the low N/P ratio (i.e., N/P = 2.0) for the efficient compaction of siRNA (Figure 2E). The AmB-loaded micelleplex platform we presented here can also be applied for simultaneous co-delivery of therapeutic siRNA and water-insoluble anticancer drugs (e.g., paclitaxel) to exploit the maximal synergy for cancer therapy.46,47
CONCLUSION
In summary, we report the development of a novel AmB-loaded micelleplex platform for siRNA delivery into mammalian cells. The micelleplexes exhibited a dual pH-responsive property to changes of physiological and endosomal pH's. Upon endocytosis, the PDMA-b-PDPA micelleplexes dissociated to release AmB into endocytic vesicles to destabilize the endosome membranes. AmB loading did not show additional toxicity or affect the intracellular micelleplex uptake. Instead, AmB increased siRNA endosomal escape and enhanced siRNA-Luc knockdown efficiency caused by the PDMA-b-PDPA micelleplexes. We proposed a synergistic membrane disruption mechanism through AmB-mediated pore formation, PDMA-b-PDPA-induced increase in osmotic pressure and membrane tension. The described method provides a useful strategy to overcome the endosomal barrier for siRNA delivery and help to realize the potential of targeted RNAi therapy.
EXPERIMENTAL PROCEDURES
Materials
Diblock copolymers PDMA-b-PDPA, PEG-b-PDPA, PEG-b-PDMA, and PSt-b-PDMA were all synthesized by the ATRP method as described in the Supporting Information. Tetramethyl rhodamine-conjugated PEG-b-PDPA copolymer (PEG114-b-P(DPA60-r-TMR6)) was synthesized following our published procedure.23 Amphotericin B and bafilomycin A1 were ordered from Sigma-Aldrich. Ultracentrifugal units (molecular weight cutoff (MWCO) 100 kDa) and dialysis tubing (MWCO 3.5 kDa) were ordered from Fisher Scientific, Inc. Diethyl pyrocarbonate-treated Milli-Q water was autoclaved and used for siRNA dissolving. All other solvents and reagents were used as received from Sigma-Aldrich or Fisher Scientific, Inc. The CellLight Reagents for early endosome (Rab5a-GFP) or late endosome/lysosome (Lamp1-GFP) labeling were purchased from Invitrogen.
Ready to use siRNA duplexes, GL3 luciferase specific siRNA (siRNA-Luc), [5′-GAU UAU GUC CGG UUA UGU AUU-3′ (sense)], and scrambled RNA (siRNA-Scr) nonspecific to any human gene, [5′-CGG UGA GCC AGG CGU GCA AUU-3′ (sense)], were custom ordered from Dharmacon (Lafayette, Co., USA). siRNA-Scr with AlexaFluor647 labeling at the 5′ end (Alexa-siRNA-Scr) was ordered from QIAGEN Inc. (Valencia, CA, USA).
Micelle Preparation
Micelle stock solution (10 mg· mL−1) in Hepes-buffer glucose solution (HBG, 20 mM Hepes-HCl buffer, 5.0 w/v % glucose, pH 7.4) was prepared by a solvent evaporation method (see Supporting Information for details). PEG114-b-P(DPA60-r-TMR6) was mixed with PDMA-b-PDPA at 20/80 weight ratios for preparation of the TMR-labeled micelles. To prepare AmB-loaded micelles, PDMA-b-PDPA copolymer and a certain amount of AmB were dissolved in THF and DMSO, respectively, and the micelles were prepared by the solvent evaporation method. The PDMA-b-PDPA micelle stock solution was diluted to 0.5 mg · mL−1 in HBG for siRNA complexation.
AmB loading percentage and encapsulation efficiency were determined by UV–Vis spectra (λ = 384 nm). The particle size and zeta-potential of the micelles or siRNA-loaded micelleplexes were determined by dynamic light scattering with a Zetasizer Nano ZS (Malvern Instruments, Worcestershire, U.K.). All the measurements were conducted using at a He–Ne laser (λ = 633 nm) at 25 °C. The zeta-potential was measured using a folded capillary cell (Malvern Instruments, Herrenberg, Germany). The presented data were averaged from three independent measurements.
Erythrocyte Lysis Assay
The erythrocyte lysis assay was conducted following a literature method with minor adaption.37 Briefly, human erythrocytes were isolated from fresh citrate-treated mouse blood, washed first in phosphorus buffer saline (PBS) solution and then in lysis assay buffers (20 mM Hepes-HCl buffer, 150 mM NaCl, pH 7.4 or pH 6.2). The erythrocyte pellets were diluted with 10-fold lysis assay buffer. A certain amount of AmB-free or AmB-loaded micelle solution was added into 135 µL of lysis assay buffer in a 96-well tissue culture plate, mixed with 15 µL of erythrocyte suspension. The tissue culture plates were then incubated at 37 °C for 1 h under constant shaking. The hemoglobin release was determined using a microplate UV–vis spectra reader (Abs450). Complete erythrocyte lysis was determined by 10% Triton X-100-treated erythrocyte solution. The Abs450 of the lysis assay buffer was set as a negative control.
Cell Culture
Cell culture medium (DMEM or Opti-MEM) was obtained from Invitrogen or Gibco. A549 human non-small-cell lung cancer (NSCLC) cells were kindly provided by Dr. John Minna (UT Southwestern Medical Center, Dallas, TX). A549 cells stably expressing CMV-driven luciferase protein (A549-Luc) were generated by lentiviral vector-mediated transfection.48 Both A549 and A549-Luc cells were tested to be mycoplasma free and cultured in DMEM with 5% FBS supplement in a humidified 10% CO2 atmosphere at 37 °C.
Live Cell Imaging
A549-Luc cells expressing Rab5a-GFP (early endosome marker) or Lamp1-GFP (late endosome/lysosome marker) were produced by transfection with the CellLight Reagents following the manufacture's protocol. The transfected A549-Luc cells were seeded in 35 mm glass-bottom dishes in 2.0 mL of DMEM medium at a density of 5 × 104 per dish. After 24 h culture, the medium was replaced with fresh Opti-MEM, and 10.0 µg of TMR-labeled micelleplexes containing 1.0 µg of siRNA was added. The cells were incubated at 37 °C and imaged at designated times using a Nikon TE2000-E confocal laser scanning microscope equipped with a far-red filter and 60× objective lens. The GFP, TMR, and Alexa dyes were excited at 488, 543, and 575 nm and detected at 515, 595, and 675 nm (far-red), respectively.
To investigate AmB-enhanced siRNA endosomal escape, A549-Luc cells were seeded in a 35 mm cell culture dish and grown for 24 h in DMEM medium. The medium was replaced by 1.0 mL of Opti-MEM containing 2.0 mg of TMR-dextran. After 15 min incubation at 37 °C, 10.0 µg of micelleplex solution containing 1.0 µg of Alexa-siRNA was added. The cells were imaged with CLSM at designated time points.
Luc Knockdown Efficiency in Vitro
The Luc knockdown studies were performed in A549-Luc cells. Cells were seeded into 96-well tissue culture plates at a density of 5000 cells per well and cultured for 24 h. The DMEM growth medium was replaced by Opti-MEM medium, and 20 µL of micelleplex HBG solution was added into each well. After 24 h incubation, Opti-MEM medium was replaced by DMEM medium. The cells were cultured for an additional 24 h and lysed. The Luc activity was measured using a microplate luminometer (Centro LB 960, Berthold Technologies GmbH., Bad Wildbad, Germany). All the measurements were done in triplicates and repeated twice.
Influence of Baf-A1 Treatment on Micelleplex Dissociation and siRNA Transfection
To investigate the influence of Baf-A1 treatment on micelleplex dissociation, A549-Luc cells expressing Rab5a-GFP or Lamp1-GFP were cultured in fresh Opti-MEM medium containing 200 nM Baf-A1 and 10 µg of TMR-labeled micelleplexes (containing 1.0 µg siRNA, 75 nM). The cells were imaged by CLSM at different time points. After live cell imaging, part of the cells were collected for flow cytometry analysis.
To investigate the Baf-A1 effect on siRNA transfection, the A549-Luc cells were treated with 200 nM Baf-A1 at different time points after micelleplex addition. After 24 h incubation, the Opti-MEM medium was replaced by DMEM medium (with 5% FBS supplement). The cells were continually cultured for an additional 24 h and lysed for Luc activity analysis.
Statistical Analysis
Data were presented as the mean ± standard deviation. The statistical significance was determined using the analysis of variance (ANOVA) and two-tail Student's t test. p values < 0.05 were considered statistically significant.
Acknowledgments
This research was supported by the NIH grants R01CA129011 (J.G.) and R01CA102792 (D.A.B.). We would like to acknowledge the Shared Live Imaging Resource at the Harold C. Simmons Cancer Center, which is supported in part by an NCI Cancer Center Support Grant, P30CA142543.
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES AND NOTES
- 1.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-Nucleotide RNAs Mediate RNA Interference in Cultured Mammalian Cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 2.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and Specific Genetic Interference by Double-Stranded RNA in Caenorhabditis Elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 3.de Fougerolles A, Vornlocher HP, Maraganore J, Lieberman J. Interfering with Disease: A Progress Report on siRNA-Based Therapeutics. Nat. Rev. Drug Discovery. 2007;6:443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hannon GJ, Rossi JJ. Unlocking the Potential of the Human Genome with RNA Interference. Nature. 2004;431:371–378. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- 5.Pecot CV, Calin GA, Coleman RL, Lopez-Berestein G, Sood AK. RNA Interference in the Clinic: Challenges and Future Directions. Nat. Rev. Cancer. 2011;11:59–67. doi: 10.1038/nrc2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Juliano R, Alam MR, Dixit V, Kang H. Mechanisms and Strategies for Effective Delivery of Antisense and siRNA Oligonucleotides. Nucleic Acids Res. 2008;36:4158–4171. doi: 10.1093/nar/gkn342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas M, Lu JJ, Chen J, Klibanov AM. Non-Viral siRNA Delivery to the Lung. Adv. Drug Delivery Rev. 2007;59:124–133. doi: 10.1016/j.addr.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whitehead KA, Langer R, Anderson DG. Knocking Down Barriers: Advances in siRNA Delivery. Nat. Rev. Drug Discovery. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Green JJ, Langer R, Anderson DG. A Combinatorial Polymer Library Approach Yields Insight into Nonviral Gene Delivery. Acc. Chem. Res. 2008;41:749–759. doi: 10.1021/ar7002336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang HC, Samsonova O, Bae YH. Trafficking Micro-environmental pHs of Polycationic Gene Vectors in Drug-Sensitive and Multidrug-Resistant MCF7 Breast Cancer Cells. Biomaterials. 2010;31:3071–3078. doi: 10.1016/j.biomaterials.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meyer M, Philipp A, Oskuee R, Schmidt C, Wagner E. Breathing Life into Polycations: Functionalization with pH-Responsive Endosomolytic Peptides and Polyethylene Glycol Enables siRNA Delivery. J. Am. Chem. Soc. 2008;130:3272–3273. doi: 10.1021/ja710344v. [DOI] [PubMed] [Google Scholar]
- 12.Mok H, Lee SH, Park JW, Park TG. Multimeric Small Interfering Ribonucleic Acid for Highly Efficient Sequence-Specific Gene Silencing. Nat. Mat. 2010;9:272–278. doi: 10.1038/nmat2626. [DOI] [PubMed] [Google Scholar]
- 13.Wang XL, Ramusovic S, Nguyen T, Lu ZR. Novel Polymerizable Surfactants with pH-Sensitive Amphiphilicity and Cell Membrane Disruption for Efficient siRNA Delivery. Bioconjugate Chem. 2007;18:2169–2177. doi: 10.1021/bc700285q. [DOI] [PubMed] [Google Scholar]
- 14.Wang XL, Xu R, Wu X, Gillespie D, Jensen R, Lu ZR. Targeted Systemic Delivery of a Therapeutic siRNA with a Multifunctional Carrier Controls Tumor Proliferation in Mice. Mol. Pharmaceutics. 2009;6:738–746. doi: 10.1021/mp800192d. [DOI] [PubMed] [Google Scholar]
- 15.Yu H, Wagner E. Bioresponsive Polymers for Nonviral Gene Delivery. Curr. Opin. Mol. Ther. 2009;11:165–178. [PubMed] [Google Scholar]
- 16.Boussif O, Lezoualc'h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. A Versatile Vector for Gene and Oligonucleotide Transfer into Cells In Culture and In Vivo: Polyethylenimine. Proc. Natl. Acad. Sci. U. S. A. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tzeng SY, Guerrero-Cazares H, Martinez EE, Sunshine JC, Quinones-Hinojosa A, Green JJ. Non-Viral Gene Delivery Nanoparticles Based on Poly(beta-amino esters) for Treatment of Glioblastoma. Biomaterials. 2011;32:5402–5410. doi: 10.1016/j.biomaterials.2011.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hatakeyama H, Akita H, Ito E, Hayashi Y, Oishi M, Nagasaki Y, Danev R, Nagayama K, Kaji N, Kikuchi H, et al. Systemic Delivery of siRNA to Tumors Using a Lipid Nanoparticle Containing a Tumor-Specific Cleavable PEG-lipid. Biomaterials. 2011;32:4306–4316. doi: 10.1016/j.biomaterials.2011.02.045. [DOI] [PubMed] [Google Scholar]
- 19.Pittella F, Zhang M, Lee Y, Kim HJ, Tockary T, Osada K, Ishii T, Miyata K, Nishiyama N, Kataoka K. Enhanced Endosomal Escape of siRNA-Incorporating Hybrid Nanoparticles fromCalciumPhosphate and PEG-Block Charge-Conversional Polymer for Efficient Gene Knockdown with Negligible Cytotoxicity. Biomaterials. 2011;32:3106–3114. doi: 10.1016/j.biomaterials.2010.12.057. [DOI] [PubMed] [Google Scholar]
- 20.Walker GF, Fella C, Pelisek J, Fahrmeir J, Boeckle S, Ogris M, Wagner E. Toward Synthetic Viruses: Endosomal pH-Triggered Deshielding of Targeted Polyplexes Greatly Enhances Gene Transfer in Vitro and in Vivo. Mol. Ther. 2005;11:418–425. doi: 10.1016/j.ymthe.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 21.Endoh T, Ohtsuki T. Cellular siRNA Delivery Using Cell-Penetrating Peptides Modified for Endosomal Escape. Adv. Drug Delivery Rev. 2009;61:704–709. doi: 10.1016/j.addr.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 22.Kwon EJ, Bergen JM, Pun SH. Application of An HIV gp41-Derived Peptide for Enhanced Intracellular Trafficking of Synthetic Gene and siRNA Delivery Vehicles. Bioconjugate Chem. 2008;19:920–927. doi: 10.1021/bc700448h. [DOI] [PubMed] [Google Scholar]
- 23.Zhou K, Wang Y, Huang X, Luby-Phelps K, Sumer BD, Gao J. Tunable, Ultrasensitive pH-Responsive Nanoparticles Targeting Specific Endocytic Organelles in Living Cells. Angew. Chem., Int. Ed. 2011;50:6109–6114. doi: 10.1002/anie.201100884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Casey JR, Grinstein S, Orlowski J. Sensors and Regulators of Intracellular pH. Nat. Rev. Mol. Cell Biol. 2010;11:50–61. doi: 10.1038/nrm2820. [DOI] [PubMed] [Google Scholar]
- 25.Won YY, Sharma R, Konieczny SF. Missing Pieces in Understanding the Intracellular Trafficking of Polycation/DNA Complexes. J. Controlled Release. 2009;139:88–93. doi: 10.1016/j.jconrel.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jain JP, Jatana M, Chakrabarti A, Kumar N. Amphotericin-B-Loaded Polymersomes Formulation (PAMBO) Based on (PEG)-PLA Copolymers: An In Vivo Evaluation in a Murine Model. Mol. Pharm. 2011;8:204–212. doi: 10.1021/mp100267k. [DOI] [PubMed] [Google Scholar]
- 27.Binet A, Bolard J. Recovery of Hepatocytes from Attack by the Pore Former Amphotericin B. Biochem. J. 1988;253:435–440. doi: 10.1042/bj2530435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozmin SA. Chemical “Knockout” Challenges the Amphotericin B Channel Model. Nat. Chem. Biol. 2008;4:19–20. doi: 10.1038/nchembio0108-19. [DOI] [PubMed] [Google Scholar]
- 29.Garcia-Chaumont C, Seksek O, Jolles B, Bolard J. A Cationic Derivative of Amphotericin B as a Novel Delivery System for Antisense Oligonucleotides. Antisense Nucleic Acid Drug Dev. 2000;10:177–184. doi: 10.1089/oli.1.2000.10.177. [DOI] [PubMed] [Google Scholar]
- 30.Gary DJ, Lee H, Sharma R, Lee JS, Kim Y, Cui ZY, Jia D, Bowman VD, Chipman PR, Wan L, et al. Influence of Nano-Carrier Architecture on In Vitro siRNA Delivery Performance and In Vivo Biodistribution: Polyplexes vs Micelleplexes. ACS Nano. 2011;5:3493–3505. doi: 10.1021/nn102540y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Du J, Armes SP. pH-Responsive Vesicles Based on a Hydrolytically Self-Cross-Linkable Copolymer. J. Am. Chem. Soc. 2005;127:12800–12801. doi: 10.1021/ja054755n. [DOI] [PubMed] [Google Scholar]
- 32.Bowman EJ, Siebers A, Altendorf K. Bafilomycins: A Class of Inhibitors of Membrane ATPases From Microorganisms, Animal Cells, and Plant Cells. Proc. Natl. Acad. Sci. U. S. A. 1988;85:7972–7976. doi: 10.1073/pnas.85.21.7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson LS, Dunn KW, Pytowski B, McGraw TE. Endosome Acidification and Receptor Trafficking: Bafilomycin A1 Slows Receptor Externalization by A Mechanism Involving the Receptor's Internalization Motif. Mol. Biol. Cell. 1993;4:1251–1266. doi: 10.1091/mbc.4.12.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emans N, Biwersi J, Verkman AS. Imaging of Endosome Fusion in BHK Fibroblasts Based on A Novel Fluorometric Avdin-Biotin Binding Assay. Biophys. J. 1995;69:716–728. doi: 10.1016/S0006-3495(95)79947-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berezhna SY, Supekova L, Supek F, Schultz PG, Deniz AA. siRNA in Human Cells Selectively Localizes to Target RNA Sites. Proc. Natl. Acad. Sci. USA. 2006;103:7682–7687. doi: 10.1073/pnas.0600148103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andreoli TE. On the Anatomy of Amphotericin B-Cholesterol Pores in Lipid Bilayer Membranes. Kidney Int. 1973;4:337–345. doi: 10.1038/ki.1973.126. [DOI] [PubMed] [Google Scholar]
- 37.Plank C, Oberhauser B, Mechtler K, Koch C, Wagner E. The Influence of Endosome-Disruptive Peptides on Gene Transfer Using Synthetic Virus-Like Gene Transfer Systems. J. Biol. Chem. 1994;269:12918–12924. [PubMed] [Google Scholar]
- 38.Papadopoulos S, Jurgens KD, Gros G. Protein Diffusion in Living Skeletal Muscle Fibers: Dependence on Protein Size, Fiber Type, and Contraction. Biophys. J. 2000;79:2084–2094. doi: 10.1016/S0006-3495(00)76456-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benoit DS, Srinivasan S, Shubin AD, Stayton PS. Synthesis of Folate-Functionalized RAFT Polymers for Targeted siRNA Delivery. Biomacromolecules. 2011;12:2708–2714. doi: 10.1021/bm200485b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yezhelyev MV, Qi L, O'Regan RM, Nie S, Gao X. Proton-Sponge Coated Quantum Dots for siRNA Delivery and Intracellular Imaging. J. Am. Chem. Soc. 2008;130:9006–9012. doi: 10.1021/ja800086u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller DK, Griffiths E, Lenard J, Firestone RA. Cell Killing by Lysosomotropic Detergents. J. Cell Biol. 1983;97:1841–1851. doi: 10.1083/jcb.97.6.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lomas H, Massignani M, Abdullah KA, Canton I, Lo Presti C, MacNeil S, Du J, Blanazs A, Madsen J, Armes SP, et al. Non-Cytotoxic Polymer Vesicles for Rapid and Efficient Intracellular Delivery. Faraday Discuss. 2008;139:143–159. doi: 10.1039/b717431d. [DOI] [PubMed] [Google Scholar]
- 43.Tseng YC, Mozumdar S, Huang L. Lipid-Based Systemic Delivery of siRNA. Adv. Drug Delivery Rev. 2009;61:721–731. doi: 10.1016/j.addr.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang HW, Chen FY, Lee MT. Molecular Mechanism of Peptide-Induced Pores in Membranes. Phys. Rev. Lett. 2004;92:198304. doi: 10.1103/PhysRevLett.92.198304. [DOI] [PubMed] [Google Scholar]
- 45.Varkouhi AK, Scholte M, Storm G, Haisma HJ. Endosomal Escape Pathways for Delivery of Biologicals. J. Controlled Release. 2011;151:220–228. doi: 10.1016/j.jconrel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 46.Cao N, Cheng D, Zou S, Ai H, Gao J, Shuai X. The Synergistic Effect of Hierarchical Assemblies of siRNA and Chemotherapeutic Drugs Co-Delivered into Hepatic Cancer Cells. Biomaterials. 2011;32:2222–2232. doi: 10.1016/j.biomaterials.2010.11.061. [DOI] [PubMed] [Google Scholar]
- 47.Qiu LY, Bae YH. Self-Assembled Polyethylenimine-Graft-Poly(epsilon-caprolactone) Micelles as Potential Dual Carriers of Genes and Anticancer Drugs. Biomaterials. 2007;28:4132–4142. doi: 10.1016/j.biomaterials.2007.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dong Y, Bey EA, Li LS, Kabbani W, Yan J, Xie XJ, Hsieh JT, Gao J, Boothman DA. Prostate Cancer Radiosensitization through Poly(ADP-Ribose) Polymerase-1 Hyperactivation. Cancer Res. 2010;70:8088–8096. doi: 10.1158/0008-5472.CAN-10-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]


