Abstract
Background
The mammalian cyclin-dependent kinase subunit (Cks) family has two members, Cks1 and Cks2. Overexpression of Cks1 and Cks2 has been reported to be associated with high aggressiveness and poor prognosis in a few malignancies, including prostate and hepatocellular carcinomas. However, whether Cks1 and Cks2 are overexpressed in esophageal carcinoma remains uncharacterized.
Aims
To investigate whether overexpression of the Cks family is clinically relevant to esophageal carcinoma, and whether expression patterns of Cks1 and Cks2 in esophageal carcinoma have diagnostic and prognostic value.
Methods
Real-time quantitative reverse transcriptase polymerase chain reaction, immunohistochemistry and Western blot analyses were applied to detect the expression of Cks1 and Cks2 at the mRNA and protein levels respectively. The associations between Cks1 / Cks2 expressions and clinical features / p27kip1 expressions in esophageal carcinoma, were analysed.
Results
Expressions of Cks1 and Cks2 at both mRNA (Cks1: 15 out of 26 cases, 58%, Cks2: 17 out of 26 cases, 65%) and protein (Cks1: 30 out of 56 cases, 54%, Cks2: 34 out of 56 cases, 61%) levels were significantly higher in esophageal carcinoma than those in the adjacent noncancerous tissues. Overexpressions of Cks1 and Cks2 in esophageal carcinoma were closely associated with poor differentiation features like histologic grade of esophageal carcinoma, regional lymph nodes and neoplastic embolus. The expressions of both Cks1 and Cks2 were negatively associated with p27kip1 at the protein level.
Conclusions
Overexpression of Cks1 and Cks2 is associated with the aggressive tumour behaviours of esophageal carcinoma, and thus has diagnostic and prognostic value. Further efforts are needed to develop novel biomarkers for esophageal carcinoma based on Cks1 and Cks2 expressions.
Keywords: Cks1, Cks2, esophageal carcinoma, p27kip1
The development of tumour is closely related to the disorder of cell cycle, the core of cell cycle regulation is a group of Cyclin-dependent kinase (CDKs). Activation and inactivation of CDKs are regulated by cell cycle proteins and cyclin-dependent kinase inhibitor (CKIs), respectively. Cyclin-dependent kinase subunit (Cks) proteins, which consist of Cks1 and Cks2 in mammalian, are highly conserved molecules in eukaryotes. [1] Cks1 and Cks2 are subunits of CDKs required for cell division. Cks1 is required for SCFSkp2-mediated ubiquitination and degradation of p27kip1 that is essential for the G1/S transition during the cell cycle. Low levels of p27kip1 protein have been found to be associated with the high aggressiveness and poor prognosis in patients with a variety of malignancies. [2–7] Consistent emerging evidence shows that Cks1 and Cks2 are overexpressed in tumours of other tissue origins. Our laboratory has reported that clinical significance and expression of Cks in hepatocellular carcinoma. [8] Furthermore, an elevated expression of Cks1 and Cks2 are found to be coincident with the reduction of p27kip1 protein in tumour cells of gastric, breast, colorectal, prostate carcinomas. [9–15]
However, whether Cks1 or Cks2 is over expressed in esophageal carcinoma has not been reported. Esophageal carcinoma is one of the most frequent human cancers. About 22 million people worldwide died of esophageal cancer each year. The highest esophageal carcinoma frequencies are found in Asia, especially in China. The incidence of esophageal carcinoma is associated with the chronic stimulation of nitrosamines, inflammation and trauma, genetic factors as well as drinking water, trace elements in food and vegetables. Because the treatment effect of radiotherapy, chemotherapy in esophageal carcinoma is not satisfactory, we hope Cks1 and Cks2 to become the diagnostic markers and therapeutic targets of esophageal carcinoma.
Materials and methods
Patient recruitments and sample collections
Fresh surgical specimens of both esophageal cancer tissues and the matched adjacent noncancerous esophageal tissues (located ≥5cm away from the tumour margins) were obtained from 56 patients. The tissue collection procedures were according to local ethical guidelines and approved beforehand by the institution’s Human Investigation Committee. All patients had undergone surgery in the Department of Chest Surgery, Zhongshan Hospital, Xiamen University, from 2009 to 2011. None of the patients had received preoperative treatment, such as radiation or chemotherapies. The pathological classifications were performed according to Edmondson’s Classification. Tumour tissues were divided into two parts: one part was quickly frozen in liquid nitrogen for RNA and protein extractions and the other part was fixed in 10% formalin and then paraffin-embedded for immunohistochemical staining.
Quantitative real-time PCR
Total RNA extraction and reverse transcription were carried out separately with the RNA Fast1000 Kit (Fastagen Biotech, Shanghai, China) and M-MuLV reverse transcriptase (Fastagen Biotech) by following the manufacturers’ instructions. A total volume of 20ul was used for the PCR reaction, which contained template cDNA, 250nM probes, 125nM forward and reverse primers and 1×PCR Master Mix (Invitrogen Corporation, Shanghai, China). The reaction was initiated by activation of the Taq polymerase at 95°C for 10 min, followed by 45 cycles at 95°C for 15s and 60°C for 1 min. The PCR product length and the PCR primers are as follows: Cks1, 170 bp, forward primer TACGACGACGAGGAGTTTGAGT, reverse primer AGCAAGATGTGAGGTTCTGGTTC and probe TCATGCTGCCCAAGGACATAGCCAAGC; Cks2, 124 bp, forward primer CAAGCAGATCTACTACTCGGACAAG, reverse primer CCTCCACTCCTCTTCAGACATCA and probe CGACGAACACTACGAGTACCGGCATGT; p27kip1, 146 bp, forward primer GTTAGCGGAGCAATGCGC, reverse primer CAGGCTTCTTGGGCGTCTG and probe AAGCGACCTGCAACCGACGATTCTTCT; and β-actin, 293 bp, forward primer TCACCCACACTGTGCCCATCTACGA, reverse primer CAGCGGAACCGCTCATTGCCAATGG and probe ATGCCCTCCCCCAT GCCATCCTGCGT. All primers were synthesized by Invitrogen Corporation. Data were normalized with the β-actin housekeeping gene and were expressed as 2−ΔΔCt. The cases were classified into two groups: high-expression (2−ΔΔCt > 1) and low-expression (2−ΔΔCt < 1) groups.
Immunohistochemical staining
Immunohistochemical (IHC) staining was performed on formalin-fixed and paraffin-embedded tissue sections. The sections were deparaffinized with xylene and stepwise rehydrated with series dilutions of ethanol as described elsewhere. For epitope retrieval, slides were incubated in 0.01M citric acid buffer (pH 6.0) at 140°C for 20 min. The dilutions of the antibodies are Cks1 (1:300), Cks2 (1:300) and p27kip1 (1:200). After being washed, the specifically bound antibodies were detected with the EliVision™ Plus Kit (Maixin Bio, Fuzhou, China) according to the manufacturer’s instructions. All sections were counterstained with haematoxylin. Nonimmune rabbit serum (10%) was used for negative controls. No detectable staining was observed in all negative control slides.
Positively stained cells per 1000 cells in random fields were scored and the averages were derived from at least five slides. The scoring criteria were as follows: ++++, > 50% of the cells are positive; +++, 20–50% positive; ++, 10–20% positive; +, < 10% positive; and −, no positive staining. Expression levels of Cks1 and Cks2 proteins were categorized as low (− to ++) or high (+++ to ++++). Expression levels of p27kip1 protein were categorized as low (− to +++) or high (++++). The scoring was carried out blindly and independently by two observers.
Western blotting
Total protein was extracted from approximately 0.5g of cryopreservative tissues with the RIPA buffer (Beyotime, Shanghai, China). The protein concentrations were determined with the BCA Kit (Beyotime). Aliquots containing 30mg of protein were subjected to SDS-polyacrylamide gel electrophoreses and electroblotted onto a PVDF membrane (Millipore Co., Billerica, U.S.A.) for Western blot analyses. For Cks1/2 and p27kip1 staining, the membrane was cut into 4 pieces for staining with anti-p27kip1, anti-Cks1, anti-Cks2 and anti-β-actin antibodies individually. The experiments were repeated for 3 times and the representative results were showed. The specifically bound antibodies were detected with the ECL-plus chemiluminescent detection reagents (Millipore Co., Billerica, U.S.A.). Anti-p27kip1, anti-Cks1, anti-Cks2 and anti-β-actin antibodies were purchased from Abcam (Hong Kong) Ltd. (Abcam, Hong Kong).
Statistical analysis
All data were processed with the SPSS 13.0 (SPSS lnc., Chicago, IL, USA) statistical software package. The Spearman rank correlation was used to study the difference between Cks1 or Cks2 mRNA and protein expression and various clinicopathological features. The cross tabulation and Pearson’s χ2 were used to analyse the data from the esophageal carcinoma. The relations between Cks1 or Cks2 and p27Kip1 expressions were analysed with the Spearman rank correlation. Differences were considered significant if the P-value was < 0.05, which was calculated by the two-tailed tests.
Results
Expressions of Cks1 and Cks2 are elevated at the mRNA and protein levels in esophageal carcinoma
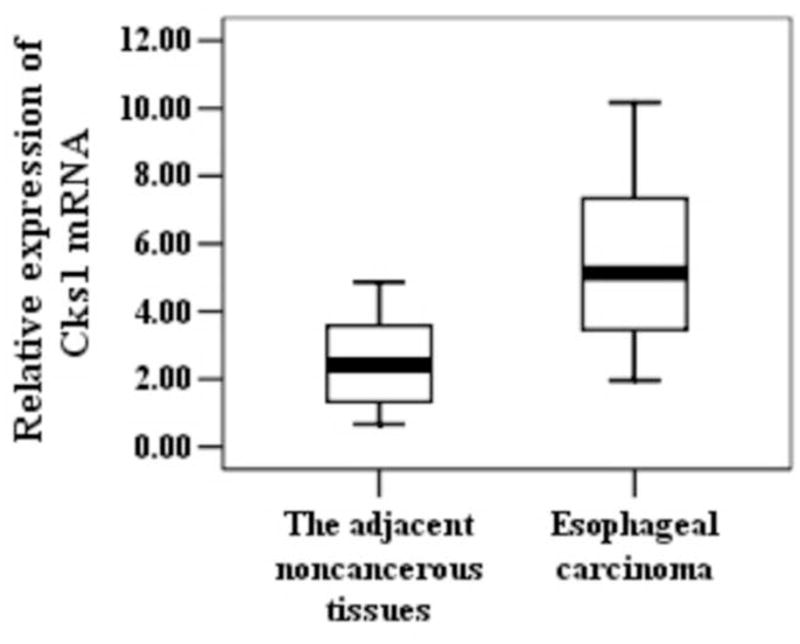
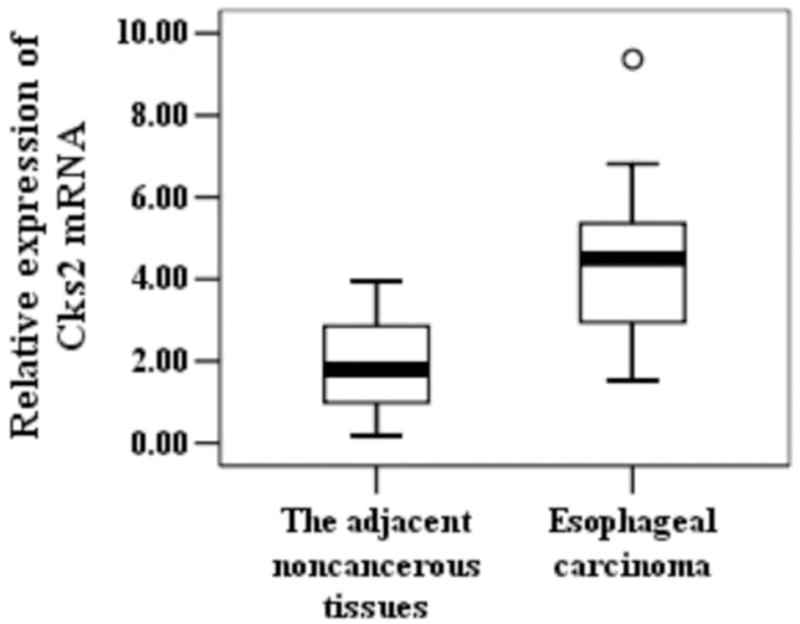
To determine whether Cks1 and Cks2 were overexpressed during the development of esophageal carcinoma, we measured their expressions at both mRNA and protein levels in esophageal carcinoma and its adjacent esophageal tissues. Real-time PCR analyses revealed that both Cks1 and Cks2 mRNA levels were significantly elevated in esophageal carcinoma (Figure. 1). The majority of esophageal carcinoma showed high Cks1 (15 out of 26 cases, 58%) and Cks2 (17 out of 26 cases, 65%) expressions. To establish whether Cks1 and Cks2 protein levels were also increased in esophageal carcinoma, immunohistochemical staining (Figure. 2) and Western blot analyses (Figure. 3) were applied. The results showed that the protein levels of Cks1 and Cks2 were not uniformly high in esophageal carcinoma. Therefore, quantitative analyses were used to assess the Cks protein expressions in these tissues. The data in Table 2 clearly demonstrated that most adjacent esophageal tissues showed lower Cks1 and Cks2 expressions. In contrast, the majority of esophageal carcinoma showed higher Cks1 (30 out of 56 cases, 54%) and Cks2 (34 out of 56 cases, 61%) expressions.
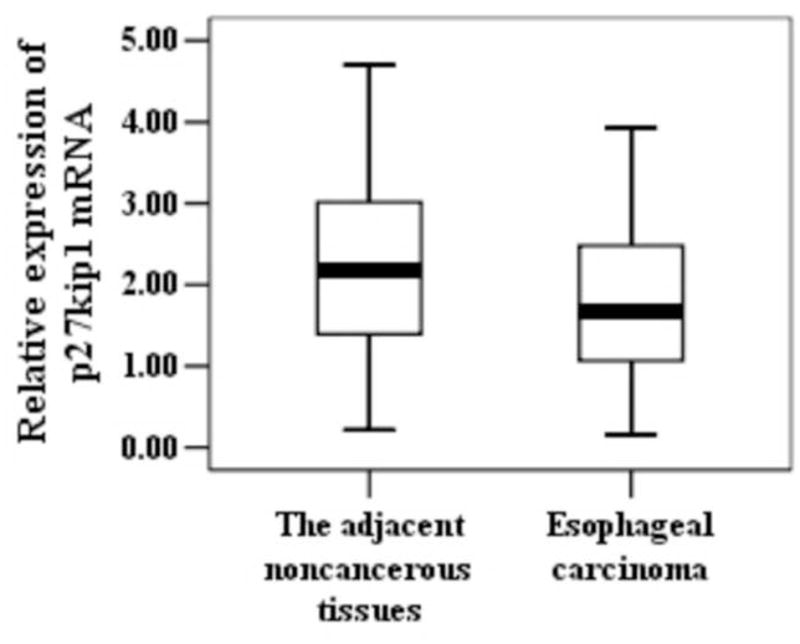
Figure 1.

Expression of Cks1, Cks2 and p27kip1 at the mRNA level in different esophageal tissues. Total RNA samples were extracted from esophageal carcinoma and the adjacent noncancerous tissues. Expressions of Cks1, Cks2 and p27kip1 were assessed with real-time PCR. Data were normalized to β-actin. P-value < 0.05 is considered statistically significant. °statistically insignificant.
Figure 2.
Immunohistochemical staining of Cks1, Cks2 and p27kip1. Note that more cells in moderately to poorly differentiated esophageal carcinoma (T) exhibited strong nuclear staining of Cks1 (A1) and Cks2 (B1), and weak staining of p27kip1 (C1). In contrast, most cells in the adjacent noncancerous tissues (N) exhibited were weak staining of Cks1 (A2) and Cks2 (B2) and strong staining of p27kip1(C2). All the above results come from the one sample.
Figure 3.

Western blot analysis of Cks1, Cks2 and p27kip1. The proteins extracted from esophageal carcinoma (T) and the adjacent noncancerous tissues (N) were subjected to Western blot analyses with anti-Cks1, Cks2 and p27kip1 as indicated. β-actin was used as the loading control. All the above results come from the one sample.
Table 2.
Expression of Cks1, Cks2 and p27kip1 proteins in different esophageal tissues
| Tissues | N | Cks1(protein) | Cks2(protein) | p27 kip1(protein) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||
| X±SX* | Low | High | P | X±SX* | Low | High | P | X±SX* | Low | High | P | ||
| The adjacent noncancerous tissues | 56 | 1.13±0.15 | 48 | 8 | 1.14±0.14 | 49 | 7 | 1.12±0.11 | 21 | 35 | |||
| Esophageal carcinoma | 56 | 2.59±0.15 | 26 | 30 | <0.05# | 2.61±0.16 | 22 | 34 | <0.05# | 2.62±0.12 | 32 | 24 | <0.05# |
Immunohistochemical results were converted to numeric values based on the estimated levels (0–4). Sx, standard error.
P values < 0.05 are considered statistically significant.
P values > 0.05 are considered statistically nonsignificant.
Overexpressions of Cks1 and Cks2 are associated with aggressive behaviour of esophageal carcinoma
To further investigate whether overexpressions of Cks1 and Cks2 in esophageal carcinoma had clinicopathological relevance, the associated Cks1 and Cks2 expressions with tumour pathological features were analysed. The results showed that overexpressions of Cks1 and Cks2 at both mRNA and protein levels were significantly associated with histologic grade of esophageal carcinoma (Table 3). In addition, overexpression of Cks proteins was also associated with regional lymph nodes and neoplastic embolus. The association of Cks overexpression with other clinicopathological features was not consistent at the mRNA and protein levels, which included tumour size and position, morphological type, neural invasion and distant metastasis, as well as patient sex. Together, the results suggest that expressions of Cks1 and Cks2 have potential value for esophageal carcinoma diagnosis.
Table 3.
Relationships between Cks1 and Cks2 expression and clinicopathological features in esophageal carcinoma
| Features | N | Cks1 (protein) | Cks2 (protein) | N | Cks1 (mRNA) | Cks2 (mRNA) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||||
| Low | High | P | Low | High | P | High | X±Sx | P | High | X±Sx | P | |||
| Age | ||||||||||||||
| >60 | 35 | 15 | 20 | 11 | 24 | 17 | 10 | 4.51±0.46 | 12 | 5.62±0.57 | ||||
| ≤60 | 21 | 11 | 10 | 0.340 | 11 | 10 | 0.346 | 9 | 5 | 3.99±0.54 | 0.288 | 5 | 4.70±0.71 | 0.314 |
| Sex | ||||||||||||||
| Male | 48 | 22 | 26 | 18 | 30 | 23 | 13 | 4.52±0.38 | 15 | 5.32±0.50 | ||||
| Female | 8 | 4 | 4 | 0.738 | 4 | 4 | 0.702 | 3 | 2 | 2.86±0.43 | 0.119 | 2 | 5.16±0.64 | 0.907 |
| Primary tumor | ||||||||||||||
| T1 | 5 | 4 | 1 | 3 | 2 | 1 | 1 | 5.36 | 1 | 8.67 | ||||
| T2 | 10 | 5 | 5 | 5 | 5 | 2 | 1 | 3.93±0.43 | 2 | 5.75±0.13 | ||||
| T3 | 35 | 14 | 21 | 11 | 24 | 20 | 12 | 4.41±0.42 | 12 | 5.10±0.55 | ||||
| T4 | 6 | 3 | 3 | 0.144 | 3 | 3 | 0.323 | 3 | 1 | 3.75±0.97 | 0.615 | 3 | 5.22±0.52 | 0.355 |
| Regional lymph nodes | ||||||||||||||
| N0 | 11 | 10 | 1 | 9 | 2 | 6 | 1 | 2.92±0.40 | 2 | 3.97±0.35 | ||||
| N1 | 45 | 16 | 29 | 0.001* | 13 | 32 | 0.003* | 20 | 14 | 4.76±0.40 | 0.014* | 15 | 5.70±0.54 | 0.067 |
| Distant metastasis | ||||||||||||||
| M0 | 50 | 22 | 28 | 20 | 30 | 25 | 14 | 4.31±0.37 | 16 | 5.30±0.46 | ||||
| M1 | 6 | 4 | 2 | 0.400 | 2 | 4 | 0.978 | 1 | 1 | 4.91 | 0.746 | 1 | 5.200 | 0.948 |
| Histologic grade | ||||||||||||||
| G1 | 13 | 11 | 2 | 9 | 4 | 4 | 1 | 2.95±0.31 | 2 | 2.83±0.56 | ||||
| G2 | 31 | 11 | 20 | 12 | 19 | 15 | 7 | 4.35±0.55 | 8 | 5.26±0.56 | ||||
| G3 | 12 | 4 | 8 | 0.003* | 1 | 11 | 0.001* | 7 | 7 | 5.08±0.36 | 0.027* | 7 | 6.80±0.69 | 0.003* |
| Neoplastic embolus | ||||||||||||||
| Positive | 29 | 8 | 21 | 10 | 29 | 19 | 14 | 4.68±0.43 | 14 | 5.84±0.55 | ||||
| Negative | 27 | 18 | 9 | 0.041* | 12 | 5 | 0.215 | 7 | 1 | 4.41±0.79 | 0.100 | 3 | 3.83±0.41 | 0.023* |
| Neural invasion | ||||||||||||||
| Positive | 39 | 16 | 23 | 14 | 25 | 18 | 12 | 4.30±0.39 | 13 | 5.76±0.52 | ||||
| Negative | 17 | 10 | 7 | 0.306 | 8 | 9 | 0.139 | 8 | 3 | 4.41±0.79 | 0.706 | 4 | 4.27±0.79 | 0.096 |
| Tumor position | ||||||||||||||
| Upper | 4 | 1 | 3 | 3 | 1 | 2 | 1 | 4.97±0.39 | 1 | 5.63±3.04 | ||||
| Middle | 38 | 18 | 20 | 16 | 22 | 17 | 8 | 4.35±0.47 | 10 | 5.20±0.57 | ||||
| Lower | 14 | 7 | 7 | 0.806 | 3 | 11 | 0.089 | 7 | 6 | 4.10±0.66 | 0.534 | 6 | 5.44±0.72 | 0.882 |
| Morphological type | ||||||||||||||
| Ulcerative type | 39 | 15 | 24 | 14 | 25 | 18 | 12 | 4.04±0.36 | 13 | 5.26±0.53 | ||||
| Medullay type | 14 | 8 | 6 | 7 | 7 | 6 | 1 | 4.28±0.68 | 2 | 4.91±1.04 | ||||
| Protruded type | 3 | 3 | 0 | 0.098 | 1 | 2 | 0.773 | 2 | 2 | 7.07±2.31 | 0.484 | 2 | 6.82±1.75 | 0.812 |
| Tumor size (cm) | ||||||||||||||
| >3 | 33 | 16 | 17 | 17 | 16 | 16 | 11 | 4.24±0.80 | 10 | 4.90±0.82 | ||||
| ≤3 | 23 | 10 | 13 | 0.277 | 5 | 18 | 0.054 | 10 | 4 | 4.39±0.31 | 0.608 | 7 | 5.55±0.53 | 0.410 |
P value < 0.05 is considered statistically significant.
P values > 0.05 are considered statistically nonsignificant.
Cks1 and Cks2 expressions are negatively associated with p27kip1 protein levels in esophageal carcinoma
Although it is a tumour suppressor, p27kip1 seldom has abnormalities at the genomic level, and downregulation of p27kip1 mRNA is rarely observed in human cancers. The abundance of p27kip1 protein in tumour cells is frequently reduced via the Cks1-mediated degradation. Consistently, we did not observe significant differences in p27kip1 mRNA expression levels in esophageal carcinoma (Table 1). To study whether the abundance of p27kip1 protein was reduced in esophageal carcinoma, immunohistochemical staining (Figure. 2) and Western blot analyses (Figure. 3) were used to assess p27kip1 protein in the aforementioned tissues. The results revealed that the abundance of p27kip1 protein was significantly reduced in esophageal carcinoma. Interestingly, statistical analyses revealed that the abundance of p27kip1 was not only negatively correlated with Cks1 expression, but also negatively correlated with Cks2 expression (Figure. 4). The biological significance of the negative correlation between Cks2 and p27kip1 abundance remains elusive.
Table 1.
Expression of Cks1, Cks2 and p27kip1 mRNA in esophageal tissues
| N | Cks1(mRNA) | Cks2(mRNA) | p27kip1(mRNA) | |||
|---|---|---|---|---|---|---|
| Low* | High* | Low | High | Low | High | |
| 26 | 11 | 15 | 9 | 17 | 14 | 12 |
high-expression (2−ΔΔCt > 1) and low-expression (2−ΔΔCt < 1)
Figure 4.

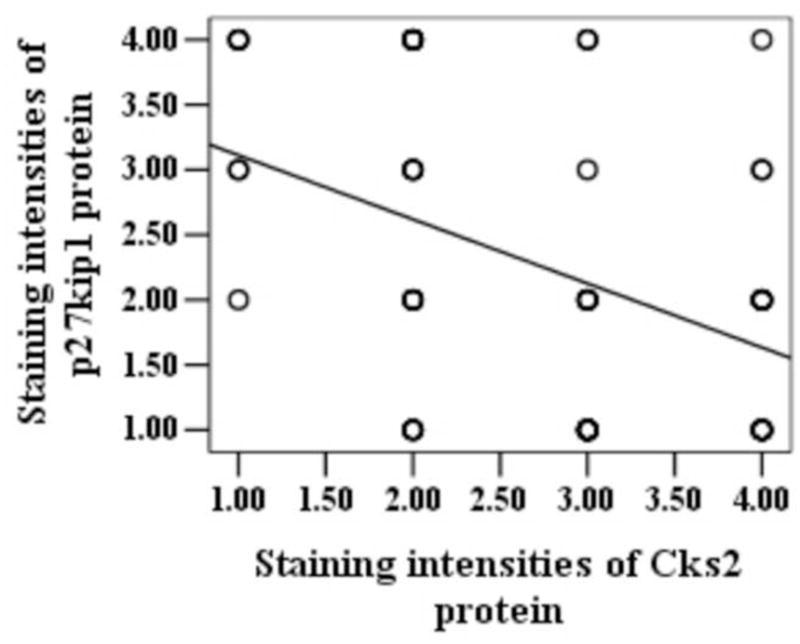
Regression analysis of the relationship between the abundance of Cks1/Cks2 and p27kip1 proteins. The circles in (a) and (b) indicates the detected patients immunostained cells in esophageal carcinoma tissue slides from 56 patients were categorized into five groups (0–4) based on the staining intensities. The two oblique lines indicated that there were negative correlations between expressions of Cks1 and p27kip1 (r = −0.447, P = 0.001), and between the expressions of Cks2 and p27kip1 (r = −0.407, P = 0.003).
The consistency between mRNA and protein levels in overexpressed Cks1 / Cks2 in esophageal carcinoma
To investigate the consistency between mRNA and protein levels in overexpressed Cks1 and Cks2, we counted 26 cases of esophageal carcinoma (Figure. 5). The percent of Cks1 high experssion at mRNA and protein levels is 53% (14 out of 26 cases). And this proportion for Cks2 is 57% (15 out of 26 cases). The percent of Cks1 low experssion at mRNA and protein levels is 4% (1 out of 26 cases). And this proportion for Cks2 is 8% (2 out of 26 cases). There were some discrepancies in Cks1/Cks2 expressions at the mRNA and protein levels.
Figure 5.

The consistency between mRNA and protein levels in overexpressed Cks1/Cks2. The percent of Cks1 high experssion at mRNA and protein levels is 53%. And this proportion for Cks2 is 57%. The percent of Cks1 low experssion at mRNA and protein levels is 4%. And this proportion for Cks2 is 8%.
Discussion
The clinical significance of overexpressed Cks1 and Cks2 in esophageal carcinoma
In this study, we observed that expressions of Cks1 and Cks2 were elevated at both mRNA and protein levels in esophageal carcinoma in comparison with the adjacent noncancerous tissues. The results showed that overexpressions of Cks1 and Cks2 at both mRNA and protein levels were significantly associated with histological grade of esophageal carcinoma, regional lymph nodes and neoplastic embolus. The association of Cks overexpression with other clinicopathological features was not consistent at the mRNA and protein levels. It is possible that the difference is tiny and that more samples are needed to determine whether the difference is significant. Nevertheless, the data clearly shows that overexpressions of Cks1 and Cks2 are strongly associated with various clinicopathological features that are commonly used to determine aggressive tumour behaviours, including histologic grade, regional lymph nodes and neoplastic embolus. The results suggest that the overexpressions of Cks1 and Cks2 may contribute to the development of esophageal carcinoma. Because of the lack of a complete course of esophageal carcinoma patients, we did not conduct the Kaplan – Meier survival analysis. Further studies are needed to clarify this issue.
Negative association between Cks1 / Cks2 expression and p27kip1
The control of p27Kip1 protein level is affected by ubiquitin dependent degradation, in an ubiquitin and Skp2-independent manner at G1. The effect of p27Kip1 on the cell cycle is regulated mainly by its stability. It has been revealed that Cks1 is required for p27Kip1 ubiquitination. [16–19] The association between Cks1 overexpression and reduction abundance of p27kip1 has been shown in several malignancies. [9, 10] In our former study, the abundance of p27kip1 was negatively correlated with Cks1 expression in hepatocellular carcinoma. [8] In this report, we also demonstrate a negative correlation between Cks1 expression and p27Kip1 protein level, which further suggests that Cks1 overexpression contributes to tumorigenesis of esophageal carcinoma by promoting the SCFSkp2-mediated degradation of p27Kip1.
The Cks family, Cks1 and Cks2, are small but essential components of cyclin/CDK complexes contributing to the cell cycle control and have 81% identical amino acids. Various tumors have revealed the dysregulation of Cks and other cell cycle-related regulators including cyclins and CDKs. Cks1 is an essential cofactor in the degradation of the p27kip1 by the SCFSkp2 ubiquitin ligase. In contrast, the function of Cks2 in mammalian somatic cells remains poorly understood, although we found that Cks2 was negatively associated with p27kip1 protein in esophageal carcinoma. Accumulating evidence demonstrates that expression of Cks2 is elevated in malignant tumours of different tissues, including bladder, prostate, cervical and hepatocellular carcinomas. [9, 11–12] Whether Cks2 also contributes to p27kip1 degradation in esophageal carcinoma remains to be investigated. Although Cks2 does not have the proteolytic activity against the p27kip1, both Cks1 and Cks2 are essential for the progression into and exit from mitosis.
The consistency between mRNA and protein levels in overexpressed Cks1/Cks2
To investigate the consistency between mRNA and protein level in Cks overexpression, we counted 26 cases of esophageal carcinoma. The analysis showed that the percent of Cks1 overexperssion at mRNA and protein levels is 53%. And this proportion for Cks2 is 57%. There were some discrepancies in Cks expressions at the mRNA and protein levels. It is possible that the difference is caused by the different translation efficiencies or stabilities of the protein in tumour tissues. In addition, several genetic and epigenetic factors, such as DNA methylation, genetic mutation and abnormal posttranscriptional regulations, may also have contributed to the variation of the results.
Conclusion
In conclusion, we found that Cks1 and Cks2 overexpressed in esophageal carcinoma. The results suggest that abnormal expression of Cks1 and Cks2 may contribute to esophageal carcinoma’s initiation and progression. Alterations in gene expression in esophageal carcinoma may yield clues to their pathogenesis. Analysis of a larger number of esophageal carcinoma specimens and correlation with biological phenotypes with gene expression patterns may identify clinically meaningful characteristics of this malignancy. This could provide important clues to develop novel diagnostic and prognostic markers as well as strategies for efficient prevention and therapy strategy for esophageal carcinoma. In this study, we first demonstrate that Cks1 and Cks2 expression is strongly correlated with the progression of esophageal carcinoma. Our comprehensive expression profiling data of esophageal carcinoma provide a new insight into the molecular biology level of esophageal carcinoma.
References
- 1.Richardson HE, Stueland CS, Thomas J, Russell P, Reed SI. Human cDNAs encoding homologs of the small p34Cdc28/Cdc2-associated protein of Saccharomyces cerevisiae and Schizosaccharomyces pombe. Genes Dev. 1990;4:1332–1344. doi: 10.1101/gad.4.8.1332. [DOI] [PubMed] [Google Scholar]
- 2.Wang X, Gorospe M, Huang Y, Holbrook NJ. P27Kip1 overexpression causes apoptotic death of mammalian cells. Oncogene. 1997;15:2991–2997. doi: 10.1038/sj.onc.1201450. [DOI] [PubMed] [Google Scholar]
- 3.Hara T, Kamura T, Nakayama K, Oshikawa K, Hatakeyama S. Degradation of p27(Kip1) at the G(0)-G(1) transition mediated by a Skp2-independent ubiquitination pathway. J Biol Chem. 2001;276:48937–48943. doi: 10.1074/jbc.M107274200. [DOI] [PubMed] [Google Scholar]
- 4.Kamura T, Hara T, Matsumoto M, Ishida N, Okumura F, Hatakeyama S, Yoshida M, Nakayama K, Nakayama KI. Cytoplasmic ubiquitin ligase KPC regulates proteolysis of p27(Kip1) at G1 phase. Nat Cell Biol. 2004;6:1229–1235. doi: 10.1038/ncb1194. [DOI] [PubMed] [Google Scholar]
- 5.Vlach J, Hennecke S, Amati B. Phosphorylation dependent degradation of the cyclin-dependent kinase inhibitor p27. EMBO J. 1997;16:5334–5344. doi: 10.1093/emboj/16.17.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sutterluty H, Chatelain E, Marti A, Wirbelauer C, Senften M, Muller U, Krek W. p45SKP2 promotes p27Kip1 degradation and induces S phase in quiescent cells. Nat Cell Biol. 1999;1:207–214. doi: 10.1038/12027. [DOI] [PubMed] [Google Scholar]
- 7.Sheaff RJ, Groudine M, Gordon M, Roberts JM, Clurman BE. Cyclin E-CDK2 is a regulator of p27Kip1. Genes Dev. 1997;11:1464–1478. doi: 10.1101/gad.11.11.1464. [DOI] [PubMed] [Google Scholar]
- 8.Shen DY, Fang ZX, You P, Liu PG, Wang F, Huang CL, Yao XB, Chen ZX, Zhang ZY. Clinical significance and expression of cyclin kinase subunits 1 and 2 in hepatocellular carcinoma. Liver Int. 2010;30(1):119–125. doi: 10.1111/j.1478-3231.2009.02106.x. [DOI] [PubMed] [Google Scholar]
- 9.Lan Y, Zhang Y, Wang J, Lin C, Ittmann MM, Wang F. Aberrant expression of Cks1 and Cks2 contributes to prostate tumorigenesis by promoting proliferation and inhibiting programmed cell death. Int J Cancer. 2008;123(3):543–551. doi: 10.1002/ijc.23548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang MA, Kim JT, Kim JH, Kim SY, Kim YH, Yeom YI, Lee Y, Lee HG. Upregulation of the cycline kinase subunit CKS2 increases cell proliferation rate in gastric cancer. J Cancer Res Clin Oncol. 2009;135(6):761–769. doi: 10.1007/s00432-008-0510-3. [DOI] [PubMed] [Google Scholar]
- 11.Wong YF, Cheung TH, Tsao GS, Lo KW, Yim SF, Wang VW, Heung MM, Chan SC, Chan LK, Ho TW, Wong KW, Li C, Guo Y, Chung TK, Smith DI. Genome-wide gene expression profiling of cervical cancer in Hong Kong women by oligonucleotide microarray. Int J Cancer. 2006;118(10):2461–2469. doi: 10.1002/ijc.21660. [DOI] [PubMed] [Google Scholar]
- 12.Lyng H, Brøvig RS, Svendsrud DH, Holm R, Kaalhus O, Knutstad K, Oksefjell H, Sundfør K, Kristensen GB, Stokke T. Gene expressions and copy numbers associated with metastatic phenotypes of uterine cervical cancer. BMC Genomics. 2006;7:268. doi: 10.1186/1471-2164-7-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawakami K, Enokida H, Tachiwada T, Gotanda T, Tsuneyoshi K, Kubo H, Nishiyama K, Takiguchi M, Nakagawa M, Seki N. Identification of differentially expressed genes in human bladder cancer though genome-wide gene expression profiling. Oncol Rep. 2006;16(3):521–531. [PubMed] [Google Scholar]
- 14.Wiese AH, Auer J, Lassmann S, Nährig J, Rosenberg R, Höfler H, Rüger R, Werner M. Identification of gene signatures for invasive colorectal tumor cells. Cancer Detect Prev. 2007;31(4):282–295. doi: 10.1016/j.cdp.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 15.Lin HM, Chatterjee A, Lin YH, Anjomshoaa A, Fukuzawa R, McCall JL, Reeve AE. Genome wide expression profiling identifies genes associated with colorectal liver metastasis. Oncol Rep. 2007;17(6):1541–1549. doi: 10.3892/or.17.6.1541. [DOI] [PubMed] [Google Scholar]
- 16.Ganoth D, Bornstein G, Ko TK, Larsen B, Tyers M, Pagano M, Hershko A. The cell-cycle regulatory protein Cks1 is required for SCFSkp2-mediated ubiquitinylation of p27. Nature Cell Biol. 2001;3:321–324. doi: 10.1038/35060126. [DOI] [PubMed] [Google Scholar]
- 17.Spruck C, Strohmaier H, Watson M, Smith AP, Ryan A, Krek TW, Reed SI. A CDK-independent function of mammalian Cks1: targeting of SCFSkp2 to the CDK inhibitor p27. Mol Cell. 2001;7:639–650. doi: 10.1016/s1097-2765(01)00210-6. [DOI] [PubMed] [Google Scholar]
- 18.Sitry D, Seeliger MA, Ko TK, Ganoth D, Breward SE, Itzhaki LS, Pagano M, Hershko A. Three different binding sites of Cks1 are required for p27-ubiquitin ligation. J Biol Chem. 2002;277:42233–42240. doi: 10.1074/jbc.M205254200. [DOI] [PubMed] [Google Scholar]
- 19.Pagano M, Tam SW, Theodoras AM, Beer-Romero P, Del Sal G, Chau V, Yew PR, Draetta GF, Rolfe M. Role of the ubiquitin – proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science. 1995;269:682–685. doi: 10.1126/science.7624798. [DOI] [PubMed] [Google Scholar]


