Abstract
The primary cilium is a protrusion from the cell surface that serves as a specialized compartment for signal transduction. Many signaling factors are known to be dynamically concentrated within cilia and to require cilia for their function. Yet protein entry into primary cilia remains poorly understood. To enable a mechanistic analysis of soluble protein entry into cilia, we developed a method for semi-permeabilization of mammalian cells in which the plasma membrane is permeabilized while the ciliary membrane remains intact. Using semi-permeabilized cells as the basis for an in vitro diffusion-to-capture assay, we uncovered a size-dependent diffusion barrier that restricts soluble protein exchange between the cytosol and the cilium. The manipulability of this in vitro system enabled an extensive characterization of the ciliary diffusion barrier and led us to show that the barrier is mechanistically distinct from those at the axon initial segment and the nuclear pore complex. Because semi-permeabilized cells enable a range of experimental perturbations that would not be easily feasible in intact cells, we believe this methodology will provide a unique resource for investigating primary cilium function in development and disease.
Keywords: primary cilia, cilium, semi-permeabilization, diffusion barrier, digitonin
Introduction
The primary cilium is a cellular compartment with a key role in signal transduction. Cilia are thought to promote signaling by concentrating signaling proteins within a specialized environment (Nachury, 2014). How proteins are targeted to cilia has therefore emerged as a central question in the field, in particular because the cilium is not a membrane-enclosed organelle. Rather, the ciliary membrane is topologically continuous with the plasma membrane, and the ciliary lumen is likewise continuous with the cytosol. The leading model for how select proteins become concentrated within cilia is that dedicated ciliary trafficking machineries transport cargos across a diffusion barrier that partitions the cilium from the cell body (for review see Garcia-Gonzalo & Reiter, 2012; Reiter, Blacque, & Leroux, 2012). However, it is also possible that biological molecules become concentrated within cilia in the absence of a barrier through diffusion-and-retention mechanisms (Francis, Sfakianos, Lo, & Mellman, 2011; Nair et al., 2005) or via local production in the case of lipids, ions, and other small molecules (Bielas et al., 2009; Decaen, Delling, Vien, & Clapham, 2013; Delling, Decaen, Doerner, Febvay, & Clapham, 2013; Jacoby et al., 2009). Thus far, technical limitations have made it difficult to achieve a mechanistic and molecular understanding of ciliary compartmentalization and transport.
The need for new tools to study these questions is highlighted by recent studies that reached opposite conclusions regarding the existence of a ciliary diffusion barrier for soluble proteins. Specifically, while a septin-based diffusion barrier at the base of cilia was shown to impede the diffusion of membrane proteins from the plasma membrane into the ciliary membrane (Chih et al., 2012; Hu et al., 2010), Calvert and colleagues found no evidence for a soluble protein diffusion barrier at the connecting cilium of frog photoreceptors (Calvert, Schiesser, & Pugh, 2010; Najafi, Maza, & Calvert, 2012). By contrast, Kee et al. found that micro-injected fluorescent probes larger than ~40kDa were not detected in primary cilia protruding from human retinal pigment epithelium cells (Kee et al., 2012). Kee et al. further proposed that nucleoporins are present at the ciliary barrier where they might function to exclude inert molecules and let in importin-bound cargoes, similar to their role at the nuclear pore complex. Alternatively, several proteins altered in ciliopathies have been localized to the transition zone at the base of the cilium, where they may form the ciliary barrier (Garcia-Gonzalo & Reiter, 2012; Reiter et al., 2012). At present, the ultra-structural, molecular, and functional properties of the ciliary diffusion barrier remain poorly understood.
We recently developed an experimental system for analysis of protein entry into primary cilia based on semi-permeabilized cells in which the plasma membrane is disrupted while the cilium remains intact (Breslow, Koslover, Seydel, Spakowitz, & Nachury, 2013). As in classic studies of other cellular organelles using semi-permeabilized cells, this approach enables a variety of experimental manipulations not possible in intact cells. Specifically, we used semi-permeabilized cells to acutely introduce recombinant probes of defined properties at specific concentrations. We then quantified the ability of these probes to enter cilia in the absence of potential confounding effects from ongoing active transport. Semi-permeabilized cells also allowed us to directly add various reagents without the need for labor-intensive microinjection and without concerns for indirect effects of the applied perturbations. Thus, our semi-permeabilized cell system provides a powerful means to define the properties of the diffusion barrier that prevents the free exchange of soluble proteins between the cytosol and the cilium (Breslow et al., 2013). Furthermore, because key physical and functional features of primary cilia remain intact following semi-permeabilization, we expect this approach to be broadly useful to understanding active ciliary trafficking and signaling. Here we provide a detailed description of this methodology and its application to studying the ciliary diffusion barrier.
Methods
Rationale
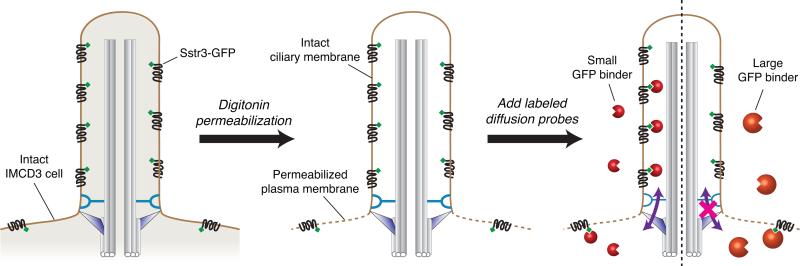
The mammalian primary cilium is roughly 1000-fold smaller than the cell body and in many cases cannot be spatially resolved from the cell body by conventional fluorescence microscopy. Thus, we found it difficult to analyze the ciliary diffusion barrier by visualizing the equilibration of fluorescently labeled inert macromolecules between cytosol and cilium. We therefore designed a diffusion-to-capture assay in which labeled probes of interest are added to the cytosolic compartment of semi-permeabilized cells and allowed to diffuse into cilia, where they are captured by effectively irreversible binding to an engineered receptor (Fig. 1). In this way, the fluorescent probes accumulate to very high levels within cilia, giving rise to a strong signal that is readily distinguished from the background and that can be monitored quantitatively and in real time. The diffusion probes we developed are GFP-binding proteins, either anti-GFP antibodies or protein fusions to an anti-GFP nanobody, which consists of a 12kDa GFP-binding protein (GBP) (Kirchhofer et al., 2010). The ciliary diffusion trap is a GFP fusion to the cytoplasmic tail of SSTR3, a G protein-coupled receptor (GPCR) that localizes to the cilium. Because the GBP can be produced recombinantly in bacteria and fused to a variety of other proteins, a range of diffusion probes can be tested for their ability to enter cilia and be captured by SSTR3-GFP.
Figure 1. Schematic of the ciliary entry assay using semi-permeabilized cells.
Mouse IMCD3 kidney epithelial cells stably expressing SSTR3-GFP are selectively permeabilized by low amounts of the detergent digitonin, disrupting the plasma membrane while leaving the primary cilium intact. Following semi-permeabilization, cells are incubated with dye-labeled GFP-binding proteins that rapidly diffuse into the cytosol. Left: For a small GFP-binding protein (GBP), diffusion across the transition zone into the cilium occurs readily, leading to GBP capture by SSTR3-GFP and to prominent GBP fluorescence in the cilium. Right: For a large GBP, diffusion into the cilium is impeded, leading to a lack of ciliary GBP fluorescence. Because SSTR3-GFP is present at low levels in the plasma membrane, a weak GBP signal is detected at the plasma membrane regardless of the GBP size.
Production and characterization of protein probes for ciliary entry
The recombinant GBP binds GFP with a dissociation constant of less than 1 nM, is readily expressed to high yields in bacteria and is amenable to fusion with other proteins (Kirchhofer et al., 2010). After purification on immobilized Ni2+, His6-GBP fusions are further purified by size-exclusion chromatography (thus enabling hydrodynamic size determination) before covalent labeling by fluorescent conjugates to enable detection in imaging assays.
1. Cloning of GBP and fusions
To express the GBP, DNA can be produced by gene synthesis to encode the sequence:
MGMADVQLVESGGALVQPGGSLRLSCAASGFPVNRYSMRWYRQAPGKEREWVAGM
SSAGDRSSYEDSVKGRFTISRDDARNTVYLQMNSLKPEDTAVYYCNVNVGFEYWG
QGTQVTVSSHHHHHH,
which includes a C-terminal 6xHis tag for purification. We recommend using the DNAWorks website (http://helixweb.nih.gov/dnaworks/) to design oligonucleotides for gene synthesis and following standard protocols for oligonucleotide assembly (Hoover & Lubkowski, 2002). After cloning the GBP-encoding DNA into a suitable bacterial expression vector such as pET28c(+), additional protein moieties can be inserted N-terminal to the GBP sequence by standard cloning methods.
2. Expression and purification of GBP fusions
Plasmids encoding GBP fusion proteins are transformed into Rosetta2(DE3)-pLysS cells and bacteria inoculated into a 10 mL Luria Broth (LB) culture with appropriate antibiotics. Following growth at 37°C to an OD600 of ~0.8, dilute the culture into 1L of 2YT media containing appropriate antibiotics. When the culture has reached OD600 of 0.6, cells are cold-shocked in an ice-water bath for 15 min, IPTG is added to a final concentration of 0.2 mM, and the culture is grown overnight at 18 °C. After 18 to 20h of growth, harvest cells by centrifugation and resuspend the cell pellet in 50 mL ice-cold lysis buffer (20 mM sodium phosphate pH 7.2, 500 mM NaCl, 10 mM imidazole, 0.5 mM DTT, 1% NP-40, and a protease inhibitor cocktail). Note: all steps are conducted at 4 °C from this point onwards. Cells are lysed by sonication (we use 3× 30s cycles of sonication – 0.8s on, 0.2s off at 70% power on a Branson Model 450 Sonifier), and the lysate is centrifuged at 20,000 x g for 45 min. Collect the supernatant, which corresponds to the cleared lysate, and add to 2 mL Ni-NTA agarose resin (Qiagen) that has been pre-equilibrated in lysis buffer. After incubation for at least 1 h, wash the resin twice with 50 mL lysis buffer and transfer the resin to a Poly-Prep column (Bio-Rad). Wash the column by gravity flow with 10 mL lysis buffer and elute with lysis buffer containing 300 mM imidazole and 0.5 mM DTT, collecting four 1 mL fractions. Determine the peak fraction by assaying for protein concentration using the Bradford assay (Bio-Rad).
3. Size-exclusion chromatography of GBP fusions
Using a FPLC system (Äkta, GE Healthcare), equilibrate a Superdex 200 HiLoad 16/60 size exclusion column (GE Healthcare) in column buffer (20 mM Hepes pH 7.7, 150 mM NaCl, 0.5 mM DTT). Load 1 to 2 mL of the peak fraction(s) eluted from the Ni-NTA column and develop the size exclusion column with 1.25 column volume of column buffer, collecting fractions of eluted protein. Using UV absorbance and/or analysis of fractions by SDS/PAGE gel electrophoresis, determine the peak fractions containing the purified GBP fusion protein. Add glycerol to the peak fractions to a final concentration of 5% and freeze the GBP fusion in 25-50 μl aliquots. To determine the Stokes radius of the GBP fusion, compare its elution volume to those of standards of known size (such as Bio-Rad Gel Filtration Standard #151-1901). Specifically, use the total column volume, VC, the void volume of the column, V0, and the elution volume, Ve, to calculate the partition coefficient, Kav, for the GBP fusion and for the standards according to the equation:
The stokes radius, Rs, can then be determined from the relationship:
4. Dye-labeling of GBP fusions
To label GBP fusions, incubate 200 to 400 μg protein with ~30 μg amine-reactive dye such as Alexa Fluor 647 succinimidyl ester (Life Technologies) in a total volume of 500 μl (dilute protein into PBS with 125 mM NaHCO3 for the reaction buffer; the dye can be dissolved and added in DMSO). After 1 h incubation at room temperature, remove unconjugated dye by applying the reaction mixture to a desalting column such as NAP-5 (GE Healthcare) that has been previously equilibrated in PBS. Elute the protein from the column per the manufacturer's instructions, collecting 250 μl fractions. Determine the peak fractions and the degree of labeling by measuring absorbance at 280 nm and 650 nm per the manufacturer's instructions. We routinely use a NanoDrop 2000c spectrophotometer (Thermo Fisher) and typically obtain a degree of labeling of 1 to 3 fluorophores per protein. Flash freeze aliquots of labeled GBP fusions in liquid nitrogen and store at −80 °C. A summary of GBP fusions that we have used is provided in Table 1.
Table 1.
Properties of GFP-binding proteins used to analyze the ciliary diffusion barrier
| GFP Binder | Molecular Weight (kDa) | Stokes Radius (nm) | Ciliary Entry* |
|---|---|---|---|
| GBP# | 13.5 | 1.4 | ++++ |
| Trx-GBP | 26 | 2.4 | ++++ |
| MBP-GBP | 57 | 3.1 | ++ |
| ZZ-GBP | 29 | 3.6 | + |
| NusA-GBP | 70 | 4.3 | +/− |
| Luciferase-GBP | 75 | 4.3 | - |
| Anti-GFP antibody | 145 | 5.1 | - |
| ZZ-GBP + IgG | 175 | >5.1 | - |
| LacZ-GBP | 520 | 6.1 | - |
Ciliary entry was assessed after incubating semi-permeabilized cells for 10 min with indicated GFP binders at 55 nM.
Abbreviations: GBP, GFP-binding protein; Trx, Thioredoxin; MBP, Maltose Binding Protein; ZZ, tandem Z domain from protein A; NusA, N-using Substance A; IgG, Immunoglobulin G; LacZ, β-galactosidase.
Adapted from Breslow et al. (2013).
Growth and semi-permeabilization of ciliated IMCD3 cells
1. Generation of an IMCD3 cell line expressing SSTR3-GFP
To date, our studies have primarily used mouse IMCD3 (inner medullary collecting duct) epithelial cells, as they are easy to work with, ciliate readily, and are compatible with semi-permeabilization. Our preliminary experiments indicate that selective permeabilization of the plasma membrane but not the ciliary membrane can be achieved in other ciliated cell lines; however we recommend rigorous validation before proceeding with other cell types (see below). We grow IMCD3 cells in DMEM/F12 medium supplemented with 10% fetal bovine serum (FBS), 2 mM glutamine, and penicillin-streptomycin (Life Technologies) and induce ciliation by changing to medium containing 0.2% FBS for 24 to 48 h. To enable GBP capture inside cilia, we stably express the ciliary GPCR somatostatin receptor 3 (SSTR3) tagged at its intracellular C-terminus with a localization and affinity purification (LAP) tag consisting of an S tag followed by a cleavage site for the HRV3C protease and GFP. To achieve single-site integration of SSTR3-GFP, we routinely use the recombination-based Flp-In system (Life Technologies) and a Flp-In IMCD3 host cell line with an FRT site integrated at a single site in the genome (Mukhopadhyay et al., 2010). Following the manufacturer's instructions, IMCD3 Flp-In cells are transfected with the Flp recombinase-expressing plasmid pOG44 and with a Blasticidin resistance plasmid expressing SSTR3-LAP under the control of the EF1α promoter. After Blasticidin selection, resistant clones are isolated and checked for expression and ciliary localization of SSTR3-GFP. Bright GFP fluorescence should be evident in all cilia (detected via immunofluorescent staining of acetylated tubulin with the 6-11B-1 antibody; Sigma Aldrich) and weaker GFP signal is typically also seen in the plasma membrane.
2. Validation of GFP-binding proteins
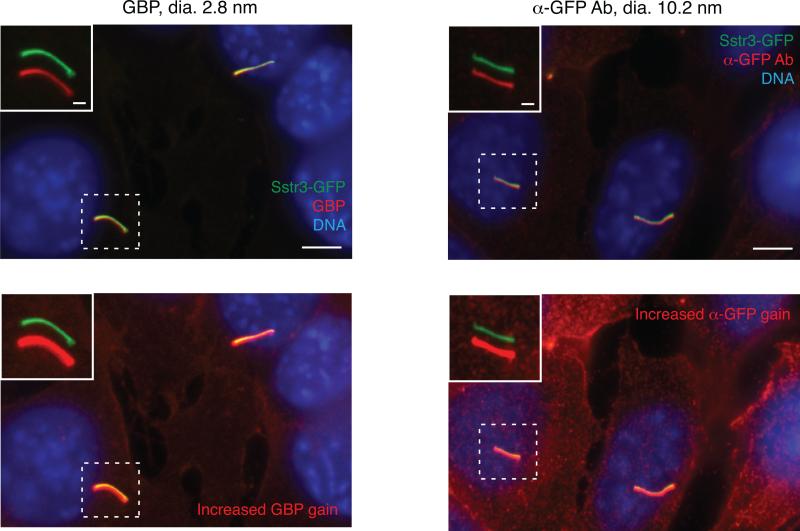
To verify that the recombinant GFP-binding proteins produced above are able to bind ciliary SSTR3-GFP, we first test them in a conventional immunofluorescence procedure. First plate 50,000 IMCD3-[SSTR3-LAP] cells per well in a 24-well plate containing 12mm #1.5 glass coverslips (Thermo Fisher Scientific) that have been previously acid-cleaned in a solution composed of 2 parts nitric acid, 1 part HCl. After growth for 24 h, replace the medium with serum starvation medium containing 0.2% FBS. After an additional 24 to 48 h, fix cells on coverslips with 4% paraformaldehyde in PBS for 10min. After 3 washes to remove fixation solution, fully permeabilize cells by adding PBS with 0.1% Triton X-100 to coverslips for 10 min. After two additional PBS washes, block coverslips for 30 min in PBS supplemented with 3% BSA (Thermo Fisher Scientific) and 5% normal donkey serum (NDS, Jackson ImmunoResearch). Dilute GBP fusions to 50 nM in PBS with 3% BSA and apply 60 μl of the diluted solution to each coverslip for 30 min at room temperature. Wash coverslips five times with PBS with 3% BSA and stain nuclei with DAPI or Hoechst stain if desired. Mount coverslips on glass slides and image on a standard epifluorescence microscope equipped with an oil-immersion objecting providing ≥60X magnification and with suitable excitation/emission filters for GFP and Alexa Fluor 647. The pattern of GBP fluorescence is expected to match that of SSTR3-GFP – specifically, there should be prominent GBP labeling of primary cilia (Figure 2, top left) and weaker staining of the SSTR3-GFP in the plasma membrane (Figure 2, bottom left). We recommend comparing the GBP staining distribution to a positive control provided by a well-validated anti-GFP antibody (e.g. Rockland #600-101-215) (Figure 2, right). We also note that some GBP fusions may display non-specific staining of cellular structures. This possibility can be assessed by staining IMCD3 cells lacking SSTR3-GFP. It may still be possible to qualitatively assess ciliary entry for such GBP fusions, but quantitative analysis of ciliary entry will be complicated by non-specific staining (see “Data Analysis” section).
Figure 2. Validation of GFP-binding proteins in fully permeabilized cells.
IMCD3 cells stably expressing SSTR3-GFP were fixed and stained using a conventional immunofluorescence protocol with Triton X-100 permeabilization. SSTR3-GFP was stained with either Alexa Fluor 647-labeled GBP nanobody (left-hand panels) or an anti-GFP antibody and indirect immunofluorescence (right-hand panels). Insets show magnified views of cilia with channels shifted to aid visualization. Bottom panels are shown with increased gain in the GBP and anti-GFP panels to illustrate weaker staining of plasma membrane SSTR3-GFP. Scale bars are 5 μm (main panels) and 1 μm (insets). Adapted from Breslow et al. (2013).
3. Semi-permeabilization with digitonin
Selective permeabilization of the plasma membrane while leaving the cilium intact is achieved by briefly exposing cells on ice to very low concentrations of the detergent digitonin. Alternatively, we have also used Perfringolysin O (Breslow et al., 2013), a cholesterol-dependent pore-forming toxin (Heuck, Moe, & Johnson, 2010); however, we typically use digitonin as the procedure is simpler. Successful semi-permeabilization with digitonin is critically dependent on the concentration of digitonin applied to the cells. We therefore only use high-purity digitonin (≥95 %; EMD Millipore #300410) and prepare a stock solution in DMSO at 20 mg/mL, freezing aliquots at −20 °C to ensure consistent results (avoid repeated freeze/thaw and use extra care in handling digitonin in powder form). It is necessary to first perform a titration of the digitonin concentration to determine the optimal conditions for semi-permeabilization. Here, the extent of permeabilization will be assessed by the accessibility of cytosolic versus ciliary epitopes to antibodies. The optimal conditions for semi-permeabilization will allow antibodies to access cytosolic epitopes while the ciliary interior will remain inaccessible to antibodies. Meanwhile, the GBP nanobody will be able to enter cilia, as it can cross the diffusion barrier at the base of the cilium. Thus, for each digitonin concentration to be tested, permeabilized cells are incubated with i) the mAb414 antibody (Covance Inc.), which detects cytosol-exposed nucleoporins at the nuclear envelope, ii) the GBP nanobody, and iii) a commercial anti-GFP antibody. Note that the protocol below describes the procedure for digitonin titration but is essentially identical to how subsequent analyses of ciliary entry are conducted once an optimal digitonin concentration has been identified.
For each digitonin concentration to be tested, seed 50,000 IMCD3-[SSTR3-GFP] cells onto 12mm coverslips in 24-well plates (2 coverslips per digitonin concentration). After 24 h, replace the media with serum starvation media, and after an additional 24h, remove the coverslips from growth medium and transfer to HKM buffer on ice (20 mM Hepes, pH 7.4, 115 mM KOAc, 1 mM MgCl2, 1 mM EGTA). To ensure temperature control, we place coverslips on a parafilm-covered aluminum block (Diversified Biotech) placed on top of a bed of ice. Gently wash coverslips once with ice-cold HKM buffer, then replace the buffer with 100 μl HKM supplemented with digitonin at concentrations ranging from 0 to 100 μg/mL (e.g. 0, 15, 20, 30, 40, 60, and 100 μg/mL), permeabilizing two coverslips for each digitonin concentration to be tested (typically ~30 μγ/μλ digitonin is optimal for semi-permeabilization). After incubation on ice for 7 min, wash coverslips twice with ice-cold digitonin-free HKM buffer. Keep coverslips on ice until the assay is about to be started.
4. Incubation with ciliary entry probes
The assay is initiated by addition to the coverslip of 60 μl of ciliary entry probe diluted in HKM buffer supplemented with 0.1% BSA (HKMB). Assays are typically conducted at room temperature but can also be performed on ice or at 37 °C with similar results. For initial titration of digitonin, add to the first coverslip 20 nM mAb414 antibody mixed with 25 to 50 nM dye-labeled GBP and to the second coverslip 20 nM mAb414 antibody mixed with 25 to 50 nM anti-GFP antibody. To analyze results of probe entry into cilia, continue with next steps “Imaging probe entry in fixed cells” or “Imaging probe entry in real-time”.
Optional and alternative steps
A variety of optional steps may be performed prior to semi-permeabilization or after semi-permeabilization but before addition of ciliary entry probes. For example, we have disrupted the actin cytoskeleton by pre-treating intact cells with Cytochalasin D and have performed RNAi against candidate components of the ciliary diffusion barrier. After semi-permeabilization, it is possible to immobilize ciliary SSTR3-GFP using the multivalent lectin wheat germ agglutinin (WGA), which binds and crosslinks glycosylated SSTR3, thereby influencing the spatial and temporal properties of GBP capture in cilia (Breslow et al., 2013). Alternatively, one can add various exogenous reagents (antibodies, recombinant proteins, nucleotide analogs, cytosolic extract etc.) to semi-permeabilized cells that may affect the properties of the ciliary diffusion barrier. Finally, one can also sequentially add several GBP fusions with different properties (size, dye-labeling) to semi-permeabilized cells.
Imaging probe entry into cilia
1. Imaging probe entry in fixed cells
After incubating coverslips with GFP binders and mAb414 for 10 min, wash coverslips twice with HKM buffer and fix coverslips in PBS with 4% paraformaldehyde for 10 min. Wash coverslips twice with HKM and once with PBS. To facilitate the indirect detection of anti-GFP or anti-nucleoporin antibodies, we recommend further permeabilization with 0.1% Triton X-100 in PBS for 10 min. After blocking in PBS with 3% BSA and 5% NDS, incubate coverslips in appropriate secondary antibodies for detection of mAb414 and/or anti-GFP antibodies. Optionally, one can also add antibodies to stain other cellular markers at this step, such as anti-γ-tubulin antibody GTU-88 (Sigma Aldrich) to mark the base of the cilium. After five washes with PBS with 3% BSA and optional DNA staining, mount coverslips on a 2 μl drop of mounting medium (80% glycerol, 10 mM Tris pH 7.4) spotted on a slide. Examine coverslips on an epifluorescence microscope, capturing images for several fields of view in the appropriate channels for GFP, GBP or anti-GFP, as well as any other proteins stained.
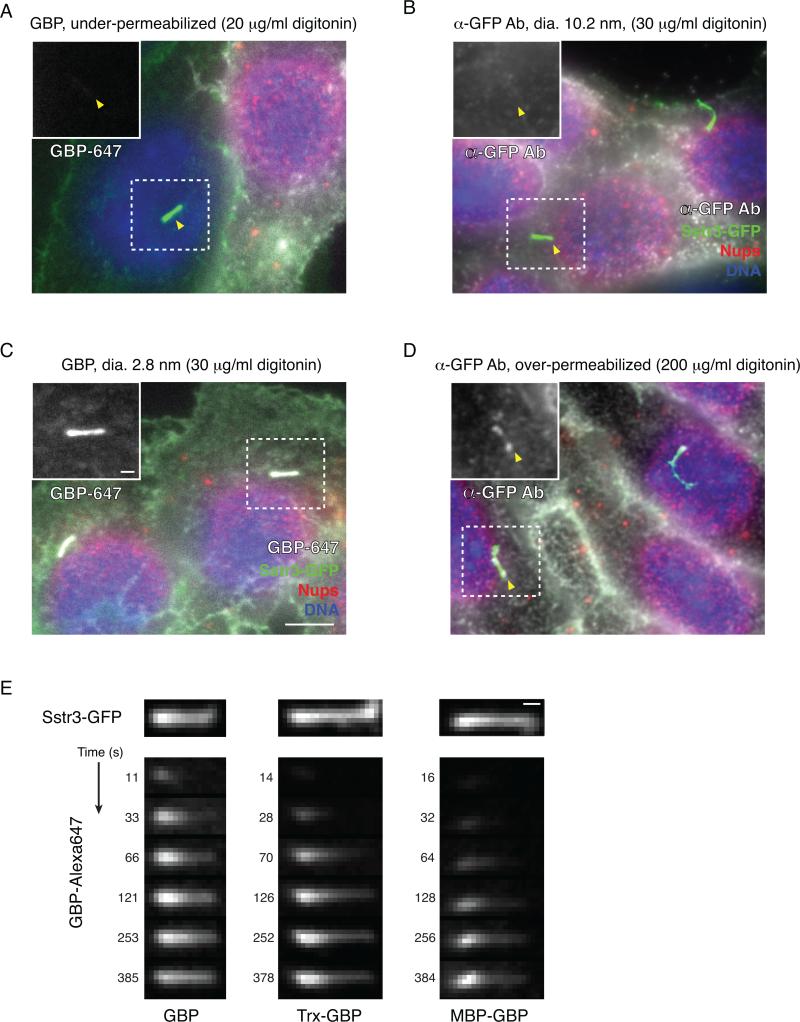
When evaluating the optimal conditions for semi-permeabilization, the plasma membrane will remain intact at the lowest concentrations of digitonin tested or if no digitonin is added. Under such conditions, cells (identified by ciliary GFP fluorescence and DNA staining) will lack mAb414 staining at the nuclear envelope; additionally, there will be no GBP or anti-GFP signal, either at the cilium or the plasma membrane (Figure 3A. As the digitonin concentration is increased, the plasma membrane of the vast majority (>90%) of the cells will become permeabilized, and cells will therefore display mAb414 staining in a nuclear rim pattern. Additionally, permeabilized cells are expected to show GBP or anti-GFP staining of SSTR3-GFP in the plasma membrane. However, only the GBP but not the anti-GFP antibody will be present in the cilium (Figure 3B,C), as the latter will be excluded by the size-dependent ciliary diffusion barrier. If too much digitonin is used, the ciliary membrane may be permeabilized, leading to anti-GFP antibody staining of ciliary SSTR3-GFP (Figure 3D). We use the lowest amount of digitonin that is sufficient to selectively permeabilize >80-90% of cells. Additionally, we recommend including a control coverslip in all subsequent experiments to assess the degree of plasma membrane permeabilization.
Figure 3. Optimization of digitonin concentration for semi-permeabilization and live imaging of ciliary entry.
A. Example of under-permeabilization with digitonin concentration too low. Here the cell to the right is permeabilized, but the one to the left with the dashed rectangle is not, as evidenced by the lack of mAb414 staining at the nuclear envelope and by the lack of staining by the GBP nanobody. The position of the cilium in the unpermeabilized cell is indicated with an arrowhead. Insets in A-D show magnified views of the cilium in the GBP channel.
B. Representative results of ciliary entry assay for a large GFP binding protein – an anti-GFP antibody – with optimal digitonin concentration (30 μg/mL digitonin in this case). Although nucleoporins are prominently detected by the mAb414 antibody, the anti-GFP antibody is only present at the plasma membrane and not in the cilium (marked by arrowhead).
C. Representative results of a ciliary entry assay for a small GFP-binding protein (GBP) when semi-permeabilization has been optimally achieved. Following addition of Alexa Fluor 647-labeled GBP to semi-permeabilized IMCD3 cells stably expressing SSTR3-GFP, a GBP signal is clearly seen in the cilium and to a lesser extent at the plasma membrane, matching the distribution of SSTR3-GFP. Nucleoporins (Nups) at the nuclear envelope are readily detected by the mAb414 antibody.
D. Example of over-permeabilization, in which the large anti-GFP antibody weakly labels SSTR3-GFP in the cilium as well as in the plasma membrane.
E. Time-series images obtained from live imaging on a confocal spinning-disk microscope show kinetics of ciliary entry for different size GBP fusion proteins. Following semi-permeabilization, the indicated GBP fusions were added at 110 nM and images were recorded at the indicated intervals. Note that for all GBP fusions, the ciliary signal increases with time and progresses from base to tip.
Scale bars are 5 μm (main panels A-D) and 1 μm (insets and panel E). Adapted from Breslow et al. (2013).
2. Imaging probe entry in real-time
To image probe entry in real-time, the assay is performed as above, but the GBP is added to semi-permeabilized cells on a coverslip placed on a microscope stage. Images are then acquired in the GFP and Alexa Fluor 647 (GBP) channels at successive time-points after GBP addition. To minimize the background signal from GBP in solution, we recommend using an inverted confocal microscope equipped with a spinning-disk head and a 60x oil-immersion objective. Alternatively, if cilia are formed on the ventral surface of cells against the glass coverslip (as is the case in IMCD3 cells; Breslow et al., 2013), a total internal fluorescence (TIRF) microscope may be used. Take particular care to ensure that the temperature of the buffer, coverslip, and microscope have fully equilibrated before adding GBP fusions to permeabilized cells, as thermal fluctuations can cause significant focal drift during the course of the experiment. For this purpose, a temperature-controlled stage or environmental chamber is highly beneficial.
For live imaging, cell growth and semi-permeabilization are done as described above. However, we recommend seeding 250,000 cells on a 25mm round #1.5 coverslip placed in a 35mm dish (Corning). Semi-permeabilize cells as above, using 600 μl digitonin buffer, then wash in HKMB pre-equilibrated at the intended imaging temperature (we often image at 37 °C). Next, place the coverslip on the microscope stage, adding 400 μl HKMB to the coverslip. Here we typically use a custom-fabricated aluminum block that fits the stage of a Nikon microscope and that uses a rubber O-ring to hold and seal a 25mm coverslip (Stanford Varian Machine shop). Alternatively, one may use a commercially available product such as the Quick Release Magnetic Imaging Chamber from Warner Instruments (QR-43C). Identify the focal plane of the cilia using bright field or GFP illumination and then wait 10 min for thermal equilibration of the imaging system and biological sample. Dilute the GBP in HKMB to 4-fold the intended final concentration. Identify a field of interest, initiate image acquisition, and then carefully add 133 μl GBP to the coverslip. We have found that it is helpful to dispense the GBP from a P1000 pipette evenly around the coverslip and to mix very gently after GFP addition by pipetting up and down. Acquire images in the GFP and GBP channels at appropriate time intervals of 8 to 30s). The time interval between images should be determined based on the entry rate of the GBP fusion of interest, the rate of photobleaching, and microscope parameters such as acquisition speed, camera sensitivity, etc. It is not strictly necessary to capture the GFP channel at every time point, however the GFP signal can be used to assess and account for focal drift or cilium movement. To keep cilia in focus and minimize the effects of focal drift, we also recommend acquiring z-stacks of 3 to 5 focal planes (0.4 μm spacing) straddling the ciliary focal plane at the time of GBP addition. If needed, cell permeabilization can be verified using a fluorescently labeled dextran or with a dye-labeled phalloidin derivative. Finally, we note that the added GBP will bind rapidly to the plasma membrane pool of SSTR3-GFP, which can complicate the analysis of ciliary signal increase over time. To reduce the impact of this issue, one may “pre-block” the plasma membrane pool of SSTR3-GFP by first incubating permeabilized cells for 5-10 min in HKMB with unlabeled LacZ-GBP (which is unable to enter cilia), and then adding the labeled GBP of interest. The unlabeled LacZ-GBP will saturate plasma membrane SSTR3-GFP but will not enter cilia and therefore will not interfere with subsequent ciliary capture of the labeled GBP. Typical results showing a time-dependent increase in ciliary GBP signal and a progression of the GBP signal from base to tip are shown in Figure 3E.
Data Analysis
1. Measuring the intensity and spatial distribution of ciliary GBP fluorescence
To measure the intensity and spatial distribution of the GBP signal within the cilium, we quantify the average ciliary GBP intensity and perform line trace analyses using the image analysis software ImageJ (National Institutes of Health). To obtain the mean ciliary GBP intensity, first use the polygon tool to draw a region of interest (ROI) around the cilium. To account for the signal of GBP captured by SSTR3-GFP in regions of the plasma membrane immediately above or below the cilium, define a similar region that corresponds to a part of the plasma membrane that does not overlap with the cilium. Determine the mean fluorescence intensity in each region using the “Measure” command, and calculate the background-corrected mean ciliary intensity of GBP by subtracting the mean intensity of the plasma membrane ROI from the mean intensity of the ciliary ROI. This procedure can be repeated for many cilia, allowing an average value and standard deviation to be determined.
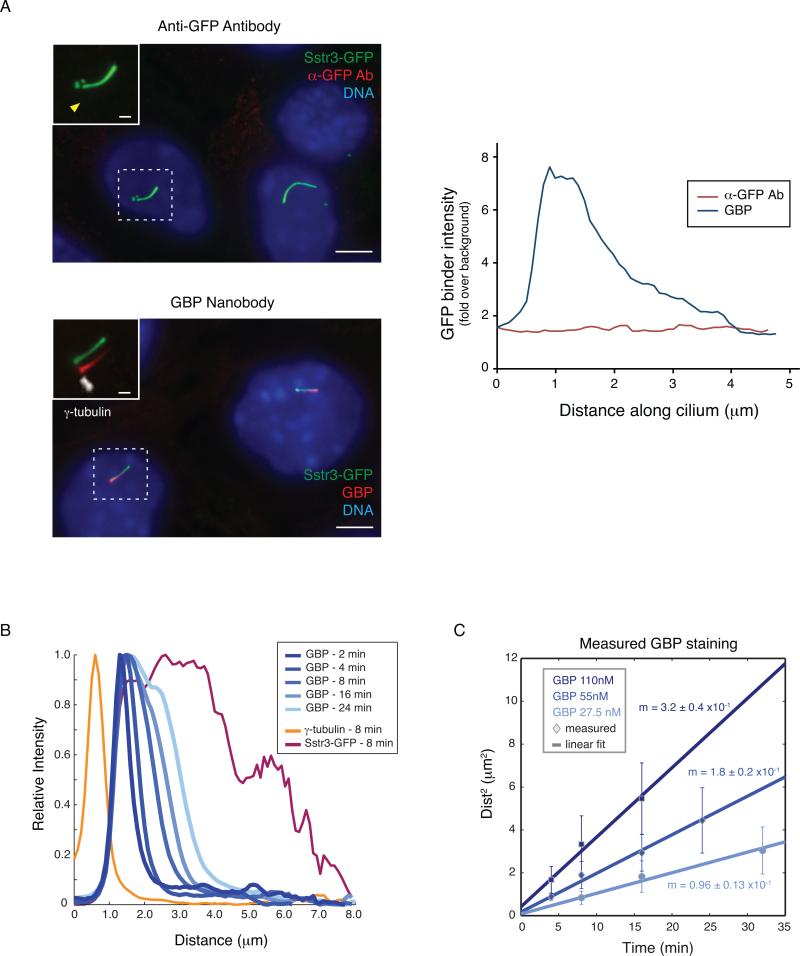
To perform a line trace analysis, first use the segmented line tool to draw connected line segments along the length of the cilium in the GFP channel. If possible, it is helpful to define the base of the cilium by staining for a centrosomal marker such as γ-tubulin (Fig. 4A, left). To obtain the fluorescence intensities, select a given channel from the image and use the “Analyze -> Plot Profile” command. The intensity values underlying the resulting plot are displayed by clicking the ‘List’ button on the plot window and can be copied to other programs for graphical representation and further analysis (Figure 4A, right). Key parameters that can be calculated are the average position-dependent ciliary signal (Figure 4B) and the time-dependent progression of GBP into the cilium (Figure 4C). To determine the distance stained by a given GBP, it is necessary to define the beginning and end of the ciliary signal; for this purpose, we use the positions where the GBP signal decreases below a threshold of 25% of the maximum ciliary signal. We also note that more consistent results may be obtained under conditions where there are sharper boundaries for the GBP signal, such as with WGA treatment. In WGA treated cells, the square of the distance stained by the GBP is linearly proportional to the time after GBP addition and to the concentration of GBP added (Figure 4C), as expected for a diffusion-controlled process.
Figure 4. Analyzing fixed images from diffusion-to-capture assays reveals a ciliary diffusion barrier.
A. Left: Semi-permeabilized IMCD3-[SSTR3-GFP] cells were incubated with either an anti-GFP antibody (top) or GBP (bottom). Image insets show magnified views of cilia with channels shifted to aid visualization, and the arrowhead shows the position of the primary cilium in the anti-GFP antibody channel. Scale bars are 5 μm (main panels) and 1 μm (insets). Right: Background-corrected intensities were determined using ImageJ software and line intensity profiles are plotted along the length of the cilium for the GBP and anti-GFP channels.
B. Average line intensity profiles reveal time-dependent progression of GBP signal from the base to the tip of the cilium. Here the lectin wheat germ agglutinin, which crosslinks and immobilizes SSTR3-GFP, has been used to facilitate detection of base-to-tip GBP progression. The data shown were obtained from samples fixed at the indicated times after incubation with labeled GBP. Line intensity profiles for individual cilia were first aligned by using γ-tubulin staining to define the base of the cilium and then averaged across ≥10 cilia per condition.
C. Measurement of the distance stained by the GBP in WGA-treated cells reveals that the square of the distance is proportional to the time after GBP addition and to the concentration of GBP applied to semi-permeabilized cells. Error bars indicate standard deviations (n ≥10). Adapted from Breslow et al. (2013).
2. Measurement of ciliary entry kinetics from live imaging
To calculate rates of ciliary entry, it is first necessary to determine the ciliary GBP intensity at each time-point. First, using the SSTR3-GFP fluorescence as a guide, identify for each time-point the z-plane that best captures the focal plane of the cilium. The plane with maximal ciliary GFP intensity corresponds to the optimal z-plane; however, if the GFP image is not in focus in any of the captured planes, the time-point in question should be discarded. The ciliary GBP signal can next be calculated either for the whole cilium (using ImageJ as described above) or for the proximal region of the cilium where the GBP signal first appears. We prefer to measure the intensity near the base of the cilium because GBP fusions enter cilia from the base and are rapidly captured by nearby SSTR3-GFP molecules. When the signal from the entire cilium is analyzed, other processes contribute to the rate of GBP fusion capture, and the observed rate of signal increase may not accurately reflect the rate of diffusion barrier crossing. Specifically, once proximal binding sites become saturated, subsequent ciliary capture is affected by diffusion of GBP fusions within cilia and by the rate at which free SSTR3-GFP molecules diffuse from distal to proximal regions of the cilium. By measuring GBP signals near the base, the potential confounding effects of these processes can be minimized. To measure GBP intensity near the base at a given time-point, first identify the region corresponding to the cilium. For this purpose, a simple threshold-based approach using the GFP fluorescence relative to background can be implemented in Matlab (Mathworks, Inc.) or ImageJ (e.g. pixels with GFP intensity ≥3x that of a plasma membrane region). The proximal region of the cilium to be analyzed is defined by determining the pixels within the cilium that have the highest GBP intensity. We typically use the average intensity of 4-9 pixels near the base and then subtract the background signal arising from GBP in solution and/or GBP bound to plasma membrane SSTR3-GFP. Plotting the average GBP intensity near the cilium base against time reveals a rapid initial increase that plateaus as available SSTR3-GFP binding sites become occupied (Figure 5). These curves are well fit by exponential equations of the form,
where IBase is the GBP intensity near the base and k is the observed entry rate. This exponential function matches the first-order kinetics expected for a process in which crossing the ciliary diffusion barrier is the initial rate-limiting step in ciliary GBP capture. The observed entry rate, k, can be obtained by fitting the data to an exponential equation and averaging fitted rate constants for all cilia analyzed (Figure 5). As expected for a size-dependent barrier, we find that the ciliary entry rate decreases significantly as GBP size increases.
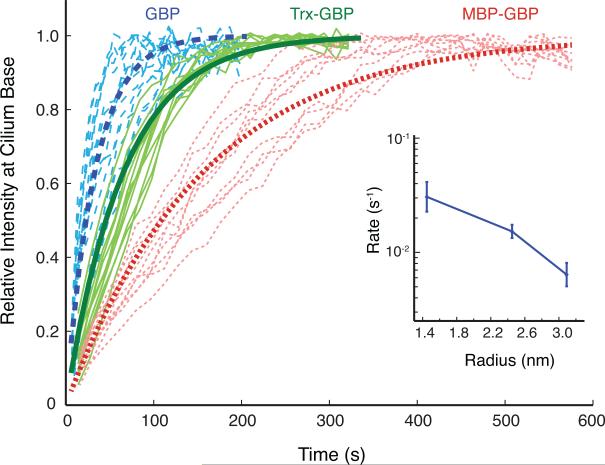
Figure 5. Kinetic analysis of GBP entry into primary cilia of semi-permeabilized cells.
Rates of GBP ciliary entry were determined by measuring the GBP signal at the proximal part of the cilium as a function of time (see Figure 3E for representative images). Traces from individual cilia are shown in thin lines for GBP (blue lines with long dashes), Trx-GBP (solid green lines), and MBP-GBP (red lines with short dashes). Fitted exponential curves corresponding to the average entry rates obtained from analyzing ≥10 cilia are shown in thick lines. The inset graph at right shows the average entry rate plotted versus the Stokes radius of the diffusion probe. Error bars indicate standard deviation. Adapted from Breslow et al. (2013).
Conclusions and Future Outlook
The semi-permeabilized cell system described here provides a sensitive and quantitative means to investigate the diffusion barrier that partitions the cilium from the cell body. Additionally, our assay enables powerful functional and mechanistic studies of the ciliary diffusion barrier in an in vitro context. In our work to date, we have used pharmacological reagents, recombinant proteins, and function-blocking antibodies in semi-permeabilized cells to acutely interfere with the actin cytoskeleton or nucleoporins in order to test their role in regulating ciliary entry (Breslow et al., 2013). Interestingly, we found that the ciliary diffusion barrier is mechanistically distinct from those at the nuclear pore complex and at the axon initial segment. In the future, we expect that the experimental system described here will provide a means to define the functional and molecular properties of the ciliary diffusion barrier and thereby understand how compartmentalization of the cilium supports ciliary signaling.
More broadly, we expect that semi-permeabilized cells will provide a unique tool to investigate many aspects of primary cilium function. For example, we recently used semi-permeabilized cells to dissect the relative contributions of active transport versus diffusion to the movement of signaling receptors within the cilium (Ye et al., 2013). The value of semi-permeabilized cells is also supported by our finding that several key features of cilia are preserved following semi-permeabilization. Specifically, the diffusion barrier that restricts membrane protein exchange between the plasma membrane and the ciliary membrane remains intact, and intraflagellar transport (IFT) particles remain inside the cilium (Breslow et al., 2013). Although IFT movement is interrupted following semi-permeabilization, addition of cytoplasmic extract and ATP can temporarily re-activate IFT train movement. Notably, it has thus far been quite difficult to assess the roles of components such as the IFT complexes in specific ciliary processes because they are required for ciliogenesis; here, in vitro assays in semi-permeabilized cells may offer a means to overcome this obstacle. As many fundamental insights into cellular processes have been made possible by semi-permeabilized cell assays, we hope that application of this classic methodology to the primary cilium will help to illuminate ciliary function in health and disease.
Acknowledgments
We thank Nancy Zhang for experimental assistance and members of the Nachury lab for helpful discussions. The methods described here were developed with support from the Damon Runyon Cancer Research Foundation to D.K.B. (DRG-2087-11 and DFS-11-14) and from the National Institutes of Health (GM089933) to M.V.N.
References
- Bielas SL, Silhavy JL, Brancati F, Kisseleva MV, Al-Gazali L, Sztriha L, Gleeson JG. Mutations in INPP5E, encoding inositol polyphosphate-5-phosphatase E, link phosphatidyl inositol signaling to the ciliopathies. Nature Genetics. 2009;41(9):1032–1036. doi: 10.1038/ng.423. doi: 10.1038/ng.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow DK, Koslover EF, Seydel F, Spakowitz AJ, Nachury MV. An in vitro assay for entry into cilia reveals unique properties of the soluble diffusion barrier. The Journal of cell biology. 2013;203(1):129–147. doi: 10.1083/jcb.201212024. doi: 10.1083/jcb.201212024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvert PD, Schiesser WE, Pugh EN. Diffusion of a soluble protein, photoactivatable GFP, through a sensory cilium. The Journal of General Physiology. 2010;135(3):173–196. doi: 10.1085/jgp.200910322. doi: 10.1085/jgp.200910322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chih B, Liu P, Chinn Y, Chalouni C, Komuves LG, Hass PE, Peterson AS. A ciliopathy complex at the transition zone protects the cilia as a privileged membrane domain. Nature Cell Biology. 2012;14(1):61–72. doi: 10.1038/ncb2410. doi: 10.1038/ncb2410. [DOI] [PubMed] [Google Scholar]
- Decaen PG, Delling M, Vien TN, Clapham DE. Direct recording and molecular identification of the calcium channel of primary cilia. Nature. 2013;504(7479):315–318. doi: 10.1038/nature12832. doi: 10.1038/nature12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delling M, Decaen PG, Doerner JF, Febvay S, Clapham DE. Primary cilia are specialized calcium signalling organelles. Nature. 2013;504(7479):311–314. doi: 10.1038/nature12833. doi: 10.1038/nature12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis SS, Sfakianos J, Lo B, Mellman I. A hierarchy of signals regulates entry of membrane proteins into the ciliary membrane domain in epithelial cells. The Journal of cell biology. 2011 doi: 10.1083/jcb.201009001. doi: 10.1083/jcb.201009001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalo FR, Reiter JF. Scoring a backstage pass: Mechanisms of ciliogenesis and ciliary access. The Journal of cell biology. 2012;197(6):697–709. doi: 10.1083/jcb.201111146. doi: 10.1083/jcb.201111146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuck AP, Moe PC, Johnson BB. The cholesterol-dependent cytolysin family of gram-positive bacterial toxins. Subcell Biochem. 2010;51:551–577. doi: 10.1007/978-90-481-8622-8_20. doi: 10.1007/978-90-481-8622-8_20. [DOI] [PubMed] [Google Scholar]
- Hoover DM, Lubkowski J. DNAWorks: an automated method for designing oligonucleotides for PCR-based gene synthesis. Nucleic acids research. 2002;30(10):e43. doi: 10.1093/nar/30.10.e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Milenkovic L, Jin H, Scott MP, Nachury MV, Spiliotis ET, Nelson WJ. A septin diffusion barrier at the base of the primary cilium maintains ciliary membrane protein distribution. Science. 2010;329(5990):436–439. doi: 10.1126/science.1191054. doi: 10.1126/science.1191054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby M, Cox JJ, Gayral S, Hampshire DJ, Ayub M, Blockmans M, Schurmans S. INPP5E mutations cause primary cilium signaling defects, ciliary instability and ciliopathies in human and mouse. Nature Genetics. 2009;41(9):1027–1031. doi: 10.1038/ng.427. doi: 10.1038/ng.427. [DOI] [PubMed] [Google Scholar]
- Kee HL, Dishinger JF, Blasius TL, Liu C-J, Margolis B, Verhey KJ. A size-exclusion permeability barrier and nucleoporins characterize a ciliary pore complex that regulates transport into cilia. Nature Cell Biology. 2012;14(4):431–437. doi: 10.1038/ncb2450. doi: 10.1038/ncb2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhofer A, Helma J, Schmidthals K, Frauer C, Cui S, Karcher A, Rothbauer U. Modulation of protein properties in living cells using nanobodies. Nature Structural & Molecular Biology. 2010;17(1):133–138. doi: 10.1038/nsmb.1727. doi: 10.1038/nsmb.1727. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S, Wen X, Chih B, Nelson CD, Lane WS, Scales SJ, Jackson PK. TULP3 bridges the IFT-A complex and membrane phosphoinositides to promote trafficking of G protein-coupled receptors into primary cilia. Genes & development. 2010;24(19):2180–2193. doi: 10.1101/gad.1966210. doi: 10.1101/gad.1966210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury MV. How do cilia organize signalling cascades? Philos Trans R Soc Lond B Biol Sci. 2014;369(1650) doi: 10.1098/rstb.2013.0465. doi: 10.1098/rstb.2013.0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair KS, Hanson SM, Mendez A, Gurevich EV, Kennedy MJ, Shestopalov VI, Slepak VZ. Light-dependent redistribution of arrestin in vertebrate rods is an energy-independent process governed by protein-protein interactions. Neuron. 2005;46(4):555–567. doi: 10.1016/j.neuron.2005.03.023. doi: 10.1016/j.neuron.2005.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafi M, Maza NA, Calvert PD. Steric volume exclusion sets soluble protein concentrations in photoreceptor sensory cilia. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(1):203–208. doi: 10.1073/pnas.1115109109. doi: 10.1073/pnas.1115109109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter JF, Blacque OE, Leroux MR. The base of the cilium: roles for transition fibres and the transition zone in ciliary formation, maintenance and compartmentalization. EMBO Reports. 2012;13(7):608–618. doi: 10.1038/embor.2012.73. doi: 10.1038/embor.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Breslow DK, Koslover EF, Spakowitz AJ, Nelson WJ, Nachury MV. Single molecule imaging reveals a major role for diffusion in the exploration of ciliary space by signaling receptors. eLife. 2013;2:e00654. doi: 10.7554/eLife.00654. doi: 10.7554/eLife.00654. [DOI] [PMC free article] [PubMed] [Google Scholar]