Abstract
Background
There is an established association between type 2 diabetes and accelerated cognitive decline. The exact mechanism linking type 2 diabetes and reduced cognitive function is less clear. The monoamine system, which is extensively involved in cognition, can be altered by type 2 diabetes status. Thus, this study hypothesized that sequence variants in genes linked to dopamine metabolism and associated pathways are associated with cognitive function as assessed by the Digit Symbol Substitution Task, the Modified Mini‐Mental State Examination, the Stroop Task, the Rey Auditory‐Verbal Learning Task, and the Controlled Oral Word Association Task for Phonemic and Semantic Fluency in the Diabetes Heart Study, a type 2 diabetes‐enriched familial cohort (n = 893).
Methods
To determine the effects of candidate variants on cognitive performance, genetic association analyses were performed on the well‐documented variable number tandem repeat located in the 3' untranslated region of the dopamine transporter, as well as on single‐nucleotide polymorphisms covering genes in the dopaminergic pathway, the insulin signaling pathway, and the convergence of both. Next, polymorphisms in loci of interest with strong evidence for involvement in dopamine processing were extracted from genetic datasets available in a subset of the cohort (n = 572) derived from Affymetrix® Genome‐Wide Human SNP Array 5.0 and 1000 Genomes imputation from this array.
Results
The candidate gene analysis revealed one variant from the DOPA decarboxylase gene, rs10499695, to be associated with poorer performance on a subset of Rey Auditory‐Verbal Learning Task measuring retroactive interference (P = 0.001, β = −0.45). Secondary analysis of genome‐wide and imputed data uncovered another DOPA decarboxylase variant, rs62445903, also associated with retroactive interference (P = 7.21 × 10−7, β = 0.3). These data suggest a role for dopaminergic genes, specifically a gene involved in regulation of dopamine synthesis, in cognitive performance in type 2 diabetes.
Keywords: Cognition, dopamine, DAT1 VNTR, DOPA decarboxylase, polymorphism, type 2 diabetes
Introduction
Although cognitive decline is a part of normal aging, the rate of cognitive decline can be influenced by factors such as lifestyle, health, and genetics. Previous epidemiological and imaging studies have established an association between type 2 diabetes (T2D) and accelerated cognitive decline (Manschot et al. 2006; Luchsinger 2012; Palta et al. 2014; Ryan et al. 2014). In a review on the relationship between diabetes and cognition, Mayeda and colleagues postulate that insulin dysregulation and hyperglycemia are causal factors in neurodegeneration and cognitive decline (2015). However, the exact mechanism linking diabetes and poor cognitive performance remains unclear.
DA (Dopamine), a catecholamine neurotransmitter, plays a major role in learning and behavior by modulating neural activity in the prefrontal cortex (Clark and Noudoost 2014). DA has been implicated in disorders that influence cognitive performance such as ADHD (attention‐deficit/hyperactivity disorder, Genro et al. 2010; Sharma and Couture 2014) and Parkinson's disease (Kehagia et al. 2013; Ko et al. 2013). Several studies have examined the influence of genetic polymorphisms of genes from the dopaminergic pathway on cognitive performance. These studies have reported significant evidence of association with a range of cognitive measures (e.g., Savitz et al. 2006; Frank and Fossella 2011; Nemoda et al. 2011; Bowirrat et al. 2012; Störmer et al. 2012). One polymorphism that has received considerable attention is the variable number tandem repeat located in the 3' untranslated region of the DA transporter gene, (DAT1 VNTR, rs28363170) which has been associated with several aspects of executive functioning (Brown et al. 2011; Wittmann et al. 2013; Gordon et al. 2015; Sambataro et al. 2015; Schneider et al. 2015).
Insulin insufficiency is a characteristic of individuals with T2D, where peripheral insulin resistance is not compensated for by adapting insulin secretion, thus resulting in chronic hyperglycemia (Wu et al. 2014). Beyond insulin's fundamental role in metabolic regulation (Plum et al. 2006), insulin dysregulation may also be centrally involved in DA homeostasis (Kleinridders et al. 2015). For example, insulin receptors are collocated on DA neurons in the substantia nigra (Henderson et al. 2000), a region involved in cognitive control (Boehler et al. 2011), decision‐making (Ding and Gold 2013), and reinforcement learning (de Berker and Rutledge 2014). Not insignificantly, pancreatic β‐cells co‐express the DA D3 receptor and transporter, DRD3 and DAT1 respectively, which are implicated in inhibitory control of glucose‐stimulated insulin release (Ustione and Piston 2012). Thus, it is reasonable to consider the impact of polymorphisms in the DA pathway in the pathogenesis of T2D complications, including cognitive decline. Based upon these observations, the purpose of this study was twofold: (1) to investigate the role of common and multi‐allelic genetic variants in cognitive performance; and (2) to perform a focused association analysis of dopaminergic pathway loci using genotyped and imputed data.
The majority of reports in the DA candidate gene literature contain relatively small sample sizes with modest coverage of the genes in question. Therefore, this study took advantage of an extensively genotyped familial cohort of T2D patients and related controls, the Diabetes Heart Study (DHS) – Mind, to investigate the role of sequence variants in genes linked to DA metabolism (e.g., DDC, TH, DRD2, DRD3, and DAT1) and associated pathways in cognitive function. We hypothesized that polymorphisms in these pathways were associated with performance on a cognitive battery in T2D‐affected individuals and unaffected siblings.
Material and Methods
Subjects
In all, 893 participants from 550 families of European ancestry (783 T2D‐affected; 110 controls) were recruited (Table 1). Recruitment and ascertainment for the study have been described elsewhere (Bowden et al. 2010; Hugenschmidt et al. 2013; Raffield et al. 2015). Briefly, siblings concordant for T2D, plus available nondiabetic siblings, were enrolled. T2D, confirmed by fasting glucose and glycated hemoglobin (HbA1C), was defined as occurring after the age of 35 years, without advanced renal insufficiency, and not initially treated exclusively by insulin therapy. The DHS – Mind sample is an ancillary study to the original DHS initiated in 2008 to examine the relationship between T2D and cognitive function (for a review of the DHS family of studies, see Bowden et al. 2010). The current analysis included a T2D enriched sample of 321 newly recruited T2D‐affected participants in addition to 573 individuals initially recruited in the original DHS investigation from 1998 to 2005.
Table 1.
Demographic characteristics of the 893 DHS–Mind participants
| Mean (SD) or % | Median (range) | |
|---|---|---|
| Demographic Information | ||
| Age (years) | 65.8 (9.7) | 66.2 (37.7–93.2) |
| Gender (% female) | 52.9 | |
| BMI (kg/m2) | 32.5 (6.7) | 31.6 (14.6–59) |
| % smoking (current and prior) | 54.6 | |
| Hypertension (%) | 86.9 | |
| Self‐reported history of prior CVD (%) | 34.8 | |
| Type 2 diabetes | ||
| % affected | 87.7 | |
| Duration (year) | 15.4 (7.7) | 13.8 (0.4–44) |
| Glucose (mg/dL) | 147.6 (55) | 135 (40–408) |
| Hemoglobin A1C (%) | 7.3 (1.4) | 7.3 (1.9–14.8) |
| Medication use (%) | ||
| Antidiabetic | 73.2 | |
| Cholesterol‐lowering | 52.3 | |
| Antihypertensive | 67.4 | |
| Education (%) | ||
| Less than high school | 14.8 | |
| High school | 47.7 | |
| Greater than high school | 37.5 | |
| Cognitive Battery | ||
| 3MSE | 90.9 (7.1) | 92 (43–100) |
| DSST | 50 (16) | 50 (10–106) |
| RAVLT‐R | 41.2 (10.2) | 41 (11–66) |
| RAVLT‐RI | 7.64 (3.2) | 8 (0–15) |
| COWA‐Semantic | 30.8 (8.4) | 30 (11–69) |
| COWA‐Phonemic | 29.7 (11.7) | 29 (2–67) |
| Stroop word card | 19.2 (5.1) | 18 (11–75) |
| Stroop color card | 26 (7.1) | 25 (14–88) |
| Stroop color‐word card | 59.2 (22.2) | 53 (26–193) |
3MSE, Modified Mini‐Mental State Examination; BMI, body‐mass index; COWA, Controlled Oral Word Association Task; CVD, cardiovascular disease; DSST, digit symbol substitution Test; RAVLT, Rey Auditory‐verbal Learning Task; R, recall; RI, retroactive interference.
All studies were approved by the Wake Forest School of Medicine Institutional Review Board and conducted in accordance with the Declaration of Helsinki. All experiments were undertaken with the understanding and written consent of subjects.
Cognitive tasks
This study includes an extension and secondary analysis of cognitive battery data described in detail in a previous study (Hugenschmidt et al. 2013). The battery assessed a number of cognitive domains by employing the Modified Mini‐Mental State Examination (3MSE; n = 883); the DSST (Digit Symbol Substitution Task; n = 889); the Rey Auditory‐Verbal Learning Task (RAVLT‐R; n = 891); the COWA (Controlled Oral Word Association Task; n = 849); and the Stroop Task (n = 883). The 3MSE is often used to screen for cognitive impairment and dementia; a series of questions were used to assess an individual's orientation to place and time, word recall and fluency, and construction (McDowell et al. 1997). The DSST measured short‐term working memory by asking individuals to match symbols to numbers as quickly as possible within a limited amount of time (Swiger et al. 2013). The RAVLT measured episodic memory and verbal learning by reading individuals a set of 15 unrelated nouns (known as list A) and asking them to repeat back as many as they could remember. Scores are the sum of correct words recalled across the first five trials. A subsection of the test required individuals to recall words from list A, directly after the introduction of a distractor list B. A measure of performance during this subsection is termed retroactive interference (RAVLT‐RI) – as recently learned words from list B interfere with the recall of words from list A (Mitrushina et al. 1991). The COWA task was used to measure phonemic (letter) and semantic (category) fluency by allotting individuals one minute to name as many words as possible starting with a given letter (F,A,S) or within a given category (animals and kitchen) (Shao et al. 2014). The Stroop task was broken down to three phases. First, individuals were shown a word card (W) and asked to read the names of colors printed in black ink. Then they were shown a color card (C) and asked to name color swatches. Finally, individuals were presented a CW (color‐word) card and asked to name the incongruent color of the print used for the color names, while trying to suppress the automatic processing of printed words. For the Stroop analysis, a group of subscores (Table S1) was chosen based on previous analyses and including two measures of the stroop effect (Jensen 1965). Color‐blind individuals were excluded from the Stroop Task (n = 2). Importantly, this study aimed to examine the functional effects of dopaminergic polymorphisms in a T2D‐affected population on cognitive performance, thus individuals whose performance was indicative of mild cognitive impairment or dementia were not excluded. Scores were natural log transformed when necessary to approximate a normal distribution.
Genotyping
Initially, SNPs (single‐nucleotide polymorphisms) were selected to form an a priori set of candidate variants covering genes in the dopaminergic pathway, the insulin signaling pathway, and the convergence of both. Genotyping was accomplished using the Sequenom MassARRAY iPLEX™ multiplexing assay. In general, the protocol included a multiplex polymerase chain reaction (PCR) followed by a single‐base primer extension reaction and MALDI‐TOF mass spectrometry‐based allele detection (Oeth et al. 2005). During quality control, both samples and SNPs were excluded with call rates less than 90% and discordant samples were removed. The final candidate gene list included 13 SNPs (Table 2).
Table 2.
Candidate variants, supporting literature, and association P‐values (β –values) with cognitive phenotypes assuming an additive model of inheritance
| Chr | Position | Rsid | Gene | Name | Function | Location | Alleles mjr/mnr | MAF | Association P‐values (β‐values); covariates – age, sex, T2D affected status, educationa | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3MSE | DSST | RAVLT‐R | RAVLT‐RI | COWA‐ Semantic | COWA‐ Phonemic | Stroop‐I | Stroop‐II | Stroop‐III | Stroop‐IV | Rationale | |||||||||
| 5 | 13,92,904 | rs28363170 | DAT1 | Dopamine Active Transporter 1 | Membrane spanning receptor; mediates reuptake of dopamine from synapse | 3' UTR | 10‐R/8, 9, 11‐R | 0.28b | 0.616 (−0.182) | 0.866 (−0.132) | 0.835 (0.105) | 0.400 (−0.129) | 0.671 (0.192) | 0.403 (0.525) | 0.860 (−0.004) | 0.337 (0.040) | 0.470 (l−0.019) | 0.253 (0.029) | Newman et al. (2014), Schneider et al. (2015), Yang et al. (2007) |
| 14 | 10,47,93,397 | rs1130214 | AKT1 | v‐akt murine thymoma viral oncogene homolog 1 | serine‐threonine protein kinase involved in metabolism and DA receptor signaling | 3' UTR | G/T | 0.24 | 0.711 (0.134) | 0.377 (0.687) | 0.256 (0.563) | 0.084 (0.256) | 0.254 (−0.516) | 0.828 (−0.138) | 0.777 (−0.006) | 0.418 (0.032) | 0.153 (−0.037) | 0.004 (0.070) | Cox et al. (2014), Mackenzie and Elliott (2014), Xia et al., 2015 |
| 4 | 11,34,58,223 | rs1880529 | CAMKIID | Calcium/Calmodulin‐ Dependent Protein Kinase II Delta | Regulates calmodulin‐dependent protein kinase activity; positive regulation of ERK signaling pathway | Intronic | T/C | 0.32 | 0.514 (0.225) | 0.666 (−0.320) | 0.516 (−0.309) | 0.570 (0.082) | 0.355 (−0.400) | 0.463 (−0.443) | 0.493 (−0.015) | 0.870 (−0.006) | 0.926 (−0.002) | 0.862 (−0.004) | Sprooten et al. (2013), Southam et al. (1999), Zhang et al. (2014) |
| 7 | 5,05,50,906 | rs10499695 | DDC | DOPA decarboxylase | Catalyzes decarboxylation of L‐DOPA to DA | Intronic | T/C | 0.46 | 0.020 (−0.761) | 0.223 (−0.855) | 0.039 (−0.928) | 0.001 (−0.448) | 0.751 (−0.130) | 0.898 (0.073) | 0.177 (−0.027) | 0.503 (−0.025) | 0.333 (−0.023) | 0.969 (−0.001) | Borelli et al. (1997), Rorsman et al. (1995), Zhu et al., (2013) |
| 7 | 5,04,77,395 | rs3887825 | DDC | DOPA decarboxylase | Catalyzes decarboxylation of L‐DOPA to DA | Intronic | G/A | 0.47 | 0.228 (0.411) | 0.806 (0.180) | 0.155 (0.670) | 0.375 (0.126) | 0.889 (0.059) | 0.938 (−0.046) | 0.773 (−0.006) | 0.602 (0.020) | 0.790 (−0.007) | 0.598 (0.012) | Borelli et al. (1997), Rorsman et al. (1995) Zhu et al., (2013) |
| 7 | 5,05,56,808 | rs6969081 | DDC | DOPA decarboxylase | Catalyzes decarboxylation of L‐DOPA to DA | Intronic | T/A | 0.5 | 0.217 (−0.431) | 0.915 (−0.080) | 0.277 (−0.524) | 0.010 (−0.376) | 0.794 (−0.114) | 0.425 (0.487) | 0.379 (−0.019) | 0.996 (0.0002) | 0.429 (−0.020) | 0.139 (−0.035) | Borelli et al. (1997), Rorsman et al. (1995), Zhu et al., (2013) |
| 3 | 11,41,71,968 | rs6280 | DRD3 | Dopamine Receptor D3 | DA receptor localized to limbic regions; inhibits AC | Exonic | T/C | 0.49 | 0.475 (0.240) | 0.853 (0.133) | 0.230 (0.554) | 0.373 (0.124) | 0.249 (−0.484) | 0.616 (−0.295) | 0.982 (−0.0004) | 0.796 (0.010) | 0.990 (0.0003) | 0.472 (0.016) | Bombin et al. (2008), Ustione and Piston (2012) |
| 10 | 6,07,92,132 | rs2456778 | FAM53B | Family With Sequence Similarity 53, Member B | Regulates β‐catenin nuclear localization | Intronic | T/A | 0.21 | 0.951 (0.023) | 0.571 (0.465) | 0.653 (0.238) | 0.0862 (−0.277) | 0.067 (0.875) | 0.301 (0.692) | 0.983 (0.0005) | 0.826 (−0.010) | 0.719 (−0.010) | 0.289 (0.028) | Gelernter et al. (2014), Kanazawa et al. (2005), Kizil et al. (2014) |
| 11 | 28,18,521 | rs2237892 | KCNQ1 | Potassium Channel, Voltage Gated KQT‐ Like Subfamily Q, Member 1 | Action potential repolarization | Intronic | C/T | 0.15 | 0.704 (0.284) | 0.492 (−1.10) | 0.453 (−0.771) | 0.758 (−0.096) | 0.697 (0.361) | 0.973 (0.044) | 0.271 (0.051) | 0.024 (−0.189) | 0.089 (0.093) | 0.799 (0.013) | Abbott (2014a), Abbott et al. (2014b), Hansen et al. (2006) Thevenod (2002), Unoki et al. (2008) |
| 11 | 28,35,964 | rs2237895 | KCNQ1 | Potassium Channel, Voltage Gated KQT‐ Like Subfamily Q, Member 1 | Action potential repolarization | Intronic | A/C | 0.32 | 0.177 (0.447) | 0.018 (1.67) | 0.398 (−0.384) | 0.371 (0.124) | 0.214 (0.509) | 0.190 (0.751) | 0.556 (−0.012) | 0.394 (−0.032) | 0.736 (−0.008) | 0.958 (0.001) | Abbott (2014a), Abbott et al. (2014b), Hansen et al. (2006) Thevenod (2002), Unoki et al. (2008) |
| 11 | 28,28,300 | rs2283228 | KCNQ1 | Potassium Channel, Voltage Gated KQT‐ Like Subfamily Q, Member 1 | Action potential repolarization | Intronic | A/C | 0.16 | 0.243 (0.810) | 0.910 (0.171) | 0.927 (−0.088) | 0.848 (0.055) | 0.183 (1.15) | 0.581 (0.668) | 0.889 (−0.006) | 0.149 (−0.112) | 0.806 (0.012) | 0.621 (−0.023) | Abbott (2014a), Abbott et al. (2014b), Hansen et al. (2006), Thevenod (2002), Unoki et al. (2008) |
| 11 | 9,29,75,544 | rs1080963 | MTNR1B | Melatonin Receptor 1B | Implicated in circadian entrainment and GPCR signaling | Intronic | C/G | 0.26 | 0.723 (−0.125) | 0.302 (−0.784) | 0.071 (−0.878) | 0.187 (−0.194) | 0.684 (−0.178) | 0.459 (−0.454) | 0.880 (0.003) | 0.038 (−0.082) | 0.293 (0.027) | 0.053 (0.046) | Bonnefond and Froguel (2015), Yokoyama et al. (2014) |
| 1 | 1,12,62,099 | rs1883965 | MTOR | Mechanistic Target Of Rapamycin | Kinase; mediates cellular response to stress | Intronic | G/A | 0.31 | 0.251 (−0.429) | 0.045 (−1.59) | 0.489 (−0.359) | 0.268 (−0.173) | 0.025 (−1.03) | 0.764 (−0.193) | 0.516 (0.014) | 0.845 (0.008) | 0.506 (0.018) | 0.993 (−0.0002) | Bowling et al. (2014), Melnik (2015), Johnson et al. (2013) |
| 11 | 21,72,610 | rs10770141 | TH | Tyrosine Hydroxylase | Rate‐limiting enzyme in the synthesis of DA | Upstream | G/A | 0.34 | 0.706 (0.116) | 0.184 (−0.871) | 0.634 (−0.202) | 0.568 (−0.074) | 0.433 (−0.302) | 0.249 (−0.621) | 0.521 (−0.012) | 0.682 (−0.014) | 0.470 (−0.016) | 0.242 (0.025) | Aliev et al. (2014), Borelli et al. (2003), Rao et al. (2010) |
AC, adenylyl cyclase; DA, dopamine; ERK, extracellular signal regulated kinases; GPCR, G‐protein coupled receptor; K(+), potassium; L‐DOPA, L‐3,4‐dihydroxyphenylalanine.
Association P‐value <0.05 in bold.
RAVLT‐RI additionally adjusted for number of words recalled after initial exposure.
MAF based on frequency of all minor alleles.
For the DAT1 VNTR, genotyping was achieved using PCR with the forward primer: 5'‐ TGTGG TGTAGGGAACGGCCTGAG‐3' and reverse primer: 5'‐CTTCC TGGAGGTCACGGCTCAAGG‐3' followed by 2% agarose gel electrophoresis. Allele determination was made based on the size of fragments compared with known genotypes (Sano et al. 1993; Kang et al. 1999) and standards.
Finally, SNPs in loci of interest with strong evidence for involvement in DA processing (DDC, TH, DRD2, DRD3, and DAT1) were extracted from genetic datasets available in a subset of the cohort (n = 572) derived from Affymetrix® Genome‐Wide Human SNP Array 5.0 and 1000 Genomes (1000G) imputation from this array. The genes included were DAT1 (SLC6A3, DA transporter), DDC (DOPA decarboxylase), DRD2 (DA D2 receptor), DRD3 (DA D3 receptor), and TH (tyrosine hydroxylase). Genotyping and quality control procedures for these data are described elsewhere (Cox et al. 2014). Briefly, for the GWAS set, exclusion criteria for SNP performance included Hardy–Weinberg equilibrium P < 1 × 10−6, call rate <95%, and minor allele frequency <0.01. Only SNPs with a confidence score >0.90 and information score >0.50 were used from the imputed dataset. Initially 649 SNPs were isolated from both datasets. After reduction due to >5% missingness and <5% minor allele frequency, 484 SNPs remained for analysis.
Statistical analyses
To determine the effect of candidate variants on cognitive performance, genetic association analyses were completed for the 13 candidate SNPs and the DAT1 VNTR. Next, targeted association analyses were performed using the GWAS and imputed data. All SNP association analyses were carried out using variance‐components analysis in SOLAR version 8.0.1 (Texas Biomedical Research Institute San Antonio, TX) to account for relatedness among subjects (Almasy and Blangero 1998). Genetic association was investigated assuming an additive model of inheritance and adjusting for age, sex, T2D‐affected status, and education. An additional covariate was added when analyzing retroactive interference in the RAVLT in order to adjust for recall performance after a single exposure to the word list. Significance levels were adjusted for multiple comparisons by Bonferroni correction. For the candidate variant analysis, P < 3.8 × 10−3; for SNPs extracted from genome‐wide and imputed data, statistical significance was accepted at 1 × 10−4.
Results
Table 1 provides summary statistics of the analyzed sample. The significant enrichment of T2D‐affected participants is reflected in prevalent comorbid indices such as participant‐reported CVD (cardiovascular disease; 34.8%), high mean BMI (body‐mass index; 32.5 kg/m2), and a significant proportion of individuals with hypertension (86.9%). In general, cognitive performance among the different tests was normally distributed and performance ranged from above average to indicative of cognitive impairment (Hoyer et al. 2004; Loonstra et al. 2001; Savage and Gouvier 1992; Roy et al. 2015; Vogel et al. 2013; Table 1). Education levels ranged from less than high school to attainment of highly trained occupations which is reflective of a community‐based cohort.
Initially, the cognitive data from examination of these subjects were evaluated for association with 14 genetic variants. Characteristics of candidate SNPs and DAT1 VNTR are summarized in Table 2 with a brief description of gene function and relevant references. Table 2 also summarizes the results of the genetic association analysis. The variant analysis was conducted in the DHS – Mind cohort to evaluate the association of dopaminergic and other literature‐supported polymorphisms with cognitive performance adjusting for age, sex, T2D status, and education
Assuming an additive model with the 10 repeat allele as the risk allele (Yang et al. 2007), the DAT1 VNTR was not significantly associated with any cognitive phenotypes (Table 2). The most suggestive association was with poorer performance (i.e., higher score) on the Stroop‐IV, which measures difficulty in color‐naming due to interference of the conflicting printed words known as the stroop effect (P = 0.253; β = 0.029).
In general, when SNPs in candidate genes were evaluated (Table 2) most associations were, at best, nominal. However, one coding variant from DDC, the DOPA decarboxylase gene, rs10499695 was significantly associated with poorer performance on a subset of RAVLT that measures retroactive interference (P = 0.001, β = −0.45). Additionally, an AKT1 gene polymorphism, rs1130214, was nominally associated with performance on Stroop‐IV (P = 0.004; β = 0.07), though this did not meet our Bonferroni‐corrected P‐value threshold.
Taking advantage of GWAS and imputed data, a targeted analysis was carried out in a subset of the cohort (n = 572). 484 SNPs in our genes of interest from GWAS and imputed data passed quality control. The top 50 SNPs associated with each of the cognitive phenotypes are included in Table S2A–J. This targeted association analysis revealed a significant association between SNP rs62445903 in DDC and retroactive interference on the RAVLT (P = 7.21 × 10−7, β = 0.3). Among the most strongly associated SNPs were several variants from DRD2 (e.g., rs73557283, P = 1.3 × 10−3, β = 0.12 and rs77195172, P = 1.85 × 10−3, β = 0.12 with Stroop‐II score) and DAT1 (e.g., rs10052016, P = 1.61 × 10−3, β = 0.11 and rs10053602, P = 3.94 × 10−3, β = 0.10 with Stroop‐IV score) genes. Figure 1 represents locus plots of the DOPA decarboxylase, DA D2 receptor, and DA D3 receptor genes in RAVLT‐RI, Stroop‐II, and COWA‐Phonemic tasks, respectively.
Figure 1.
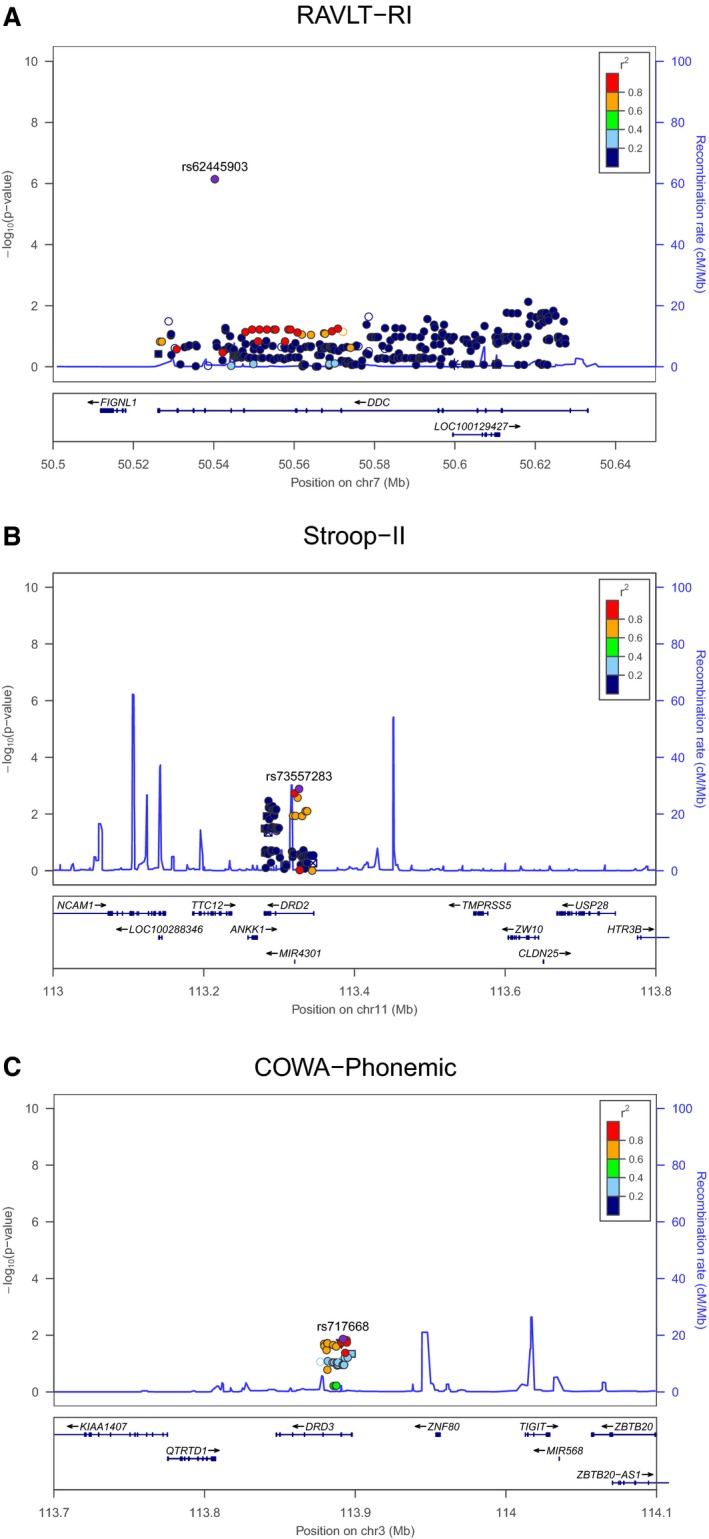
Locus plots for GWAS and Imputed associations with (A) RAVLT‐RI (B) Stroop‐II and (C) Stroop‐IV. Association analyses were performed assuming an additive model of inheritance with adjustment for age, sex, T2D affected status, and education; additional adjustment for recall performance after first exposure to the word list was included for RAVLT‐RI. Abbreviations: GWAS, genome‐wide association study; RAVLT‐RI, Rey Auditory‐verbal Learning Task—retroactive interference; T2D, type 2 diabetes.
Discussion
After searching the literature, candidate genetic variants were selected based on studies investigating specific components of the dopaminergic pathway, cognitive function, T2D pathogenesis, and particularly combinations thereof. In the T2D‐enriched DHS sample, the majority of candidate SNPs were not strongly associated with measures of cognitive function at conservative levels of significance. However, of special interest is SNP rs10499695 located intronically in the gene DDC, (DOPA decarboxylase) which was significantly associated with retroactive interference on the RAVLT. In this section of the RAVLT task, a distractor list is read between presentation and recall of the original word list; disruption of recall of the retained words is termed retroactive interference. Interestingly, this DDC SNP has previously been shown to be associated with alerting attention, which specifically measures an individual's ability to maintain a state of alertness and readiness to respond to stimuli (Zhu et al. 2013). This is congruent considering it is theoretically a diverted attention, e.g., the distractor list, which underpins the phenomena of retroactive interference (Dewar et al. 2007).
An extensive candidate gene literature surrounds the DAT1 VNTR, a 40‐base pair repeat unit with 9 and 10‐repeat units occurring most commonly. For example, Schneider and colleagues found that 10‐repeat carriers were less susceptible to the “Stroop effect” (2015) that arises from discordance between words and color words, and is scored similar to Stroop‐III in this study. However, DAT1 genotype was not associated with Stroop‐III performance or any other cognitive measure in the DHS – Mind sample. This is not surprising given that replication in both genetic and cognitive literature is a challenge due to variations in sample size, misinterpretation of associations (Hirschhorn 2005), and heterogeneity of phenotyping procedures (e.g., see Hart et al. 2013). However, the importance of the DAT1 VNTR should not be discarded in light of a recent meta‐analysis of in vivo imaging studies which found that DAT1 VNTR genotype has functional consequences on DA transporter activity (Faraone et al. 2014). Thus, future studies are needed to completely understand the role of the DAT1 VNTR in cognitive performance in T2D‐affected individuals.
Next, to explore the genetic consequence of the dopaminergic system on cognitive capacity in T2D, association analyses of both genotyped and imputed SNPs from regions of interest were completed. The most notable finding was that a significant association between a polymorphism from the DOPA decarboxylase gene and retroactive interference in the RAVLT was revealed. This intronic variant, rs62445903, is located approximately 78 kb upstream of the significant DDC SNP, rs10499695, uncovered in the candidate gene analysis. A pair‐wise linkage disequilibrium test using 1000G as the reference panel suggested the two SNPs from DDC are most likely in linkage disequilibrium, with a reduced correlation coefficient due to a MAF less than 10% for the imputed SNP, rs62445903 (D' = 1; r 2 = 0.12). Furthermore, rs62445903 is no longer significant when the model is adjusted for the candidate variant rs10499695. Although rs10499695 was also included on the Affymetrix® array, it failed to reach significance in this secondary analysis. This could be due to the reduced sample size in the GWAS and imputed analysis compared to the candidate SNP analysis (n = 572 vs. n = 893, respectively) especially since the two genotyping procedures were 99.1% concordant. Importantly, the appearance of significant SNPs on the DDC locus in both analyses highlights this gene as a locus of interest for cognitive capacity in T2D.
Interestingly, the GWAS/Imputation analysis revealed DOPA decarboxylase polymorphisms among the 50 strongest associations for each of the cognitive indices. The DDC gene undergoes alternative splicing creating two distinct mRNAs – neural and nonneuronal (Ichinose et al. 1992). Although it is most well‐known for synthesis of DA and serotonin in the central nervous system, the enzyme is also located in pancreatic β cells where it is theorized to be involved in local, dopaminergic regulation of insulin secretion (Rubi et al. 2005; Ustione et al. 2013). The abundance of nominal associations in the DDC gene in a T2D population has biological relevance with regard to potential roles in cognitive function as DDC deficiency due to frame‐shift mutations or substitutions leads to impaired cognitive function (Bertoldi 2014).
Intriguingly, the most significant SNP associations in the GWAS and imputation analysis (refer to Table S2) were generally segregated by cognitive phenotype. For instance, 100% of the top 50 SNPs associated with Stroop‐I score were from DDC, while 43 of the top 50 associations for Stroop‐II score were polymorphisms from the DRD2 gene. This is compelling since Stroop‐I scores represent an individual's personal tempo, while Stroop‐II scores represent a color‐naming factor (Jensen 1965). Thus, this study tenably uncovered a genetic component that further validates Jensen's statistical conclusion of a cluster of Stroop subscores representing different cognitive dimensions (1965).
Although the present analysis was able to reveal polymorphisms significantly and suggestively associated with cognitive phenotypes in a T2D‐rich familial population, there are limitations to the study. Firstly, as with the majority of studies investigating the genetic contribution to complex disorders, sample size is a concern; the GWAS and imputed association analysis in particular was limited to fewer than 500 individuals (Hong and Park, 2012). Thus, a future study closely investigating the influence of the dopaminergic pathway on cognition in a larger cohort is warranted. Secondly, candidate genes for this study were selected by gathering information from previous genetic studies and knowledge of biologic pathways in the pursuit of SNPs that harbor functional significance. However, the majority of the candidate SNPs as well as the current significant associations included intronic variants. Intronic SNPs may act as regulatory elements through interaction with enhancers to affect transcription (Stadhouders et al. 2012), or may affect chromatin structure by changes in DNA methylation patterns, thus modulating gene activity (Zaina et al. 2010). SNP rs10499695 is associated with retroactive interference and is located within an ETS‐family transcription factor binding site (Boyle et al. 2012). Moreover, these variants may not be protein‐coding but the field of medical genetics requires a more thorough understanding of polymorphisms outside protein‐coding regions, especially for those that may affect RNA splicing (Xiong et al. 2015). For instance, the other DDC SNP significantly associated with retroactive interference, rs62445903, is located in intron 9. Interestingly, there is an isoform of human DDC that includes an alternative exon 10 that is located within intron 9 and this particular isoform is expressed in nonneural kidney tissue (Vassilacopoulou et al. 2004). Effects on alternative splicing are one way intronic SNPs modulate mRNA expression and translation and this is perhaps the mechanism of action by which DDC affects cognition. Finally, it is possible that the DDC SNPs tagged in this study may be in LD with untyped putatively functional variants, which was not investigated in this study.
In conclusion, this study offers evidence of the involvement of dopaminergic genes in cognitive performance among T2D‐affected individuals, especially through regulation of DA synthesis. Identification of such pathways is critical in order to unveil new avenues of treatment as well as garner a more thorough understanding of the detrimental cognitive effects of T2D. Future studies will be needed to validate the findings and expand knowledge on the role of the DDC gene in individual cognitive ability.
Conflict of Interest
None declared.
Supporting information
Table S1. Formulas for derived Stroop scores.
Table S2. Top 50 association P‐values of genome‐wide and imputed data with cognitive phenotypes adjusted for age, sex, T2D‐affected status, and education. RAVLT‐RI model additionally adjusted for recall performance after first exposure to the word list. Abbreviations: RAVLT‐RI, Rey Auditory‐verbal Learning Task—retroactive interference; T2D, type 2 diabetes.
Acknowledgments
This study was supported in part by the National Institutes of Health through R01 HL67348, R01 HL092301, R01 NS058700 (to DWB), F32 DK083214‐01 (to CEH), and F31 AG044879 (to LMR). The authors thank the other investigators, the staff, and the Diabetes Heart Study participants for their valuable contributions.
Martelle S. E., Raffield L. M., Palmer N. D., Cox A. J., Freedman B. I., Hugenschmidt C. E., Williamson J. D., Bowden D. W.. Dopamine pathway gene variants may modulate cognitive performance in the DHS – Mind Study, Brain and Behavior, 2016; 6(4), e00446, doi: 10.1002/brb3.446
References
- Abbott, G. W. 2014a. Biology of the KCNQ1 potassium channel. New J. Sci. 2014:1–26. [Google Scholar]
- Abbott, G. W. , Tai K. K., Neverisky D. L., Hansler A., Hu Z., Roepke T. K., et al. 2014b. KCNQ1, KCNE2, and Na+‐coupled solute transporters form reciprocally regulating complexes that affect neuronal excitability. Sci. Signal. 7:ra22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aliev, G. , Shahida K., Gan S. H., Firoz C., Khan A., Abuzenadah A. M., et al. 2014. Alzheimer disease and type 2 diabetes mellitus: the link to tyrosine hydroxylase and probable nutritional strategies. CNS Neurol. Disord. Drug Targets 13:467–477. [DOI] [PubMed] [Google Scholar]
- Almasy, L. , and Blangero J.. 1998. Multipoint quantitative‐trait linkage analysis in general pedigrees. Am. J. Hum. Genet. 62:1198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Berker, A. O. , and Rutledge R. B.. 2014. A role for the human substantia nigra in reinforcement learning. J. Neurosci. 34:12947–12949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoldi, M. 2014. Mammalian Dopa decarboxylase: structure, catalytic activity and inhibition. Arch. Biochem. Biophys. 546:1–7. [DOI] [PubMed] [Google Scholar]
- Boehler, C. N. , Bunzeck N., Krebs R. M., Noesselt T., Schoenfeld M. A., Heinze H. J., et al. 2011. Substantia nigra activity level predicts trial‐to‐trial adjustments in cognitive control. J. Cogn. Neurosci. 23:362–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bombin, I. , Arango C., Mayoral M., Castro‐Fornieles J., Gonzalez‐Pinto A., Gonzalez‐Gomez C., et al. 2008. DRD3, but not COMT or DRD2, genotype affects executive functions in healthy and first‐episode psychosis adolescents. Am. J. Med. Genet. B Neuropsychiatr. Genet. 147b:873–879. [DOI] [PubMed] [Google Scholar]
- Bonnefond, A. , and Froguel P.. 2015. Rare and common genetic events in type 2 diabetes: what should biologists know? Cell Metab. 21:357–368. [DOI] [PubMed] [Google Scholar]
- Borelli, M. I. , Villar M. J., Orezzoli A., and Gagliardino J. J.. 1997. Presence of DOPA decarboxylase and its localisation in adult rat pancreatic islet cells. Diabetes Metab. 23:161–163. [PubMed] [Google Scholar]
- Borelli, M. I. , Rubio M., Garcia M. E., Flores L. E., and Gagliardino J. J.. 2003. Tyrosine hydroxylase activity in the endocrine pancreas: changes induced by short‐term dietary manipulation. BMC Endocr. Disord. 3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden, D. W. , Cox A. J., Freedman B. I., Hugenschimdt C. E., Wagenknecht L. E., Herrington D., et al. 2010. Review of the Diabetes Heart Study (DHS) family of studies: a comprehensively examined sample for genetic and epidemiological studies of type 2 diabetes and its complications. Rev. Diabet. Stud. 7:188–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowirrat, A. , Chen T. J., Oscar‐Berman M., Madigan M., Chen A. L., Bailey J. A., et al. 2012. Neuropsychopharmacology and neurogenetic aspects of executive functioning: should reward gene polymorphisms constitute a diagnostic tool to identify individuals at risk for impaired judgment? Mol. Neurobiol. 45:298–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling, H. , Zhang G., Bhattacharya A., Perez‐Cuesta L. M., Deinhardt K., Hoeffer C. A., et al. 2014. Antipsychotics activate mTORC1‐dependent translation to enhance neuronal morphological complexity. Sci. Signal. 7:ra4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle, A. P. , Hong E. L., Hariharan M., Cheng Y., Schaub M. A., Kasowski M., et al. 2012. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 22:1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, A. B. , Biederman J., Valera E., Makris N., Doyle A., Whitfield‐Gabrieli S., et al. 2011. Relationship of DAT1 and adult ADHD to task‐positive and task‐negative working memory networks. Psychiatry Res. 193:7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, K. L. , and Noudoost B.. 2014. The role of prefrontal catecholamines in attention and working memory. Front. Neural. Circuits. 8:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, A. J. , Hugenschmidt C. E., Raffield L. M., Langefeld C. D., Freedman B. I., Williamson J. D., et al. 2014. Heritability and genetic association analysis of cognition in the Diabetes Heart Study. Neurobiol. Aging 35:1958 e1953‐1958 e1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar, M. T. , Cowan N., and Sala S. D.. 2007. Forgetting due to retroactive interference: a fusion of Muller and Pilzecker's (1900) early insights into everyday forgetting and recent research on anterograde amnesia. Cortex 43:616–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, L. , and Gold J. I.. 2013. The basal ganglia's contributions to perceptual decision making. Neuron 79:640–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraone, S. V. , Spencer T. J., Madras B. K., Zhang‐James Y., and Biederman J.. 2014. Functional effects of DA transporter gene genotypes on in vivo DA transporter functioning: a meta‐analysis. Mol. Psychiatry 19:880–889. [DOI] [PubMed] [Google Scholar]
- Frank, M. J. , and Fossella J. A.. 2011. Neurogenetics and pharmacology of learning, motivation, and cognition. Neuropsychopharmacology 36:133–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter, J. , Sherva R., Koesterer R., Almasy L., Zhao H., Kranzler H. R., et al. 2014. Genome‐wide association study of cocaine dependence and related traits: FAM53B identified as a risk gene. Mol. Psychiatry 19:717–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genro, J. P. , Kieling C., Rohde L. A., and Hutz M. H.. 2010. Attention‐deficit/hyperactivity disorder and the DArgic hypotheses. Expert Rev. Neurother. 10:587–601. [DOI] [PubMed] [Google Scholar]
- Gordon, E. M. , Devaney J. M., Bean S., and Vaidya C. J.. 2015. Resting‐state striato‐frontal functional connectivity is sensitive to DAT1 genotype and predicts executive function. Cereb. Cortex 25:336–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, H. H. , Ebbesen C., Mathiesen C., Weikop P., Ronn L. C., Waroux O., et al. 2006. The KCNQ channel opener retigabine inhibits the activity of mesencephalic dopaminergic systems of the rat. J. Pharmacol. Exp. Ther. 318:1006–1019. [DOI] [PubMed] [Google Scholar]
- Hart, A. B. , de Wit H., and Palmer A. A.. 2013. Candidate gene studies of a promising intermediate phenotype: failure to replicate. Neuropsychopharmacology 38:802–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson, D. C. , Cagliero E., Gray C., Nasrallah R. A., Hayden D. L., Schoenfeld D. A., et al. 2000. Clozapine, diabetes mellitus, weight gain, and lipid abnormalities: a five‐year naturalistic study. Am. J. Psychiatry 157:975–981. [DOI] [PubMed] [Google Scholar]
- Hirschhorn, J. N. 2005. Genetic approaches to studying common diseases and complex traits. Pediatr. Res. 57:74r–77r. [DOI] [PubMed] [Google Scholar]
- Hong, E. P. , and Park J. W.. 2012. Sample size and statistical power calculation in genetic association studies. Genomics Inform. 10:117–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyer, W. J. , Stawski R. S., Wasylyshyn C., and Verhaeghen P.. 2004. Adult age and digit symbol substitution performance: a meta‐analysis. Psychol. Aging 19:211–214. [DOI] [PubMed] [Google Scholar]
- Hugenschmidt, C. E. , Hsu F. C., Hayasaka S., Carr J. J., Freedman B. I., Nyenhuis D. L., et al. 2013. The influence of subclinical cardiovascular disease and related risk factors on cognition in type 2 diabetes mellitus: the DHS‐Mind study. J. Diabetes Complications 27:422–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinose, H. , Sumi‐Ichinose C., Ohye T., Hagino Y., Fujita K., and Nagatsu T.. 1992. Tissue‐specific alternative splicing of the first exon generates two types of mRNAs in human aromatic L‐amino acid decarboxylase. Biochemistry 31:11546–11550. [DOI] [PubMed] [Google Scholar]
- Jensen, A. R. 1965. Scoring the Stroop test. Acta Psychol. (Amst) 24:398–408. [DOI] [PubMed] [Google Scholar]
- Johnson, S. C. , Rabinovitch P. S., and Kaeberlein M.. 2013. mTOR is a key modulator of ageing and age‐related disease. Nature 493:338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazawa, A. , Tsukada S., Kamiyama M., Yanagimoto T., Nakajima M., and Maeda S.. 2005. Wnt5b partially inhibits canonical Wnt/beta‐catenin signaling pathway and promotes adipogenesis in 3T3‐L1 preadipocytes. Biochem. Biophys. Res. Commun. 330:505–510. [DOI] [PubMed] [Google Scholar]
- Kang, A. M. , Palmatier M. A., and Kidd K. K.. 1999. Global variation of a 40‐bp VNTR in the 3'‐untranslated region of the DA transporter gene (SLC6A3). Biol. Psychiatry 46:151–160. [DOI] [PubMed] [Google Scholar]
- Kehagia, A. A. , Barker R. A., and Robbins T. W.. 2013. Cognitive impairment in Parkinson's disease: the dual syndrome hypothesis. Neurodegener. Dis. 11:79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizil, C. , Kuchler B., Yan J. J., Ozhan G., Moro E., Argenton F., et al. 2014. Simplet/Fam53b is required for Wnt signal transduction by regulating beta‐catenin nuclear localization. Development 141:3529–3539. [DOI] [PubMed] [Google Scholar]
- Kleinridders, A. , Cai W., Cappellucci L., Ghazarian A., Collins W. R., Vienberg S. G., et al. 2015. Insulin resistance in brain alters DA turnover and causes behavioral disorders. Proc. Natl Acad. Sci. USA 112:3463–3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko, J. H. , Antonelli F., Monchi O., Ray N., Rusjan P., Houle S., et al. 2013. Prefrontal DArgic receptor abnormalities and executive functions in Parkinson's disease. Hum. Brain Mapp. 34:1591–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loonstra, A. S. , Tarlow A. R., and Sellers A. H.. 2001. COWAT metanorms across age, education, and gender. Appl. Neuropsychol. 8:161–166. [DOI] [PubMed] [Google Scholar]
- Luchsinger, J. A. 2012. Type 2 diabetes and cognitive impairment: linking mechanisms. J. Alzheimers Dis. 30(Suppl 2):S185–S198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie, R. W. , and Elliott B. T.. 2014. Akt/PKB activation and insulin signaling: a novel insulin signaling pathway in the treatment of type 2 diabetes. Diabetes Metab. Syndr. Obes. 7:55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manschot, S. M. , Brands A. M., van der Grond J., Kessels R. P., Algra A., Kappelle L. J., et al. 2006. Brain magnetic resonance imaging correlates of impaired cognition in patients with type 2 diabetes. Diabetes 55:1106–1113. [DOI] [PubMed] [Google Scholar]
- Mayeda, E. R. , Whitmer R. A., and Yaffe K.. 2015. Diabetes and cognition. Clin. Geriatr. Med. 31:101–115, ix. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell, I. , Kristjansson B., Hill G. B., and Hebert R.. 1997. Community screening for dementia: the Mini Mental State Exam (MMSE) and Modified Mini‐Mental State Exam (3MS) compared. J. Clin. Epidemiol. 50:377–383. [DOI] [PubMed] [Google Scholar]
- Melnik, B. C. 2015. The pathogenic role of persistent milk signaling in mTORC1‐ and milk‐microRNA‐driven type 2 diabetes mellitus. Curr. Diabetes Rev. 11:46–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrushina, M. , Satz P., Chervinsky A., and D'Elia L.. 1991. Performance of four age groups of normal elderly on the Rey Auditory‐Verbal Learning Test. J. Clin. Psychol. 47:351–357. [DOI] [PubMed] [Google Scholar]
- Nemoda, Z. , Szekely A., and Sasvari‐Szekely M.. 2011. Psychopathological aspects of DArgic gene polymorphisms in adolescence and young adulthood. Neurosci. Biobehav. Rev. 35:1665–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, D. P. , Cummins T. D., Tong J. H., Johnson B. P., Pickering H., Fanning P., et al. 2014. Dopamine transporter genotype is associated with a lateralized resistance to distraction during attention selection. J. Neurosci. 34:15743–15750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeth, P. , Beaulieu M., Park C., Kosman D., del Mistro G., van den Boom D., et al. 2005. iPLEX™ assay: increased plexing efficiency and flexibility for MassARRAY system through single base primer extension with mass‐modified terminators. Sequenom San Diego (CA). Sequenom application note.
- Palta, P. , Schneider A. L., Biessels G. J., Touradji P., and Hill‐Briggs F.. 2014. Magnitude of cognitive dysfunction in adults with type 2 diabetes: a meta‐analysis of six cognitive domains and the most frequently reported neuropsychological tests within domains. J. Int. Neuropsychol. Soc. 20:278–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plum, L. , Belgardt B. F., and Bruning J. C.. 2006. Central insulin action in energy and glucose homeostasis. J. Clin. Invest. 116:1761–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffield, L. M. , Cox A. J., Hugenschmidt C. E., Freedman B. I., Langefeld C. D., Williamson J. D., et al. 2015. Heritability and genetic association analysis of neuroimaging measures in the Diabetes Heart Study. Neurobiol. Aging 36:1602 e1607–1602 e1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, F. , Zhang K., Zhang L., Rana B. K., Wessel J., Fung M. M., et al. 2010. Human tyrosine hydroxylase natural allelic variation: influence on autonomic function and hypertension. Cell. Mol. Neurobiol. 30:1391–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorsman, F. , Husebye E. S., Winqvist O., Bjork E., Karlsson F. A., and Kampe O.. 1995. Aromatic‐L‐amino‐acid decarboxylase, a pyridoxal phosphate‐dependent enzyme, is a beta‐cell autoantigen. Proc. Natl Acad. Sci. USA 92:8626–8629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, S. , Kim N., Desai A., Komaragiri M., Baxi N., Jassil N., et al. 2015. Cognitive function and control of type 2 diabetes mellitus in young adults. N. Am. J. Med. Sci. 7:220–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubi, B. , Ljubicic S., Pournourmohammadi S., Carobbio S., Armanet M., Bartley C., et al. 2005. DA D2‐like receptors are expressed in pancreatic beta cells and mediate inhibition of insulin secretion. J. Biol. Chem. 280:36824–36832. [DOI] [PubMed] [Google Scholar]
- Ryan, J. P. , Fine D. F., and Rosano C.. 2014. Type 2 diabetes and cognitive impairment: contributions from neuroimaging. J. Geriatr. Psychiatry Neurol. 27:47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambataro, F. , Podell J. E., Murty V. P., Das S., Kolachana B., Goldberg T. E., et al. 2015. Dopamine transporter 3' UTR VNTR modulates striatal function during working memory updating across the adult age span. Eur. J. Neurosci. 42:1912–1918. [DOI] [PubMed] [Google Scholar]
- Sano, A. , Kondoh K., Kakimoto Y., and Kondo I.. 1993. A 40‐nucleotide repeat polymorphism in the human DA transporter gene. Hum. Genet. 91:405–406. [DOI] [PubMed] [Google Scholar]
- Savage, R. M. , and Gouvier W. D.. 1992. Rey Auditory‐Verbal Learning Test: the effects of age and gender, and norms for delayed recall and story recognition trials. Arch. Clin. Neuropsychol. 7:407–414. [PubMed] [Google Scholar]
- Savitz, J. , Solms M., and Ramesar R.. 2006. The molecular genetics of cognition: DA, COMT and BDNF. Genes Brain Behav. 5:311–328. [DOI] [PubMed] [Google Scholar]
- Schneider, K. K. , Schote A. B., Meyer J., and Frings C.. 2015. Genes of the DArgic system selectively modulate top‐down but not bottom‐up attention. Cogn. Affect. Behav. Neurosci. 15:104–116. [DOI] [PubMed] [Google Scholar]
- Shao, Z. , Janse E., Visser K., and Meyer A. S.. 2014. What do verbal fluency tasks measure? Predictors of verbal fluency performance in older adults. Front. Psychol. 5:772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, A. , and Couture J.. 2014. A review of the pathophysiology, etiology, and treatment of attention‐deficit hyperactivity disorder (ADHD). Ann. Pharmacother. 48:209–225. [DOI] [PubMed] [Google Scholar]
- Southam, L. , Ashfield R., Cox R., Lathrop M., and Ashcroft S. J.. 1999. Human islets of Langerhans express the delta(C) isoform of calcium/calmodulin‐dependent protein kinase II. Diabetes Metab. Res. Rev. 15:243–246. [DOI] [PubMed] [Google Scholar]
- Sprooten, E. , Fleming K. M., Thomson P. A., Bastin M. E., Whalley H. C., Hall J., et al. 2013. White matter integrity as an intermediate phenotype: exploratory genome‐wide association analysis in individuals at high risk of bipolar disorder. Psychiatry Res. 206:223–231. [DOI] [PubMed] [Google Scholar]
- Stadhouders, R. , van den Heuvel A., Kolovos P., Jorna R., Leslie K., Grosveld F., et al. 2012. Transcription regulation by distal enhancers: who's in the loop? Transcription 3:181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Störmer, V. S. , Passow S., Biesenack J., and Li S. C.. 2012. Dopaminergic and cholinergic modulations of visual‐spatial attention and working memory: insights from molecular genetic research and implications for adult cognitive development. Dev. Psychol. 48:875–889. [DOI] [PubMed] [Google Scholar]
- Swiger, K. J. , Manalac R. J., Blumenthal R. S., Blaha M. J., and Martin S. S.. 2013. Statins and cognition: a systematic review and meta‐analysis of short‐ and long‐term cognitive effects. Mayo Clin. Proc. 88:1213–1221. [DOI] [PubMed] [Google Scholar]
- Thevenod, F. 2002. Ion channels in secretory granules of the pancreas and their role in exocytosis and release of secretory proteins. Am. J. Physiol. Cell Physiol. 283:C651–C672. [DOI] [PubMed] [Google Scholar]
- Unoki, H. , Takahashi A., Kawaguchi T., Hara K., Horikoshi M., Andersen G., et al. 2008. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat. Genet. 40:1098–1102. [DOI] [PubMed] [Google Scholar]
- Ustione, A. , and Piston D. W.. 2012. DA synthesis and D3 receptor activation in pancreatic beta‐cells regulates insulin secretion and intracellular [Ca(2+)] oscillations. Mol. Endocrinol. 26:1928–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustione, A. , Piston D. W., and Harris P. E.. 2013. Minireview: Dopaminergic regulation of insulin secretion from the pancreatic islet. Mol. Endocrinol. 27:1198–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilacopoulou, D. , Sideris D. C., Vassiliou A. G., and Fragoulis E. G.. 2004. Identification and characterization of a novel form of the human L‐dopa decarboxylase mRNA. Neurochem. Res. 29:1817–1823. [DOI] [PubMed] [Google Scholar]
- Vogel, A. , Stokholm J., and Jorgensen K.. 2013. Performances on Symbol Digit Modalities Test, Color Trails Test, and modified Stroop test in a healthy, elderly Danish sample. Neuropsychol. Dev. Cogn. B Aging Neuropsychol. Cogn. 20:370–382. [DOI] [PubMed] [Google Scholar]
- Wittmann, B. C. , Tan G. C., Lisman J. E., Dolan R. J., and Duzel E.. 2013. DAT genotype modulates striatal processing and long‐term memory for items associated with reward and punishment. Neuropsychologia 51:2184–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y. , Ding Y., Tanaka Y., and Zhang W.. 2014. Risk factors contributing to type 2 diabetes and recent advances in the treatment and prevention. Int. J. Med. Sci. 11:1185–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, S. F. , Xie Z. X., Qiao Y., Li L. R., Cheng X. R., Tang X., et al. 2015. Differential effects of quercetin on hippocampus‐dependent learning and memory in mice fed with different diets related with oxidative stress. Physiol. Behav. 138:325–331. [DOI] [PubMed] [Google Scholar]
- Xiong, H. Y. , Alipanahi B., Lee L. J., Bretschneider H., Merico D., Yuen R. K., et al. 2015. RNA splicing. The human splicing code reveals new insights into the genetic determinants of disease. Science 347:1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, B. , Chan R. C., Jing J., Li T., Sham P., and Chen R. Y.. 2007. A meta‐analysis of association studies between the 10‐repeat allele of a VNTR polymorphism in the 3'‐UTR of DA transporter gene and attention deficit hyperactivity disorder. Am. J. Med. Genet. B Neuropsychiatr. Genet. 144b:541–550. [DOI] [PubMed] [Google Scholar]
- Yokoyama, J. S. , Evans D. S., Coppola G., Kramer J. H., Tranah G. J., and Yaffe K.. 2014. Genetic modifiers of cognitive maintenance among older adults. Hum. Brain Mapp. 35:4556–4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaina, S. , Perez‐Luque E. L., and Lund G.. 2010. Genetics talks to epigenetics? The interplay between sequence variants and chromatin structure. Curr. Genomics 11:359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Wang Y., Wang J., Zhou X., Shu N., and Zhang Z.. 2014. White matter integrity disruptions associated with cognitive impairments in type 2 diabetic patients. Diabetes 63:3596–3605. [DOI] [PubMed] [Google Scholar]
- Zhu, B. , Chen C., Moyzis R. K., Dong Q., He Q., Li J., et al. 2013. The DOPA decarboxylase (DDC) gene is associated with alerting attention. Prog. Neuropsychopharmacol. Biol. Psychiatry 43:140–145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Formulas for derived Stroop scores.
Table S2. Top 50 association P‐values of genome‐wide and imputed data with cognitive phenotypes adjusted for age, sex, T2D‐affected status, and education. RAVLT‐RI model additionally adjusted for recall performance after first exposure to the word list. Abbreviations: RAVLT‐RI, Rey Auditory‐verbal Learning Task—retroactive interference; T2D, type 2 diabetes.


