Abstract
All biological information, since the last common ancestor of all life on earth, has been encoded by a genetic alphabet consisting of only four nucleotides that form two base pairs. Long standing efforts to develop two synthetic nucleotides that form a third, unnatural base pair (UBP) have recently yielded three promising candidates, one based on alternate hydrogen bonding, and two based on hydrophobic and packing forces. All three UBPs are replicated and transcribed with remarkable efficiency and fidelity, and the latter two thus demonstrate that hydrogen bonding is not unique in its ability to underlie the storage and retrieval of genetic information. This review highlights these recent developments as well as the applications enabled by the UBPs, including the expansion of the evolution process to include new functionality and the creation of semi-synthetic life that stores increased information.
Keywords: Unnatural, DNA, base pair, hydrophobic, semi-synthetic
Graphical Abstract
1. Introduction and Overview
DNA encodes life, and no other molecule has captured the attention and imagination of scientists and the general public more. However, as a polymer DNA is chemically and physically rather unremarkable, consisting of nothing more than a linear sequence of the deoxyribonucleotides, dG, dC, dA, and dT. What makes DNA remarkable, and absolutely unique, is its sequence specific duplex formation and replication, as well as its sequence specific transcription into RNA, which consists of the ribonucleotides G, C, A and U. This sequence specificity is made possible by the selective pairing of the four letters of the genetic alphabet to form two base pairs; (d)G pairs with (d)C and (d)A pairs with dT or U. The same base pairing between mRNA and tRNA enables decoding of the nucleotide sequences into protein sequences. It is the selective formation of these two base pairs that allows this otherwise unremarkable molecule to encode the diversity of life.
The development of synthetic nucleotides that form a third, unnatural base pair (UBP), and thus expand the genetic alphabet, has been a goal of chemists since the idea was suggested over half a century ago.[1] A UBP would have wide ranging in vitro applications, especially if one or both of the constituent nucleotides could be derivatized with linkers for the site-specific attachment of groups with interesting physicochemical properties. Moreover, the development of a UBP that functions within a cell has emerged as a central goal of the developing field of synthetic biology. Such a UBP would allow for the creation of semi-synthetic organisms, which store increased information in their DNA, and thus form the foundation of semi-synthetic life with the potential to possess new and useful attributes or functions.
While nucleotides with modified sugars[2-5] or phosphates[6,7] have been explored extensively and have important applications, the generation of a UBP requires nucleotides with modified nucleobases, as it is the nucleobases that mediate base pair formation. A wide variety of synthetic nucleotides bearing nucleobase analogs have been reported,[8-10] and several have been identified that stably pair with one another within an otherwise natural duplex.[11-19] However, efficient and high fidelity replication of DNA containing a UBP has proven much more challenging, and early work from the Benner laboratory yielded what were for over a decade the only promising candidates.[20,21] These early UBPs were designed to interact via complementary hydrogen bonding (H-bonding) patterns not employed by the natural nucleotides, but there is no reason to assume a priori that H-bonding is the only force sufficient to underlie the storage and retrieval of genetic information. For example, we[22] and others[12,23-26] have explored metal-dependent pairing. However, hydrophobic and packing (including ring stacking) forces have been explored most intensively, and along with optimized versions of the original alternate H-bonding UBPs, several of these predominantly hydrophobic UBPs now form a small group of candidates that have been developed to a high level of proof-of-concept (Figure 1). Progress in the development of these UBPs through the middle of the last decade has already been reviewed.[27-30] Here, we focus on more recent developments, with earlier studies included only for context. Also for context, we include a discussion of the forces that likely underlie the replication of DNA containing predominantly hydrophobic UBPs. Lastly, we present a discussion of currently emerging applications, culminating in the recent creation of the first semi-synthetic organism that stably harbors a UBP in its DNA.
Figure 1.
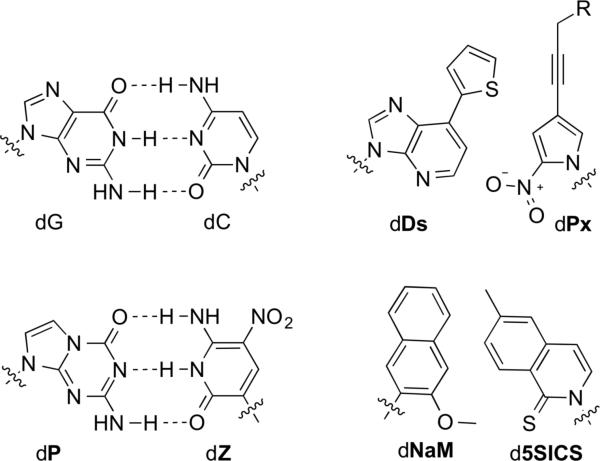
Natural dG-dC base pair and dZ-dP, dDs-dPx (R = H or -CH(OH)-CH2OH), and dNaM-d5SICS UBPs. Sugar and phosphate are omitted for clarity.
2. Alternate H-Bonding UBPs
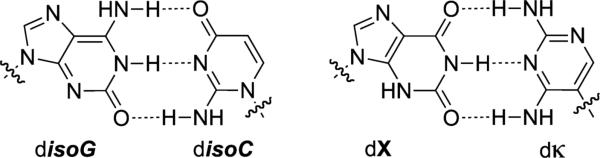
In 1962, Alexander Rich proposed that disoC and disoG, constitutional isomers of dC and dG (Figure 2), should be capable of selective pairing via complementary H-bonding patterns similar to, but distinct from, those employed by the natural base pairs.[1] However, it was not until three decades later that the Rappaport[31,32] and Benner[20,21] laboratories independently explored the development of such UBPs. In particular, the Benner laboratory's early progress, initially with disoC-disoG, and then with dκ-dX, remained state-of-the art for over a decade (Figure 2). Highlights of this seminal work include the demonstration that DNA containing disoC-disoG was at least reasonably well replicated by the Klenow fragment of E. coli DNA polymerase I (Kf)[20] and the later demonstration with cell extracts and synthetic tRNA, that they could be decoded at the ribosome.[33]
Figure 2.
disoC-disoG and dX-dκ UBPs. Sugars and phosphates are omitted for clarity.
The major limitations of these early UBP candidates were the deamination and epimerization of disoC and dκ; the misparing of disoG and dT/U, due to the tautomerization of disoG; and the poor transcription of DNA containing disoG into RNA containing isoC.[34,35] The mispairing problem was later partially circumvented by the use of 2-thiothymidine deoxyribonucleoside triphosphate instead of natural dTTP, which made possible the first PCR-mediated amplification of DNA containing a UBP, a landmark in the development of UBPs.[36] Nonetheless, the fidelity of UBP replication was only 98%[36] (throughout, fidelity refers to the retention of the UBP per doubling of the DNA), which translates into a third of the UBPs being lost after 20 PCR doublings (0.9820 = 0.668 retention) and is too low for most applications. More recently, Benner reported the dZ and dP nucleotides (Figure 1),[37,38] which are more stable, do not epimerize, and form a UBP that when incorporated into DNA is PCR amplified with a fidelity 99.8%.[39] Very recently, the transcription and reverse transcription of dZ-dP was reported.[40]
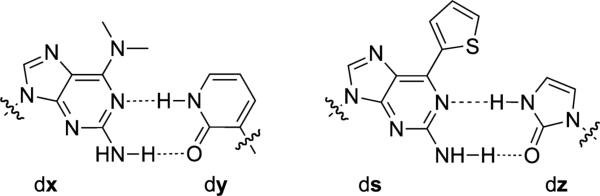
In 2000, the Hirao group reported the development of an orthogonal H-bonding UBP formed between the purine analog dx and the pyridone analog dy (Figure 3).[41,42] To decrease the mispairing of dx with natural nucleotides, they developed the dsdy pair the following year by replacing the dimethylamino group of dx with the more bulky and aromatic thiophenyl moiety of ds (Figure 3).[43] While mispairing of dy, mostly with dA, precluded PCR amplification, DNA containing ds was successfully transcribed into mRNA containing y, which was then translated with the aid of a synthetic tRNA containing s, into a protein with the unnatural amino acid 3-chlorotyrosine.[44] Conversely, DNA containing dy was transcribed into RNA containing s with low fidelity. To reduce mispairing of dy with dA, dz was developed to pair with ds by replacing the pyridone scaffold with a smaller cyclic dehydroimidazolone scaffold (Figure 3), however, the resulting triphosphates, both dzTP and zTP, were recognized inefficiently by DNA and RNA polymerases, respectively.[45]
Figure 3.
dx-dy and ds-dz UBPs. Sugars and phosphates are omitted for clarity.
3. Conceptual Interlude
The forces underlying the replication of natural DNA clearly include the complementary H-bonds that link the pairing natural nucleobases. However, more than 15 years ago the Kool group reported the remarkable observation that Kf polymerase selectively pairs dA with dF, whose difluorotoluene nucleobase is a shape mimic of thymine in which the H-bond donor N-H has been replaced with a C-H group and the H-bond accepting carbonyl groups have been replaced with C-F bonds.[46,47] Although the ability of the fluorine atom to participate in H-bonding was and still is actively debated,[48,49] it is clear that the F···H hydrogen bond is much weaker than the bonds it replaces (3.8 kcal mol−1 less stable according to recent estimates[49]). While DNA primers terminating with the dF–dA pair are not efficiently extended, the selective pairing of dF with dA was interpreted as evidence of a “geometrical selection” mechanism of DNA replication (which was also independently proposed by Goodman[50,51]) and it clearly demonstrates that forces other than H-bonding also contribute to DNA synthesis. Such forces might be harnessed to control the replication of a UBP.
It has long been known that H-bonding is not the sole force contributing to duplex stability.[52-56] However, caution must be taken when considering how these forces might contribute to replication because the polymerase active site provides a unique (and incompletely understood) environment. For example, it was noted decades ago that the destabilization associated with forming a mispair in duplex DNA could not account for the fidelity of DNA replication.[57,58] Nonetheless, given that DNA synthesis involves triphosphates leaving their aqueous environment and developing at least some of the same intrabase interactions that stabilize a DNA duplex, it seems likely that some of the same forces may contribute to DNA synthesis.[59]
One prominent force that favors duplex formation is that associated with the hydrophobic effect.[54,60,61] Qualitatively, the hydrophobic effect may be understood as an interaction that causes hydrophobic moieties to aggregate or cluster in water, and it plays a central role in micelle formation, as well as in many biological processes such as membrane formation, protein folding, and molecular recognition. The “water ordering” or “iceberg” model, first proposed by Frank and Evans in 1945[62] still constitutes an important part of our understanding of the hydrophobic effect. This model posits that a non-polar molecule has a tendency to induce the ordering of nearby waters into a crystalline- or clathrate-like cage structure such that a maximal number of H-bonds may be retained despite the production of the requisite cavity. This mechanism nicely explained the initially surprising result that the hydrophobic effect could be driven by entropy. However, with further studies it became clear that increasing the temperature or the size of the hydrophobic solute could cause the hydrophobic effect to be driven predominantly by enthalpy changes. Thus, the original iceberg model was extended to allow for varying clathrate stability.[63-66] Specifically, it has been postulated that the H-bonds of the clathrate structure are disrupted before those of bulk water as the temperature is increased, and H-bonding networks cannot be maintained around large, hydrophobic molecules, especially those with low surface curvature. Thus, at high temperatures or with sufficiently large non-polar molecules, less order is induced and the positive entropy of mixing or the negative enthalpy associated with H-bond rupture are more likely to dominate the associated change in free energy. While some recent work has suggested that the binding of hydrophobic molecules to ordered environments such as proteins or DNA might also be favored by the displacement of high energy waters,[67] the revised iceberg model is currently the only widely accepted model of the hydrophobic effect.
A variety of studies have demonstrated that the hydrophobic effect may be harnessed for the stable pairing of nucleotides bearing predominantly hydrophobic nucleobases. The Kool laboratory used a “dangling end” assay, wherein the nucleotide of interest is attached to the end of one strand of a duplex where it can interact with its flanking nucleobase but has no H-bonding partner, to demonstrate that duplex stabilization was uniquely correlated with nucleobase surface area, which led them to conclude that it was mediated predominantly by the hydrophobic effect.[68] In another study by Leumann and co-workers, an entropic origin for the stability of DNA containing pairs formed between nucleotides bearing fluorinated biphenyl rings was also interpreted as evidence of the hydrophobic effect.[69] The importance of the hydrophobic effect in the pairing of nucleotides with hydrophobic nucleobases has been further supported by computational studies of Hobza and co-workers.[70,71]
While there is a consensus that the hydrophobic effect favors the sequestration of the aromatic faces of the nucleobases within the hydrophobic core of the duplex, the exact nature and role of the non-H-bonding forces that arise between the nucleobases have been more controversial. Early views centered around two opposing perspectives. The first emphasized nucleobase overlap and dispersive forces (i.e. electron correlation),[72] while the second focused on interactions between the permanent dipoles associated with the exocyclic groups of one nucleobase and the delocalized electron density of the other (i.e. induction).[73] The latter appeared consistent with structural data (which showed that polar exocyclic groups tend to be positioned above the rings of neighboring nucleobases),[73,74] but increasingly sophisticated theoretical approaches have consistently identified dispersive interactions as the most important.[75-77] For example, computational studies from Hobza and co-workers suggest that while the hydrophobic effect is the dominant force driving nucleobase association, dispersion interactions determine the specific structure adopted.[78] In something of a compromise between the original perspectives, recent theoretical studies based on the topological analyses of calculated electron density have suggested that the bonding interactions between flanking nucleobases are manifest between specific atoms, especially the ring nitrogen atoms and the polar amino and carbonyl substituents.[79-81] These results refocus attention on local interactions between specific ring atoms and substituents of the nucleobases.
The overlap of nucleobases within a DNA duplex, which are at least partially developed during replication, are an example of a broader class of molecular interactions observed between aromatic rings. These interactions have been studied intensively for decades, and are typically referred to as π-π stacking, although recent studies have suggested that this term is overused, and even potentially misused.[82,83] The conventional model of Hunter and Sanders emphasized the repulsion between π clouds and focused on electrostatic quadrupole-quadrupole interactions that result in favorable interactions between the negatively charged π-electrons of one ring and the positively charged σ-framework of the other.[84] These studies neatly explained the otherwise perplexing edge-to-face geometry observed with benzene, and the offset, or parallel-displaced geometry observed with many other aromatics. However, more recent theoretical studies have again emphasized the importance of the dispersion interactions that result from the close contact of the interacting rings, which were difficult to capture in early studies due to the high levels of theory required.[85-88]
The effects of derivatizing the stacking aromatic rings with different substituents have also been studied intensively with small molecules. The traditional view, exemplified by the Hunter-Sanders model[84,89-92] and the polar/π model of Cozzi and Siegel[93-97] describes the effects of substituents in terms of their non-local effect on the repulsive aromatic electron density via polarization and resonance effects within the ring to which they are attached, thus electron withdrawing substituents are predicted to stabilize the interaction between two rings and electron donating substituents are predicted to destabilize it. However, high-level calculations have now decisively demonstrated that both electron withdrawing and donating substituents can stabilize the interaction of two aromatic rings.[98-101] Calculations from the Wheeler and Houk laboratories[102-105] explained these results by suggesting that substituent effects are not manifested via the aromatic rings and associated resonance effects, but instead arise from direct interactions of the local dipole associated with the substituent and the electric field of the other ring, as well as additional local dispersive interactions mediated by the polarized bond. The Wheeler-Houk model was recently supported and refined by high-level calculations from Sherrill and co-workers.[106] Most recently, experimental studies have suggested that substituent effects are mostly additive, which seems inconsistent with their being mediated via resonance effects, and strongly supports the Wheeler-Houk model.[107]
The interactions of heterocyclic aromatics have also been studied. Calculations have suggested that the addition of nitrogen atoms favors electrostatic interactions, but that this stabilization is offset by increases in exchange-repulsion, which along with only small changes in induction terms, make dispersion interactions dominant.[108] In contrast, Sherrill reported calculations that suggest that aza substitution weakens both dispersion exchange-repulsion forces, and that electrostatic effects of aza substitution are governed by direct interactions between the localized electron density of the aza substituent and an electron deficient hydrogen atom of the interacting ring.[109] Finally, Liedl and co-workers recently reported the effects of heteroatom substitution on the stability of aromatic rings interacting via a parallel-displaced geometry.[110] Generally, their results predict that the position of substitution has a large effect on the stability of the complex, as placing an electronegative atom, especially a nitrogen, more proximal to the center of high electron density of the interacting ring was destabilizing. The stabilities and the specific geometries of the favored structures were interpreted in terms of non-directional dispersive interactions and more directional electrostatics, and the differences in stabilities and structures were strongly correlated with the magnitude of the dipole moment associated with the heteroatom.
Finally, experimental studies employing model systems have also been used to study the interactions between aromatic rings.[111-115] Generally, these studies are consistent with the current perspective provided by theory: a dominant role for the hydrophobic effect, which is fine-tuned by dispersive and local electrostatic interactions.
Collectively, these studies suggest that in addition to complementary H-bonding, the hydrophobic effect, desolvation, dispersive, and electrostatic interactions between nucleobases can contribute to the stability of a DNA duplex. What remained unclear, was whether these forces can mediate efficient replication. Moreover, because the contribution of these forces to duplex stability is largely independent of sequence, it was even less clear if these forces could contribute to sequence-specific replication. For example, while Kool's results with dA with dF illustrate that, at least in some cases, these forces are sufficient to mediate triphosphate insertion, they also suggest that they may not be sufficient to prevent mispairing with a natural nucleotide (from the perspective of developing a UBP, the dF-dA pair is a mispair). Thus, it was unclear whether hydrophobic and packing forces would be sufficiently strong and specific to underlie the development of a functional UBP.
4. Predominantly Hydrophobic UBPs
Since 1999, our group has pursued the idea of using hydrophobic and packing forces to create a UBP. Our development strategy has been inspired by medicinal chemistry and involves the synthesis and evaluation of a wide variety of analogs and the elucidation of structure-activity relationship (SAR) data to guide optimization. During early development efforts, the most useful SAR data was generated via steady-state kinetics, which was used to determine the efficiency (i.e. second-order rate constant) and fidelity (i.e. the ratio of second-order rate constants for correct and incorrect pairings) of UBP synthesis (i.e. incorporation of the unnatural triphosphate opposite its cognate unnatural nucleotide in a template), as well as the subsequent extension step (i.e. incorporation of the next correct nucleotide). However, as the UBPs were optimized, their replication became too efficient to be characterized by steady-state methods (due to rate-limiting product dissociation, a step that is not relevant to processive synthesis), and thus we turned to pre-steady-state kinetics and the efficiency and fidelity with which DNA containing the UBPs is PCR amplified. During later stages of development, these studies employed multiple polymerases and sequence contexts to ensure the generality of the SAR data.
Our first generation analogs, ultimately including 37 different synthetic nucleotides, were designed to explore the ability of the hydrophobic effect and packing interactions to replace interstrand H-bonding. The nucleobase analogs were based largely on the relatively large isocarbostyril-, napthyl-, and indole-like scaffolds (we note that in many cases, the “nucleobases” are not actually basic, but the designation is still used for simplicity). The details of these early efforts have already been reviewed,[28,29] but in general, a variety of pairs were identified that were capable of relatively stable and selective pairing in duplex DNA.[116] More importantly, several were identified that were synthesized with rates approaching those of a natural pair,[117] or with at least modest selectivity, suggesting that hydrophobic and packing forces could be harnessed for this step of replication.[117,118] However, once synthesized, all of the first generation candidates were extended with very low efficiency. In fact, this pattern of large nucleobase analogs and efficient synthesis but poor extension emerged as the most pronounced SAR from the first generation analogs.
Structural studies of duplex DNA suggested that these first generation UBPs do not pair in an edge-to-edge manner, but instead pair via cross-strand intercalation of the large aromatic nucleobases (see Section 5). We speculated that this mode of pairing might also favor UBP synthesis, due to hydrophobic and dispersion forces, but result in a structure refractory to continued primer extension, due to the associated mispositioning of the primer terminus. Thus, our second generation analogs, ultimately consisting of 55 synthetic nucleotides, were mostly based on benzene,[119-121] pyridine,[122,123] or pyridone[124] scaffolds to test whether smaller nucleobases that should be less prone to intercalate could be optimized for replication. Perhaps surprisingly, several of these second generation UBPs formed stably and selectively in duplex DNA, despite the absence of H-bonding and extended aromatic surface area.[121]
Among these second generation UBPs, several were identified that are synthesized and/or extended with at least reasonable efficiency.[119,125-127] However, optimization efforts yielded only flat SAR, and problematically, the SAR identified an apparent conflict among the required physicochemical properties. The conflict revolved around the nature of the substituent positioned ortho to the glycosidic linkage, and which is expected to be disposed into the developing minor groove during replication. Efficient UBP synthesis required this substituent to be hydrophobic in both the incoming and the templating nucleobase. However, efficient extension appeared to have contradictory requirements, as it required the substituent in the template to be hydrophobic and the substituent at the primer terminus to be able to accept an H-bond.[122,124,125] Indeed, H-bond acceptors are positioned at the analogous positions of each natural nucleobase, and at the primer terminus they are known to accept H-bonds from the polymerase,[128] and biochemical studies have shown that these interactions are critical for primer elongation.[34,128-130]
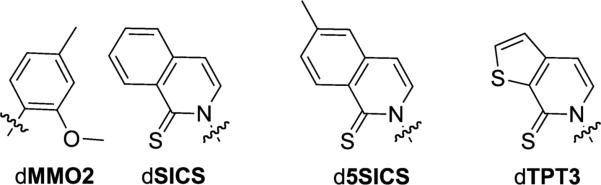
The different physicochemical properties required for UBP synthesis and extension challenged the notion that a solution could be designed rationally. Thus, we developed and implemented two different screens to evaluate all possible pairs formed between 60 diverse nucleotides culled from our first and second generation efforts (resulting in ~1,800 possible UBPs).[131] Remarkably, the top hit identified by both screens was the UBP formed between dMMO2 and dSICS (Figure 4). A particularly interesting aspect of dMMO2-dSICS is the nature of the substituents at the position ortho to the glycosidic linkage, which appears to resolve the above mentioned conflict. The sulfur atom of dSICS is softer and more polarizable than an oxygen atom, making it more hydrophobic, but it is still capable of accepting an H-bond. A simple rotation of the O-methyl group of dMMO2 allows this ortho substituent to present either a hydrophobic methyl group or an oxygen H-bond acceptor.
Figure 4.
dMMO2-dSICS UBP and d5SICS and dTPT3 unnatural nucleotides.
Thus, the dMMO2-dSICS UBP became the founding member of a third generation of candidate UBPs, reinvigorating development efforts and rapidly resulting in the identification of dMMO2-d5SICS (Figure 4). With dMMO2-d5SICS, the problematic self-pairing of dSICS-dSICS was eliminated,[131] and in contrast to our first and second generation UBPs, this UBP is recognized with high fidelity by a variety of A, B and X family DNA polymerases.[132] The least efficient step in the replication of DNA containing dMMO2-d5SICS is the incorporation of dMMO2TP against d5SICS, and optimization of this step eventually yielded dNaM-d5SICS (Figure 1),[133,134] which is synthesized an order of magnitude more efficiently.
Both dMMO2-d5SICS and dNaM-d5SICS represented landmarks in our development program, as they were the first of our UBPs that when incorporated into DNA are efficiently PCR amplified.[135] Indeed, massive amplification of DNA containing dNaM-d5SICS is possible with no significant loss of the UBP (i.e. 1024-fold amplification with >99.9% fidelity), and importantly, with no significant sequence bias.[136] This efficient, high fidelity, and sequence-independent replication makes dNaM-d5SICS especially attractive for applications that require massive amplification of a randomized template, such as SELEX (see Section 6). Importantly, using T7 RNA polymerase, both dMMO2-d5SICS and dNaM-d5SICS are efficiently transcribed into RNA (i.e. dMMO2 or dNaM directs the incorporation of 5SICS into RNA, and d5SICS directs the incorporation of either MMO2 or especially NaM into RNA) with efficiencies only ~30- fold reduced relative to natural nucleotides and with a fidelity of 93% to 99%.[137]
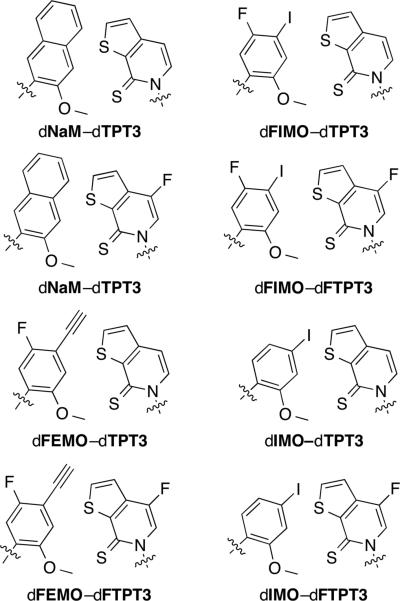
Despite the high efficiency and high fidelity replication and transcription of DNA containing dNaM-d5SICS, the overall rate of its replication is still slower than that of natural base pairs. After efforts to further optimize dNaM resulted in only flat SAR,[134,138-140] our efforts turned to the optimization of d5SICS, since continued primer extension after the incorporation of this unnatural nucleotide limits replication. These efforts eventually yielded dTPT3 (Figure 4).[141] We then supplemented this optimization effort with another screen of 111 unnatural nucleotides (many of them newly synthesized since our original screening efforts) forming ~6,000 candidate UBPs.[142] This screen yielded an entire family of well replicated UBPs (Figure 5). While dNaM-dTPT3 remains the most efficiently replicated UBP identified to date, each member of this family of UBPs is replicated more efficiently than dNaM-d5SICS, which as described below (Section 7), is sufficiently well replicated for in vivo use. This suggests that all of these UBPs should be suitable for practical applications.
Figure 5.
dNaM–dTPT3-like family of UBPs
Since 2003, the Hirao group has also focused on the development of predominantly hydrophobic UBPs. Their approach was inspired by Kool's work with dQ and dF, which are hydrophobic isosteres of dA and dT (Figure 6).[143,144] To reduce mispairing with dA, the Hirao group first developed dPa as a partner for dQ,[145] and then further decorated dQ with a bulky thienyl moiety, resulting in dDs (Figure 6). The resulting dDs-dPa UBP was the first that could be PCR amplified with an error rate below 1% (corresponding to >99% fidelity),[146] although this required the use of the γ-amidotriphosphates of dDs and dA (dDsTPN and dATPN, respectively) to reduce formation of the dDs-dDs and dA-dPa mispairs. A high fidelity of transcription (>95%) of the biotinylated analog of Pa was also demonstrated.
Figure 6.
dQ-dF UBP and dDs, dPa, and dPn unnatural nucleotides.
Further development of the dDs-dPa pair resulted from a substitution of the aldehyde group of dPa with a nitro group, yielding the dPn nucleoside (Figure 6).[147] The dDs-dPn UBP exhibited improved fidelity during PCR and bypassed the need to use dATPN. Finally, the addition of a propynyl moiety to dPn resulted in the dDs-dPx UBP (Figure 1, with R = H), which further improved the fidelity (>99.9%) and eliminated the need to use either dDsTPN or dATPN.[148]
The efficient and selective replication of d5SICS-dNaM, its related analogs, and dDs-dPx, clearly indicate that forces other than H-bonding can also be harnessed to mediate stability and high fidelity replication through the optimization of predominantly hydrophobic nucleobases. Although these studies largely proceeded empirically, several general trends are apparent. Most generally, the data are consistent with the primacy of the hydrophobic effect and local electrostatic and dispersive interactions between the nucleobases (natural and unnatural), subject to the steric constraints imposed by the polymerase active site. The specific positions of heteroatoms within the nucleobase scaffolds can have substantial positive or negative effects, largely due to electrostatic interactions and/or desolvation. The effects of modifications were typically greater with the triphosphates than with the templates. As already discussed, efficient UBP synthesis required the substituent ortho to the glycosidic linkage to mediate hydrophobic packing interactions, while UBP extension required it to be capable of H-bonding, and efficient replication required substituents capable of both. At the meta and para positions, oriented toward the developing major groove, increased aromatic surface area and other substituents that appear to mediate favorable local electrostatic and dispersive interactions were found to optimize UBP synthesis, but to disfavor extension, and efficient replication required an intermediate level of these forces. Indeed, many modifications were found to have opposite effects on UBP synthesis and extension, and optimization required compromises.
5. Structural Studies
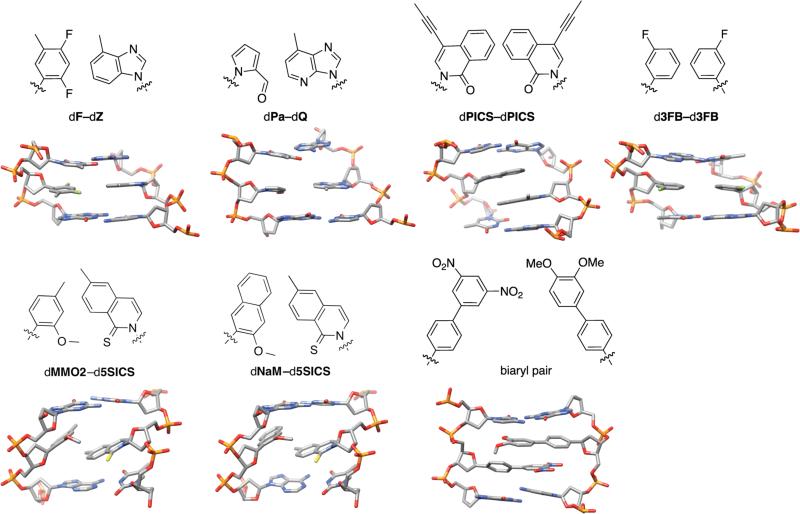
The structures of duplex DNA containing nucleotides with synthetic nucleobases have provided invaluable information regarding the forces that stabilize UBP formation and which might contribute to their replication. The first structure of a predominantly hydrophobic UBP in duplex DNA was that of dF paired with a hydrophobic isostere of dA called dZ (not to be confused with Benner's analog of the same name) (Figure 7).[149] The duplex adopted a regular B-form structure with the nucleobases packed within the duplex interior and paired in an edge-to-edge, planar manner. The C1’-C1’ distance was found to be slightly increased (by 0.8 Å), compared to a natural base pair, presumably to accommodate the additional hydrogen atom in the interface between the nucleobase analogs. The structure of the dQ-dPa pair, which was also designed to structurally mimic a natural purine-pyrimidine pair, is also well accommodated within a B-form duplex with a structure similar to that of a natural base pair (Figure 7).[145] The structure of DNA containing the pyrene nucleobase paired against the abasic site revealed that even this large nucleobase analog did not result in major structural perturbations of the duplex.[150] Several other structures of DNA[25,151-154] and RNA[155] containing at least partially hydrophobic nucleobase analogs have been reported with similar results, demonstrating that the DNA duplex is able to accommodate these nucleobases, with more or less natural-like geometries.
Figure 7.
Structures of UBPs.
The structures of duplexes containing dPICS-dPICS (one of our first generation UBPs, specifically a “self pair” formed between two identical dPICS analogs, which was efficiently synthesized, but not extended, see above),[154] d3FB-d3FB (a second generation UBP),[119] dMMO2-d5SICS,[138] and dNaM-d5SICS[156] have all been reported (Figure 7). Unlike previously reported structures, the NMR structure of DNA containing the dPICS self pair showed that the nucleobases did not pair in an edge-to-edge, planar manner, but instead paired via cross-strand intercalation. While the X-ray crystal structure of the d3FB-d3FB second generation UBP revealed a planar, edge-to-edge mode of pairing, the NMR structures of dMMO2-d5SICS and dNaM-d5SICS again revealed intercalative modes of pairing. The ortho sulfur and methoxy groups are oriented into the minor groove of the duplex, with the latter rotated out of planarity with the aromatic ring allowing for the establishment of van der Waals contacts with the sulfur group of d5SICS. The stacking interface between the nucleobases is formed by the methyl group and proximal portion of the aromatic ring of d5SICS and the para substituent of dMMO2 or dNaM. However, relative to the dPICS self pair, there is somewhat less overlap between the nucleobases, due to their geometry and/or reduced size. Based on molecular modeling, 1H NMR analysis, and circular dichroism studies, Leumann and co-workers had previously proposed such an intercalative mode of pairing between biaryl derivatized nucleotides,[13,14,157] which was later confirmed by NMR structural studies (Figure 7).[158] Finally, duplexes containing nucleotides bearing multiple aromatic chromophores in lieu of nucleobases associate in a zipper-like motif wherein each individual chromophore interstrand intercalates in a similar fashion (for reviews, see refs [159,160]). Thus, this intercalative mode of pairing appears to be general with large aromatic nucleobase analogs, and is consistent with an important role for hydrophobicity and dispersion forces.
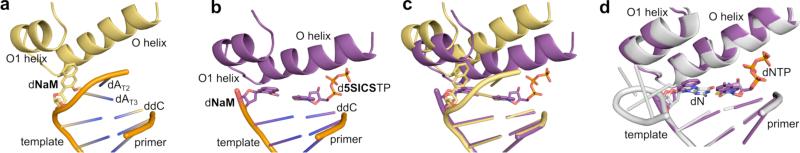
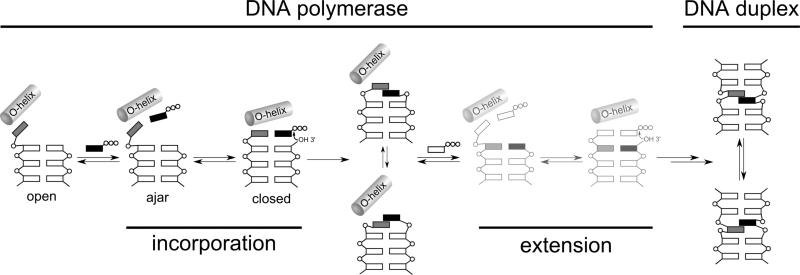
While the intercalative mode of pairing in duplex DNA clearly contributes to UBP stability, it results in a structure that is more similar to a mispair than a correct natural pair. It was thus difficult to reconcile the efficient and high fidelity replication of these UBPs with the accepted mechanism of polymerase recognition, in which binding of the correct (natural) triphosphate induces a large conformational change involving closure of the fingers domain over the palm and thumb domains to produce a closed and rigid complex that geometrically selects for the conserved Watson-Crick structure of a natural base pair.[50,51,161-164] To address this apparent contradiction and elucidate the mechanism of UBP synthesis, in collaboration with the Marx laboratory, we used X-ray crystallography to characterize the structure of several KlenTaq complexes (KlenTaq is the large fragment of Taq DNA polymerase I; Figure 8).[156] Comparison of the binary and ternary complexes revealed that with dNaM in the templating position, the addition of d5SICSTP induces the same large conformational changes in the polymerase induced by the addition of a cognate, natural triphosphate[163,165,166] (Figure 8a–c). In fact, comparison of the ternary complexes with natural and unnatural substrates reveals remarkable similarities, including the interactions between triphosphate and polymerase, the orientations of the active site side chains, and even the bound metal ions (Figure 8d). Most remarkably, unlike the intercalated structure formed in a free duplex, the nucleobases of the developing UBP in the polymerase active site of the closed complex adopt a co-planar structure with nearly optimal edge-to-edge packing and a C1’-C1’ internucleotide distance that is roughly the same as that of a natural base pair (11.0 Å versus 10.6 Å, respectively, compared to 9.1 Å for dNaM-d5SICS in a free duplex). In contrast, addition of dNaMTP to a complex of KlenTaq bound to template containing d5SICS does not fully induce the formation of the catalytically competent closed state.[167] However, it does induce the formation of a partially closed state, in which d5SICS moves toward the insertion site and dNaMTP is bound to the O-helix via its triphosphate moiety. This state is similar to the ajar state, which is thought to allow the polymerase to test for correct base pairing during the synthesis of natural DNA.[164,168,169]
Figure 8.
KlenTaq polymerase induces the dNaM-d5SICS UBP to adopt a natural, Watson-Crick-like structure. Structure of the polymerase-template binary complex (a), polymerase-template-d5SICSTP ternary complex (b), a superposition of binary and ternary complexes (c), and a superposition of the ternary complex where d5SICSTP is paired with dNaM and the ternary complex where dGTP is paired with dC (d). Reproduced from Betz, et al.[156]
The structures of several post-incorporation complexes were also characterized, wherein the UBP strand context and flanking sequences were varied.[167] While structural differences were observed, in each case the UBP was positioned in the correct post-insertion site, where it again assumed an intercalated structure as it does in the free duplex.
These structural studies, along with the analysis of UBP replication in different sequence contexts,[133] allowed us to propose a mechanism for UBP replication (Figure 9). Upon binding of an unnatural triphosphate to the O-helix, the polymerase samples different conformations, and with sufficient UBP stability, populates the catalytically active closed conformation. With d5SICSTP incorporation, the intermediate states are populated only transiently, but with dNaMTP incorporation, the series of conformational changes are halted at the ajar-like state, either due to the stability of this complex or the instability of the corresponding closed complex, and further progress towards the incorporation of dNaMTP requires thermal fluctuations to populate the closed state. In either case, with the population of the closed complex, the developing UBP adopts a Watson-Crick-like structure that facilitates covalent attachment of the incoming unnatural nucleotide. After incorporation, the polymerase returns to the open conformation, a molecule of pyrophosphate is released, and the UBP translocates to the post-insertion site, where it cross-strand intercalates. UBP extension then requires thermal fluctuations to both deintercalate the UBP and reorganize the polymerase active site. This intercalation/de-intercalation mechanism explains the balance of electrostatic and dispersive interactions that was required for the simultaneous optimization of UBP synthesis and extension. In fact, more than just being consistent with their mediating replication, this mechanism suggests that hydrophobic and packing forces may be ideal due to their relatively strong but non-directional and thus plastic nature.
Figure 9.
Proposed mechanism of replication of d5SICS-dNaM UBP. Intermediates based on solved structures are shown in darker color and those not yet validated by structural studies are shown in lighter color. The steps corresponding to incorporation of the unnatural triphosphate and subsequent extension of the nascent UBP are indicated. Phosphates are indicated with open circles, natural nucleotides are indicated with open rectangles, and the unnatural nucleotides are indicated with grey and black boxes. Reproduced from Betz, et al.[167]
6. In vitro applications
With the development of replicable UBPs, interesting applications soon followed. Many early applications took advantage of the inherent increase in hybridization specificity associated with three base pairs, relative to two (reviewed in Kimoto, 2011[170]). A more recent and potentially revolutionary application is the use of UBPs in SELEX for the evolution of aptamers or nucleic acid catalysts with unnatural functionality. This important milestone was first reached by the Hirao group, who used a derivative of the dDs-dPx UBP (Figure 1, with R = CH(OH)CH2OH) to evolve aptamers that use the unique functionality of dDs to recognize VEGF-165 or IFN-γ.[171] Since no straightforward sequencing method exists for determining the position of multiple UBPs, one or multiple dDs analogs were incorporated into defined positions of a 43-nt randomized region, with their positions identified by a two- or three-nucleotide barcode. After multiple rounds of selection and PCR amplification, all unnatural nucleotides were converted to natural nucleotides by PCR, and the resulting population was analyzed by Ion Torrent deep sequencing. The positions of the dDs nucleotide were then reconstituted from the identity of the barcodes. Both selections with the UBP yielded aptamers that bound their target with ~100-fold greater affinity than aptamers evolved from selections with only natural nucleotides. The highest affinity unnatural aptamer for VEGF-165 had two dDs nucleotides, neither of which could be replaced with dA without a decrease in affinity, and the highest affinity aptamer for IFN-γ had three, two of which could not be replaced with dA.
Soon after Hirao's work, the Benner laboratory evolved aptamers containing the dP and dZ unnatural nucleotides that recognize breast cancer cells with a dissociation constant of 30 nM.[172] To identify specific binders after 12 rounds of in vitro selection, the authors sequenced the resulting unnatural library via controllable mutation of dZ into dC and dT (developed in Ref. [39]), followed by standard deep sequencing with an Ion Torrent instrument. Importantly, as in the previous example, aptamer affinity for the target cells was significantly reduced when unnatural nucleotides were mutated to a natural counterpart thus highlighting the importance of the unnatural functionality.
While the functionality made available by the inclusion of the dDs-dPx or dP-dZ UBPs was clearly utilized in the evolved aptamers, a much greater range of physicochemical properties would be accessible by using the UBPs to site-specifically attach other functionality to DNA or RNA. Attachment of such cargo would allow for an almost limitless range of functionality to be subjected to the evolution process. For this to be accomplished, the unnatural nucleotides must be modified with a linker that does not interfere with replication or transcription and that can be used to attach the cargo of interest to the triphosphate or after incorporation of the unnatural nucleotide into DNA or RNA.
The first example of such site-specific labeling of an oligonucleotide was reported by the Dervan laboratory who used a template containing disoC to direct the incorporation of 6-aminohexylisoG into RNA, which was followed by post-transcriptional labeling with N-hydroxysuccinimidobiotin (NHS-biotin) or EDTA dianhydride.[173] More recently, the dDsdPa/dPn/dPx family of UBPs has been used by Hirao and coworkers to site-specifically label DNA[146,148] and RNA.[146,174,175] Several of the RNA labeling experiments relied on the direct incorporation during transcription of the unnatural ribonucleoside triphosphate covalently attached to the functional group of interest. However, the attachment of larger moieties, such as fluorophores, sometimes interfered with transcription. To overcome this limitation, dDs was used to template the transcription of the alkyne-modified ribonucleoside triphosphate, Eth-C4-PaTP, with the resulting RNA then modified with a biotin tag or a fluorophore via Click chemistry.[176] The Hirao group has also reported a series of linker-modified dPx analogs that are well replicated,[177] one of which was used in the above mentioned selection, and the use of other linkers or linkers with interesting functionality attached will no doubt increase the range of activities that can be sampled.
We synthesized and evaluated UBP replication and transcription with a variety of (d)5SICSTP, (d)MMO2TP, and (d)NaMTP derivatives modified with linkers bearing free or protected amines for coupling to cargo via NHS esters[178] or alkynyl groups for coupling to cargo via Click chemistry.[179] With the d5SICS scaffold, addition of a propynyl ether linker at the free meta position is accommodated better than its aliphatic analogue, but the protected propargyl amine linker is best tolerated. Interestingly, the dTPT3 scaffold is more tolerant of linker derivatization than the d5SICS scaffold.[141] While we have not yet identified linker modifications that are well tolerated by the dNaM scaffold, the para and meta positions of dMMO2 are tolerant of linker attachment, and while aliphatic and ether-based linkers are accommodated, the direct attachment of an ethynyl group to the nucleobase core is best tolerated. In general, these results may be rationalized based on the ternary structure of KlenTaq polymerase bound to 5’ propargylamido dUTP.[180] The structure revealed an H-bond between Arg660 and the nitrogen atom of the propargylamido linker in the developing major groove. In the absence of the linker, Arg660 interacts with the phosphate backbone of the primer terminus,[181,182] and thus the linkers must be either small enough not to perturb this interaction, or capable of replacing it with a stabilizing interaction.
To demonstrate the application of the linker-modified UBPs, DNA containing dNaM or d5SICS was used to direct the transcription of Methanococcus jannaschii tRNATyr with the amino-linker modified variants of 5SICS or MMO2, respectively, at the third position of the anticodon, which in the latter case was used to site-specifically biotinylate the tRNA.[178] Towards material-like applications, DNA containing the UBP formed between dNaM and propynyl ether-derivatized d5SICS was amplified and used to site-specifically array two nSH3 domains (Src homology 3 domain from the human CrkII adaptor protein), with the expected topology confirmed by atomic force microscopy.[179] We also synthesized the α-phosphorothioate variant of d5SICSTP and demonstrated its use in DNA backbone thiolation and postamplification labeling.[178] Together, these linker-modified nucleotides allow for the site-specific modification of nucleic acids with up to three different functional groups with unprecedented spatial control.
7. UBPs and semi-synthetic organisms
Perhaps the ultimate goal of developing UBPs is for their use in vivo as the foundation of semi-synthetic organisms that store and retrieve increased genetic information (Figure 10). With the demonstration that dNaM-d5SICS is replicated with high efficiency and fidelity in any sequence context, and that it is also efficiently transcribed into RNA, we initiated efforts towards this goal. With the wealth of knowledge and tools available, Escherichia coli was the obvious candidate for the creation of the first semi-synthetic organism. However, the two membranes possessed by this organism complicated the first challenge of how to make the requisite unnatural triphosphates available within the cell. We first explored a strategy employed by many nucleoside drugs, namely, the passive diffusion of the free nucleoside into the cell followed by its triphosphorylation via the sequential action of the kinases of the nucleoside salvage pathway.[183] While we found that a variety of nucleosides with predominantly hydrophobic nucleobases were monophosphorylated by the nucleoside kinase from D. melanogaster, we were unable to identify monophosphate kinases that catalyzed conversion to the diphosphates, which are more specific for their natural substrates.[184] In addition, overexpression of kinases in an attempt to compensate for their low activity resulted in significant levels of toxicity, presumably due to misregulation of the natural triphosphate pool.
Figure 10.
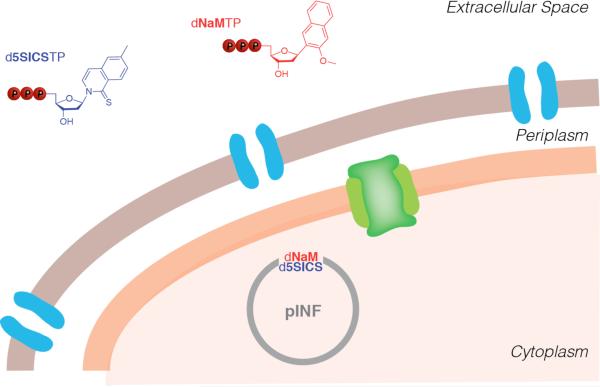
Recently developed semi-synthetic organism. Porins in the outer membrane and PtNTT2 in the inner membrane are illustrated in blue and green, respectively.
Challenged with having to identify a novel route to make the unnatural triphosphates available intracellularly, we noted that the genomes of a variety of intracellular bacteria and algal plastids do not encode the enzymes required for nucleoside triphosphate synthesis. Instead, they encode dedicated nucleoside triphosphate transporters (NTTs) and scavenge the requisite triphosphates from their cellular milieu.[185-192] We examined the plasmid-based expression of eight different NTTs in E. coli, and found that one from Phaeodactylum tricornutum (PtNTT2)[191] was active and able to import both d5SICSTP and dNaMTP (Figure 10). The addition of 250 μM of d5SICSTP and dNaMTP to the media resulted in steady state intracellular levels of approximately 90 μM and 30 μM, respectively. These concentrations are significantly above the sub-micromolar KM values of the unnatural triphosphates for DNA polymerases,[134] setting the stage for replication of the UBP in a living organism.
To determine whether E. coli equipped with PtNTT2 could use the imported unnatural triphosphates to replicate DNA containing the UBP, we used solid phase synthesis and circular extension PCR to replace a single dA-dT in the pUC19 plasmid with the dNaM-dTPT3 UBP, resulting in pINF (information plasmid). E. coli cells expressing a plasmid-borne PtNTT2 were grown in media supplemented with dNaMTP and d5SICSTP and transformed with pINF (Figure 10). As controls, cells were also transformed with the parent pUC19 plasmid, or grown either without induction of the transporter or without the unnatural triphosphates. After overnight growth, pINF, which had been amplified ~107-fold, was recovered, digested, and dephosphorylated to free nucleosides, and analysis by LCMS/MS demonstrated the presence of approximately one d5SICS per plasmid. In contrast, no d5SICS was detected when cells were transformed with the fully natural pUC19 plasmid, when they were transformed with pINF but the transporter was not induced, or when the unnatural triphosphates were not provided. Importantly, since the synthetic pINF contained dNaM-dTPT3, and d5SICS was only provided as triphosphate to the media, the detection of d5SICS in the recovered plasmid unambiguously demonstrated in vivo replication. This conclusion was confirmed by sequencing and by using a biotin shift assay after amplification of the recovered pINF with d5SICS and a biotinylated analog of dNaM. Based on the lower limit of detection provided by the analysis of UBP retention and the amplification level, the fidelity of replication is at least 99.4%. Thus, this modified E. coli represents the first semi-synthetic organism that propagates a UBP in its DNA.
8. Implications and future directions
It has been more than 50 years since Alex Rich first proposed the development of UBPs, and 25 years since Steve Benner's lab produced the first viable candidates. Today, three classes of UBPs have been developed and validated to a high level of proof-of-concept: one based on alternate H-bonding, exemplified by dZ-dP, and two based on hydrophobic and packing forces, exemplified by dNaM-d5SICS and dDs-dPx. The data demonstrate that at least for a single UBP embedded within an otherwise natural environment, H-bonds are not unique in their ability to mediate stable and selective pairing, replication, and transcription into RNA, and that they may be replaced by the hydrophobic effect and dispersive interactions mediated by nucleobase scaffolds with complementary structures, combined with desolvation and electrostatic interactions mediated by judiciously positioned heteroatoms. Continued optimization of all three classes is expected to yield UBPs that are replicated with rates and fidelities that are indistinguishable from those of a natural base pair.
Future efforts should also focus on developing methodologies to sequence DNA containing the UBPs, especially DNA containing multiple UBPs. This could include modification of the nucleobase with fluorophores for Sanger sequencing, modification of the nucleobase and sugar with fluorophores and reversible terminators, respectively, for sequencing with the Illumina platform,[193] or modification of the triphosphate moiety with a fluorophore for sequencing with the Pacific Biosciences methodology.[194] Sequencing with the Ion Torrent[195] or Oxford Nanopore[196] platforms would also be attractive as they would not require any nucleotide modification. In any case, modifications to the hardware and software will be required. Efforts should also include an increased focus on the systematic exploration and optimization of linkers for the site-specific attachment of different cargo to DNA. Such efforts will continue to facilitate the application of the UBPs in different technologies that draw upon the potential physicochemical properties of the cargo, and importantly, in an evolvable context. The use of an UBP for the evolution of novel devices or materials represents perhaps their most significant in vitro application.
It is our opinion that the most exciting application of UBPs is their use for the creation of semi-synthetic organisms that store and retrieve increased information. Again drawing upon well established tenets of medicinal chemistry, the availability of the dNaM-dTPT3 family of UBPs is especially important, as the different nucleotides possess a range of different pharmacokinetic-like properties, such as triphosphate uptake, stability, and off-target activity. Correspondingly, SAR studies should move toward including these parameters, in addition to in vitro and in vivo replication efficiency and fidelity.
The next step towards the creation of semi-synthetic organisms capable of retrieving increased information is the in vivo transcription of DNA containing the UBP into RNA. In conjunction with the orthogonal tRNA/aminoacyl synthetase pairs developed by Schultz and co-workers,[197-199] this will set the stage to explore the retrieval of the information encoded by the UBP in the form of novel proteins. There is no reason such efforts should be limited to bacterial cells, and the engineering of eukaryotic cells to store and retrieve increased information brings with it its own potentially transformative applications. Such semi-synthetic bacterial and eukaryotic organisms will form the foundation of semi-synthetic life with the potential to possess and evolve new and useful attributes or functions. DNA underlies all that life is, has been, and might evolve to be, and it would appear that UBPs now promise to dramatically increase the potential of this already remarkable molecule.
Acknowledgements
The work from our laboratory described in this review was made possible through the continuous support of the National Institute of General Medical Sciences (GM060005). We also thank all of the current and former Romesberg laboratory members who contributed to this project.
Biography
 Denis Malyshev received his MS degree from the Higher Chemical College of the Russian Academy of Sciences in 2008. He obtained his PhD from The Scripps Research Institute in 2013 working under the direction of Prof. Floyd Romesberg. Currently, He is a scientist at Synthorx, Inc., working to develop novel biomolecules containing unnatural building blocks for life science and biotechnology applications.
Denis Malyshev received his MS degree from the Higher Chemical College of the Russian Academy of Sciences in 2008. He obtained his PhD from The Scripps Research Institute in 2013 working under the direction of Prof. Floyd Romesberg. Currently, He is a scientist at Synthorx, Inc., working to develop novel biomolecules containing unnatural building blocks for life science and biotechnology applications.
 Floyd Romesberg studied chemistry at The Ohio State University and later obtained his PhD at Cornell University under the direction of David Column. He was then a postdoctoral fellow at UC Berkeley with Peter Schultz. Since 1998 he has been on the faculty of The Scripps Research Institute, working to expand the genetic alphabet, discover new approaches to combating bacteria, and understand the role of dynamics in protein evolution and function.
Floyd Romesberg studied chemistry at The Ohio State University and later obtained his PhD at Cornell University under the direction of David Column. He was then a postdoctoral fellow at UC Berkeley with Peter Schultz. Since 1998 he has been on the faculty of The Scripps Research Institute, working to expand the genetic alphabet, discover new approaches to combating bacteria, and understand the role of dynamics in protein evolution and function.
References
- 1.Rich A. Horizons in Biochemistry. Academic Press; New York: 1962. [Google Scholar]
- 2.Fa M, Radeghieri A, Henry AA, Romesberg FE. J. Am. Chem. Soc. 2004;126:1748–1754. doi: 10.1021/ja038525p. [DOI] [PubMed] [Google Scholar]
- 3.Veedu RN, Wengel J. Chem. Biodiver. 2010;7:536–542. doi: 10.1002/cbdv.200900343. [DOI] [PubMed] [Google Scholar]
- 4.Chaput JC, Yu HY, Zhang S. Chem. Biol. 2012;19:1360–1371. doi: 10.1016/j.chembiol.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 5.Pinheiro VB, Holliger P. Curr. Opin. Chem. Biol. 2012;16:245–252. doi: 10.1016/j.cbpa.2012.05.198. [DOI] [PubMed] [Google Scholar]
- 6.Nielsen PE. Mol. Biotechnol. 2004;26:233–248. doi: 10.1385/MB:26:3:233. [DOI] [PubMed] [Google Scholar]
- 7.Guga P, Koziolkiewicz M. Chem. Biodiver. 2011;8:1642–1681. doi: 10.1002/cbdv.201100130. [DOI] [PubMed] [Google Scholar]
- 8.Simons C. Nucleoside Mimetics: Their Chemistry and Biological Properties. Gordon and Breach Science Publishers; Amsterdam: 2001. [Google Scholar]
- 9.Stambasky J, Hocek M, Kocovsky P. Chem. Rev. 2009;109:6729–6764. doi: 10.1021/cr9002165. [DOI] [PubMed] [Google Scholar]
- 10.Merino P. Chemical Synthesis of Nucleoside Analogs. Wiley; New York: 2013. [Google Scholar]
- 11.Guckian KM, Morales JC, Kool ET. J. Org. Chem. 1998;63:9652–9656. doi: 10.1021/jo9805100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weizman H, Tor Y. J. Am. Chem. Soc. 2001;123:3375–3376. doi: 10.1021/ja005785n. [DOI] [PubMed] [Google Scholar]
- 13.Brotschi C, Haberli A, Leumann CJ. Angew. Chem. Int. Ed. 2001;40:3012–3014. doi: 10.1002/1521-3773(20010817)40:16<3012::AID-ANIE3012>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 14.Brotschi C, Mathis G, Leumann CJ. Chemistry. 2005;11:1911–1923. doi: 10.1002/chem.200400858. [DOI] [PubMed] [Google Scholar]
- 15.Paul N, Nashine VC, Hoops G, Zhang P, Zhou J, Bergstrom DE, Davisson VJ. Chem. Biol. 2003;10:815–825. doi: 10.1016/j.chembiol.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 16.Kim TW, Kool ET. J. Org. Chem. 2005;70:2048–2053. doi: 10.1021/jo048061t. [DOI] [PubMed] [Google Scholar]
- 17.Krueger AT, Lu H, Lee AH, Kool ET. Acc. Chem. Res. 2007;40:141–150. doi: 10.1021/ar068200o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuramoto K, Tarashima N, Hirama Y, Kikuchi Y, Minakawa N, Matsuda A. Chem. Comm. 2011;47:10818–10820. doi: 10.1039/c1cc13805g. [DOI] [PubMed] [Google Scholar]
- 19.Wojciechowski F, Leumann CJ. Chem. Soc. Rev. 2011;40:5669–5679. doi: 10.1039/c1cs15027h. [DOI] [PubMed] [Google Scholar]
- 20.Switzer C, Moroney SE, Benner SA. J. Am. Chem. Soc. 1989;111:8322–8323. [Google Scholar]
- 21.Piccirilli JA, Krauch T, Moroney SE, Benner SA. Nature. 1990;343:33–37. doi: 10.1038/343033a0. [DOI] [PubMed] [Google Scholar]
- 22.Meggers E, Holland PL, Tolman WB, Romesberg FE, Schultz PG. J. Am. Chem. Soc. 2000;122:10714–10715. [Google Scholar]
- 23.Tanaka K, Tasaka M, Cao H, Shionoya M. Eur. J. Pharm. Sci. 2001;13:77–83. doi: 10.1016/s0928-0987(00)00210-4. [DOI] [PubMed] [Google Scholar]
- 24.Czlapinski JL, Sheppard TL. J. Am. Chem. Soc. 2001;123:8618–8619. doi: 10.1021/ja0162212. [DOI] [PubMed] [Google Scholar]
- 25.Kaul C, Muller M, Wagner M, Schneider S, Carell T. Nat. Chem. 2011;3:794–800. doi: 10.1038/nchem.1117. [DOI] [PubMed] [Google Scholar]
- 26.Switzer C, Sinha S, Kim PH, Heuberger BD. Angew. Chem. Int. Ed. 2005;44:1529–1532. doi: 10.1002/anie.200462047. [DOI] [PubMed] [Google Scholar]
- 27.Benner SA. Acc. Chem. Res. 2004;37:784–797. doi: 10.1021/ar040004z. [DOI] [PubMed] [Google Scholar]
- 28.Henry AA, Romesberg FE. Curr. Opin. Chem. Biol. 2003;7:727–733. doi: 10.1016/j.cbpa.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 29.Leconte AM, Romesberg FE. In: Protein Eng. RajBhandary C. K. a. U. L., editor. Springer-Verlag; Berlin: 2009. pp. 291–314. [Google Scholar]
- 30.Hirao I, Kimoto M, Yamashige R. Acc. Chem. Res. 2012;45:2055–2065. doi: 10.1021/ar200257x. [DOI] [PubMed] [Google Scholar]
- 31.Rappaport HP. Nucleic Acids Res. 1988;16:7253–7267. doi: 10.1093/nar/16.15.7253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rappaport HP. Biochemistry. 1993;32:3047–3057. doi: 10.1021/bi00063a016. [DOI] [PubMed] [Google Scholar]
- 33.Bain JD, Switzer C, Chamberlin AR, Benner SA. Nature. 1992;356:537–539. doi: 10.1038/356537a0. [DOI] [PubMed] [Google Scholar]
- 34.Switzer CY, Moroney SE, Benner SA. Biochemistry. 1993;32:10489–10496. doi: 10.1021/bi00090a027. [DOI] [PubMed] [Google Scholar]
- 35.Horlacher J, Hottiger M, Podust VN, Hübscher U, Benner SA. Proc. Natl. Acad. Sci. USA. 1995;92:6329–6333. doi: 10.1073/pnas.92.14.6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sismour AM, Benner SA. Nucleic Acids Res. 2005;33:5640–5646. doi: 10.1093/nar/gki873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Z, Sismour AM, Sheng P, Puskar NL, Benner SA. Nucleic Acids Res. 2007;35:4238–4249. doi: 10.1093/nar/gkm395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hutter D, Benner SA. J. Org. Chem. 2003;68:9839–9842. doi: 10.1021/jo034900k. [DOI] [PubMed] [Google Scholar]
- 39.Yang Z, Chen F, Alvarado JB, Benner SA. J. Am. Chem. Soc. 2011;133:15105–15112. doi: 10.1021/ja204910n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leal NA, Kim HJ, Hoshika S, Kim MJ, Carrigan MA, Benner SA. ACS Synth. Biol. 2014 doi: 10.1021/sb500268n. [DOI] [PubMed] [Google Scholar]
- 41.Ishikawa M, Hirao I, Yokoyama S. Tetrahedron Lett. 2000;41:3931–3934. [Google Scholar]
- 42.Ohtsuki T, Kimoto M, Ishikawa M, Mitsui T, Hirao I, Yokoyama S. Proc. Natl. Acad. Sci. USA. 2001;98:4922–4925. doi: 10.1073/pnas.091532698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fujiwara T, Kimoto M, Sugiyama H, Hirao I, Yokoyama S. Bioorg. Med. Chem. Lett. 2001;11:2221–2223. doi: 10.1016/s0960-894x(01)00415-2. [DOI] [PubMed] [Google Scholar]
- 44.Hirao I, et al. Nat. Biotechnol. 2002;20:177–182. doi: 10.1038/nbt0202-177. [DOI] [PubMed] [Google Scholar]
- 45.Hirao I, Harada Y, Kimoto M, Mitsui T, Fujiwara T, Yokoyama S. J. Am. Chem. Soc. 2004;126:13298–13305. doi: 10.1021/ja047201d. [DOI] [PubMed] [Google Scholar]
- 46.Moran S, Ren RXF, Rumney S, Kool ET. J. Am. Chem. Soc. 1997;119:2056–2057. doi: 10.1021/ja963718g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moran S, Ren RX, Kool ET. Proc. Natl. Acad. Sci. USA. 1997;94:10506–10511. doi: 10.1073/pnas.94.20.10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kool ET, Sintim HO. Chem. Comm. 2006:3665–3675. doi: 10.1039/b605414e. [DOI] [PubMed] [Google Scholar]
- 49.Khakshoor O, Wheeler SE, Houk KN, Kool ET. J. Am. Chem. Soc. 2012;134:3154–3163. doi: 10.1021/ja210475a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goodman MF. Proc. Natl. Acad. Sci. USA. 1997;94:10493–10495. doi: 10.1073/pnas.94.20.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kool ET. Ann. Rev. Biochem. 2002;71:191–219. doi: 10.1146/annurev.biochem.71.110601.135453. [DOI] [PubMed] [Google Scholar]
- 52.Sturtevant JM, Rice SA, Geiduschek EP. Discuss. Faraday Soc. 1958;25:138–149. [Google Scholar]
- 53.Ts'o POP, I.S. M, Olson AC. J. Am. Chem. Soc. 1963;85:1289–1296. [Google Scholar]
- 54.Saenger W. Principles of Nucleic Acid Structure. Springer; New York: 1984. [Google Scholar]
- 55.Yakovchuk P, Protozanova E, Frank-Kamenetskii MD. Nucleic Acids Res. 2006;34:564–574. doi: 10.1093/nar/gkj454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Petersheim M, Turner DH. Biochemistry. 1983;22:256–263. doi: 10.1021/bi00271a004. [DOI] [PubMed] [Google Scholar]
- 57.Petruska J, Goodman MF, Boosalis MS, Sowers LC, Cheong C, Tinoco I., Jr. Proc. Natl. Acad. Sci. USA. 1988;85:6252–6256. doi: 10.1073/pnas.85.17.6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Loeb LA, Kunkel TA. Annu Rev Biochem. 1982;51:429–457. doi: 10.1146/annurev.bi.51.070182.002241. [DOI] [PubMed] [Google Scholar]
- 59.Kool ET. Annu. Rev. Biophys. Biomol. Struct. 2001;30:1–22. doi: 10.1146/annurev.biophys.30.1.1. [DOI] [PubMed] [Google Scholar]
- 60.Herskovits T. Arch. Biochem. Biophys. 1962;97:474–484. [Google Scholar]
- 61.Friedman RA, Honig B. Biophys. J. 1995;69:1528–1535. doi: 10.1016/S0006-3495(95)80023-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Frank HS, Evans MW. J. Chem. Phys. 1945;13:507–532. [Google Scholar]
- 63.Southall NT, Dill KA, Haymet ADJ. J. Phys. Chem. B. 2002;106:521–533. [Google Scholar]
- 64.Chandler D. Nature. 2005;437:640–647. doi: 10.1038/nature04162. [DOI] [PubMed] [Google Scholar]
- 65.Luksic M, Urbic T, Hribar-Lee B, Dill KA. J. Phys. Chem. A. 2012;116:6177–6186. doi: 10.1021/jp300743a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Blokzijl W, Engberts JBFN. Angew. Chem. Int. Ed. 1993;32:1545–1579. [Google Scholar]
- 67.Biedermann F, Nau WM, Schneider HJ. Angew. Chem. Int. Ed. 2014;53:11158–11171. doi: 10.1002/anie.201310958. [DOI] [PubMed] [Google Scholar]
- 68.Guckian KM, Schweizer BA, Ren RXF, Sheils CJ, Tahmassebi DC, Kool ET. J. Am. Chem. Soc. 2000;122:2213–2222. doi: 10.1021/ja9934854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zahn A, Brotschi C, Leumann CJ. Chem. Eur. J. 2005;11:2125–2129. doi: 10.1002/chem.200401128. [DOI] [PubMed] [Google Scholar]
- 70.Reha D, Hocek M, Hobza P. Chem. Eur. J. 2006;12:3587–3595. doi: 10.1002/chem.200501126. [DOI] [PubMed] [Google Scholar]
- 71.Zendlova L, Reha D, Hocek M, Hobza P. Chem. Eur. J. 2009;15:7601–7610. doi: 10.1002/chem.200802170. [DOI] [PubMed] [Google Scholar]
- 72.Hobza P, Sponer J, Polasek M. J. Am. Chem. Soc. 1995;117:792–798. [Google Scholar]
- 73.Bugg CE, Thomas JM, Sundaralingam M, Rao ST. Biopolymers. 1971;10:175–219. doi: 10.1002/bip.360100113. [DOI] [PubMed] [Google Scholar]
- 74.Sarai A, Mazur J, Nussinov R, Jernigan RL. Biochemistry. 1988;27:8498–8502. doi: 10.1021/bi00422a030. [DOI] [PubMed] [Google Scholar]
- 75.Sedlak R, Jurecka P, Hobza P. J. Chem. Phys. 2007;127:075104. doi: 10.1063/1.2759207. [DOI] [PubMed] [Google Scholar]
- 76.Fiethen A, Jansen G, Hesselmann A, Schutz M. J. Am. Chem. Soc. 2008;130:1802–1803. doi: 10.1021/ja076781m. [DOI] [PubMed] [Google Scholar]
- 77.Hobza P, Sponer J. J. Am. Chem. Soc. 2002;124:11802–11808. doi: 10.1021/ja026759n. [DOI] [PubMed] [Google Scholar]
- 78.Kolar M, Kubar T, Hobza P. J. Phys. Chem. A. 2011;115:8038–8046. doi: 10.1021/jp202878d. [DOI] [PubMed] [Google Scholar]
- 79.Waller MP, Robertazzi A, Platts JA, Hibbs DE, Williams PA. J. Comput. Chem. 2006;27:491–504. doi: 10.1002/jcc.20363. [DOI] [PubMed] [Google Scholar]
- 80.Matta CF, Castillo N, Boyd RJ. J. Phys. Chem. A. 2006;110:563–578. doi: 10.1021/jp054986g. [DOI] [PubMed] [Google Scholar]
- 81.Kamya PR, Muchall HM. J. Phys. Chem. A. 2011;115:12800–12808. doi: 10.1021/jp203918z. [DOI] [PubMed] [Google Scholar]
- 82.Grimme S. Angew. Chem. Int. Ed. 2008;47:3430–3434. doi: 10.1002/anie.200705157. [DOI] [PubMed] [Google Scholar]
- 83.Martinez CR, Iverson BL. Chem. Sci. 2012;3:2191–2201. [Google Scholar]
- 84.Hunter CA, Sanders JKM. J. Am. Chem. Soc. 1990;112:5525–5534. [Google Scholar]
- 85.Andersson Y, Langreth DC, Lundqvist BI. Phys. Rev. Lett. 1996;76:102–105. doi: 10.1103/PhysRevLett.76.102. [DOI] [PubMed] [Google Scholar]
- 86.Hobza P, Sponer J. Chem. Rev. 1999;99:3247–3276. doi: 10.1021/cr9800255. [DOI] [PubMed] [Google Scholar]
- 87.Shishkin OV, Gorb L, Hobza P, Leszczynski J. Int. J. Quantum Chem. 2000;80:1116–1124. [Google Scholar]
- 88.Sponer J, Leszczynski J, Hobza P. Biopolymers. 2001;61:3–31. doi: 10.1002/1097-0282(2001)61:1<3::AID-BIP10048>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 89.Hunter CA, Lawson KR, Perkins J, Urch CJ. J. Chem. Soc. Perkins Trans. 2. 2001:651–669. [Google Scholar]
- 90.Cockroft SL, Hunter CA, Lawson KR, Perkins J, Urch CJ. J. Am. Chem. Soc. 2005;127:8594–8595. doi: 10.1021/ja050880n. [DOI] [PubMed] [Google Scholar]
- 91.Cockroft SL, Hunter CA. Chem. Soc. Rev. 2007;36:172–188. doi: 10.1039/b603842p. [DOI] [PubMed] [Google Scholar]
- 92.Cockroft SL, et al. Org. Biomol. Chem. 2007;5:1062–1080. doi: 10.1039/b617576g. [DOI] [PubMed] [Google Scholar]
- 93.Cozzi F, Cinquini M, Annunziata R, Dwyer T, Siegel JS. J. Am. Chem. Soc. 1992;114:5729–5733. [Google Scholar]
- 94.Cozzi F, Cinquini M, Annuziata R, Siegel JS. J. Am. Chem. Soc. 1993;115:5330–5331. [Google Scholar]
- 95.Cozzi F, Siegel JS. Pure Appl. Chem. 1995;67:683–689. [Google Scholar]
- 96.Cozzi F, Annunziata R, Benaglia M, Cinquini M, Raimondi L, Baldridge KK, Siegel JS. Org. Biomol. Chem. 2003;1:157–162. doi: 10.1039/b208871a. [DOI] [PubMed] [Google Scholar]
- 97.Cozzi F, Annunziata R, Benaglia M, Baldridge KK, Aguirre G, Estrada J, Sritana-Anant Y, Siegel JS. Phys. Chem. Chem. Phys. 2008;10:2686–2694. doi: 10.1039/b800031j. [DOI] [PubMed] [Google Scholar]
- 98.Sinnokrot MO, Sherrill CD. J. Phys. Chem. A. 2003;107:8377–8379. [Google Scholar]
- 99.Sinnokrot MO, Sherrill CD. J. Phys. Chem. A. 2006;110:10656–10668. doi: 10.1021/jp0610416. [DOI] [PubMed] [Google Scholar]
- 100.Sinnokrot MO, Sherrill CD. J. Am. Chem. Soc. 2004;126:7690–7697. doi: 10.1021/ja049434a. [DOI] [PubMed] [Google Scholar]
- 101.Watt M, Hardebeck LK, Kirkpatrick CC, Lewis M. J. Am. Chem. Soc. 2011;133:3854–3862. doi: 10.1021/ja105975a. [DOI] [PubMed] [Google Scholar]
- 102.Wheeler SE, Houk KN. J. Am. Chem. Soc. 2008;130:10854–10855. doi: 10.1021/ja802849j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wheeler SE. J. Am. Chem. Soc. 2011;133:10262–10274. doi: 10.1021/ja202932e. [DOI] [PubMed] [Google Scholar]
- 104.Wheeler SE. Acc. Chem. Res. 2013;46:1029–1038. doi: 10.1021/ar300109n. [DOI] [PubMed] [Google Scholar]
- 105.Wheeler SE, Bloom JW. J. Phys. Chem. A. 2014;118:6133–6147. doi: 10.1021/jp504415p. [DOI] [PubMed] [Google Scholar]
- 106.Parrish RM, Sherrill CD. J. Am. Chem. Soc. 2014;136:17386–17389. doi: 10.1021/ja5101245. [DOI] [PubMed] [Google Scholar]
- 107.Hwang J, Li P, Carroll WR, Smith MD, Pellechia PJ, Shimizu KD. J. Am. Chem. Soc. 2014;136:14060–14067. doi: 10.1021/ja504378p. [DOI] [PubMed] [Google Scholar]
- 108.Wang W, Hobza P. ChemPhysChem. 2008;9:1003–1009. doi: 10.1002/cphc.200700587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sherrill CD. Acc. Chem. Res. 2013;46:1020–1028. doi: 10.1021/ar3001124. [DOI] [PubMed] [Google Scholar]
- 110.Huber RG, Margreiter MA, Fuchs JE, von Grafenstein S, Tautermann CS, Liedl KR, Fox T. J. Chem. Inf. Model. 2014;54:1371–1379. doi: 10.1021/ci500183u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shetty AS, Zhang J, Moore JS. J. Am. Chem. Soc. 1996;118:1019–1027. [Google Scholar]
- 112.Gardner RR, McKay SL, Gellman SH. Org. Lett. 2000;2:2335–2338. doi: 10.1021/ol006096j. [DOI] [PubMed] [Google Scholar]
- 113.Cubberley MS, Iverson BL. J. Am. Chem. Soc. 2001;123:7560–7563. doi: 10.1021/ja015817m. [DOI] [PubMed] [Google Scholar]
- 114.Goodman AJ, Breinlinger EC, McIntosh CM, Grimaldi LN, Rotello VM. Org. Lett. 2001;3:1531–1534. doi: 10.1021/ol015838l. [DOI] [PubMed] [Google Scholar]
- 115.Rashkin MJ, Waters ML. J. Am. Chem. Soc. 2002;124:1860–1861. doi: 10.1021/ja016508z. [DOI] [PubMed] [Google Scholar]
- 116.Berger M, Ogawa AK, McMinn DL, Wu Y, Schultz PG, Romesberg FE. Angew. Chem. Int. Ed. 2000;39:2940–2942. doi: 10.1002/1521-3773(20000818)39:16<2940::aid-anie2940>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 117.Ogawa AK, Wu YQ, McMinn DL, Liu JQ, Schultz PG, Romesberg FE. J. Am. Chem. Soc. 2000;122:3274–3287. [Google Scholar]
- 118.McMinn DL, Ogawa AK, Wu Y, Liu J, Schultz PG, Romesberg FE. J. Am. Chem. Soc. 1999;121:11585–11586. [Google Scholar]
- 119.Henry AA, Olsen AG, Matsuda S, Yu CZ, Geierstanger BH, Romesberg FE. J. Am. Chem. Soc. 2004;126:6923–6931. doi: 10.1021/ja049961u. [DOI] [PubMed] [Google Scholar]
- 120.Hwang GT, Romesberg FE. Nucleic Acids Res. 2006;34:2037–2045. doi: 10.1093/nar/gkl049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Matsuda S, Romesberg FE. J. Am. Chem. Soc. 2004;126:14419–14427. doi: 10.1021/ja047291m. [DOI] [PubMed] [Google Scholar]
- 122.Kim Y, Leconte AM, Hari Y, Romesberg FE. Angew. Chem. Int. Ed. 2006;45:7809–7812. doi: 10.1002/anie.200602579. [DOI] [PubMed] [Google Scholar]
- 123.Hari Y, Hwang GT, Leconte AM, Joubert N, Hocek M, Romesberg FE. Chembiochem. 2008;9:2796–2799. doi: 10.1002/cbic.200800577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Leconte AM, Matsuda S, Hwang GT, Romesberg FE. Angew. Chem. Int. Ed. 2006;45:4326–4329. doi: 10.1002/anie.200601272. [DOI] [PubMed] [Google Scholar]
- 125.Leconte AM, Matsuda S, Romesberg FE. J. Am. Chem. Soc. 2006;128:6780–6781. doi: 10.1021/ja060853c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Matsuda S, Henry AA, Romesberg FE. J. Am. Chem. Soc. 2006;128:6369–6375. doi: 10.1021/ja057575m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Matsuda S, Leconte AM, Romesberg FE. J. Am. Chem. Soc. 2007;129:5551–5557. doi: 10.1021/ja068282b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Meyer AS, Blandino M, Spratt TE. J. Biol. Chem. 2004;279:33043–33046. doi: 10.1074/jbc.C400232200. [DOI] [PubMed] [Google Scholar]
- 129.Morales JC, Kool ET. J. Am. Chem. Soc. 2000;122:1001–1007. doi: 10.1021/ja993464+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Spratt TE. Biochemistry. 2001;40:2647–2652. doi: 10.1021/bi002641c. [DOI] [PubMed] [Google Scholar]
- 131.Leconte AM, Hwang GT, Matsuda S, Capek P, Hari Y, Romesberg FE. J. Am. Chem. Soc. 2008;130:2336–2343. doi: 10.1021/ja078223d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hwang GT, Romesberg FE. J. Am. Chem. Soc. 2008;130:14872–14882. doi: 10.1021/ja803833h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Seo YJ, Hwang GT, Ordoukhanian P, Romesberg FE. J. Am. Chem. Soc. 2009;131:3246–3252. doi: 10.1021/ja807853m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lavergne T, Malyshev DA, Romesberg FE. Chem. Eur. J. 2012;18:1231–1239. doi: 10.1002/chem.201102066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Malyshev DA, Seo YJ, Ordoukhanian P, Romesberg FE. J. Am. Chem. Soc. 2009;131:14620–14621. doi: 10.1021/ja906186f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Malyshev DA, Dhami K, Quach HT, Lavergne T, Ordoukhanian P, Torkamani A, Romesberg FE. Proc. Natl. Acad. Sci. USA. 2012;109:12005–12010. doi: 10.1073/pnas.1205176109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Seo YJ, Matsuda S, Romesberg FE. J. Am. Chem. Soc. 2009;131:5046–5047. doi: 10.1021/ja9006996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Malyshev DA, Pfaff DA, Ippoliti SI, Hwang GT, Dwyer TJ, Romesberg FE. Chem. Eur. J. 2010;16:12650–12659. doi: 10.1002/chem.201000959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Seo YJ, Romesberg FE. Chembiochem. 2009;10:2394–2400. doi: 10.1002/cbic.200900413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lavergne T, Degardin M, Malyshev DA, Quach HT, Dhami K, Ordoukhanian P, Romesberg FE. J. Am. Chem. Soc. 2013;135:5408–5419. doi: 10.1021/ja312148q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Li L, Degardin M, Lavergne T, Malyshev DA, Dhami K, Ordoukhanian P, Romesberg FE. J. Am. Chem. Soc. 2014;136:826–829. doi: 10.1021/ja408814g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Dhami K, Malyshev DA, Ordoukhanian P, Kubelka T, Hocek M, Romesberg FE. Nucleic Acids Res. 2014;42:10235–10244. doi: 10.1093/nar/gku715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Morales JC, Kool ET. Nat. Struct. Biol. 1998;5:950–954. doi: 10.1038/2925. [DOI] [PubMed] [Google Scholar]
- 144.Morales JC, Kool ET. J. Am. Chem. Soc. 1999;121:2323–2324. doi: 10.1021/ja983502+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mitsui T, Kitamura A, Kimoto M, To T, Sato A, Hirao I, Yokoyama S. J. Am. Chem. Soc. 2003;125:5298–5307. doi: 10.1021/ja028806h. [DOI] [PubMed] [Google Scholar]
- 146.Hirao I, Kimoto M, Mitsui T, Fujiwara T, Kawai R, Sato A, Harada Y, Yokoyama S. Nat. Methods. 2006;3:729–735. doi: 10.1038/nmeth915. [DOI] [PubMed] [Google Scholar]
- 147.Hirao I, Mitsui T, Kimoto M, Yokoyama S. J. Am. Chem. Soc. 2007;129:15549–15555. doi: 10.1021/ja073830m. [DOI] [PubMed] [Google Scholar]
- 148.Kimoto M, Kawai R, Mitsui T, Yokoyama S, Hirao I. Nucleic Acids Res. 2009;37:e14. doi: 10.1093/nar/gkn956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Guckian KM, Krugh TR, Kool ET. J. Am. Chem. Soc. 2000;122:6841–6847. doi: 10.1021/ja994164v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Smirnov S, Matray TJ, Kool ET, de los Santos C. Nucleic Acids Res. 2002;30:5561–5569. doi: 10.1093/nar/gkf688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Atwell S, Meggers E, Spraggon G, Schultz PG. J. Am. Chem. Soc. 2001;123:12364–12367. doi: 10.1021/ja011822e. [DOI] [PubMed] [Google Scholar]
- 152.Klewer DA, Hoskins A, Zhang PM, Davisson VJ, Bergstrom DE, LiWang AC. Nucleic Acids Res. 2000;28:4514–4522. doi: 10.1093/nar/28.22.4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Johar Z, Zahn A, Leumann CJ, Jaun B. Chem. Eur. J. 2008;14:1080–1086. doi: 10.1002/chem.200701304. [DOI] [PubMed] [Google Scholar]
- 154.Matsuda S, et al. J. Am. Chem. Soc. 2007;129:10466–10473. doi: 10.1021/ja072276d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Minasov G, Matulic-Adamic J, Wilds CJ, Haeberli P, Usman N, Beigelman L, Egli M. RNA. 2000;6:1516–1528. doi: 10.1017/s1355838200001114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Betz K, Malyshev DA, Lavergne T, Welte W, Diederichs K, Dwyer TJ, Ordoukhanian P, Romesberg FE, Marx A. Nat. Chem. Biol. 2012;8:612–614. doi: 10.1038/nchembio.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Brotschi C, Leumann CJ. Angew. Chem. Int. Ed. 2003;42:1655–1658. doi: 10.1002/anie.200250516. [DOI] [PubMed] [Google Scholar]
- 158.Johar Z, Zahn A, Leumann CJ, Jaun B. Chem. Eur. J. 2008;14:1080–1086. doi: 10.1002/chem.200701304. [DOI] [PubMed] [Google Scholar]
- 159.Malinovskii VL, Wenger D, Haner R. Chem. Soc. Rev. 2010;39:410–422. doi: 10.1039/b910030j. [DOI] [PubMed] [Google Scholar]
- 160.Teo YN, Kool ET. Chem. Rev. 2012;112:4221–4245. doi: 10.1021/cr100351g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Echols H, Goodman MF. Ann. Rev. Biochem. 1991;60:477–511. doi: 10.1146/annurev.bi.60.070191.002401. [DOI] [PubMed] [Google Scholar]
- 162.Kunkel TA. J. Biol. Chem. 2004;279:16895–16898. doi: 10.1074/jbc.R400006200. [DOI] [PubMed] [Google Scholar]
- 163.Rothwell PJ, Waksman G. Adv. Prot. Chem. 2005;71:401–440. doi: 10.1016/S0065-3233(04)71011-6. [DOI] [PubMed] [Google Scholar]
- 164.Wu EY, Beese LS. J. Biol. Chem. 2011;286:19758–19767. doi: 10.1074/jbc.M110.191130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Li Y, Korolev S, Waksman G. EMBO J. 1998;17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Doublié S, Tabor S, Long AM, Richardson CC, Ellenberger T. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 167.Betz K, Malyshev DA, Lavergne T, Welte W, Diederichs K, Romesberg FE, Marx A. J. Am. Chem. Soc. 2013;135:18637–18643. doi: 10.1021/ja409609j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hohlbein J, Aigrain L, Craggs TD, Bermek O, Potapova O, Shoolizadeh P, Grindley ND, Joyce CM, Kapanidis AN. Nat. Commun. 2013;4:2131. doi: 10.1038/ncomms3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Berezhna SY, Gill JP, Lamichhane R, Millar DP. J. Am. Chem. Soc. 2012;134:11261–11268. doi: 10.1021/ja3038273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kimoto M, Cox RS, 3rd, Hirao I. Expert. Rev. Mol. Diagn. 2011;11:321–331. doi: 10.1586/erm.11.5. [DOI] [PubMed] [Google Scholar]
- 171.Kimoto M, Yamashige R, Matsunaga K, Yokoyama S, Hirao I. Nat. Biotechnol. 2013;31:453–457. doi: 10.1038/nbt.2556. [DOI] [PubMed] [Google Scholar]
- 172.Sefah K, et al. Proc. Natl. Acad. Sci. USA. 2014;111:1449–1454. doi: 10.1073/pnas.1311778111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Tor Y, Dervan PB. J. Am. Chem. Soc. 1993;115:4461–4467. [Google Scholar]
- 174.Kimoto M, Sato A, Kawai R, Yokoyama S, Hirao I. Nucleic Acids Symp. Ser. 2009:73–74. doi: 10.1093/nass/nrp037. [DOI] [PubMed] [Google Scholar]
- 175.Morohashi N, Kimoto M, Sato A, Kawai R, Hirao I. Molecules. 2012;17:2855–2876. doi: 10.3390/molecules17032855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ishizuka T, Kimoto M, Sato A, Hirao I. Chem. Comm. 2012;48:10835–10837. doi: 10.1039/c2cc36293g. [DOI] [PubMed] [Google Scholar]
- 177.Yamashige R, Kimoto M, Takezawa Y, Sato A, Mitsui T, Yokoyama S, Hirao I. Nucleic Acids Res. 2012;40:2793–2806. doi: 10.1093/nar/gkr1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Seo YJ, Malyshev DA, Lavergne T, Ordoukhanian P, Romesberg FE. J. Am. Chem. Soc. 2011;133:19878–19888. doi: 10.1021/ja207907d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Li Z, et al. Chem. Eur. J. 2013;19:14205–14209. [Google Scholar]
- 180.Obeid S, Baccaro A, Welte W, Diederichs K, Marx A. Proc. Natl. Acad. Sci. USA. 2010;107:21327–21331. doi: 10.1073/pnas.1013804107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Li Y, Kong Y, Korolev S, Waksman G. Protein Sci. 1998;7:1116–1123. doi: 10.1002/pro.5560070505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Li Y, Waksman G. Protein Sci. 2001;10:1225–1233. doi: 10.1110/ps.250101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Wu Y, Fa M, Tae EL, Schultz PG, Romesberg FE. J. Am. Chem. Soc. 2002;124:14626–14630. doi: 10.1021/ja028050m. [DOI] [PubMed] [Google Scholar]
- 184.Yan H, Tsai MD. Adv. Enzymol. Relat. Areas Mol. Biol. 1999;73:103–134. doi: 10.1002/9780470123195.ch4. [DOI] [PubMed] [Google Scholar]
- 185.Winkler HH, Neuhaus HE. Trends Biochem. Sci. 1999;24:64–68. doi: 10.1016/s0968-0004(98)01334-6. [DOI] [PubMed] [Google Scholar]
- 186.Amiri H, Karlberg O, Andersson SG. J. Mol. Evol. 2003;56:137–150. doi: 10.1007/s00239-002-2387-0. [DOI] [PubMed] [Google Scholar]
- 187.Hatch TP, Al-Hossainy E, Silverman JA. J. Bacteriol. 1982;150:662–670. doi: 10.1128/jb.150.2.662-670.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Winkler HH. J. Biol. Chem. 1976;251:389–396. [PubMed] [Google Scholar]
- 189.Horn M, Wagner M. J. Eukaryot. Microbiol. 2004;5:509–514. doi: 10.1111/j.1550-7408.2004.tb00278.x. [DOI] [PubMed] [Google Scholar]
- 190.Haferkamp I, Schmitz-Esser S, Wagner M, Neigel N, Horn M, Neuhaus HE. Mol. Microbiol. 2006;60:1534–1545. doi: 10.1111/j.1365-2958.2006.05193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Ast M, Gruber A, Schmitz-Esser S, Neuhaus HE, Kroth PG, Horn M, Haferkamp I. Proc. Natl. Acad. Sci. USA. 2009;106:3621–3626. doi: 10.1073/pnas.0808862106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Haferkamp I, Linka N. Plant Biol. 2012;14:675–690. doi: 10.1111/j.1438-8677.2012.00591.x. [DOI] [PubMed] [Google Scholar]
- 193.Bentley DR, et al. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Eid J, et al. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 195.Rothberg JM, et al. Nature. 2011;475:348–352. doi: 10.1038/nature10242. [DOI] [PubMed] [Google Scholar]
- 196.Oxford Nanopore Technologies Introduction to nanopore sensing. 2014 can be found under http://nanoporetech.com/technology.
- 197.Liu CC, Schultz PG. Annu. Rev. Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 198.Young TS, Schultz PG. J. Biol. Chem. 2010;285:11039–11044. doi: 10.1074/jbc.R109.091306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Wang L, Xie J, Schultz PG. Annu. Rev. Biophys. Biomol. Struct. 2006;35:225–249. doi: 10.1146/annurev.biophys.35.101105.121507. [DOI] [PubMed] [Google Scholar]