Abstract
Aims
IDOL (inducible degrader of the low-density lipoprotein receptor, LDLR) is an E3 ubiquitin ligase that promotes the ubiquitination and degradation of the LDLR. IDOL is a potential therapeutic target for the development of a novel class of low-density lipoprotein cholesterol (LDL-C)-lowering therapies. In an attempt to develop a mouse model for testing IDOL inhibitors, we examined the effects of adeno-associated virus (AAV)-mediated stable expression of human IDOL in the livers of mice ‘humanized’ with regard to lipoprotein metabolism.
Methods and results
Using a liver-specific AAV serotype 8 (AAV8)-mediated delivery, AAV-hIDOL produced a dose-dependent increase in LDL-C levels and a decrease in liver LDLR protein. Furthermore, we expressed hIDOL in a ‘humanized’ mouse model of heterozygous familial hypercholesterolaemia (LDLR+/−/Apobec1−/−/hApoB-Tg, LAhB). In this model, total cholesterol (TC) and LDL-C levels were increased by ∼60% starting from 1 week and were sustainable for at least 3 weeks post-injection. Finally, we demonstrate that the effects caused by hIDOL expression are LDLR- dependent given the unchanged plasma lipids in LAhB mice lacking LDLR.
Conclusion
In conclusion, our study demonstrates a dose-dependent physiological effect of human IDOL on LDL metabolism in mice. This provides a potential model for preclinical testing of IDOL inhibitors for reduction of LDL-C levels.
Keywords: IDOL, LDLR, Lipoproteins, Hypercholesterolaemia, Therapeutic targets
1. Introduction
IDOL (inducible degrader of LDLR), originally known as MYLIP, is an E3 ubiquitin ligase that promotes the ubiquitination and degradation of the LDLR in the lysosome through the interaction with its cytoplasmic domain.1,2 Transient adenoviral overexpression of mouse IDOL in wild-type mice induced a marked decrease in LDLR protein and an increase in LDL-C.1 Other targets of IDOL might include VLDLR and ApoER2.3 IDOL was originally described as an LXR target gene.1
Human genetics support a role for genetic variation in IDOL influencing LDL-C levels. Common genetic variants near the IDOL/MYLIP gene are associated with LDL-C levels.4–6 In addition, a missense variant in IDOL, N342S, was found to be associated with lower plasma cholesterol levels in Mexicans,7 and a complete loss-of-function IDOL allele was reported to be associated with lower LDL levels in Danish population.8 Thus, IDOL is a potential novel therapeutic target for pharmacologic inhibition to reduce LDL-C levels. Indeed, pharmacologic inhibition of IDOL is starting to attract some attention. Novel IDOL inhibitors that modulate cholesterol levels are being identified. Some of them were found to increase LDLR levels in cell lines.9 Furthermore, a new study found that inhibition of IDOL in non-human primates (NHP), using antisense oligonucleotide (ASO)-mediated knockdown, might reduce LDL-C levels.10
The mouse has been widely used as a model for drug discovery. However, the contribution of hepatic IDOL to the regulation of LDLR and LDL-C levels in mice is unclear.1,10,11 Furthermore, wild-type C57BL/6 mice do not represent an optimal model to study the regulation of LDL, as mouse lipoproteins are predominantly HDL animals. In this study, we used an adeno-associated virus serotype 8 (AAV8) vector to stably express human IDOL in the livers of humanized hypercholesterolaemic mice12 and observed substantial reduction in hepatic LDLR protein and increase in plasma LDL-C levels. We suggest that this model may be useful for testing the efficacy of IDOL inhibitors.
2. Materials and methods
2.1. Animals
Animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Pennsylvania and conform the NIH guidelines as stated in the Guide for the care and use of laboratory animals. Mice were anaesthetized by continuous inhalation of isoflurane (3–5%) and euthanized at the end of the study by cervical dislocation under the same anaesthesia conditions.
2.2. hIDOL stable expression using AAV
AAV8-hIDOL was obtained from the Penn Vector Core (Philadelphia, PA, USA) and was described.13 Briefly, the cDNA of hIDOL was purchased (Origene, MD) and cloned into AAV8 vector under the liver-specific thyroxine-binding globulin (TBG) promoter. AAV vectors containing an empty expression cassette (AAV-Null) or the indicated dose of AAV-hIDOL were administered by intra-peritoneal (i.p.) injection.
2.3. Plasma lipids (Mira and FPLC)
At the indicated time points, blood was collected from retro-orbital cavity, and plasma lipids were analysed using clinical chemistry analyzer (Cobas Mira Autoanalyzer; Roche Diagnostic Systems, Indianapolis, IN, USA). To examine the different lipoprotein fractions, plasma was separated by FPLC using a Superose™ 6 10/300 column (GE Life Sciences, Marlborough, MA, USA). Individual fractions were assayed for cholesterol or triglycerides (TGs) concentrations by using commercially available assay kits (Infinity™ cholesterol and triglycerides Reagents; Thermo Scientific, Waltham, MA, USA).
2.4. Gene expression (RT–PCR)
Total RNA was isolated from frozen liver tissue using the TRIzol method according to the manufacturer’s instructions (Life Technologies, Waltham, MA, USA). cDNA was prepared using High Capacity cDNA Reverse Transcription kit (Applied Biosystems). Real-time RT–PCR was performed in an ABI 7900 Real-Time PCR System (Applied Biosystems, Waltham, MA, USA) using SYBR green detection. The relative quantity of each mRNA was calculated using the delta CT method with β-actin as the housekeeping gene.
2.5. SDS–PAGE/western blot
Samples were subjected to SDS–PAGE using 4–12% polyacrylamide gels (life technologies) and transferred to nitrocellulose membranes. Primary antibodies for LDLR were from Medimabs (Canada) and for Actin were from Santa Cruz. Secondary antibodies conjugated to horseradish peroxidase (HRP) were from Jackson ImmunoResearch. Membranes were detected on Imagequant LAS 4000 digital imaging system (GE Healthcare) following addition of Luminata HRP Substrate detection reagent (Millipore).
2.6. Statistics
All experiments were repeated three times and analysed using Student's t-test or one-way ANOVA as indicated in the figure legends for different experiments.
3. Results
3.1. Expression of human IDOL in livers of wild-type mice produces a dose-dependent reduction in LDLR protein and increase in plasma non-HDL cholesterol
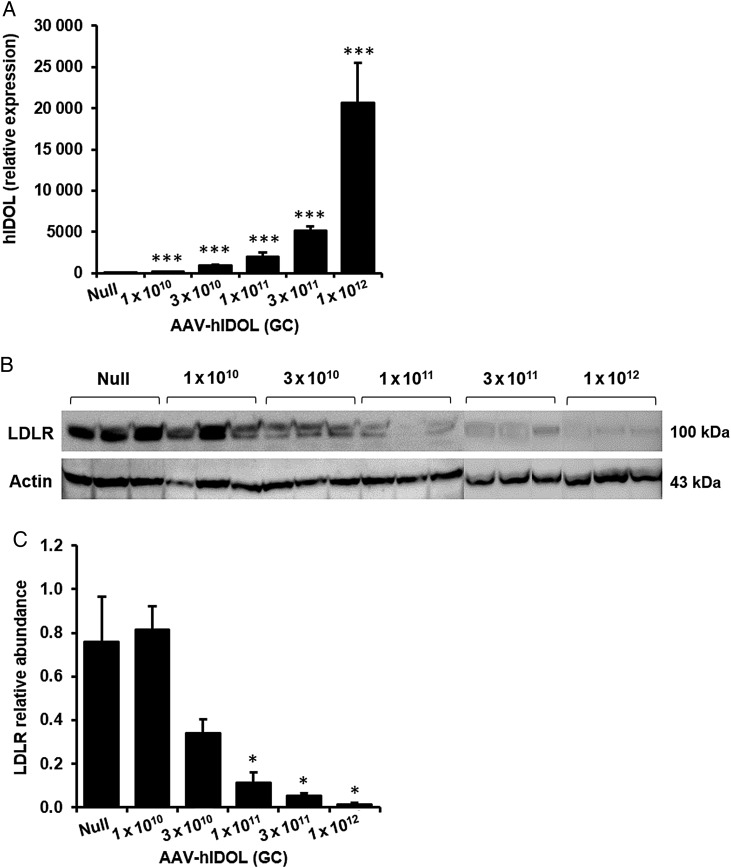
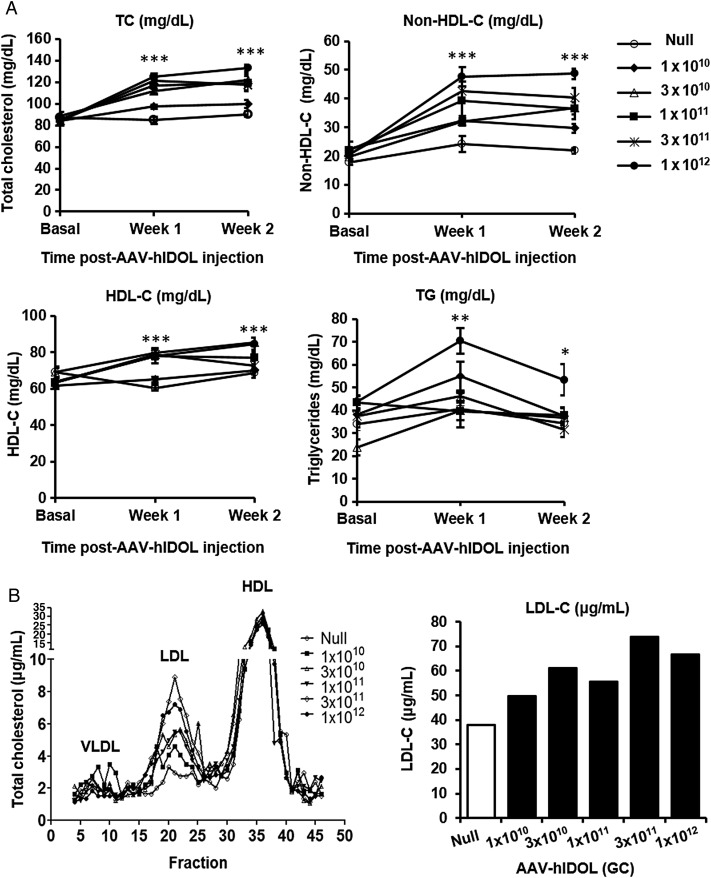
We used an AAV8 vector to perform a dose-ranging study of human IDOL expression in the livers of wild-type C57BL/6 mice. Five doses of AAV-hIDOL at half-log increases were administered and compared with a sixth group of control mice injected with AAV-Null. Dose-dependent hepatic expression of human IDOL mRNA was noted (Figure 1A). Hepatic expression of human IDOL produced a dose-dependent decrease of the mouse LDLR protein, with little protein detected at the two highest doses (Figure 1B). Significant dose-dependent increases in total and non-HDL cholesterol were observed (Figure 2A). At the highest vector dose, non-HDL-C levels increased more than two-fold. A dose-dependent increase in LDL-C was observed by FPLC (Figure 2B). Plasma TG levels were unchanged through the dose range except at the highest dose where they increased by ∼75% (Figure 2A).
Figure 1.
Dose-dependent decrease of the mouse liver LDLR by hIDOL in C57BL/6 mice. Five doses of AAV-hIDOL (1 × 1010, 3 × 1010, 1 × 1011, 3 × 1011, 1 × 1012 GC) were administered to male C57BL/6 mice (n = 5) by intraperitoneal injection and compared with a group injected with AAV-Null as control. (A) hIDOL gene expression as measured by RT–PCR. (B) LDLR protein levels examined in total liver lysates by western blot (n = 3 mice per group) and (C) quantified by densitometry. Data were analysed using Student's t-test (***P < 0.001; *P < 0.05 vs. ‘Null’).
Figure 2.
LDL-C levels increased in a dose-dependent fashion due to the expression of hIDOL in C57BL/6 mice. (A) Plasma total cholesterol, non-HDL-C, HDL-C, and triglycerides (n = 5). Data were from a representative experiment and analysed using one-way ANOVA at each time point (***P < 0.001; **P < 0.01; *P < 0.05 vs. ‘Null’). (B) FPLC profile from pooled plasma showing the distribution of plasma cholesterol in the different lipoprotein subsets. Total LDL-C was isolated and calculated in each group as shown in the bar graph.
3.2. Expression of human IDOL substantially raises LDL-C in a humanized mouse model of heterozygous familial hypercholesterolaemia but not homozygous familial hypercholesterolaemia
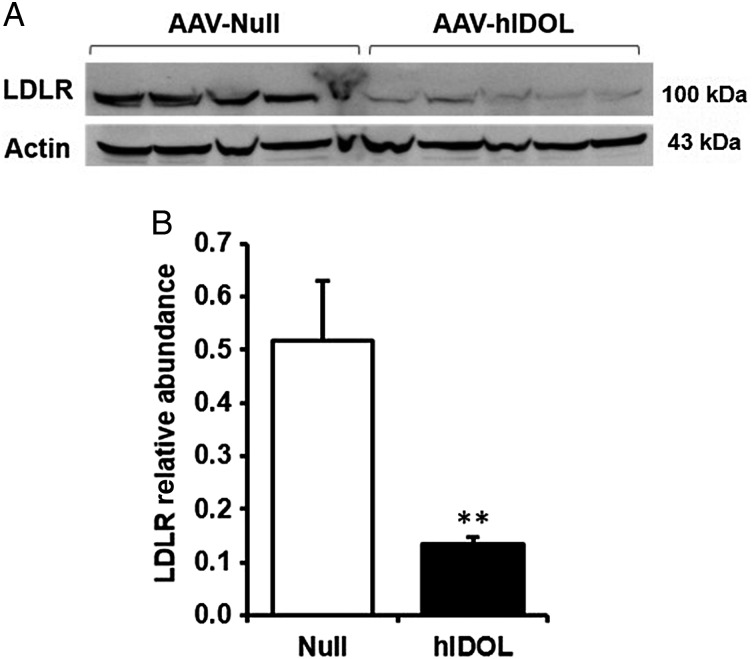
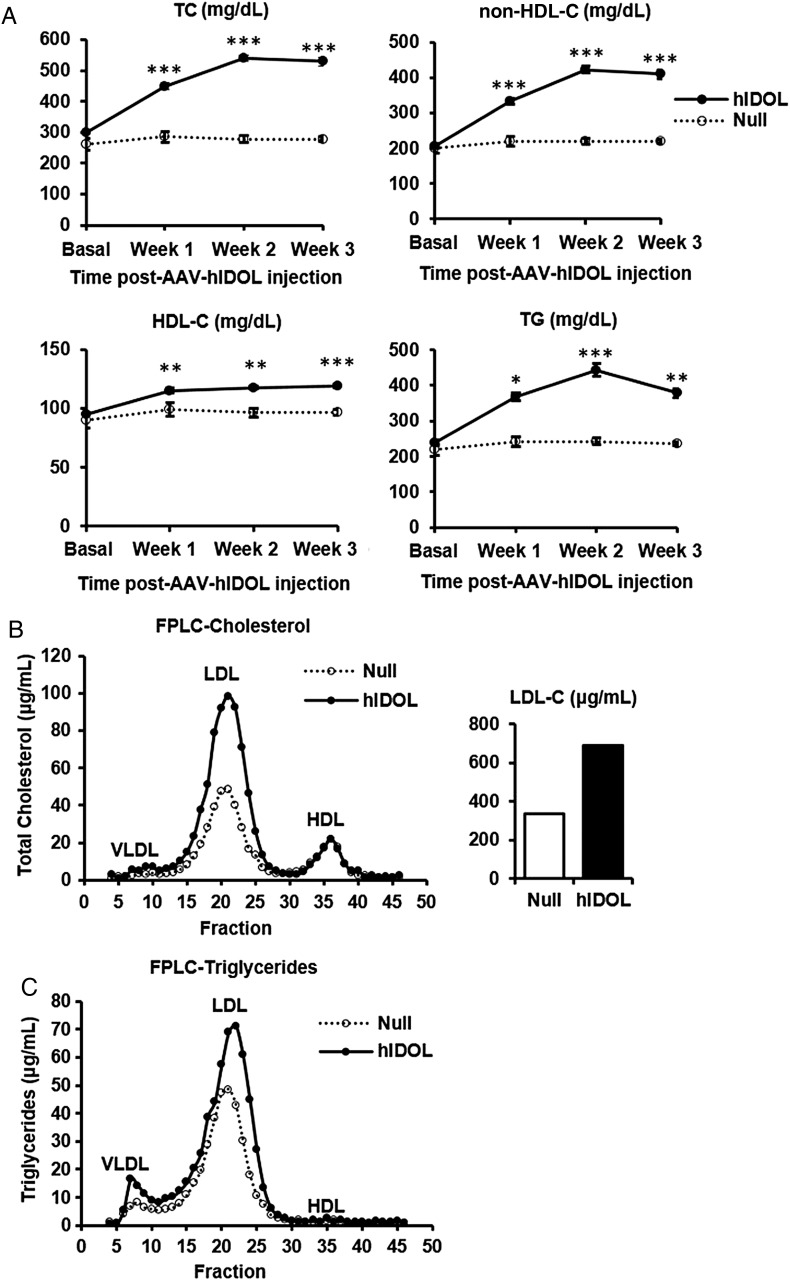
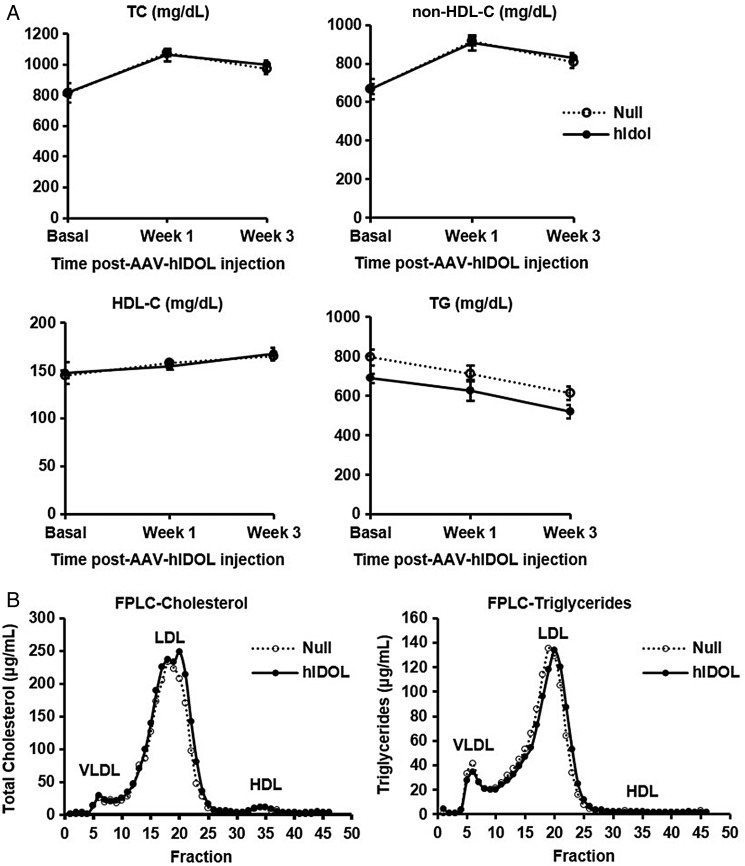
We then turned to the LDLR+/−/Apobec1−/−/hApoB-Tg mouse model,12 a ‘humanized’ mouse model in which the apoB-containing lipoproteins particles contain only apoB100, most of it human apoB. We injected AAV-hIDOL or AAV-null at a dose of 1 × 1011 GC. Hepatic LDLR protein was markedly reduced (Figure 3). TC and LDL-C levels were increased by ∼60% starting from 1 week and were sustainable for at least 3 weeks post-injection (Figure 4A). A marked two-fold increase of the LDL fraction was seen upon separation of lipoproteins by FPLC (Figure 4B).
Figure 3.
Hepatic LDLR protein was markedly reduced by hIDOL in a humanized mouse model of heterozygous familial hypercholesterolaemia (LDLR+/−/Apobec1−/−/hApoB-Tg). Male mice (n = 5) were injected with AAV-hIDOL or AAV-Null (1 × 1011 GC). (A) LDLR protein levels examined in total liver lysates by western blot and (B) quantified by densitometry. Data were analysed using Student's t-test (**P < 0.01 vs. ‘Null’).
Figure 4.
LDL-C levels increased by hIDOL in LDLR+/−/Apobec1−/−/hApoB-Tg or LAhB-het mice. (A) Plasma total cholesterol, non-HDL-C, HDL-C, and triglycerides (n = 5). Data were from a representative experiment and analysed using one-way ANOVA at each time point (***P < 0.001; **P < 0.01; *P < 0.05 vs. ‘Null’). (B) FPLC profile from pooled plasma showing the distribution of plasma cholesterol and (C) triglycerides in the different lipoprotein subsets. Total LDL-C was isolated and calculated in each group as shown in the bar graph.
To establish the dependence of the LDL-raising effect of human IDOL on the LDL receptor, we turned to a humanized mouse model of homozygous familial hypercholesterolaemia (LDLR−/−/Apobec1−/−/hApoB-Tg) lacking the LDL receptor. We again injected AAV-hIDOL or AAV-null at a dose of 1 × 1011 GC. Compared with control vector injected mice, no significant changes in plasma TC or non-HDL-C (Figure 5A) or LDL-C (Figure 5B) were seen. This confirms that the LDL-raising effects of human IDOL expression are dependent on the LDLR.
Figure 5.
hIDOL effects are LDLR dependent. hIDOL was expressed in LDLR−/−/Apobec1−/−/hApoB-Tg or LAhB-KO mice, and plasma lipid profile was examined. (A) Plasma total cholesterol, non-HDL-C, HDL-C, and triglycerides (n = 5). (B) FPLC profile from pooled plasma showing the distribution of plasma cholesterol and triglycerides in the different lipoprotein subsets.
4. Discussion
Human genetics suggest that IDOL is a potential therapeutic target for inhibition to reduce LDL-C levels. Common variants at the Mylip/IDOL locus found by GWAS were associated with LDL-C levels. And a rare variant leading to a complete loss of function was associated with lower LDL levels. Thus, IDOL is a validated target for inhibition to reduce plasma LDL-C levels. In this study, in an attempt to develop a mouse model for testing IDOL inhibitors, we examined the effects of stable expression of human IDOL in mouse liver using AAV8-mediated delivery. We demonstrated that hIDOL causes a dose-dependent decrease in LDLR protein expression in mouse liver. This was translated into a dose-dependent increase in plasma total cholesterol (TC) and LDL-C levels in C57BL/6 mice. By FPLC, we were able to detect a moderate but nonetheless dose-dependent increase in the fraction corresponding to LDL particles that are present at a modest level in control WT C57BL/6 mice.
Wild-type mice are largely HDL animals. Unlike humans, the circulating lipoproteins are mostly HDL in the fasting state.14 Due to the hepatic and intestinal expression of apolipoprotein B mRNA-editing enzyme catalytic polypeptide 1 (Apobec-1) in mice, their non-HDL particles are essentially ApoB48-containing lipoproteins that are cleared mainly by LDLR-independent mechanisms.15,16 To better understand the role of IDOL in the liver in the regulation of liver LDLR and plasma LDL-C in settings that are relevant to humans, we used a ‘humanized’ mouse model of heterozygous familial hypercholesterolaemia (LDLR+/−/Apobec1−/−/hApoB-Tg aka LAhB)12 in which we expressed the human version of IDOL (hIDOL).
A medium range dose (1 × 1011 GC) was chosen to be administered to LAhB mice. In these conditions, we found a significant decrease in LDLR protein expression in mouse liver. This was translated into a significant increase in plasma TC and LDL-C levels. The fraction corresponding to LDL particles that are relatively abundant in controls in this model was increased by two-fold. Moreover, we demonstrated that the effects of hIDOL expression seen in this study are LDLR dependent as shown by the lack of effect of hIDOL in mice that are LDLR deficient.
In mice, IDOL is constitutively expressed in the liver and other tissues like macrophages and intestine. Its regulation by LXR agonists seems to be most effective in non-hepatic cells. This might be of particular interest when IDOL is targeted for therapy given its potential off-target effect due to non-hepatic expression. In contrast, LXR activation induces IDOL expression in liver and other tissues in NHP, highlighting a species-specific regulation of IDOL in the liver.1,10 Further studies are required to fully understand the regulation of IDOL in mouse liver. Nonetheless, this study demonstrates that under conditions where IDOL expression is increased in the liver, one would expect an increase in circulating LDL and plasma cholesterol likely through the degradation of liver LDLR. Indeed, it was shown that IDOL could be up-regulated by the increase of endogenous LXR ligands in the liver of Apoe−/−Npc1−/− mice that displayed elevated intracellular cholesterol levels.17 It would certainly be interesting to examine whether the addition of cholesterol to chow diet would increase IDOL expression. Moreover, it would be interesting to identify variants in the MYLIP locus that are associated with increased expression of IDOL.
On the other hand, human IDOL might be associated with a lower LDL clearance rates than in mice. For instance, unlike human IDOL, the mouse version of the protein has a Ser at position 342 (S342) instead of Asn (N342). Mutation of Ser342 in mouse IDOL to Asn reflecting the human IDOL polymorphism at this residue shows that mouse N342 IDOL was more efficient than S342 at degrading total LDLR protein.7
In conclusion, this study demonstrates that hIDOL is capable of degrading murine LDLR in mouse liver in vivo. Here we show a physiological effect of hIDOL on LDL metabolism in humanized hypercholesterolaemic mice and provide a potential model for preclinical testing of IDOL inhibitors for reduction of LDL-C levels.
Conflict of interest: none declared.
Funding
This work was supported by National Heart, Lung, and Blood Institute (P01 HL59407-15).
References
- 1.Zelcer N, Hong C, Boyadjian R, Tontonoz P. LXR regulates cholesterol uptake through Idol-dependent ubiquitination of the LDL receptor. Science 2009;325:100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang L, Reue K, Fong LG, Young SG, Tontonoz P. Feedback regulation of cholesterol uptake by the LXR-IDOL-LDLR axis. Arterioscler Thromb Vasc Biol 2012;32:2541–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hong C, Duit S, Jalonen P, Out R, Scheer L, Sorrentino V, Boyadjian R, Rodenburg KW, Foley E, Korhonen L, Lindholm D, Nimpf J, van Berkel TJ, Tontonoz P, Zelcer N. The E3 ubiquitin ligase IDOL induces the degradation of the low density lipoprotein receptor family members VLDLR and ApoER2. J Biol Chem 2010;285:19720–19726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, Johansen CT, Fouchier SW, Isaacs A, Peloso GM, Barbalic M, Ricketts SL, Bis JC, Aulchenko YS, Thorleifsson G, Feitosa MF, Chambers J, Orho-Melander M, Melander O, Johnson T, Li X, Guo X, Li M, Shin Cho Y, Jin Go M, Jin Kim Y, Lee JY, Park T, Kim K, Sim X, Twee-Hee Ong R, Croteau-Chonka DC, Lange LA, Smith JD, Song K, Hua Zhao J, Yuan X, Luan J, Lamina C, Ziegler A, Zhang W, Zee RY, Wright AF, Witteman JC, Wilson JF, Willemsen G, Wichmann HE, Whitfield JB, Waterworth DM, Wareham NJ, Waeber G, Vollenweider P, Voight BF, Vitart V, Uitterlinden AG, Uda M, Tuomilehto J, Thompson JR, Tanaka T, Surakka I, Stringham HM, Spector TD, Soranzo N, Smit JH, Sinisalo J, Silander K, Sijbrands EJ, Scuteri A, Scott J, Schlessinger D, Sanna S, Salomaa V, Saharinen J, Sabatti C, Ruokonen A, Rudan I, Rose LM, Roberts R, Rieder M, Psaty BM, Pramstaller PP, Pichler I, Perola M, Penninx BW, Pedersen NL, Pattaro C, Parker AN, Pare G, Oostra BA, O'Donnell CJ, Nieminen MS, Nickerson DA, Montgomery GW, Meitinger T, McPherson R, McCarthy MI, McArdle W, Masson D, Martin NG, Marroni F, Mangino M, Magnusson PK, Lucas G, Luben R, Loos RJ, Lokki ML, Lettre G, Langenberg C, Launer LJ, Lakatta EG, Laaksonen R, Kyvik KO, Kronenberg F, König IR, Khaw KT, Kaprio J, Kaplan LM, Johansson A, Jarvelin MR, Janssens AC, Ingelsson E, Igl W, Kees Hovingh G, Hottenga JJ, Hofman A, Hicks AA, Hengstenberg C, Heid IM, Hayward C, Havulinna AS, Hastie ND, Harris TB, Haritunians T, Hall AS, Gyllensten U, Guiducci C, Groop LC, Gonzalez E, Gieger C, Freimer NB, Ferrucci L, Erdmann J, Elliott P, Ejebe KG, Döring A, Dominiczak AF, Demissie S, Deloukas P, de Geus EJ, de Faire U, Crawford G, Collins FS, Chen YD, Caulfield MJ, Campbell H, Burtt NP, Bonnycastle LL, Boomsma DI, Boekholdt SM, Bergman RN, Barroso I, Bandinelli S, Ballantyne CM, Assimes TL, Quertermous T, Altshuler D, Seielstad M, Wong TY, Tai ES, Feranil AB, Kuzawa CW, Adair LS, Taylor HA, Borecki IB, Gabriel SB, Wilson JG, Holm H, Thorsteinsdottir U, Gudnason V, Krauss RM, Mohlke KL, Ordovas JM, Munroe PB, Kooner JS, Tall AR, Hegele RA, Kastelein JJ, Schadt EE, Rotter JI, Boerwinkle E, Strachan DP, Mooser V, Stefansson K, Reilly MP, Samani NJ, Schunkert H, Cupples LA, Sandhu MS, Ridker PM, Rader DJ, van Duijn CM, Peltonen L, Abecasis GR, Boehnke M, Kathiresan S. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 2010;466:707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waterworth DM, Ricketts SL, Song K, Chen L, Zhao JH, Ripatti S, Aulchenko YS, Zhang W, Yuan X, Lim N, Luan J, Ashford S, Wheeler E, Young EH, Hadley D, Thompson JR, Braund PS, Johnson T, Struchalin M, Surakka I, Luben R, Khaw KT, Rodwell SA, Loos RJ, Boekholdt SM, Inouye M, Deloukas P, Elliott P, Schlessinger D, Sanna S, Scuteri A, Jackson A, Mohlke KL, Tuomilehto J, Roberts R, Stewart A, Kesäniemi YA, Mahley RW, Grundy SM, McArdle W, Cardon L, Waeber G, Vollenweider P, Chambers JC, Boehnke M, Abecasis GR, Salomaa V, Järvelin MR, Ruokonen A, Barroso I, Epstein SE, Hakonarson HH, Rader DJ, Reilly MP, Witteman JC, Hall AS, Samani NJ, Strachan DP, Barter P, van Duijn CM, Kooner JS, Peltonen L, Wareham NJ, McPherson R, Mooser V, Sandhu MS, Wellcome Trust Case Control Consortium. Genetic variants influencing circulating lipid levels and risk of coronary artery disease. Arterioscler Thromb Vasc Biol 2010;30:2264–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J, Buchkovich ML, Mora S, Beckmann JS, Bragg-Gresham JL, Chang HY, Demirkan A, Den Hertog HM, Do R, Donnelly LA, Ehret GB, Esko T, Feitosa MF, Ferreira T, Fischer K, Fontanillas P, Fraser RM, Freitag DF, Gurdasani D, Heikkilä K, Hyppönen E, Isaacs A, Jackson AU, Johansson A, Johnson T, Kaakinen M, Kettunen J, Kleber ME, Li X, Luan J, Lyytikäinen LP, Magnusson PK, Mangino M, Mihailov E, Montasser ME, Müller-Nurasyid M, Nolte IM, O'Connell JR, Palmer CD, Perola M, Petersen AK, Sanna S, Saxena R, Service SK, Shah S, Shungin D, Sidore C, Song C, Strawbridge RJ, Surakka I, Tanaka T, Teslovich TM, Thorleifsson G, Van den Herik EG, Voight BF, Volcik KA, Waite LL, Wong A, Wu Y, Zhang W, Absher D, Asiki G, Barroso I, Been LF, Bolton JL, Bonnycastle LL, Brambilla P, Burnett MS, Cesana G, Dimitriou M, Doney AS, Döring A, Elliott P, Epstein SE, Eyjolfsson GI, Gigante B, Goodarzi MO, Grallert H, Gravito ML, Groves CJ, Hallmans G, Hartikainen AL, Hayward C, Hernandez D, Hicks AA, Holm H, Hung YJ, Illig T, Jones MR, Kaleebu P, Kastelein JJ, Khaw KT, Kim E, Klopp N, Komulainen P, Kumari M, Langenberg C, Lehtimäki T, Lin SY, Lindström J, Loos RJ, Mach F, McArdle WL, Meisinger C, Mitchell BD, Müller G, Nagaraja R, Narisu N, Nieminen TV, Nsubuga RN, Olafsson I, Ong KK, Palotie A, Papamarkou T, Pomilla C, Pouta A, Rader DJ, Reilly MP, Ridker PM, Rivadeneira F, Rudan I, Ruokonen A, Samani N, Scharnagl H, Seeley J, Silander K, Stancáková A, Stirrups K, Swift AJ, Tiret L, Uitterlinden AG, van Pelt LJ, Vedantam S, Wainwright N, Wijmenga C, Wild SH, Willemsen G, Wilsgaard T, Wilson JF, Young EH, Zhao JH, Adair LS, Arveiler D, Assimes TL, Bandinelli S, Bennett F, Bochud M, Boehm BO, Boomsma DI, Borecki IB, Bornstein SR, Bovet P, Burnier M, Campbell H, Chakravarti A, Chambers JC, Chen YD, Collins FS, Cooper RS, Danesh J, Dedoussis G, de Faire U, Feranil AB, Ferrières J, Ferrucci L, Freimer NB, Gieger C, Groop LC, Gudnason V, Gyllensten U, Hamsten A, Harris TB, Hingorani A, Hirschhorn JN, Hofman A, Hovingh GK, Hsiung CA, Humphries SE, Hunt SC, Hveem K, Iribarren C, Järvelin MR, Jula A, Kähönen M, Kaprio J, Kesäniemi A, Kivimaki M, Kooner JS, Koudstaal PJ, Krauss RM, Kuh D, Kuusisto J, Kyvik KO, Laakso M, Lakka TA, Lind L, Lindgren CM, Martin NG, März W, McCarthy MI, McKenzie CA, Meneton P, Metspalu A, Moilanen L, Morris AD, Munroe PB, Njølstad I, Pedersen NL, Power C, Pramstaller PP, Price JF, Psaty BM, Quertermous T, Rauramaa R, Saleheen D, Salomaa V, Sanghera DK, Saramies J, Schwarz PE, Sheu WH, Shuldiner AR, Siegbahn A, Spector TD, Stefansson K, Strachan DP, Tayo BO, Tremoli E, Tuomilehto J, Uusitupa M, van Duijn CM, Vollenweider P, Wallentin L, Wareham NJ, Whitfield JB, Wolffenbuttel BH, Ordovas JM, Boerwinkle E, Palmer CN, Thorsteinsdottir U, Chasman DI, Rotter JI, Franks PW, Ripatti S, Cupples LA, Sandhu MS, Rich SS, Boehnke M, Deloukas P, Kathiresan S, Mohlke KL, Ingelsson E, Abecasis GR, Consortium Global Lipids Genetics Consortium. Discovery and refinement of loci associated with lipid levels. Nat Genet 2013;45:1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weissglas-Volkov D, Calkin AC, Tusie-Luna T, Sinsheimer JS, Zelcer N, Riba L, Tino AM, Ordoñez-Sánchez ML, Cruz-Bautista I, Aguilar-Salinas CA, Tontonoz P, Pajukanta P. The N342S MYLIP polymorphism is associated with high total cholesterol and increased LDL receptor degradation in humans. J Clin Invest 2011;121:3062–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sorrentino V, Fouchier SW, Motazacker MM, Nelson JK, Defesche JC, Dallinga-Thie GM, Kastelein JJ, Kees Hovingh G, Zelcer N. Identification of a loss-of-function inducible degrader of the low-density lipoprotein receptor variant in individuals with low circulating low-density lipoprotein. Eur Heart J 2013;34:1292–1297. [DOI] [PubMed] [Google Scholar]
- 9.Marblestone J, Wu J, LaRocque J, Agarwal S, Kodrasov M, Weinstock J, Kumar S, Mattern M, Sterner D. Ubiquitin ligase Idol inhibitors for treating hypercholesterolemia. FASEB J 2015;29:715.49 Abstract. [Google Scholar]
- 10.Hong C, Marshall SM, McDaniel AL, Graham M, Layne JD, Cai L, Scotti E, Boyadjian R, Kim J, Chamberlain BT, Tangirala RK, Jung ME, Fong L, Lee R, Young SG, Temel RE, Tontonoz P. The LXR-Idol axis differentially regulates plasma LDL levels in primates and mice. Cell Metab 2014;20:910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim GH, Oh GS, Yoon J, Lee GG, Lee KU, Kim SW. Hepatic TRAP80 selectively regulates lipogenic activity of liver X receptor. J Clin Invest 2015;125:183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kassim SH, Li H, Bell P, Somanathan S, Lagor W, Jacobs F, Billheimer J, Wilson JM, Rader DJ. Adeno-associated virus serotype 8 gene therapy leads to significant lowering of plasma cholesterol levels in humanized mouse models of homozygous and heterozygous familial hypercholesterolemia. Hum Gene Ther 2013;24:19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Somanathan S, Jacobs F, Wang Q, Hanlon AL, Wilson JM, Rader DJ. AAV vectors expressing LDLR gain-of-function variants demonstrate increased efficacy in mouse models of familial hypercholesterolemia. Circ Res 2014;115:591–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tall AR. Plasma cholesteryl ester transfer protein. J Lipid Res 1993;34:1255–1274. [PubMed] [Google Scholar]
- 15.Innerarity TL, Borén J, Yamanaka S, Olofsson SO. Biosynthesis of apolipoprotein B48-containing lipoproteins. Regulation by novel post-transcriptional mechanisms. J Biol Chem 1996;271:2353–2356. [DOI] [PubMed] [Google Scholar]
- 16.Chan L. Apolipoprotein B, the major protein component of triglyceride-rich and low density lipoproteins. J Biol Chem 1992;267:25621–25624. [PubMed] [Google Scholar]
- 17.Ishibashi M, Masson D, Westerterp M, Wang N, Sayers S, Li R, Welch CL, Tall AR. Reduced VLDL clearance in Apoe(-/-)Npc1(-/-) mice is associated with increased Pcsk9 and Idol expression and decreased hepatic LDL-receptor levels. J Lipid Res 2010;51:2655–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]