Abstract
Background
Anti-EGFR antibody–based treatment is an important therapeutic strategy for advanced colorectal cancer (CRC); despite this, several mutations—including KRAS, BRAF, and PIK3CA mutations, and HER2 amplification—are associated with the mechanisms underlying the development of resistance to anti-EGFR therapy. The aim of our study was to investigate the frequencies and clinical implications of these genetic alterations in advanced CRC.
Methods
KRAS, BRAF, and PIK3CA mutations were determined by Cobas real-time polymerase chain reaction (PCR) in 191 advanced CRC patients with distant metastasis. Microsatellite instability (MSI) status was determined by a fragmentation assay and HER2 amplification was assessed by silver in situ hybridization. In addition, KRAS mutations were investigated by the Sanger sequencing method in 97 of 191 CRC cases.
Results
Mutations in KRAS, BRAF, and PIK3CA were found in 104 (54.5%), 6 (3.1%), and 25 (13.1%) cases of advanced CRC, respectively. MSI-high status and HER2 amplification were observed in 3 (1.6%) and 16 (8.4%) cases, respectively. PIK3CA mutations were more frequently found in KRAS mutant type (18.3%) than KRAS wild type (6.9%) (P = 0.020). In contrast, HER2 amplifications and BRAF mutations were associated with KRAS wild type with borderline significance (P = 0.052 and 0.094, respectively). In combined analyses with KRAS, BRAF and HER2 status, BRAF mutations or HER2 amplifications were associated with the worst prognosis in the wild type KRAS group (P = 0.004). When comparing the efficacy of detection methods, the results of real time PCR analysis revealed 56 of 97 (57.7%) CRC cases with KRAS mutations, whereas Sanger sequencing revealed 49 cases (50.5%).
Conclusions
KRAS mutations were found in 54.5% of advanced CRC patients. Our results support that subgrouping using PIK3CA and BRAF mutation or HER2 amplification status, in addition to KRAS mutation status, is helpful for managing advanced CRC patients.
Introduction
Colorectal cancer (CRC) is the third most common cancer and the incidence of CRC is still increasing worldwide annually. Despite of early detection and therapeutic advances, regional or distant metastatic disease accounts for almost 50% of newly diagnosed CRC patients and the overall survival rates of advanced CRC patients still remain unsatisfactory. The recent identification of molecular genetics has enabled considerable advancements in the management of patients with advanced CRC. The development of targeted therapies directed against specific mutations such as those in the epidermal growth factor receptor (EGFR) tyrosine kinase gene has improved treatment efficacy and clinical outcome in advanced CRC patients [1–4]. However, CRCs are molecularly heterogeneous tumors that harbor various gene alterations including mutations in KRAS, BRAF, and PIK3CA, as well as HER2 amplification; many patients with these mutations therefore experience resistance against anti-EGFR drugs and exhibit poor prognosis [2,3,5,6]. Therefore, it is important to explore the molecular mechanism underlying the response and resistance to anti-EGFR treatment in advanced CRC.
KRAS mutations, which are commonly detected in approximately 40% of CRC cases, are thought to be associated with resistance to anti-EGFR treatment in CRC. The evaluation of KRAS mutations is thus essential prior to the use of anti-EGFR drugs to select patients who may benefit from anti-EGFR therapies [7,8]. Furthermore, recent studies suggest that additional gene mutations such as BRAF mutations, PIK3CA mutations, and HER2 amplification are implicated in resistance to EGFR-targeted drugs for CRC patients with wild type KRAS [5,9]. BRAF, which is a member of RAF family, plays an important role in the MAP kinase/ERK-signaling pathway [10]. Many previous studies have revealed that mutations in BRAF are a biomarker for poor prognosis in advanced CRC. In addition, BRAF mutant tumors show a poor response to anti-EGFR treatment, especially in CRC patients with wild type KRAS [5,11]. PIK3CA is mutated in various human cancers; in CRC, it is mutated in approximately 20% of cases. Currently, patients harboring PIK3CA mutations in exon 20 and no mutations in KRAS may show resistance to anti-EGFR treatment. Moreover, PIK3CA mutations in exon 9 and KRAS mutations tend to be found together [3,12]. Finally, HER2 amplifications are present in a small number of CRCs, and a few studies have reported the association between HER2 amplification and poor response to anti-EGFR drugs [13].
Despite these previous findings, knowledge of the frequencies and clinical implications of these genetic alterations in Korean patients is still limited. In the present study, we evaluated the prevalence of these genetic alterations in patients with advanced CRC, and assessed the relationship of these genetic alterations with the clinicopathological factors and outcome of the patients. In addition, we compared the efficacy of using Cobas real-time polymerase chain reaction (PCR) tests with that of using Sanger sequencing tests as detection methods for KRAS mutations.
Materials and Methods
Patients and tissue samples
A total of 191 advanced CRC patients with synchronous or metachronous distant metastases who underwent surgical treatment at Seoul National University Bundang Hospital between 2003 and 2009 were enrolled in this study. All patients were treated with surgical resection of the primary CRCs at the initial diagnosis and distant metastasis resected when detected. None of the patients were treated with preoperative chemo- or radiotherapy. Clinicopathologic information and follow up data were obtained from the patients’ medical records and pathology reports. Overall survival (OS) was calculated as the time between the date of surgery and the date of death.
The histopathology and classification of the tumors were determined according to WHO classification. The use of medical record data and tissue samples for this study was approved by the Institutional Review Board of Seoul National University Bundang Hospital (reference: B-1210/174-301). All samples and medical record data were anonymized before use in this study and the participants did not provide written informed consent. The Institutional Review Board waived the need for written informed consent under the condition of anonymization and no additional intervention to the participants.
KRAS, BRAF, and PIK3CA mutation analyses using the real-time PCR test
Tumor samples were collected from surgical resection specimens of the primary CRC. Hematoxylin-Eosin (HE) stained slides were reviewed by a pathologist (H.S.L). Tumor areas were identified and microscopically dissected more than a 1 x 1 cm area, which consisted of more than 60% tumor cells. One or two 8-μm-thick formalin-fixed paraffin-embedded (FFPE) tumor tissue sections were deparaffinized with xylene for 5 min at room temperature (RT), dehydrated in absolute alcohol for 5 min at RT, and allowed to air dry completely for 10 min. DNA was isolated using the Cobas DNA Sample Preparation Kit (Roche, Branchburg, NJ, USA) and the same preparation protocol for all Cobas mutation kits was used in this study. The concentration of the isolated DNA was measured using a NanoDrop UV spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA) and the DNA was diluted with DNA Specimen Diluent from the Cobas 4800 Mutation Test kit (Roche) to the optimal concentration for each gene (KRAS 4 ng/μL, BRAF 5 ng/μL, and PIK3CA 2 ng/μL). Amplification and detection were performed with an Automated Cobas X480 analyzer instrument. The real-time PCR test could detect codon 12, 13, and 61 of KRAS mutation, V600E BRAF mutation, and exon 1, 4, 7, 9, and 20 of PIK3CA mutation.
KRAS mutation analysis using the Sanger sequencing method
Tumor samples were collected from the same primary CRC specimens that had used for the real-time PCR tests. All specimens were microdissected manually and > 60% of the sample area was shown to contain tumor cells as estimated from the H&E-stained slides. Sanger sequencing analysis of KRAS mutations in codon 12, 13, and 61 was performed in 97 of the 191 FFPE tissue samples from CRC patients, as previously described [14].
HER2 analysis by dual-color silver in-situ hybridization (SISH)
HER analysis was performed on tissue array blocks from the same cohort. Construction of tissue array blocks was performed as previously described [5,15]. Briefly, a representative area of the 191 CRC case specimens was extracted, and two cores from central and peripheral area measuring 2 mm in diameter for each case was used for tissue array block construction. Bright-field dual-color SISH analysis was performed using an automatic SISH staining device (BenchMark XT, Ventana Medical Systems) according to the manufacturer’s protocols for the INFORM HER2 DNA and INFORM Chromosome 17 (CEP17) probes (Ventana Medical Systems). We interpreted HER2/CEP17 SISH signals according to the interpretive guide accompanying the INFORM HER2 DNA probe for staining gastric cancer cells (Ventana Medical systems). Tumor tissue was evaluated for hot spots of positive HER2/CEP17 signals using 20X or 40X objectives. Signals were enumerated in 20 non-overlapping tumor cell nuclei per core with 60X or 100X objectives. Small clusters were defined as 6 signals, and larger clusters as 12 signals. HER2 gene amplification was defined as a HER2/CEP17 ratio of ≥ 2.0 in central or peripheral area. Those equivocal cases with a HER2/CEP17 ratio between 1.8 and 2.2 were recounted in 20 additional non-overlapping tumor cell nuclei; the ratio was recalculated based on these results.
Microsatellite instability (MSI) analysis
Sections were prepared from FFPE tissue samples and hematoxylin and eosin–stained slides were evaluated to identify the representative tumor area and normal area in each section. These selected areas were microdissected. MSI analysis was performed as previously described [16,17]. Briefly, MSI status was determined by analyzing five microsatellite loci (BAT-26, BAT-25, D5S346, D17S250, and S2S123) using DNA auto-sequencer (ABI 3731 genetic analyzer; Applied Biosystems, Foster City, CA). According to the Bethesda guideline on MSI, tumors were classified as MSI-H when at least two of the five markers displayed novel bands, MSI-L when additional alleles were observed with one of the five markers, and MSS when all microsatellite markers examined displayed identical patterns in both tumor and normal tissues.
Statistical analysis
Statistical analyses were performed with the SPSS Statistics 18 software package (Chicago, IL, USA). The association between the clinicopathologic parameters and genetic alterations were analyzed using the Chi-square test or Fisher’s exact test. The chi-square test was performed only if at least 80% of the cells have an expected frequency of 5 or greater, and no cell has an expected frequency smaller than 1.0. If not, Fisher’s exact test was used. Age was treated as a continuous variable and compared by using independent T test because of p>0.05 by Shapiro-Wilk normality test. Kaplan-Meier survival curves were plotted, and statistically significant differences in survival curves were analyzed using the log-rank test. Multivariate survival analysis using a Cox proportional hazards model was conducted with mutational status, age, and stage at initial diagnosis. The hazard ratio (HR) and its 95% confidence interval (CI) were evaluated. In all cases, P values less than 0.05 were considered statistically significant.
Results
Patient characteristics
The clinicopathologic features of the patients are summarized in S1 Table. Patients consisted of 103 men (53.9%) and 88 women (46.1%) with a median age of 60 years (range: 28–93 years). Of the 191 cases, 49 (25.7%) tumors were located in the right colon, 71 (37.2%) tumors in the left colon, and 71 (37.2%) tumors in the rectum. Regarding the histologic differentiation grade, 165 (86.4%) tumors were low grade, and 26 (13.6%) tumors were high grade. Regarding treatments, 176 (92.1%) patients received 5-fluorouracil (5-FU)–based adjuvant chemotherapy with or without anti-EGFR treatment (cetuximab) after surgical resection; 150 (85.2%) patients received 5-FU based chemotherapy only; and 26 (14.8%) patients received 5-FU with anti-EGFR drugs.
Genetic alterations associated with EGFR signaling pathway in advanced CRCs
All the basic data are presented in S2 Table. Of the tumor cases examined, 87 (45.5%) had wild type KRAS and 104 (54.5%) had KRAS mutations. Among the tumors with KRAS mutations, mutations in codon 12 or 13 were observed in 97 (93.3%), whereas mutations in codon 61 were observed in 7 (6.7%) patients. BRAF (V600E) mutations were observed in 6 (3.1%) tumors. PIK3CA mutations were identified in 25 (13.1%) tumors. The two most common PIK3CA mutations were located in exon 9 (17 cases, 68.0%) and exon 20 (5 cases, 20.0%). Other rare mutations were located in exons 1 and 4 (2 cases, 8.0%). One case harbored a PIK3CA exon 4 mutation as well as PIK3CA exon 9 mutation. SISH analysis demonstrated HER2 gene amplification in 16 (8.4%) tumors. Three cases from this cohort were MSI–H (1.6%), and the remaining 188 (98.4%) cases were classified as MSS/MSI-L.
Out of 104 KRAS mutant type CRC cases, 23 (22.1%) had PIK3CA mutations, HER2 amplifications, or BRAF mutations (Table 1). Eighteen cases showed PIK3CA mutation, 4 cases showed HER2 amplification, one case had both BRAF and PIK3CA mutations, and one case had both PIK3CA mutation and HER2 amplification. Out of 87 KRAS wild type CRCs, BRAF mutations, PIK3CA mutations, and HER2 amplifications were found in 5 (5.7%), 6 (6.9%), and 11 (12.6%) cases, respectively; overall, 21 of 87 KRAS wild type cases (24.1%) had BRAF mutations, PIK3CA mutations, or HER2 amplifications.
Table 1. The frequencies of genetic alterations for the entire cohort of 191 advanced CRC patients.
| Gene alteration | No. | % |
|---|---|---|
| KRAS mutation (n = 104) | ||
| KRAS only | 81 | 42.4 |
| KRAS and PIK3CA | 17 | 8.9 |
| KRAS and HER2 | 4 | 2.1 |
| KRAS, BRAF, and PIK3CA | 1 | 0.5 |
| KRAS, HER2, and PIK3CA | 1 | 0.5 |
| KRAS wild type (n = 87) | ||
| All negative | 66 | 34.6 |
| HER2 only | 10 | 5.3 |
| PIK3CA only | 5 | 2.6 |
| BRAF only | 5 | 2.6 |
| HER2 and PIK3CA | 1 | 0.5 |
| Total | 191 | 100 |
Interestingly, the presence of PIK3CA mutations was significantly associated with the presence of KRAS mutations (P = 0.020; Table 2). Mutations in KRAS and BRAF were nearly mutually exclusive; however, one case harbored concomitant KRAS and BRAF mutations. HER2 amplifications and BRAF mutations tended to be more frequently observed in KRAS wild type tumors than in KRAS mutant type tumors with borderline statistical significance (P = 0.052 and P = 0.094, respectively). MSI status did not show any association with these genetic alterations in this cohort.
Table 2. Association of each genetic alteration.
| Gene alteration | Total | KRAS wild type | KRAS mutant type | P |
|---|---|---|---|---|
| BRAF mutation | 0.094* | |||
| Wild type | 185 | 82 (44.3%) | 103 (55.7%) | |
| Mutant type | 6 | 5 (83.3%) | 1 (16.7%) | |
| PIK3CA mutation | 0.020 | |||
| Wild type | 166 | 81 (48.8%) | 85 (51.2%) | |
| Mutant type | 25 | 6 (24.0%) | 19 (76.0%) | |
| HER2 amplification | 0.052 | |||
| Negative | 175 | 76 (43.4%) | 99 (56.6%) | |
| Positive | 16 | 11 (68.8%) | 5 (31.2%) | |
| MSI status | 0.592* | |||
| MSS/MSI-L | 188 | 85 (45.2%) | 103 (54.8%) | |
| MSI-H | 3 | 2 (66.7%) | 1 (33.3%) | |
| Total | 191 | 87 (45.5%) | 104 (54.5%) |
KRAS, Kirsten rat sarcoma viral oncogene homolog; BRAF, v-raf murine sarcoma viral oncogene homolog B1; PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha; HER2, human epidermal growth factor receptor 2; MSI, microsatellite instability; MSS/MSI-L, microsatellite stable/MSI-low; MSI-H, MSI-high
*P-values are calculated by using Fisher’s exact test because less than 80% of the cells have an expected frequency of 5 or greater, or any cell has an expected frequency smaller than 1.0.
Association of genetic alterations with clinicopathologic features
Table 3 demonstrates the relationship between genetic alterations and clinicopathologic characteristics. KRAS mutant tumors were more likely to be located in the right colon (P = 0.021). These tumors were also associated with low-grade histology (P = 0.029). BRAF mutant tumors were significantly associated with T4 depth of invasion (P = 0.033). Although it did not reach the statistical significance, BRAF mutant tumors tended to be located in the right colon (P = 0.127) and to have lymphatic invasion (P = 0.097) compared to the same features in BRAF wild type tumors. Tumors with HER2 amplifications were significantly correlated with a distal location (P = 0.006). HER2 amplifications also showed an association with younger, but this difference was not statistically significant (P = 0.081). There were no other significant associations between PIK3CA mutations or MSI status with clinicopathologic factors.
Table 3. Clinicopathologic characteristics according to mutational status of each gene.
| KRAS mutation | BRAF mutation | PIK3CA mutation | HER2 amplification | MSI status | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Characteristic | Total | Mutant type | P | Mutant type | P | Mutant type | P | Positive | P | MSI-H | P |
| Age | 0.503 | 0.676 | 0.347 | 0.081 | 0.131 | ||||||
| Mean ± SD | 60.32 ± 11.64 | 61.83 ± 14.69 | 61.92 ± 10.16 | 54.69 ± 11.31 | 70.33 ± 5.51 | ||||||
| Sex | 0.752 | 0.688* | 0.130 | 0.742 | 0.596* | ||||||
| Male | 103 | 55 (53.4%) | 4 (3.9%) | 17 (16.5%) | 8 (7.8%) | 1 (1.0%) | |||||
| Female | 88 | 49 (55.7%) | 2 (2.3%) | 8 (9.1%) | 8 (9.1%) | 2 (2.3%) | |||||
| Location | 0.021 | 0.127* | 0.282 | 0.006* | 0.262* | ||||||
| Right | 49 | 35 (71.4%) | 4 (8.2%) | 9 (18.4%) | 2 (4.1%) | 2 (4.1%) | |||||
| Left | 71 | 35 (49.3%) | 1 (1.4%) | 10 (14.1%) | 2 (2.8%) | 1 (1.4%) | |||||
| Rectum | 71 | 34 (47.9%) | 1 (1.4%) | 6 (8.5%) | 12 (16.9%) | 0 (0%) | |||||
| Histologic grade | 0.029 | 0.189* | 0.538* | 0.242* | 0.357* | ||||||
| Low | 165 | 95 (57.6%) | 4 (2.4%) | 23 (13.9%) | 12 (7.3%) | 2 (1.2%) | |||||
| High | 26 | 9 (34.6%) | 2 (7.7%) | 2 (7.7%) | 4 (15.4%) | 1 (3.8%) | |||||
| T stage | 0.833 | 0.033* | 0.563 | 0.520 | 0.561* | ||||||
| T1-T3 | 117 | 63 (53.8%) | 1 (0.9%) | 14 (12.0%) | 11 (9.4%) | 1 (0.9%) | |||||
| T4 | 74 | 41(55.4%) | 5 (6.8%) | 11 (14.9%) | 5 (6.8%) | 2 (2.7%) | |||||
| pTNM stage† | 0.179* | 1.000* | 0.422* | 0.306* | 0.224* | ||||||
| I- | 2 | 1 (50.0%) | 1 (50%) | 0 (0%) | 0 (0%) | 0 (0%) | |||||
| II | 19 | 7 (52.9%) | 0 (0%) | 4 (21.1%) | 1 (5.3%) | 0 (0%) | |||||
| III | 43 | 28 (65.1%) | 1 (2.3%) | 3 (7.0%) | 1 (2.3%) | 2 (4.7%) | |||||
| IV | 127 | 68 (53.5%) | 5 (3.9%) | 18 (14.2%) | 14 (11.0%) | 1 (0.8%) | |||||
| Lymphatic invasion | 0.622 | 0.097* | 0.499 | 0.806 | 1.000* | ||||||
| Absent | 65 | 37 (56.9%) | 0 (0%) | 10 (15.4%) | 5 (7.7%) | 1 (1.5%) | |||||
| Present | 126 | 67 (53.2%) | 6 (4.8%) | 15 (11.9%) | 11 (8.7%) | 2 (1.6%) | |||||
| Venous invasion | 0.148 | 1.000* | 0.458 | 0.780* | 1.000* | ||||||
| Absent | 133 | 77 (57.9%) | 4 (3.0%) | 19 (14.3%) | 12 (9.0%) | 2 (1.5%) | |||||
| Present | 58 | 27 (46.6%) | 2 (3.4%) | 6 (10.3%) | 4 (6.9%) | 1 (1.7%) | |||||
| Perineural invasion | 0.896 | 0.685* | 0.640 | 0.844 | 1.000* | ||||||
| Absent | 91 | 50 (54.9%) | 2 (2.2%) | 13 (14.3%) | 8 (8.8%) | 1 (1.1%) | |||||
| Present | 100 | 54 (54.0%) | 4 (4.0%) | 12 (12.0%) | 8 (8.0%) | 2 (2.0%) | |||||
KRAS, Kirsten rat sarcoma viral oncogene homolog; BRAF, v-raf murine sarcoma viral oncogene homolog B1; PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha; HER2, human epidermal growth factor receptor 2; MSS, microsatellite stable; MSI-L, microsatellite instability-low; MSI-H, microsatellite instability-high; SD, standard deviation
Age was compared between two groups by using independent T test.
*P-values are calculated by using Fisher’s exact test because less than 80% of the cells have an expected frequency of 5 or greater, or any cell has an expected frequency smaller than 1.0.
†Stage is the stage at initial diagnosis.
Prognostic significance of genetic alterations
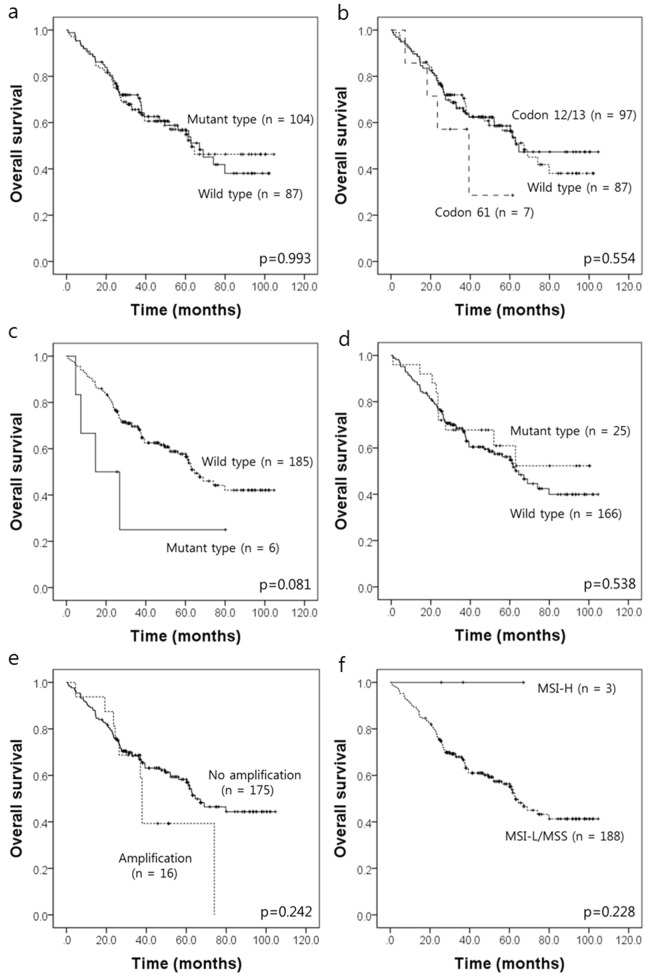
To determine the prognostic significance of these genetic alterations, survival analyses were performed using the Kaplan-Meier method for OS (Fig 1). Follow up data from all 191 CRC patients were included in the survival analysis. There were 84 CRC-related deaths, and the median follow up time was 37.9 months (range, 0.8–104.6 months). Patients with BRAF mutations showed a tendency for unfavorable outcome for OS, but this result did not reach statistical significance (P = 0.081). KRAS mutations, PIK3CA mutations, HER2 amplifications, and MSI status did not show any association with the patients’ OS (P = 0.993, P = 0.538, P = 0.368, and P = 0.538, respectively). Mutation of KRAS codon 61 tended to be associated with shorter overall survival, but it did not reach statistical significance (P = 0.554).
Fig 1.
Kaplan-Meier survival estimate graphs of overall survival (OS) in 191 advanced CRC patients according to KRAS mutations status (a), locations of KRAS mutations in advanced CRC patients (b), BRAF mutations (c), PIK3CA mutations (d), HER2 amplifications (e), and MSI status (f).
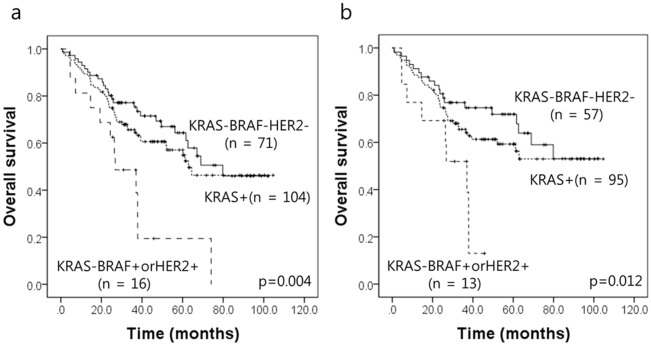
Interestingly, the KRAS wild type subgroup with BRAF mutations or HER2 amplifications showed the worst prognosis in combined analyses (P = 0.004; Fig 2A). By using Cox proportional hazards model, this subgroup was poor prognostic factor (HR, 2.055; CI, 1.093–3.861; P = 0.025) independently of age and stage at initial diagnosis (S3 Table). Among 191 advanced CRCs, 165 patients were not treated with anti-EGFR drugs patients, in whom the KRAS wild type subgroup with BRAF or HER2 alterations also showed the worst prognosis (P = 0.012; Fig 2B). By using Cox proportional hazards model, this subgroup was poor prognostic factor with borderline statistical significance (HR, 1.984; CI, 0.963–4.085; P = 0.063; data not shown). However, in 26 patients treated with 5-FU with anti-EGFR drugs, the KRAS wild type subgroup with BRAF or HER2 alterations was not associated with poor prognosis (P = 0.305, data not shown), which may be because of small number of cases. In the KRAS wild type subgroup, BRAF or HER2 alterations was associated with high grade histologic differentiation, advanced stage, lymphatic invasion and perineural invasion, but with borderline statistical significance (S4 Table).
Fig 2.
Results of combined analysis in advanced CRC patients with the KRAS wild type subgroup according to BRAF mutation and HER2 amplification regardless of anti-EGFR treatment status (n = 191) (a), in whom were not treated with anti-EGFR drugs (n = 165) (b).
Comparison of Cobas real-time PCR and Sanger sequencing methods for KRAS mutations
Of 191 CRC samples, the tissues of 97 patients were available for analysis to compare the detection of KRAS mutations with the Sanger sequencing test and Cobas real-time PCR test. Of the 97 tumors included, KRAS mutations were detected in 49 cases (50.5%) by the Sanger sequencing test. Mutations in KRAS codon 12 or 13 and KRAS codon 61 were detected in 47 (48.5%) and 2 (2.1%) cases, respectively. On the other hand, 56 cases (57.7%) of KRAS mutations were detected by the real-time PCR test; the test located 52 (53.6%) mutations in codon 12 or 13 and 4 (4.1%) mutations in codon 61. The real-time PCR test showed a higher sensitivity than that of the Sanger sequencing test.
Discussion
KRAS is a well-known driver oncogene in CRCs and the presence of KRAS mutation predicts poor response to anti-EGFR targeted therapy in metastatic CRC patients. We evaluated the frequencies and clincopathologic significance of mutations in KRAS, BRAF, and PIK3CA, and HER2 amplification, as well as the relationship of these genetic alterations in advanced CRC patients who were candidates for anti-EGFR treatment in daily practice. Mutations in KRAS, BRAF, and PIK3CA were found in 104 (54.5%), 6 (3.1%), and 25 (13.1%) cases of advanced CRC, respectively. In addition, MSI-H phenotype and HER2 amplification were observed in 3 (1.6%) and 16 (8.4%) cases, respectively.
The development of targeted therapies against specific molecular alterations has contributed to the management of advanced CRC patients, and anti-EGFR drugs are used in these patients. Although the presence of KRAS mutation is useful to exclude patients who will not benefit from anti-EGFR treatment, many patients with wild type KRAS CRC show negative responses to this treatment. To date, it is considered that BRAF mutations, PIK3CA mutations, and HER2 amplifications are associated with the underlying mechanisms of these poor responses [4,5]. In our cohort, 16 of 87 KRAS wild type cases (18.4%) had BRAF mutations or HER2 amplifications; furthermore, combined analysis showed that KRAS wild type patients with BRAF mutations or HER2 amplifications had the worst prognosis. Because the targeted therapies to BRAF mutations and HER2 amplifications have significant survival benefit in various human cancers [10,18,19], treatment with anti-BRAF and anti-HER2 agents may be a good therapeutic strategy to improve survival in CRC patients with wild type KRAS harboring BRAF mutations or HER2 amplifications and who also have primary or secondary resistance to anti-EGFR treatment.
Despite a small number of tumors with PIK3CA mutations in our cohort, we found that PIK3CA mutations largely overlapped with KRAS mutations, which was consistent with the previous studies in European [20] and Japanese [21] CRC patients with metastasis. Currently, several inhibitors targeting the PIK3CA signaling pathway have been developed, and these agents are being tested in preclinical and clinical trials of patients with CRC [22–24]. Considering our result and the previous studies [20,21] that KRAS mutations often coexisted with PIK3CA mutations, inhibiting the PIK3CA signaling pathway might be a useful therapeutic strategy to treat CRC patients with KRAS mutations.
Overall, 24 of 191 cases (12.6%) had two or more oncogenic alterations in this study. It may be interpreted that these genetic alterations occur at the same time in the same tumor, but it may be due to tumor heterogeneity. During tumor progression, oncogenic alterations develop in a subclone, which contribute to cancer metastasis and drug resistance. We used more sensitive detection methods, thus mutations in minor tumor cell population could be detected. In managing CRC patients, sensitive molecular diagnosis is helpful to detect minor oncogenic alterations, which can be the next target in advanced CRC patients with primary or secondary resistance to the first-line targeted treatment. In our cohort, one case harbored concomitant KRAS and BRAF mutations. It is well known that BRAF mutations are usually detected in KRAS wild type tumors, and that they are almost mutually exclusive with KRAS mutations in CRC. Several studies have reported that rare cases harbor combined KRAS and BRAF mutations, which occurs in less than 0.02% [25,26]. Even though the tumor biology and prognosis of patients with concomitant KRAS and BRAF mutations have been still uncertain, previous research suggests that these concomitant mutations are associated with tumor progression such as lymph node metastasis and higher T stage [25,27]. Further large-scale studies are needed to clarify the incidence and biologic function of concomitant KRAS and BRAF mutations.
Previous studies with advanced CRC patients with metastasis reported that approximately 34~45% of the patients had KRAS mutations [20,21,28]. This study with Korean CRC patients demonstrated that the frequency of KRAS mutations was 54.5%, and that these mutations were seen mainly in codons 12 or 13 (93.3%). The frequency of KRAS mutations in this cohort was slightly higher than that in previously published reports of metastatic CRC patients [20,21,28]. There are several possible explanations for it. First, we enrolled only advanced CRC patients in this cohort, which may account for the higher frequency of KRAS mutations. Second, there were some differences in mutation detection methods. We analyzed mutation status using Cobas real-time PCR, which is considered to show higher sensitivity than that of other detection methods.
It has been reported that most of KRAS mutations in CRC patients occur in codon 12 and 13. This study also demonstrated that KRAS mutations were seen mainly in codons 12 or 13 (93.3%). In our KRAS mutation subgroup analysis, mutation of KRAS codon 61 tended to be associated with shorter overall survival, but it did not reach statistical significance. The prognostic impact of KRAS codon 61 mutations has been reported in several previous studies, though with controversial results [29]. Because the incidence of KRAS codon 61 mutations is rare, further large-scale studies may help clarify the relationship between these mutations and clinical outcome.
Mutations of BRAF and PIK3CA were detected in 3.1% and 13.1% of cases, respectively. Although the frequencies of these mutations were considered to be low, they were similar to those of previous studies in advanced CRC [20,21,28]. In agreement with previous study [30], BRAF mutations were associated with aggressive CRC histology, such as higher T stage and presence of lymphatic invasion. Though the results did not reach statistical significance, patients harboring BRAF mutations also showed shorter OS.
We evaluated two detection methods for KRAS mutations in CRC samples: Cobas real-time PCR and Sanger sequencing. There was a good correlation between KRAS mutation detection by real-time PCR and that of Sanger sequencing, and real-time PCR showed higher sensitivity than that of Sanger sequencing. In our cohort, 7 cases of KRAS mutations were detected by the real-time test that were not detected by the Sanger sequencing test, specifically 5 cases for codon 12 or 13 and 2 cases for codon 61. The Sanger sequencing method, developed in 1975, was considered one of the basic mutation detection methods; however, this method appears to have limited sensitivity—a low level of the mutant allele may be undetectable by this method [31]. Conversely, the real-time PCR test including various commercial kits is considered to be a highly sensitive method that shows advantages in detecting KRAS mutations [32]. To date, the considerable intratumoral heterogeneity of molecular alterations and their clinical impact on the targeted therapy have been described in various tumors. In CRC, several studies reported the clinical significance of KRAS heterogeneity in anti-EGFR treatment [5,33,34]. Normanno et al. suggested that a low content of KRAS mutant alleles was sufficient to produce resistance to EGFR monoclonal antibodies, and the threshold in their study was 3~10% of mutant KRAS allele frequency which was unlikely to be detected by using Sanger sequencing [33]. Although further studies are needed to validate these results and clarify the role of the molecular alterations to resistance to anti EGFR treatment, the accurate detection of these mutations has great clinical significance.
In conclusion, we found that the prevalence of KRAS mutations was 54.5% in Korean advanced CRC patients, which was more frequent than that reported in other populations. BRAF mutations or HER2 amplifications were found in 16.1% of KRAS wild type patients, and furthermore, combined analysis showed that KRAS wild type patients with BRAF or HER2 amplifications had the worst prognosis. PIK3CA mutations were more frequently observed in KRAS mutant type than in wild type KRAS CRC patients. Therefore, subgrouping depending on the status of PIK3CA and BRAF mutation or HER2 amplification, in addition to KRAS mutation status, is helpful to determine CRC patient management strategies. We also demonstrated that a real time PCR method had high sensitivity for detecting KRAS mutations in CRC patients.
Supporting Information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability
All relevant data are within the Supporting Information files.
Funding Statement
This research was supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI14C1813, https://www.khidi.or.kr/eps). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Walther A, Johnstone E, Swanton C, Midgley R, Tomlinson I, et al. (2009) Genetic prognostic and predictive markers in colorectal cancer. Nat Rev Cancer 9: 489–499. 10.1038/nrc2645 [DOI] [PubMed] [Google Scholar]
- 2.Martin V, Landi L, Molinari F, Fountzilas G, Geva R, et al. (2013) HER2 gene copy number status may influence clinical efficacy to anti-EGFR monoclonal antibodies in metastatic colorectal cancer patients. Br J Cancer 108: 668–675. 10.1038/bjc.2013.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leto SM, Trusolino L (2014) Primary and acquired resistance to EGFR-targeted therapies in colorectal cancer: impact on future treatment strategies. J Mol Med (Berl) 92: 709–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Er TK, Chen CC, Bujanda L, Herreros-Villanueva M (2014) Current approaches for predicting a lack of response to anti-EGFR therapy in KRAS wild-type patients. Biomed Res Int 2014: 591867 10.1155/2014/591867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Misale S, Di Nicolantonio F, Sartore-Bianchi A, Siena S, Bardelli A (2014) Resistance to anti-EGFR therapy in colorectal cancer: from heterogeneity to convergent evolution. Cancer Discov 4: 1269–1280. 10.1158/2159-8290.CD-14-0462 [DOI] [PubMed] [Google Scholar]
- 6.Mao C, Yang ZY, Hu XF, Chen Q, Tang JL (2012) PIK3CA exon 20 mutations as a potential biomarker for resistance to anti-EGFR monoclonal antibodies in KRAS wild-type metastatic colorectal cancer: a systematic review and meta-analysis. Ann Oncol 23: 1518–1525. 10.1093/annonc/mdr464 [DOI] [PubMed] [Google Scholar]
- 7.Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, et al. (2012) Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 486: 532–536. 10.1038/nature11156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Krieken JH, Jung A, Kirchner T, Carneiro F, Seruca R, et al. (2008) KRAS mutation testing for predicting response to anti-EGFR therapy for colorectal carcinoma: proposal for an European quality assurance program. Virchows Arch 453: 417–431. 10.1007/s00428-008-0665-y [DOI] [PubMed] [Google Scholar]
- 9.Shaib W, Mahajan R, El-Rayes B (2013) Markers of resistance to anti-EGFR therapy in colorectal cancer. J Gastrointest Oncol 4: 308–318. 10.3978/j.issn.2078-6891.2013.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cantwell-Dorris ER, O'Leary JJ, Sheils OM (2011) BRAFV600E: implications for carcinogenesis and molecular therapy. Mol Cancer Ther 10: 385–394. 10.1158/1535-7163.MCT-10-0799 [DOI] [PubMed] [Google Scholar]
- 11.Yokota T (2012) Are KRAS/BRAF mutations potent prognostic and/or predictive biomarkers in colorectal cancers? Anticancer Agents Med Chem 12: 163–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, et al. (2010) Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol 11: 753–762. 10.1016/S1470-2045(10)70130-3 [DOI] [PubMed] [Google Scholar]
- 13.Yonesaka K, Zejnullahu K, Okamoto I, Satoh T, Cappuzzo F, et al. (2011) Activation of ERBB2 signaling causes resistance to the EGFR-directed therapeutic antibody cetuximab. Sci Transl Med 3: 99ra86 10.1126/scitranslmed.3002442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim MJ, Lee HS, Kim JH, Kim YJ, Kwon JH, et al. (2012) Different metastatic pattern according to the KRAS mutational status and site-specific discordance of KRAS status in patients with colorectal cancer. BMC Cancer 12: 347 10.1186/1471-2407-12-347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seo AN, Kwak Y, Kim DW, Kang SB, Choe G, et al. (2014) HER2 status in colorectal cancer: its clinical significance and the relationship between HER2 gene amplification and expression. PLoS One 9: e98528 10.1371/journal.pone.0098528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oh JR, Kim DW, Lee HS, Lee HE, Lee SM, et al. (2012) Microsatellite instability testing in Korean patients with colorectal cancer. Fam Cancer 11: 459–466. 10.1007/s10689-012-9536-4 [DOI] [PubMed] [Google Scholar]
- 17.Kim JH, Bae JM, Kim KJ, Rhee YY, Kim Y, et al. (2014) Differential Features of Microsatellite-Unstable Colorectal Carcinomas Depending on EPCAM Expression Status. Korean J Pathol 48: 276–282. 10.4132/KoreanJPathol.2014.48.4.276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumler I, Tuxen MK, Nielsen DL (2014) A systematic review of dual targeting in HER2-positive breast cancer. Cancer Treat Rev 40: 259–270. 10.1016/j.ctrv.2013.09.002 [DOI] [PubMed] [Google Scholar]
- 19.Ingold Heppner B, Behrens HM, Balschun K, Haag J, Kruger S, et al. (2014) HER2/neu testing in primary colorectal carcinoma. Br J Cancer 111: 1977–1984. 10.1038/bjc.2014.483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neumann J, Wehweck L, Maatz S, Engel J, Kirchner T, et al. (2013) Alterations in the EGFR pathway coincide in colorectal cancer and impact on prognosis. Virchows Arch 463: 509–523. 10.1007/s00428-013-1450-0 [DOI] [PubMed] [Google Scholar]
- 21.Kawazoe A, Shitara K, Fukuoka S, Kuboki Y, Bando H, et al. (2015) A retrospective observational study of clinicopathological features of KRAS, NRAS, BRAF and PIK3CA mutations in Japanese patients with metastatic colorectal cancer. BMC Cancer 15: 258 10.1186/s12885-015-1276-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J, Roberts TM, Shivdasani RA (2011) Targeting PI3K signaling as a therapeutic approach for colorectal cancer. Gastroenterology 141: 50–61. 10.1053/j.gastro.2011.05.010 [DOI] [PubMed] [Google Scholar]
- 23.Temraz S, Mukherji D, Shamseddine A (2015) Dual Inhibition of MEK and PI3K Pathway in KRAS and BRAF Mutated Colorectal Cancers. Int J Mol Sci 16: 22976–22988. 10.3390/ijms160922976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Courtney KD, Corcoran RB, Engelman JA (2010) The PI3K pathway as drug target in human cancer. J Clin Oncol 28: 1075–1083. 10.1200/JCO.2009.25.3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sahin IH, Kazmi SM, Yorio JT, Bhadkamkar NA, Kee BK, et al. (2013) Rare Though Not Mutually Exclusive: A Report of Three Cases of Concomitant KRAS and BRAF Mutation and a Review of the Literature. J Cancer 4: 320–322. 10.7150/jca.3619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Cutsem E, Kohne CH, Lang I, Folprecht G, Nowacki MP, et al. (2011) Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol 29: 2011–2019. 10.1200/JCO.2010.33.5091 [DOI] [PubMed] [Google Scholar]
- 27.Oliveira C, Velho S, Moutinho C, Ferreira A, Preto A, et al. (2007) KRAS and BRAF oncogenic mutations in MSS colorectal carcinoma progression. Oncogene 26: 158–163. [DOI] [PubMed] [Google Scholar]
- 28.Ma BB, Mo F, Tong JH, Wong A, Wong SC, et al. (2015) Elucidating the prognostic significance of KRAS, NRAS, BRAF and PIK3CA mutations in Chinese patients with metastatic colorectal cancer. Asia Pac J Clin Oncol 11: 160–169. 10.1111/ajco.12342 [DOI] [PubMed] [Google Scholar]
- 29.Imamura Y, Lochhead P, Yamauchi M, Kuchiba A, Qian ZR, et al. (2014) Analyses of clinicopathological, molecular, and prognostic associations of KRAS codon 61 and codon 146 mutations in colorectal cancer: cohort study and literature review. Mol Cancer 13: 135 10.1186/1476-4598-13-135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim B, Park SJ, Cheon JH, Kim TI, Kim WH, et al. (2014) Clinical meaning of BRAF mutation in Korean patients with advanced colorectal cancer. World J Gastroenterol 20: 4370–4376. 10.3748/wjg.v20.i15.4370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee S, Brophy VH, Cao J, Velez M, Hoeppner C, et al. (2012) Analytical performance of a PCR assay for the detection of KRAS mutations (codons 12/13 and 61) in formalin-fixed paraffin-embedded tissue samples of colorectal carcinoma. Virchows Arch 460: 141–149. 10.1007/s00428-011-1180-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arcila M, Lau C, Nafa K, Ladanyi M (2011) Detection of KRAS and BRAF mutations in colorectal carcinoma roles for high-sensitivity locked nucleic acid-PCR sequencing and broad-spectrum mass spectrometry genotyping. J Mol Diagn 13: 64–73. 10.1016/j.jmoldx.2010.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Normanno N, Rachiglio AM, Lambiase M, Martinelli E, Fenizia F, et al. (2015) Heterogeneity of KRAS, NRAS, BRAF and PIK3CA mutations in metastatic colorectal cancer and potential effects on therapy in the CAPRI GOIM trial. Ann Oncol 26: 1710–1714. 10.1093/annonc/mdv176 [DOI] [PubMed] [Google Scholar]
- 34.Baldus SE, Schaefer KL, Engers R, Hartleb D, Stoecklein NH, et al. (2010) Prevalence and heterogeneity of KRAS, BRAF, and PIK3CA mutations in primary colorectal adenocarcinomas and their corresponding metastases. Clin Cancer Res 16: 790–799. 10.1158/1078-0432.CCR-09-2446 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the Supporting Information files.