CD146 expression is gradually decreased during the long-term expansion of human umbilical cord blood-derived mesenchymal stem cells (hUCB-MSCs) in culture, and enforced CD146 downregulation accelerated senescence in hUCB-MSCs, which affected their multilineage differentiation potential, growth, and stemness. These data indicate a possible role for CD146 in inhibiting senescence in hUCB-MSCs, which may occur via the regulation of Bmi-1, Id1, and Twist1 expression. CD146 may be a novel marker for predicting senescence in hUCB-MSCs, and it could be valuable in quality-control assessments and for improving the therapeutic potential of hUCB-MSC-based therapy.
Keywords: Cellular senescence, Mesenchymal stem cell-based therapy, Cell surface marker, Melanoma cell adhesion molecule (CD146), Human umbilical cord blood-derived mesenchymal stem cells, Bmi-1, Id1, Twist1
Abstract
Therapeutic applications of mesenchymal stem cells (MSCs) for treating various diseases have increased in recent years. To ensure that treatment is effective, an adequate MSC dosage should be determined before these cells are used for therapeutic purposes. To obtain a sufficient number of cells for therapeutic applications, MSCs must be expanded in long-term cell culture, which inevitably triggers cellular senescence. In this study, we investigated the surface markers of human umbilical cord blood-derived MSCs (hUCB-MSCs) associated with cellular senescence using fluorescence-activated cell sorting analysis and 242 cell surface-marker antibodies. Among these surface proteins, we selected the melanoma cell adhesion molecule (MCAM/CD146) for further study with the aim of validating observed expression differences and investigating the associated implications in hUCB-MSCs during cellular senescence. We observed that CD146 expression markedly decreased in hUCB-MSCs following prolonged in vitro expansion. Using preparative sorting, we found that hUCB-MSCs with high CD146 expression displayed high growth rates, multilineage differentiation, expression of stemness markers, and telomerase activity, as well as significantly lower expression of the senescence markers p16, p21, p53, and senescence-associated β-galactosidase, compared with that observed in hUCB-MSCs with low-level CD146 expression. In contrast, CD146 downregulation with small interfering RNAs enhanced the senescence phenotype. In addition, CD146 suppression in hUCB-MSCs caused downregulation of other cellular senescence regulators, including Bmi-1, Id1, and Twist1. Collectively, our results suggest that CD146 regulates cellular senescence; thus, it could be used as a therapeutic marker to identify senescent hUCB-MSCs.
Significance
One of the fundamental requirements for mesenchymal stem cell (MSC)-based therapies is the expansion of MSCs during long-term culture because a sufficient number of functional cells is required. However, long-term growth inevitably induces cellular senescence, which potentially causes poor clinical outcomes by inducing growth arrest and the loss of stem cell properties. Thus, the identification of markers for evaluating the status of MSC senescence during long-term culture may enhance the success of MSC-based therapy. This study provides strong evidence that CD146 is a novel and useful marker for predicting senescence in human umbilical cord blood-derived MSCs (hUCB-MSCs), and CD146 can potentially be applied in quality-control assessments of hUCB-MSC-based therapy.
Introduction
Mesenchymal stem cells derived from human umbilical cord blood (hUCB-MSCs) are characterized by self-renewal [1], a potential for differentiation into multiple cell lineages [2], low immunogenicity [3], and paracrine functions [4–6], suggesting that they may be beneficial in allogeneic MSC-based therapy. However, enabling MSC-based therapy requires expanding MSCs in large-scale production via long-term in vitro cultivation. During this process, MSCs inevitably enter cellular senescence, which adversely affects the biological properties of stem cells associated with therapeutic outcomes, including their stemness, proliferation, differentiation potency, migration, and cytokine-production profiles [7, 8]. Thus, the senescence process of MSCs is a major issue that needs to be addressed to achieve improved outcomes in MSC-based therapy.
Typically, cellular senescence induces morphological changes that result in enlarged, multinucleated cells with a fried-egg morphology [9, 10]. Along with these morphological changes, growth arrest occurs during senescence, which is mediated by the inhibition of cell cycle procession. The senescence pathway underlying cell cycle progression proceeds via two major pathways that involve the p53/p21 and p16/retinoblastoma (Rb) proteins [11, 12]. When senescence is well advanced, the level of activated (phosphorylated) p53 increases, leading to phosphorylation of its downstream target, p21, which in turn mediates cell cycle arrest during senescence [13]. Indeed, several previous studies have demonstrated that activation of the p53/p21 pathway inhibits the growth of bone marrow-derived MSCs (BM-MSCs) [14, 15]. Phosphorylated Rb drives cell cycle progression by releasing E2F transcription factors that mediate the transcription of a variety of genes important for progression from G1 to S phase [16, 17], and p16 inhibits Rb phosphorylation, which consequently induces senescence [18]. A recent report showed that the level of phosphorylated Rb was high in early-passage MSCs, but that phosphorylated Rb levels decreased during later passages [19]. Such characteristic molecular phenotypes are often used as markers for cellular senescence. In addition, another commonly employed senescence-associated (SA) biomarker is β-galactosidase (SA β-gal) [20]. Many research groups have endeavored to establish a standard set of criteria for confirming senescence in MSCs and in doing so have focused on investigating the mechanisms underlying the senescence process in MSCs. However, despite some advances made in recent studies, knowledge regarding MSC senescence and associated biomarkers is currently limited.
Here, we sought to identify senescence-related factors in hUCB-MSCs. Because the analysis of cell surface-expression phenotypes is widely used to rapidly identify or characterize cell statuses and identify senescence factors, we analyzed the expression of surface proteins in hUCB-MSCs during long-term culture by using fluorescence-activated cell sorting (FACS) with 242 different cell surface antibodies. Among the surface proteins studied, we found that the expression of melanoma cell adhesion molecule (MCAM/CD146) gradually decreased after multiple passages of hUCB-MSCs. Furthermore, we demonstrated that CD146 suppression accelerated cellular senescence in hUCB-MSCs. To our knowledge, this study is the first to demonstrate a possible role for CD146 in determining the senescence fate of hUCB-MSCs.
Materials and Methods
Cell Culture
The Institutional Review Board of MEDIPOST Co., Ltd., approved this study (MP-2014-07-1). We collected hUCB from umbilical veins after neonatal delivery after first obtaining informed maternal consent. The hUCB harvests were processed within 24 hours of collection. The hUCB was isolated by separating mononuclear cells (MNCs) with Ficoll-Hypaque solution (density = 1.077 g/cm3; Sigma-Aldrich, St. Louis, MO, http://www.sigmaaldrich.com). The separated MNCs were washed and suspended in minimum essential medium α (Gibco/Invitrogen/ThermoFisher Scientific, Grand Island, NY, https://www.thermofisher.com), supplemented with 10% fetal bovine serum (Gibco/ThermoFisher Scientific). Cultures were maintained at 37°C in a humidified atmosphere containing 5% CO2, wherein the culture medium was changed twice a week [21]. The expansion of live cells was analyzed by using the trypan blue exclusion method. For each passage (P), MSCs were cultured for 5 days, harvested with trypsin-EDTA (Gibco), counted, and then reseeded at a cell density of 2,000 cells/cm2. Cumulative population doubling (PD) was calculated for each passage on the basis of the total number of cells at each passage [22]. This procedure was repeated until the cells stopped proliferating. Cell areas were analyzed by using ImageJ software (National Institutes of Health, Bethesda, MD, http://imagej.net) by drawing cell margins on T75 flasks from images of cultured cells and measuring total cell areas from three fields [10]. The pictures shown are representative images of cultured cells. A total of 27 lots of hUCB-MSCs was used in our experiments. Basic information related to the hUCB-MSCs is summarized in supplemental online Table 1.
Cell Surface Antibody Screening With Lysoplates
Antibodies against 242 human cell surface markers, which were lyophilized in 96-well plates at 0.5 µg/well, were acquired from BD Biosciences (San Diego, CA, http://www.bdbiosciences.com). For screening, we selected cells from two different donors, which showed typical MSC features, including the potential for multilineage differentiation, representative MSC marker expression, and proliferation (supplemental online Fig. 1). For screening, MSCs were dispensed into 96-well round-bottom plates (BD Lysoplates, BD Biosciences) at 500,000 cells/well. Subsequently, the antibodies were reconstituted and the cells were stained on ice for 20 minutes. Next, cells were washed and stained for 20 minutes with an Alexa Fluor 647-conjugated goat-anti-mouse IgG secondary antibody (Molecular Probes/ThermoFisher Scientific, Eugene, OR, https://www.thermofisher.com). Surface markers were measured by flow cytometry on a FACSCalibur instrument (BD Biosciences), and the percentage of cells expressing each cell surface antigen was calculated for 10,000 cell events. Results were analyzed in Excel 2013 (Microsoft, Redmond, WA, http://www.microsoft.com) to generate heat maps.
Flow Cytometry and Sorting
For cytometric analysis of cultured cell phenotypes, cells were stained for 15 minutes at room temperature with fluorescein isothiocyanate-conjugated antibodies against human CD14, CD45, CD47, CD71, and CD146 (BD Biosciences); phycoerythrin-conjugated antibodies against human CD29, CD44, CD73, CD90, CD106, CD165, CD274, the epidermal growth factor receptor (EGFR; BD Biosciences), and CD105 (Serotec, Kidlington, U.K., https://www.abdserotec.com); and an Alexa 647-conjugated antibody against human CD49f (BD Biosciences). Corresponding isotype-matched mouse antibodies were used as controls. The cells were washed with phosphate-buffered saline (Gibco) and fixed with 1% (vol/vol) paraformaldehyde (Sigma-Aldrich). MSC immunotypes were determined by flow cytometry on a FACSCalibur instrument, and then the percentage of expressed cell surface antigens was calculated for 10,000 gated-cell events. For sorting analyses, MSCs were incubated with a murine monoclonal primary antibody against CD146 for 10 minutes at 4°C. Both CD146-positive (CD146+) and CD146-negative (CD146−) cells were sorted to 95% purity by using a FACSVantage cell sorting system (BD Biosciences).
In Vitro Multilineage Differentiation
To assess multilineage potential, cells were incubated under specific conditions to induce differentiation into osteocytes or adipocytes. After differentiation, multilineage potential was evaluated as previously described [23]. Briefly, osteoblast or osteocyte formation was assessed by measuring the level of alkaline phosphatase (ALP; Sigma-Aldrich) activity or by von Kossa staining (Sigma-Aldrich). Quantitation of ALP or von Kossa staining was analyzed by calculating the overall percentage of cells that were positively stained. Assessment of adipocyte formation was based on the staining of accumulated lipid vacuoles with Oil Red O (Sigma-Aldrich). Lipid vacuole accumulation was quantified by calculating the percentage of stained cells in the total population. All quantitation of stained cells was performed by using i-Solution software (IMT i-Solution Inc., Daejeon, Republic of Korea, http://imt-isolution-lite.software.informer.com).
SA β-gal Staining
SA β-gal staining was used as a biomarker for senescence. A histochemical staining kit (Sigma-Aldrich) was used according to the manufacturer’s instructions to qualitatively assess SA β-gal activity, and cells were examined on an inverted microscope. To assess senescent-cell formation, the overall percentage of stained cells in the cell populations was averaged from four fields [24].
Telomerase Activity
Telomerase activity was analyzed by a telomeric repeat amplification protocol (TRAP) assay using a telomerase polymerase chain reaction (PCR) enzyme-linked immunosorbent assay kit (Roche, Mannheim, Germany, http://www.roche.com), according to manufacturer’s protocol [24]. Briefly, cells (2 × 105) were harvested for each reaction and centrifuged at 3,000g for 10 minutes at 4°C, washed twice with PBS, incubated for 20 minutes at 4°C with 200 μl lysis buffer, and centrifuged at 16,000g for 20 minutes. Telomeric repeats were added to a biotin-labeled primer during the first reaction. The PCR product was denatured, hybridized to a digoxigenin-labeled telomeric repeat-specific probe, and immobilized on a microplate. Finally, the immobilized PCR product was incubated with an anti-digoxigenin peroxidase antibody and visualized by colored-reaction product formation after substrate addition. Absorbances for the final products were measured at 450 nm by using a microplate reader. Cellular extract from 293 cells was used as a positive control (included in the kit), and the lysis reagent served as a negative control.
Western Blotting
Cell extracts were prepared in buffer containing 9.8 M urea, 4% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid, 130 mM dithiothreitol, 40 mM Tris-HCl, and 0.1% sodium dodecyl sulfate (SDS). Protein concentrations were measured by using the bicinchoninic acid kit (Sigma-Aldrich). Protein extracts (10 μg) were separated by SDS-polyacrylamide gel electrophoresis, and the resolved proteins were transferred to nitrocellulose membranes. Each membrane was incubated with antibodies against phospho-p53 (pho-p53), p21, p16, and Rb (Cell Signaling Technology, Danvers, MA, http://www.cellsignal.com); p53 and phospho-Rb (pho-Rb, Abcam, Cambridge, U.K., http://www.abcam.com); and β-actin (Sigma-Aldrich).
Quantitative Real-Time PCR and Small Interfering RNA Experiments
Quantitative real-time PCR (qPCR) was performed by using a LightCycler 480 (Roche). TaqMan probes were designed with the Universal Probe Library Assay Design Center (Roche) (supplemental online Table 2) and used to quantitatively detect mRNA for the following genes: Bmi-1, p16, Oct4, Nanog, Sox2, inhibitor of DNA binding 1 (Id1), Id2, Id3, Id4, Twist1, Twist2, and CD146. Relative expression levels of mRNAs of interest were calculated by using the comparative threshold cycle method (2-ΔΔCt), with normalization to β-actin mRNA expression. GE Dharmacon (Chicago, IL, http://dharmacon.gelifesciences.com/) designed a CD146 siRNA and scrambled siRNA for use in the siRNA experiments. siRNAs were transfected using the Dharmafect Reagent (GE Dharmacon) according to the manufacturer’s instructions. The siRNA pools consisted of four different siRNA duplexes (supplemental online Table 2).
Statistical Analysis
All data are reported as mean ± SD and were analyzed in SPSS software, version 18 (IBM, Chicago, IL, http://www-01.ibm.com). Significant differences were verified by one-way analysis of variance followed by the least-significant-difference post hoc test. p values less than .05 were considered to represent statistically significant differences.
Results
Expansion of hUCB-MSC Induced Cellular Senescence
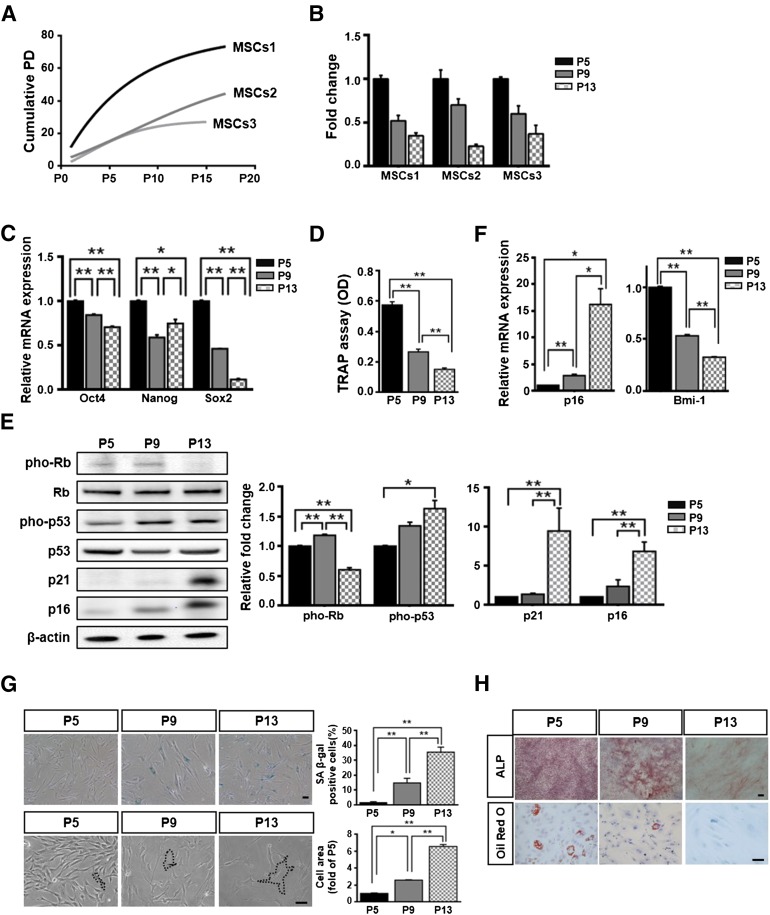
To assess the growth rate of hUCB-MSCs (n = 3), we continuously monitored cumulative PD until the cells stopped proliferating for individual lots of hUCB-MSCs. All cells eventually ceased proliferating in culture, with the number of passages being dependent on the donor (Fig. 1A). During the process of expansion, we analyzed fold-increases in cell counts at P5, P9, and P13. The fold-increases in cell growth gradually diminished from P5 to P13 (Fig. 1B). The expression of stemness markers in hUCB-MSCs, including Oct4, Nanog, and Sox2, was demonstrated at the mRNA level, which significantly decreased from P5 to P13 (Fig. 1C). The telomerase activities of hUCB-MSCs also significantly decreased during passaging (Fig. 1D). The levels of pho-p53, p16, and p21 significantly increased during passages P5 to P13, whereas the level of pho-Rb decreased between P5 and P13 (Fig. 1E). In parallel, the expression of senescence-related markers was further confirmed by quantitative real-time PCR (qPCR) analysis of p16 and Bmi-1 mRNA (Fig. 1F). Moreover, to assess the general cellular-senescence phenotypes, we monitored SA β-gal activities and cell areas during passaging from P5 or P9 to P13. SA β-gal staining and cell areas gradually increased from P5 to P13, which was associated with an enlarged and flattened cell type corresponding to the typical morphology of senescent cells (Fig. 1G). We also tested for multilineage differentiation with passaged hUCB-MSCs. ALP activity, a marker of osteoblast-associated differentiation, decreased in culture between P5 and P13. Adipogenesis, which is characterized by a decrease in lipid vacuole formation, was demonstrated by Oil Red O staining (Fig. 1H). Collectively, these data showed that the expansion of hUCB-MSCs induced the senescent phenotype.
Figure 1.
Senescence phenotype of cells following expansion. (A): Cumulative PD values from three different donors were analyzed. (B): Cell proliferation was checked by measuring fold-increases, with results normalized to the growth observed at passage (P) 5 (set as one-fold; mean ± SD; n = 2). (C): Stemness markers were quantified by quantitative real-time polymerase chain reaction (q-PCR; mean ± SD; n = 3; ∗, p < .05; ∗∗p, < .01). (D): Telomerase activity was analyzed in telomeric repeat amplification protocol assays (mean ± SD; n = 4; ∗, p < .05; ∗∗, p < .01). (E, F): Senescence-related proteins were measured by immunoblotting (E) or qPCR (F) (mean ± SD; n = 3; ∗p, < .05; ∗∗, p < .01). (C, E–F): Expression levels were normalized to β-actin, with the expression levels at P5 defined as 1. (G): The cells were stained to measure SA β-gal) expression, and quantitation was achieved by determining the percentage of SA β-gal-positive cells (upper panel; mean ± SD; n = 4; ∗, p < .05; ∗∗, p < .01). Cell areas at three passages were compared. The black lines indicate the cell margins that were drawn on the T75 flask, with the results normalized to the mean area at P5, which was defined as 1 (lower panel; mean ± SD; n = 20; ∗∗, p < .01). (H): Osteogenic and adipogenic lineages were measured by staining for alkaline phosphatase (ALP) or Oil Red O, respectively. (G, H): Scale bar = 50 μm. Abbreviations: MSC, mesenchymal stem cell; OD, optical density; PD, population doubling; pho-p53, phospho-p53; pho-Rb, phospho-retinoblastoma; SA β-gal, senescence-associated β-galactosidase; TRAP, telomeric repeat amplification protocol.
Passaging of hUCB-MSCs Downregulated Cell-Surface CD146 Expression
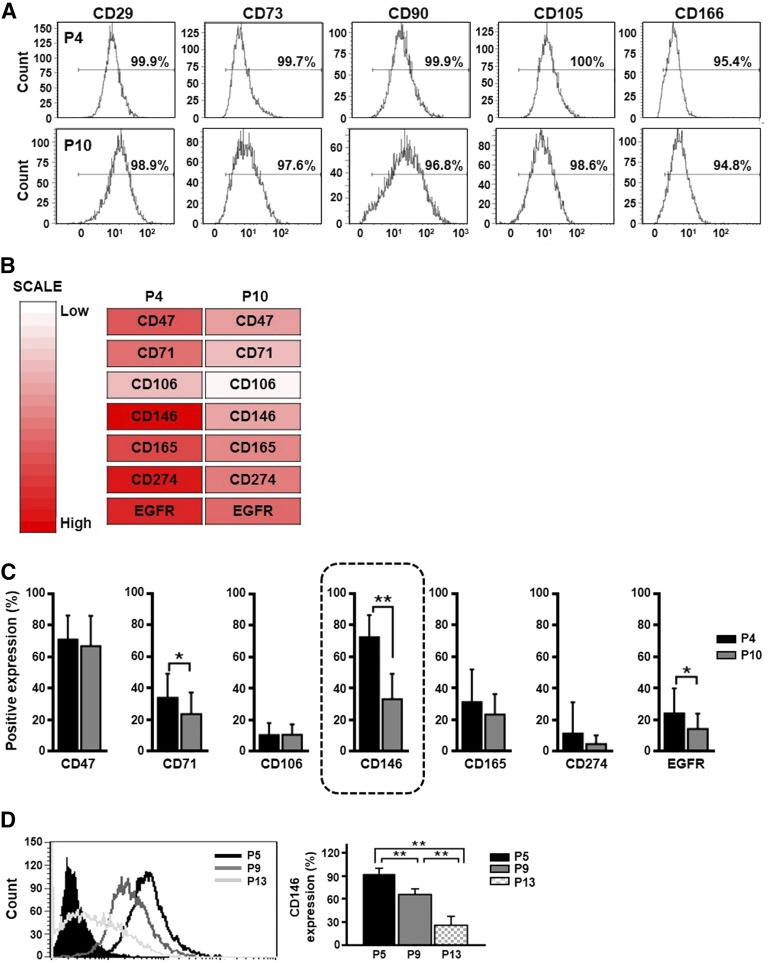
A variety of surface proteins have been proposed to govern MSC features, such as stemness, differentiation potency, and primitive selection [25]. Thus, we hypothesized that senescent hUCB-MSCs may use cell-surface proteins to actively control the aging process. To test this hypothesis, we first analyzed the cell-surface expression of CD29, CD73, CD90, CD105, and CD166 in early-stage (P4) and late-stage (P10) hUCB-MSCs, which are the minimal criteria established by the International Society for Cellular Therapy (ISCT) [26]. No change in the expression of these surface markers was observed in hUCB-MSCs between P4 and P10 (Fig. 2A). We also used a surface marker array containing antibodies against 242 CD markers to screen for expression differences during the passaging of hUCB-MSCs (n = 2) (supplemental online Table 3). As a result, we identified 7 cell-surface proteins that were markedly downregulated after passaging in culture: CD47, CD71, CD106, CD146, CD165, CD274, and EFGR (Fig. 2B). To verify these screening results, we measured the expression levels of the 7 surface proteins in early- and late-stage hUCB-MSCs from an additional 25 different donors by flow cytometry (Fig. 2C; supplemental online Table 4). The expression level of 3 markers (CD47, CD106, and CD165) did not show significant differences between the early and late stages (Fig. 2C). Although CD274 was not decreased with statistical significance, 5 lots of hUCB-MSCs showed decreased CD274 expression at P10. In particular, CD274 expression of hUCB-MSCs showed relative variation among 25 lots of hUCB-MSCs depending on the donors. CD71, CD146, and EGFR expression significantly decreased at a late stage (Fig. 2C). Notably, CD146 was the most significantly downregulated surface marker as the number of passages increased. The expression of CD146 was reduced at P10 in all hUCB-MSCs lots tested (Fig. 2C; supplemental online Table 4). In fact, the percentage of CD146-positive hUCB-MSCs gradually decreased during long-term culture, with the following percentages observed: P5 (92.3% ± 7.1%), P9 (66.0% ± 7.1%), and P13 (26.1% ± 11.4%), as shown in Figure 2D. Thus, we selected CD146 as a marker to further investigate the involvement of CD146 in the senescence of hUCB-MSCs.
Figure 2.
Screening for cell surface proteins in human umbilical cord blood-derived mesenchymal stem cells (hUCB-MSCs) with altered expression during senescence. (A): Flow cytometric analysis of both early-stage (passage [P] 4) and late-stage (P10) hUCB-MSCs based on the cell surface expression of putative MSCs markers. (B): Heat map analysis showing downregulated cell surface proteins at a late passage (P10) compared with those observed at an early passage (P4). (C): To confirm the downregulation of cell surface proteins shown in panel B, the protein expression levels of CD47, CD71, CD106, CD146, CD165, CD274, and EGFR were measured by flow cytometry at early (P4) and late (P10) stages in hUCB-MSCs from 25 different donors (mean ± SD; n = 25; ∗, p < .05; ∗∗, p < .01). CD146 expression showed the most significant decline in late-stage cells (black box). (D): CD146 expression was quantified by using flow cytometry at the indicated passages (mean ± SD; n = 3; ∗∗, p < .01). Abbreviation: EGFR, epidermal growth factor receptor.
Senescence Increased in CD146− hUCB-MSCs Compared With That Observed in CD146+ hUCB-MSCs
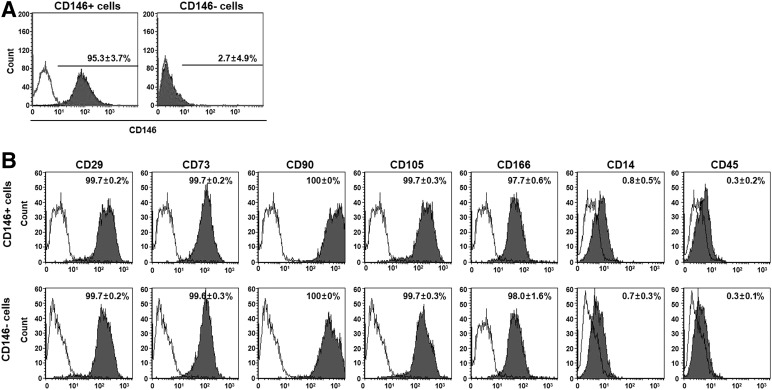
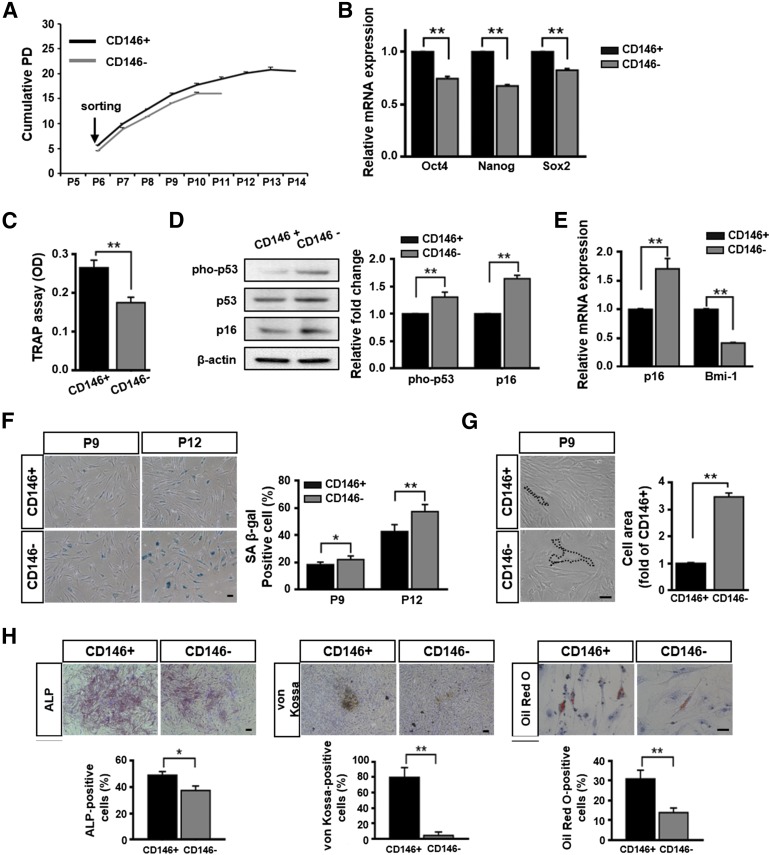
To determine whether CD146 expression is related to senescence in hUCB-MSCs, we sorted CD146+ and CD146− hUCB-MSCs by flow cytometry with an anti-CD146 antibody at P6. The sorted cells (i.e., CD146+ or CD146−) were purified to ≥95% (Fig. 3A). To analyze whether MSC surface markers were altered by CD146 expression, we tested the expression of MSC-specific antigens (CD29, CD73, CD90, CD105, CD166, CD14, and CD45) in sorted cells. Surface antigen staining showed that no difference occurred between the two populations (Fig. 3B). We also analyzed several cell-senescence phenotypes, including proliferation, stemness gene-expression levels, telomerase activity, expression levels of senescence-related proteins, SA β-gal activities, cell areas, and differentiation potentials. A comparative analysis between both populations showed that the growth rate of CD146+ cells was faster than that of CD146− cells. Specifically, the growth of CD146− cells ceased at P12, whereas CD146+ cells continued to grow until P15 (Fig. 4A). In qPCR analysis, CD146+ cells showed high expression levels of stemness genes compared with those observed in CD146− cells (Fig. 4B). Similarly, telomerase activities of CD146+ cells were significantly higher than those of CD146− cells (Fig. 4C). Western blot analysis indicated that CD146− cells showed enhanced expression of pho-p53 and p16 relative to that observed in CD146+ cells (Fig. 4D), as well as significantly augmented SA β-gal activity (Fig. 4F) and cell areas (Fig. 4G). In addition, CD146+ cells showed relatively low p16 expression but high Bmi-1 expression compared with that found in CD146− cells (Fig. 4E).
Figure 3.
Immunophenotyping results of sorted in human umbilical cord blood-derived mesenchymal stem cells (hUCB-MSCs), based on CD146 expression. (A): The percentage of CD146+ hUCB-MSCs was analyzed, and CD146+ and CD146− populations were sorted by flow cytometry. The plots shown indicate the CD146 staining profile (filled graph) versus isotype-control staining profile (unfilled graph; mean ± SD; n = 3). (B): The immunophenotypic characteristics of CD146+ and CD146− cells were examined by flow cytometry. Both CD146+ and CD146− cells were strongly positive for the MSC-specific surface markers CD29, CD73, CD90, CD105, and CD166, and they were negative for CD14 and CD45 (mean ± SD; n = 3).
Figure 4.
Senescence phenotypes of cells, based on CD146 expression. (A): Cell growth was measured by determining the cumulative PD. (B): Stemness genes were assessed by quantitative real-time polymerase chain reaction (qPCR) at passage (P) 9 (mean ± SD; n = 3; ∗∗, p < .01). (C): Telomerase activities was measured at P9 by using the TRAP assay (mean ± SD; n = 4; ∗∗, p < .01). (D, E): Expression of cell cycle inhibitors was measured by immunoblotting (D) and qPCR at P9 (E) (mean ± SD; n = 3; ∗∗, p < .01). (B, D, E): Expression levels were normalized to β-actin, with the expression levels in CD146+ cells defined as 1. (F): CD146− cells showed strong SA β-gal staining at P9 and P12 (right panel; mean ± SD; n = 4; ∗, p < .05; ∗∗, p < .01). (G): Cell areas were normalized to the mean area in CD146+ cells, which was defined as 1 at P9 (right panel; mean ± SD; n = 30; ∗∗, p < .01). The black lines indicate the cell margins. (H): In each population, multilineage differentiation was examined by ALP staining, von Kossa staining, and Oil Red O staining. Quantitative results was significantly reduced in CD146− cells at P9 (lower panel; mean ± SD; n = 3; ∗, p < .05; ∗∗, p < .01). (F–H): Scale bar = 50 μm. Abbreviations: ALP, alkaline phosphatase; OD, optical density; PD, population doubling; pho-p53, phospho-p53; SA β-gal, senescence-associated β-galactosidase; TRAP, telomeric repeat amplification protocol.
The functional effects of CD146 expression on the multilineage differentiation of hUCB-MSCs were evaluated by assessing CD146+ and CD146− cells under osteogenic- and adipogenic-specific conditions. As expected, under identical osteogenic conditions, ALP activity was significantly higher in CD146+ cells than in CD146− cells. Moreover, the level of calcium observed after von Kossa staining was also significantly higher in CD146+ cells than the levels observed in CD146− cells. Under adipogenic conditions, significantly larger lipid drops were formed within CD146+ cells than in CD146− cells, as determined by Oil Red O staining (Fig. 4H). Collectively, our results implied that CD146− cells have a higher potential for developing the senescence phenotype than do CD146+ cells.
Knockdown of CD146 Expression Accelerates Cellular Senescence in hUCB-MSCs
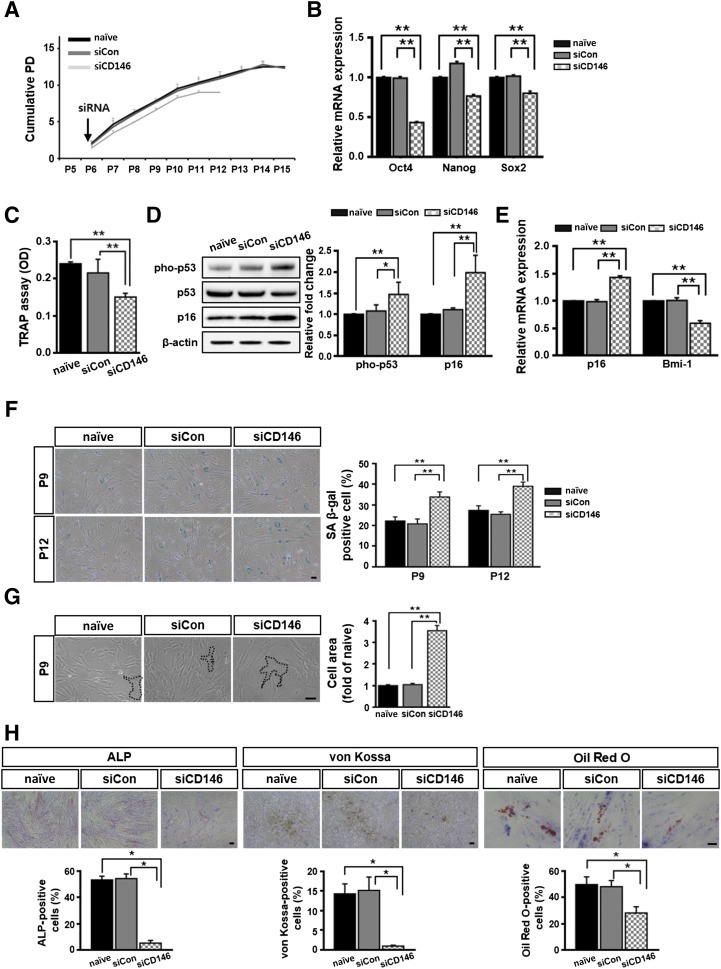
To confirm that CD146 functionally contributed to the senescence phenotype of hUCB-MSCs, we blocked CD146 expression using a siRNA (n = 3). Control experiments showed that treatment with the target siRNA also effectively inhibited CD146 expression at the protein level, as shown by flow cytometry (supplemental online Fig. 2A), with suppression being maintained for up to 28 days (supplemental online Fig. 2B). Cells with silenced CD146 expression exhibited a reduced duration of proliferation during the culture period and lower growth rates, compared with the respective measures made in naïve cells or scrambled siRNA-transfected cells (Fig. 5A). For example, the growth of CD146-silenced cells ceased at P12, whereas the other two cell populations continued to grow until P15. Thus, under the same conditions, the cumulative PD of CD146 siRNA transfectants (9.8 ± 0.3) was significantly shorter than that of naïve (15.0 ± 0.7) or scrambled siRNA (14.8 ± 0.5) transfectants (Fig. 5A). CD146 knockdown in hUCB-MSCs also resulted in decreased expression levels of stemness genes, such as Oct-4, Nanog, and Sox2 (Fig. 5B). In particular, CD146 suppression significantly reduced the telomerase activity of hUCB-MSCs to 40% of the level found in the control groups (Fig. 5C). At the protein level, CD146 silencing significantly enhanced pho-p53 production and p16 expression compared with their relative expression levels in naïve or scrambled siRNA transfectants at P9 (Fig. 5D). As expected, CD146 siRNA treated cells showed increased p16 mRNA expression levels and inhibited Bmi-1 mRNA expression (Fig. 5E). In addition, SA β-gal activities (Fig. 5F) and cell areas (Fig. 5G) were significantly increased in CD146 siRNA transfectants. CD146 silencing significantly reduced both osteogenesis and adipogenesis, as confirmed by ALP staining, von Kossa staining, and Oil Red O staining (Fig. 5H). Taken together, these data demonstrate that inhibiting CD146 expression accelerated the senescence phenotype in hUCB-MSCs.
Figure 5.
CD146 knockdown in human umbilical cord blood-derived mesenchymal stem cells accelerates the senescence process. (A): Cell growth was monitored by measuring cumulative PD. (B): Cells were assessed for their expression of stemness genes by quantitative real-time polymerase chain reaction (qPCR) at passage (P) 9 (mean ± SD; n = 3; ∗∗, p < .01). (C): Telomerase activities was measured at P9 using the telomerase PCR enzyme-linked immunosorbent assay kit (mean ± SD; n = 4; ∗∗, p < .01). (D): Expression of the cell cycle inhibitors was measured by immunoblotting at P9, with β-actin serving as a loading control fold (right panel; mean ± SD; n = 4; ∗, p < .05; ∗∗, p < .01). (E): qPCR data showing the mRNA expression levels (p16 and Bmi-1) at P9 (mean ± SD; n = 3; ∗∗, p < .01). (B, D, E): Expression levels were normalized to β-actin, with the expression levels in naïve defined as 1. (F): SA β-gal-positive cells were measured at P9 and P12. Results are shown as mean ± SD (n = 4; ∗∗, p < .01). (G): Cell areas compared at P9, which was normalized to the mean area in naïve cells, defined as 1 (mean ± SD; n = 25; ∗∗, p < .01). The black lines indicate the cell margins. (H): Multilineage differentiation was assessed by quantifying the percentage of positively stained cells at P9 (mean ± SD; n = 3; ∗, p < .05). (F–H): Scale bar = 50 μm. Abbreviations: OD, optical density; PD, population doubling; pho-p53, phospho-p53; SA β-gal, senescence-associated β-galactosidase; siCD146, CD146 small interfering RNA; siCon, small interfering scrambled RNA; siRNA, small interfering RNA; TRAP, telomeric repeat amplification protocol.
CD146 Suppression in hUCB-MSCs Downregulated Senescence Regulators
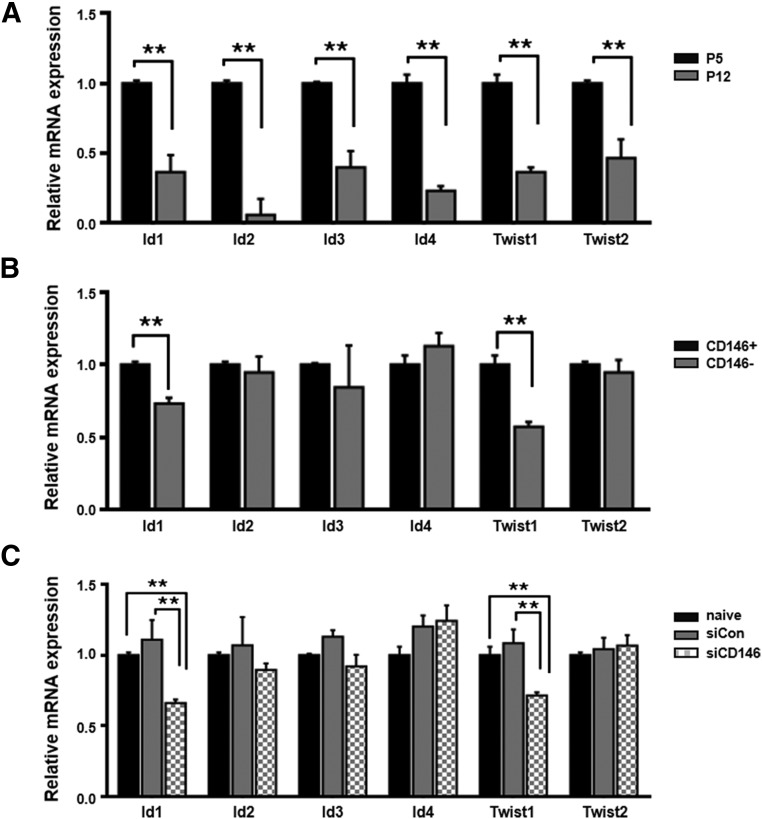
To further examine the mechanistic relationship between CD146 expression and cellular senescence, we investigated whether CD146 expression regulates the expression of Id1- or Twist-family genes, which can regulate the cellular senescence process [27]. During the onset of cellular senescence in hUCB-MSCs, the expression of Id- and Twist-family genes significantly decreased (0.2–0.6-fold lower expression at P12 than at P5), which was confirmed by qPCR (Fig. 6A). After sorting of hUCB-MSCs for CD146 expression, Id1 and Twist1 expression was lower in the CD146− subset than in CD146+ cells (Fig. 6B). To confirm these observations, we also analyzed the expression of Id- and Twist-family genes, with or without CD146 siRNA treatment. We found that Id1 and Twist1 expression levels were significantly decreased only in CD146 siRNA transfectants and not in the control groups (naïve and scramble siRNA; Fig. 6C). Collectively, these data imply that CD146 expression might be involved in maintaining expression of the Id1 and Twist1 transcription factors that are related to cellular senescence regulation.
Figure 6.
Involvement of CD146 in Id and Twist-family gene expression. (A): The expression of Id- and Twist-family genes was analyzed after human umbilical cord blood-derived mesenchymal stem cell (hUCB-MSC) expansion (both at passage [P] 5 and P12, mean ± SD; n = 3; ∗∗, p < .01). The expression levels of all genes were normalized to that of β-actin in hUCB-MSCs at P5, which was defined as 1-fold expression. (B): CD146+ and CD146− hUCB-MSCs were sorted by flow cytometry and examined for Id- or Twist-family gene expression at P9 (mean ± SD; n = 3; ∗∗, p < .01). The expression levels of all genes were normalized to that of β-actin in CD146+ cells, which was defined as 1-fold expression. (C): The gene-expression levels of Id1 and Twist1 significantly decreased siCD146 compared with the expression levels in naïve and siCon-treated cells (mean ± SD; n = 3; ∗∗, p < .01). The expression levels of all genes were normalized to that of β-actin in naïve cells, which was defined as 1-fold expression. Abbreviations: siCD146, CD146-specific siRNA; siCon, control siRNA; siRNA, small-interfering RNA.
Discussion
Currently, the cell-surface markers suggested by the ISCT are the most widely used markers, not only as the minimal criteria for defining or characterizing MSCs but also as quality-control markers for producing functional MSCs [26, 28]. Indeed, growing evidence has suggested that these surface proteins are useful biomarkers for predicting various biological characteristics of MSCs. For instance, our previous results indicated that endoglin (CD105) is a useful marker for characterizing the differentiation status of hUCB-MSCs [29]. Another report demonstrated that neural ganglioside (GD2) is associated with neural differentiation of hUCB-MSCs [23]. Moreover, several surface markers, including CD140b [30, 31] and CD271 [25, 32, 33], have been suggested as new positive markers for identifying and isolating MSCs. However, there have been few reports of markers that are functionally related to the senescence status of MSCs, and no direct evidence has previously been provided that CD146 expression is related to the cellular senescence of MSCs.
In the present study, we sought to identify cell-surface senescence-marker proteins in hUCB-MSCs during long-term culture through a large-scale, FACS-screening approach. Because we were interested in negative regulators of cellular senescence, we mainly focused on surface proteins with decreased expression during long-term culturing of hUCB-MSCs. Consequently, during screening we identified 7 surface markers that were downregulated at a late passage (P10). Because individual variations can result from unidentified factors in each hUCB-MSC lot used for screening, we validated the results with these candidate markers with 25 lots of hUCB-MSCs. After validation, we found that CD71, CD146, and EGFR significantly decreased during passaging. In particular, among these 3 validated markers, only CD146 was markedly and reproducibly decreased after passaging all lots of hUCB-MSCs to P10; hence, CD146 was selected for further investigation, specifically for its role in the senescence process. Although the passage-dependent reduction of CD274 expression in hUCB-MSCs did not reach statistical significance, 5 of 25 lots of hUCB-MSCs showed dramatically decreased CD274 expression at a late passage (supplemental online Table 4). Because CD274 has been reported to be involved in the hypoimmunogenic features of MSCs [34, 35], these late-passage cells may not be suitable for transplantation as a result of the lack of these properties. However, the overall expression levels of CD274 were in fact relatively low both in early- and late-passage naïve hUCB-MSCs. Similarly, the results from several studies demonstrated that naïve MSCs express low levels of CD274 but that proinflammatory factors such as interferon-γ stimulated CD274 expression in MSCs [36–38]. Thus, the induction potential of CD274 expression may decrease in late passages of hUCB-MSCs after stimulation with proinflammatory conditions, a possibility that future studies should address.
Here, we provide evidence that CD146 expression is markedly decreased in hUCB-MSCs passaged through senescence. Similarly, results from a recent study showed that the expression of vascular cell adhesion molecule-1 (CD106) is markedly reduced in senescent BM-MSCs, which results in low homing activity [39]. Although CD106 is also expressed in hUCB-MSCs, its expression did not change significantly during long-term culture. We found for the first time that CD146 siRNA-transfected hUCB-MSCs and the natural CD146− subset of hUCB-MSCs exhibit accelerated cellular senescence phenotypes. In addition, we further demonstrated the direct role of CD146 in regulating the cellular senescence of hUCB-MSCs. These results indicate that CD146 acts as a negative regulator of cellular senescence. Collectively, our results suggest that CD146 can be a useful alternative marker for predicting the senescence status of hUCB-MSCs.
To use CD146 as a marker for evaluating the senescence status of hUCB-MSCs, it is important to define the level of CD146 expression that occurs at senescence. Thus, we determined the cutoff CD146 expression level for defining nonsenescent hUCB-MSCs. When the proliferation of each of the 25 lots of hUCB-MSCs ceased, we measured CD146 expression at that passage by FACS analysis (supplemental online Fig. 3). Although the hUCB-MSC lots showed different passage numbers before proliferation stopped or senescence was induced, we suggest that predictions for senescence of hUCB-MSCs are possible according to this cutoff value. In addition, because freezing and thawing processes are necessary for MSC-based therapy, understanding the stability of markers during these processes is also required. Thus, we also examined the effects of freezing and thawing on CD146 expression in hUCB-MSCs, and we found that CD146 expression in hUCB-MSCs was not altered (supplemental online Fig. 4) by either process. Collectively, these results suggest the potential for using CD146 in evaluating quality-control assessments of MSC-based therapy.
Originally, CD146 was characterized as a cell-adhesion molecule that was useful as a biomarker for endothelial lineages, and it was found to play key roles in various biological processes, including cell growth, migration, and angiogenesis [40, 41]. Results from several studies have shown that CD146 augments motility, metastasis, and tumorigenesis of cancer cells, including melanoma and breast cancer cells [42, 43]. Recently, growing evidence has indicated that CD146 expression is related to the multilineage differentiation potential, proliferation, and stemness of MSCs [44–49]. Indeed, we also observed that CD146 downregulation led to decreases in the osteogenic and adipogenic differentiation potential of hUCB-MSCs, implying that such multilineage differentiation potential depends on CD146 expression levels in hUCB-MSCs (supplemental online Figs. 5 and 6). Furthermore, we showed that CD146 regulates hUCB-MSC proliferation, and stemness markers in hUCB-MSCs were decreased by CD146 suppression. Our data demonstrated that cellular senescence following long-term culture reduced the biological potentials of hUCB-MSCs, which were further accelerated by CD146 suppression. Thus, we conclude that CD146 regulates the senescence of hUCB-MSCs, which affects the osteogenic and adipogenic differentiation, proliferation, and stemness of hUCB-MSCs.
Because unsorted hUCB-MSCs have a subpopulation of CD146− cells and CD146 plays a role in stem cell phenotypes, sorted CD146+ hUCB-MSCs showed high levels of stemness markers and differentiation potentials compared with unsorted naïve hUCB-MSCs (supplemental online Fig. 7). These results suggest that hUCB-MSCs contain a subpopulation of cells expressing stemness-related factors such as CD146 that show increased stem cell characteristics compared with other cells within the population, which further indicates that hUCB-MSCs are heterogeneous. In particular, according to the results of several studies, CD146 expression distinguishes MSCs from non-MSCs in BM cells, such as fibroblasts, as well as bone cells that express several MSC markers, but not CD146 [25, 50]. Furthermore, several research groups have suggested that CD146 not only is an enrichment marker for MSCs but also serves as an additional positive marker for defining MSCs [25]. In the context of MSC heterogeneity, the overall population of MSCs includes the more committed cells with basic features of MSCs; ES-like cells with high expression of Oct4 and Sox2; and non-MSCs, such as fibroblast cells [51]. In fact, this MSC heterogeneity has led to limited and variable therapeutic outcomes in clinical trials [52–54]. Several research groups have endeavored to enrich MSCs using factors associated with stemness or multipotential differentiation to increase their therapeutic effects [55, 56]. Thus, CD146 can probably be used to enrich MSCs and improve their therapeutic potentials by reducing MSC senescence and heterogeneity.
In this study, we clearly demonstrated that CD146 expression is closely related to the cellular senescence of hUCB-MSCs. To investigate the downstream CD146-dependent mechanisms underlying cellular senescence, we also examined the expression of helix-loop-helix transcription factors, such Id1 and Twist1, during long-term culturing of hUCB-MSCs. Results from a previous study demonstrated that Id1 and Twist1 expression are downregulated during the expansion of BM-MSCs [27]. Similarly, we observed that the expression of Id- and Twist-family proteins decreased during passaging through senescence. These data indicate that Id- and Twist-family proteins may govern the senescence process of hUCB-MSCs. In agreement, data from previous reports showed that Id1 and Twist1 serve inhibitory functions during senescence in melanoma cells or MSCs by regulating key cell-cycle regulators, such as p53 and p16 [57–60].
Importantly, we also found that CD146 downregulation resulted in decreased Id1 and Twist1 mRNA levels. Although a previous report showed that CD146 regulates Id1 expression during melanoma progression [61], a link between CD146 and Twist1 expression had not been previously demonstrated. Thus, our results provide new, indirect evidence that Twist1 is a downstream target protein of CD146 during the development of cellular senescence in hUCB-MSCs. Interestingly, the results of a recent study suggested a post-translational mechanism for Id1 regulation that involved Smurf-2-mediated ubiquitination and Id1 degradation, which promoted cellular senescence by increasing p16 activity [62]. Furthermore, recent data showed that high Bmi-1 expression in adult stem cells protects them against senescence by suppressing senescence-related genes, including p16 [63, 64]. We observed a novel correlation between Bmi-1 and CD146, in that the expression level of Bmi-1 decreased following CD146 suppression. Collectively, our findings support an alternative mechanism whereby CD146 may delay cellular senescence via the transcriptional regulation of Bmi-1, Id1, or Twist1. However, further studies, which are in progress, are necessary to clarify the downstream mechanism(s) underlying the negative regulation of cellular senescence by CD146.
Conclusion
Our data showed that CD146 expression is gradually decreased during the long-term expansion of hUCB-MSCs in culture and that enforced CD146 downregulation accelerated senescence in hUCB-MSCs, which affected their multilineage differentiation potential, growth, and stemness. Collectively, these data indicate a possible role for CD146 in inhibiting senescence in hUCB-MSCs, which may occur via the regulation of Bmi-1, Id1, and Twist1 expression. Thus, we suggest that CD146 is a novel marker for predicting senescence in hUCB-MSCs, and it could be valuable in quality-control assessments and for improving the therapeutic potential of hUCB-MSC-based therapy.
Supplementary Material
Acknowledgment
This work was supported by Grant HI12C1821 (A121968) from the Korean Healthcare Technology R&D Project sponsored by the Ministry of Health, Welfare & Family Affairs, Republic of Korea.
Author Contributions
H.J.J.: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing; J.H.K., M.K., and Y.K.B.: collection and/or assembly of data; S.J.C. and W.O.: data analysis and interpretation; Y.S.Y.: data analysis and interpretation, financial support; H.B.J.: conception and design, data analysis and interpretation, manuscript writing, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
S.J.C. has compensated employment. W.O. has compensated employment, is an intellectual property and patent holder, and has stock options with MEDIPOST, Co., Ltd. The other authors indicated no potential conflicts of interest.
References
- 1.Lee OK, Kuo TK, Chen WM, et al. Isolation of multipotent mesenchymal stem cells from umbilical cord blood. Blood. 2004;103:1669–1675. doi: 10.1182/blood-2003-05-1670. [DOI] [PubMed] [Google Scholar]
- 2.Yang SE, Ha CW, Jung M, et al. Mesenchymal stem/progenitor cells developed in cultures from UC blood. Cytotherapy. 2004;6:476–486. doi: 10.1080/14653240410005041. [DOI] [PubMed] [Google Scholar]
- 3.Lee M, Jeong SY, Ha J, et al. Low immunogenicity of allogeneic human umbilical cord blood-derived mesenchymal stem cells in vitro and in vivo. Biochem Biophys Res Commun. 2014;446:983–989. doi: 10.1016/j.bbrc.2014.03.051. [DOI] [PubMed] [Google Scholar]
- 4.Kim JY, Kim DH, Kim DS, et al. Galectin-3 secreted by human umbilical cord blood-derived mesenchymal stem cells reduces amyloid-beta42 neurotoxicity in vitro. FEBS Lett. 2010;584:3601–3608. doi: 10.1016/j.febslet.2010.07.028. [DOI] [PubMed] [Google Scholar]
- 5.Kim JY, Kim DH, Kim JH, et al. Soluble intracellular adhesion molecule-1 secreted by human umbilical cord blood-derived mesenchymal stem cell reduces amyloid-β plaques. Cell Death Differ. 2012;19:680–691. doi: 10.1038/cdd.2011.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeong SY, Kim DH, Ha J, et al. Thrombospondin-2 secreted by human umbilical cord blood-derived mesenchymal stem cells promotes chondrogenic differentiation. Stem Cells. 2013;31:2136–2148. doi: 10.1002/stem.1471. [DOI] [PubMed] [Google Scholar]
- 7.Sethe S, Scutt A, Stolzing A. Aging of mesenchymal stem cells. Ageing Res Rev. 2006;5:91–116. doi: 10.1016/j.arr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 8.Bustos ML, Huleihel L, Kapetanaki MG, et al. Aging mesenchymal stem cells fail to protect because of impaired migration and antiinflammatory response. Am J Respir Crit Care Med. 2014;189:787–798. doi: 10.1164/rccm.201306-1043OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lans H, Hoeijmakers JH. Cell biology: Ageing nucleus gets out of shape. Nature. 2006;440:32–34. doi: 10.1038/440032a. [DOI] [PubMed] [Google Scholar]
- 10.Kim YM, Byun HO, Jee BA, et al. Implications of time-series gene expression profiles of replicative senescence. Aging Cell. 2013;12:622–634. doi: 10.1111/acel.12087. [DOI] [PubMed] [Google Scholar]
- 11.Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432:307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- 12.Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 13.Brown JP, Wei W, Sedivy JM. Bypass of senescence after disruption of p21CIP1/WAF1 gene in normal diploid human fibroblasts. Science. 1997;277:831–834. doi: 10.1126/science.277.5327.831. [DOI] [PubMed] [Google Scholar]
- 14.Zhou S, Greenberger JS, Epperly MW, et al. Age-related intrinsic changes in human bone-marrow-derived mesenchymal stem cells and their differentiation to osteoblasts. Aging Cell. 2008;7:335–343. doi: 10.1111/j.1474-9726.2008.00377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu Z, Jiang J, Tan W, et al. p53/p21 Pathway involved in mediating cellular senescence of bone marrow-derived mesenchymal stem cells from systemic lupus erythematosus patients. Clin Dev Immunol. 2013;2013:134243. doi: 10.1155/2013/134243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harbour JW, Dean DC. The Rb/E2F pathway: Expanding roles and emerging paradigms. Genes Dev. 2000;14:2393–2409. doi: 10.1101/gad.813200. [DOI] [PubMed] [Google Scholar]
- 17.Ito T, Sawada R, Fujiwara Y, et al. FGF-2 suppresses cellular senescence of human mesenchymal stem cells by down-regulation of TGF-beta2. Biochem Biophys Res Commun. 2007;359:108–114. doi: 10.1016/j.bbrc.2007.05.067. [DOI] [PubMed] [Google Scholar]
- 18.Ben-Porath I, Weinberg RA. The signals and pathways activating cellular senescence. Int J Biochem Cell Biol. 2005;37:961–976. doi: 10.1016/j.biocel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 19.Lin SP, Chiu FY, Wang Y, et al. RB maintains quiescence and prevents premature senescence through upregulation of DNMT1 in mesenchymal stromal cells. Stem Cell Rep. 2014;3:975–986. doi: 10.1016/j.stemcr.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurz DJ, Decary S, Hong Y, et al. Senescence-associated (beta)-galactosidase reflects an increase in lysosomal mass during replicative ageing of human endothelial cells. J Cell Sci. 2000;113:3613–3622. doi: 10.1242/jcs.113.20.3613. [DOI] [PubMed] [Google Scholar]
- 21.Jin HJ, Bae YK, Kim M, et al. Comparative analysis of human mesenchymal stem cells from bone marrow, adipose tissue, and umbilical cord blood as sources of cell therapy. Int J Mol Sci. 2013;14:17986–18001. doi: 10.3390/ijms140917986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christodoulou I, Kolisis FN, Papaevangeliou D, et al. Comparative evaluation of human mesenchymal stem cells of fetal (Wharton's jelly) and adult (adipose tissue) origin during prolonged in vitro expansion: Considerations for cytotherapy. Stem Cells Int. 2013;2013:246134. doi: 10.1155/2013/246134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin HJ, Nam HY, Bae YK, et al. GD2 expression is closely associated with neuronal differentiation of human umbilical cord blood-derived mesenchymal stem cells. Cell Mol Life Sci. 2010;67:1845–1858. doi: 10.1007/s00018-010-0292-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeoung JY, Nam HY, Kwak J, et al. A decline in Wnt3a signaling is necessary for mesenchymal stem cells to proceed to replicative senescence. Stem Cells Dev. 2015;24:973–982. doi: 10.1089/scd.2014.0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lv FJ, Tuan RS, Cheung KM, et al. Concise review: the surface markers and identity of human mesenchymal stem cells. Stem Cells. 2014;32:1408–1419. doi: 10.1002/stem.1681. [DOI] [PubMed] [Google Scholar]
- 26.Dominici M, Le Blanc K, Mueller I, et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 27.Isenmann S, Arthur A, Zannettino AC, et al. TWIST family of basic helix-loop-helix transcription factors mediate human mesenchymal stem cell growth and commitment. Stem Cells. 2009;27:2457–2468. doi: 10.1002/stem.181. [DOI] [PubMed] [Google Scholar]
- 28.Sensebé L, Bourin P, Tarte K. Good manufacturing practices production of mesenchymal stem/stromal cells. Hum Gene Ther. 2011;22:19–26. doi: 10.1089/hum.2010.197. [DOI] [PubMed] [Google Scholar]
- 29.Jin HJ, Park SK, Oh W, et al. Down-regulation of CD105 is associated with multi-lineage differentiation in human umbilical cord blood-derived mesenchymal stem cells. Biochem Biophys Res Commun. 2009;381:676–681. doi: 10.1016/j.bbrc.2009.02.118. [DOI] [PubMed] [Google Scholar]
- 30.Bühring HJ, Battula VL, Treml S, et al. Novel markers for the prospective isolation of human MSC. Ann N Y Acad Sci. 2007;1106:262–271. doi: 10.1196/annals.1392.000. [DOI] [PubMed] [Google Scholar]
- 31.Baer PC, Kuçi S, Krause M, et al. Comprehensive phenotypic characterization of human adipose-derived stromal/stem cells and their subsets by a high throughput technology. Stem Cells Dev. 2013;22:330–339. doi: 10.1089/scd.2012.0346. [DOI] [PubMed] [Google Scholar]
- 32.Battula VL, Treml S, Bareiss PM, et al. Isolation of functionally distinct mesenchymal stem cell subsets using antibodies against CD56, CD271, and mesenchymal stem cell antigen-1. Haematologica. 2009;94:173–184. doi: 10.3324/haematol.13740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones E, English A, Churchman SM, et al. Large-scale extraction and characterization of CD271+ multipotential stromal cells from trabecular bone in health and osteoarthritis: implications for bone regeneration strategies based on uncultured or minimally cultured multipotential stromal cells. Arthritis Rheum. 2010;62:1944–1954. doi: 10.1002/art.27451. [DOI] [PubMed] [Google Scholar]
- 34.Tipnis S, Viswanathan C, Majumdar AS. Immunosuppressive properties of human umbilical cord-derived mesenchymal stem cells: role of B7-H1 and IDO. Immunol Cell Biol. 2010;88:795–806. doi: 10.1038/icb.2010.47. [DOI] [PubMed] [Google Scholar]
- 35.Yan Z, Zhuansun Y, Liu G, et al. Mesenchymal stem cells suppress T cells by inducing apoptosis and through PD-1/B7-H1 interactions. Immunol Lett. 2014;162(1 Pt A):248–255. doi: 10.1016/j.imlet.2014.09.013. [DOI] [PubMed] [Google Scholar]
- 36.Sheng H, Wang Y, Jin Y, et al. A critical role of IFNgamma in priming MSC-mediated suppression of T cell proliferation through up-regulation of B7-H1. Cell Res. 2008;18:846–857. doi: 10.1038/cr.2008.80. [DOI] [PubMed] [Google Scholar]
- 37.Jang IK, Yoon HH, Yang MS, et al. B7-H1 inhibits T cell proliferation through MHC class II in human mesenchymal stem cells. Transplant Proc. 2014;46:1638–1641. doi: 10.1016/j.transproceed.2013.12.059. [DOI] [PubMed] [Google Scholar]
- 38.Wang H, Qi F, Dai X, et al. Requirement of B7-H1 in mesenchymal stem cells for immune tolerance to cardiac allografts in combination therapy with rapamycin. Transpl Immunol. 2014;31:65–74. doi: 10.1016/j.trim.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 39.Jung EM, Kwon O, Kwon KS, et al. Evidences for correlation between the reduced VCAM-1 expression and hyaluronan synthesis during cellular senescence of human mesenchymal stem cells. Biochem Biophys Res Commun. 2011;404:463–469. doi: 10.1016/j.bbrc.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 40.Kang Y, Wang F, Feng J, et al. Knockdown of CD146 reduces the migration and proliferation of human endothelial cells. Cell Res. 2006;16:313–318. doi: 10.1038/sj.cr.7310039. [DOI] [PubMed] [Google Scholar]
- 41.Ouhtit A, Gaur RL, Abd Elmageed ZY, et al. Towards understanding the mode of action of the multifaceted cell adhesion receptor CD146. Biochim Biophys Acta. 2009;1795:130–136. doi: 10.1016/j.bbcan.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 42.Mills L, Tellez C, Huang S, et al. Fully human antibodies to MCAM/MUC18 inhibit tumor growth and metastasis of human melanoma. Cancer Res. 2002;62:5106–5114. [PubMed] [Google Scholar]
- 43.Zeng G, Cai S, Liu Y, et al. METCAM/MUC18 augments migration, invasion, and tumorigenicity of human breast cancer SK-BR-3 cells. Gene. 2012;492:229–238. doi: 10.1016/j.gene.2011.10.024. [DOI] [PubMed] [Google Scholar]
- 44.Russell KC, Phinney DG, Lacey MR, et al. In vitro high-capacity assay to quantify the clonal heterogeneity in trilineage potential of mesenchymal stem cells reveals a complex hierarchy of lineage commitment. Stem Cells. 2010;28:788–798. doi: 10.1002/stem.312. [DOI] [PubMed] [Google Scholar]
- 45.Espagnolle N, Guilloton F, Deschaseaux F, et al. CD146 expression on mesenchymal stem cells is associated with their vascular smooth muscle commitment. J Cell Mol Med. 2014;18:104–114. doi: 10.1111/jcmm.12168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang X, Hirai M, Cantero S, et al. Isolation and characterization of mesenchymal stem cells from human umbilical cord blood: reevaluation of critical factors for successful isolation and high ability to proliferate and differentiate to chondrocytes as compared to mesenchymal stem cells from bone marrow and adipose tissue. J Cell Biochem. 2011;112:1206–1218. doi: 10.1002/jcb.23042. [DOI] [PubMed] [Google Scholar]
- 47.Hagmann S, Frank S, Gotterbarm T, et al. Fluorescence activated enrichment of CD146+ cells during expansion of human bone-marrow derived mesenchymal stromal cells augments proliferation and GAG/DNA content in chondrogenic media. BMC Musculoskelet Disord. 2014;15:322. doi: 10.1186/1471-2474-15-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsang WP, Shu Y, Kwok PL, et al. CD146+ human umbilical cord perivascular cells maintain stemness under hypoxia and as a cell source for skeletal regeneration. PLoS One. 2013;8:e76153. doi: 10.1371/journal.pone.0076153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ulrich C, Abruzzese T, Maerz JK, et al. Human placenta-derived CD146-positive mesenchymal stromal cells display a distinct osteogenic differentiation potential. Stem Cells Dev. 2015;24:1558–1569. doi: 10.1089/scd.2014.0465. [DOI] [PubMed] [Google Scholar]
- 50.Sacchetti B, Funari A, Michienzi S, et al. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell. 2007;131:324–336. doi: 10.1016/j.cell.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 51.Kuroda Y, Kitada M, Wakao S, et al. Unique multipotent cells in adult human mesenchymal cell populations. Proc Natl Acad Sci USA. 2010;107:8639–8643. doi: 10.1073/pnas.0911647107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Phinney DG. Biochemical heterogeneity of mesenchymal stem cell populations: Clues to their therapeutic efficacy. Cell Cycle. 2007;6:2884–2889. doi: 10.4161/cc.6.23.5095. [DOI] [PubMed] [Google Scholar]
- 53.Wagner W. Senescence is heterogeneous in mesenchymal stromal cells: Kaleidoscopes for cellular aging. Cell Cycle. 2010;9:2923–2924. doi: 10.4161/cc.9.15.12741. [DOI] [PubMed] [Google Scholar]
- 54.Phinney DG. Functional heterogeneity of mesenchymal stem cells: Implications for cell therapy. J Cell Biochem. 2012;113:2806–2812. doi: 10.1002/jcb.24166. [DOI] [PubMed] [Google Scholar]
- 55.Gang EJ, Bosnakovski D, Figueiredo CA, et al. SSEA-4 identifies mesenchymal stem cells from bone marrow. Blood. 2007;109:1743–1751. doi: 10.1182/blood-2005-11-010504. [DOI] [PubMed] [Google Scholar]
- 56.Baustian C, Hanley S, Ceredig R. Isolation, selection and culture methods to enhance clonogenicity of mouse bone marrow derived mesenchymal stromal cell precursors. Stem Cell Res Ther. 2015;6:151. doi: 10.1186/s13287-015-0139-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee TK, Man K, Ling MT, et al. Over-expression of Id-1 induces cell proliferation in hepatocellular carcinoma through inactivation of p16INK4a/RB pathway. Carcinogenesis. 2003;24:1729–1736. doi: 10.1093/carcin/bgg145. [DOI] [PubMed] [Google Scholar]
- 58.Liu H, Jia D, Li A, et al. p53 regulates neural stem cell proliferation and differentiation via BMP-Smad1 signaling and Id1. Stem Cells Dev. 2013;22:913–927. doi: 10.1089/scd.2012.0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Piccinin S, Tonin E, Sessa S, et al. A “twist box” code of p53 inactivation: Twist box: p53 interaction promotes p53 degradation. Cancer Cell. 2012;22:404–415. doi: 10.1016/j.ccr.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 60.Cakouros D, Isenmann S, Cooper L, et al. Twist-1 induces Ezh2 recruitment regulating histone methylation along the Ink4A/Arf locus in mesenchymal stem cells. Mol Cell Biol. 2012;32:1433–1441. doi: 10.1128/MCB.06315-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zigler M, Villares GJ, Dobroff AS, et al. Expression of Id-1 is regulated by MCAM/MUC18: A missing link in melanoma progression. Cancer Res. 2011;71:3494–3504. doi: 10.1158/0008-5472.CAN-10-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kong Y, Cui H, Zhang H. Smurf2-mediated ubiquitination and degradation of Id1 regulates p16 expression during senescence. Aging Cell. 2011;10:1038–1046. doi: 10.1111/j.1474-9726.2011.00746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Molofsky AV, Pardal R, Iwashita T, et al. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Park IK, Morrison SJ, Clarke MF. Bmi1, stem cells, and senescence regulation. J Clin Invest. 2004;113:175–179. doi: 10.1172/JCI20800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.