The potential effects of stem cell-released microvesicles (MVs) in proangiogenic therapy were explored. MVs were released from adipose-derived stem cells (ASCs) and were able to increase the migration and tube formation of human umbilical vein endothelial cells. MVs from ASCs, particularly from endothelial differentiation medium-preconditioned ASCs, were found to have elevated levels of microRNA-31 and to promote angiogenesis.
Keywords: Angiogenesis, miRNA, Adipose stem cell, Endothelial cell, Microvesicle
Abstract
Cell secretion is an important mechanism for stem cell-based therapeutic angiogenesis, along with cell differentiation to vascular endothelial cells or smooth muscle cells. Cell-released microvesicles (MVs) have been recently implicated to play an essential role in intercellular communication. The purpose of this study was to explore the potential effects of stem cell-released MVs in proangiogenic therapy. We observed for the first time that MVs were released from adipose-derived stem cells (ASCs) and were able to increase the migration and tube formation of human umbilical vein endothelial cells (HUVECs). Endothelial differentiation medium (EDM) preconditioning of ASCs upregulated the release of MVs and enhanced the angiogenic effect of the released MVs in vitro. RNA analysis revealed that microRNA was enriched in ASC-released MVs and that the level of microRNA-31 (miR-31) in MVs was notably elevated upon EDM-preconditioning of MV-donor ASCs. Further studies exhibited that miR-31 in MVs contributed to the migration and tube formation of HUVECs, microvessel outgrowth of mouse aortic rings, and vascular formation of mouse Matrigel plugs. Moreover, factor-inhibiting HIF-1, an antiangiogenic gene, was identified as the target of miR-31 in HUVECs. Our findings provide the first evidence that MVs from ASCs, particularly from EDM-preconditioned ASCs, promote angiogenesis and the delivery of miR-31 may contribute the proangiogenic effect.
Significance
This study provides the evidence that microvesicles (MVs) from adipose-derived stem cells (ASCs), particularly from endothelial differentiation medium (EDM)-preconditioned ASCs, promote angiogenesis. An underlying mechanism of the proangiogenesis may be the delivery of microRNA-31 via MVs from ASCs to vascular endothelial cells in which factor-inhibiting HIF-1 is targeted and suppressed. The study findings reveal the role of MVs in mediating ASC-induced angiogenesis and suggest a potential MV-based angiogenic therapy for ischemic diseases.
Introduction
Ischemic cardio- and cerebrovascular diseases continue to represent a critical and growing cause of morbidity and mortality despite the significant advances in medical, interventional, and surgical therapies for these cases. To address these concerns, the science of therapeutic angiogenesis has been evolving for the past 2 decades [1]. The protein/gene approach, including growth factors and cytokines, and the subsequent stem/progenitor cell approach have been extensively investigated in the therapy for ischemic heart disease [2, 3]. Adipose-derived stem cells (ASCs) are an attractive vehicle for cell-based therapeutics, with advantages of easy acquisition, as well as more proliferative potential and less histocompatibility antigen [4]. It has been reported that ASCs either originate from [5] or can assume the role of [6, 7] pericytes during treatment of the microvasculature. Implantation of ASCs, particularly endothelial differentiation medium (EDM)-preconditioned ASCs, is beneficial to angiogenesis in ischemic limb and heart models. Mechanistic studies have revealed that a significant proportion of beneficial effects of ASC administration may be due to secretion rather than endothelial differentiation of ASC [8–10]. Apart from soluble growth factors, cell-released microvesicles (MVs)/exosomes have been recognized recently as a new mechanism of intercellular communication [11].
MVs are plasma membrane-derived vesicles that are released into the microenvironment by various cell types, including stem cells and progenitors. Unlike protein molecules or living cells, MVs are submicron particles that target recipient cells to deliver proteins and lipids, and, therefore, trigger downstream signaling events at distant locations throughout the body [12]. An emerging MV/exosome approach of therapeutic angiogenesis may present a promising outlook for ischemic diseases [13–17]. This approach, in comparison with the stem/progenitor cell approach, avoids the possibility of tumor formation due to uncontrolled cell proliferation and microvasculature occlusion upon intra-arterial administration of implanted cells [3]. Additionally, in the treatment of ischemic heart diseases, MV-based strategy may avoid the proarrhythmic risk due to immature phenotype and syncytium formation, and may avoid myocardial ossification and/or calcification resulting from chondrocyte and osteocyte differentiation caused by cell-based paradigms [18]. Recent studies have shown that MVs deliver genetic material, mRNA and microRNA (miRNA), and thus play a critical role in modulating the function of target cells [19–24]. Indeed, secretion of MVs has been proved to mediate the angiogenic potential of tumor cells [25]. Further investigations have provided the evidence that MVs released from endothelial progenitor cells or bone marrow mesenchymal stem cells can directly promote angiogenesis in vitro and in vivo [20, 26–28]. However, the underlying molecular mechanisms remain poorly understood. In the current study, we demonstrate that ASC-released MVs promote angiogenesis. The MVs are enriched with miRNA, in which miR-31 may contribute to the induction of angiogenesis by targeting FIH1 (factor-inhibiting HIF-1) in recipient endothelial cells.
Materials and Methods
Cell Culture
Human ASCs (Thermo Fisher Scientific, Carlsbad, CA, https://www.thermofisher.com) and human umbilical vein endothelial cells (HUVECs) (Thermo Fisher Scientific) were maintained in ASC growth medium and endothelial cell growth medium (both Thermo Fisher Scientific), respectively, in a humidified atmosphere of 5% CO2 and 95% air at 37°C, according to the manufacturer’s instructions. The medium was replaced every 2–3 days, and passages 3–4 were used for all experiments.
EDM Preconditioning and Conditioned Medium Preparation of ASCs
ASCs were pretreated with EDM (PromoCell, Heidelberg, Germany, http://www.promocell.com) containing endothelial basal medium supplemented with 2% fetal bovine serum (FBS), 5 ng/ml epidermal growth factor, 10 ng/ml basic fibroblast growth factor, 20 ng/ml insulin-like growth factor, 1 µg/ml ascorbic acid, 22.5 µg/ml heparin, 0.2 µg/ml hydrocortisone, 50 ng/ml vascular endothelial growth factor, 100 U/ml penicillin, and 100 µg/ml streptomycin. The same medium was changed once during the 4-day preconditioning period. Cells were then washed with phosphate-buffered saline (PBS) and incubated in fresh endothelial basal medium/1% MV-free FBS with or without GW4869 (Thermo Fisher Scientific), an inhibitor of MV formation [29], for an additional 2 days. The culture medium was collected and centrifuged at 500g for 10 minutes at 4°C to remove untouched cells. Supernatant was used as conditioned medium (CdM). The CdM with removal of MVs via sequential centrifugations of CdM at 12,000g for 30 minutes and ultracentrifugation (Beckman Coulter L8-80M Ultracentrifuge; Brea, CA, https://www.beckmancoulter.com) at 100,000g for 60 minutes at 4°C was used as CdM-MV free. CdM or CdM-MV free, mixed with an equal volume of fresh endothelial basal medium/1% FBS, was used for HUVEC angiogenic analysis. Endothelial basal medium/1% FBS without cells was incubated in parallel for 2 days, mixed with equal amounts of fresh endothelial basal medium/1% FBS, and used as the control. All of the FBS used in this study was MV-free FBS, achieved by ultracentrifugation of FBS at 100,000g for 60 minutes at 4°C.
Preparation of MVs
MVs were isolated by sequential centrifugations of the ASC culture medium at 500g for 10 minutes, 12,000g for 30 minutes, and 100,000g for 60 minutes at 4°C. The supernatant was discarded. To remove residual soluble factors, pelleted MVs were then washed with PBS once by centrifugation at 100,000g for 60 minutes at 4°C, as described previously [30]. MVs were resuspended in indicated buffer or endothelial basal medium/1% FBS for subsequent analysis or angiogenic assay. The protein concentration of MVs was measured using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific). In this report, MVs from ASCs preconditioned with EDM are designated as MV-P.
Cell Migration Assay
HUVEC migration was estimated by a quantitative cell-migration assay that combined the use of membrane-based Boyden chambers, propidium iodide (EMD Chemicals, Gibbstown, NJ, http://www.emdmillipore.com) staining, and software-assisted counting of nuclei of migrated cells, as described in our previous report with slight modification [31]. In brief, HUVECs were treated with various CdM, or with MVs at 30 μg/ml (protein concentration) for 24 hours, or transfected as described below. The treated HUVECs in endothelial basal medium/1% FBS were then seeded in a 96-well transwell plate (BD Biosciences) at 1.25 × 104 per insert (3-µm pores). Endothelial basal medium/1% MV-free FBS was added to the upper and lower compartment of the well, followed by incubation at 37°C in 5% CO2 for 24 hours. The HUVECs were then fixed in absolute ethanol and stained with propidium iodide. The cells that migrated to the lower side of the inserts were counted.
Tube Formation Assay
HUVECs were treated with various CdM, or with MVs at 30 μg/ml (protein concentration) for 24 hours, or transfected as described below. The treated HUVECs in endothelial basal medium/1% FBS were seeded in a 96-well plate at 1 × 104 per well precoated with growth factor-reduced Matrigel (BD Biosciences). The plate was then incubated at 37°C in 5% CO2 for 4 hours. The cells were stained with 10 µM Calcein AM (Thermo Fisher Scientific) at 37°C in 5% CO2 for 30 minutes. Tube formation was checked with an inverted fluorescence microscope (Olympus IX71; Olympus, Tokyo, Japan, http://www.olympus-global.com), and tube length was calculated using ImagePro Plus software (MediaCybernetics, Rockville, MD, http://www.mediacy.com).
Electron Microscopy
MVs were fixed in 2.5% glutaraldehyde and 1% osmium tetroxide, dehydrated in alcohol, dried on a glass surface, and coated with gold using sputter coating. MV slides were then examined in a scanning electron microscope (JSM-820; JEOL, Tokyo, Japan, http://www.jeol.co.jp). Images were obtained by secondary electron microscopy at a working distance of 8 mm and an accelerating voltage of 10 kV.
Immunoblotting
Western blot for cells or MVs was performed as previously described [32]. Cells were lysed in mammalian protein extraction reagent (Thermo Fisher Scientific) supplemented with a protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO, http://www.sigmaaldrich.com). The cell lysates were resolved by electrophoresis on 4%–12% precast Bis-Tris gel (Thermo Fisher Scientific). Proteins were transferred from the gel to a nitrocellulose membrane using an iBlot Dry Blotting System (Thermo Fisher Scientific). Specific proteins were detected using primary antibodies, anti-Alix (ab88743; Abcam, Cambridge, MA, http://www.abcam.com), anti-FIH1 (AV32903; Sigma-Aldrich), and anti-β-actin (sc-47778; Santa Cruz Biotechnology, Santa Cruz, CA, http://www.scbt.com). Alix is an exosome-associated protein involved in multivesicular endosome biogenesis [12, 33].
Cell Proliferation Assay
HUVECs were seeded in a 96-well plate at 7.5 × 103 cells per well and were quiesced with endothelial basal medium/1% FBS for 24 hours. Cells were then treated with various MVs at 30 μg/ml (protein concentration) for 3 days. Untreated cells were used as a control. A Cell Counting Kit-8 (Dojindo Molecular Technologies, Rockville, MD, http://www.dojindo.com) was used for the cell proliferation assay. An absorbance spectrum at 450 nm was recorded with a spectrophotometer (SpectraMAX 190; Molecular Devices; Sunnyvale, CA, http://www.moleculardevices.com). Endothelial basal medium/1% FBS incubated in parallel without cells was used as the background.
Extraction of RNA
Extraction of total RNA, small RNA (fewer than 200 nucleotides), and large RNA from ASCs or MVs was performed using a mirVana isolation kit (Thermo Fisher Scientific) according to the manufacturer's instructions and our previous description [32]. The concentration of RNA was determined using the NanoDrop 1000 spectrophotometer.
RNA Analysis With Bioanalyzer
An Agilent 2100 Bioanalyzer (Agilent Technologies; Santa Clara, CA, http://www.agilent.com) was used following the manufacturer’s protocol. Total RNA (20 ng) from MVs and from their donor ASCs was analyzed with an Agilent RNA 6000 Nano kit and an RNA 6000 ladder, and 20 ng of small RNA from MVs was analyzed with an Agilent Small RNA kit (all Agilent Technologies).
TaqMan miRNA Assay
Reverse transcription of 10 ng of small RNA from MVs was performed using a specific miRNA primer from the corresponding TaqMan MicroRNA Assay and reagents from a TaqMan MicroRNA Reverse Transcription Kit (Thermo Fisher Scientific). Transcribed complementary DNAs (cDNAs) were subsequently preamplified using the TaqMan MicroRNA Assay and the TaqMan PreAmp Master Mix (Thermo Fisher Scientific) according to the manufacture’s protocol. Reverse-transcriptase-polymerase chain reaction (RT-PCR) was carried out using the diluted preamplified products, the specific TaqMan MicroRNA Assays, and a TaqMan Universal PCR Master Mix (Thermo Fisher Scientific), as described previously [32]. The relative miRNA levels were normalized to endogenous U6 small nuclear RNA, a highly conserved small nuclear RNA in vertebrate genomes, for each sample.
Creation and Transduction of Recombinant Lentivirus
The procedures practiced here followed the National Institutes of Health guidelines for recombinant DNA research. To deplete the expression of miR-31 in ASCs, a lentiviral Lenti/ZIPmiR-31 (ZIPmiR-31) and a control, Lenti/ZIPmiR-Cont (ZIPmiR-Cont), were created using commercial plasmids pLenti/ZIPmiR-31 and pLenti/ZIPmiR-Cont (System Biosciences, Mountain View, CA, https://www.systembio.com), respectively. To test the targeting activity of miR-31, a full-length (1–5,787 base pairs) and a half-length (1–2,681 base pairs) of 3′ untranslated region (3′-UTR) of FIH1 were each cloned into a luciferase reporter plasmid (Applied Biological Materials, Richmond, British Columbia, Canada, https://www.abmgood.com), and designated pLenti/UTR-Luc/FIH1-FL and pLenti/UTR-Luc/FIH1-HL, respectively. Meanwhile, a plasmid containing a mutated half-length of FIH1 3′-UTR, pLenti/UTR-Luc/FIH1-HLmut, was generated by replacing the nucleotides UCUUGC with AAGCUU at the predicted miR-31 target site 117–124 [34]. Then, the lentiviral Lenti/UTR-Luc/FIH1-FL (Luc/FIH1-FL), Lenti/UTR-Luc/FIH1-HL (Luc/FIH1-HL), and Lenti/UTR-Luc/FIH1-HLmut (Luc/FIH1-HLmut) were created using the above plasmids. A lentiviral Lenti/UTR-Luc/Blank (Luc/Blank) was used as a control. ASCs were transduced with ZIPmiR-Cont or ZIPmiR-31 at a multiplicity of infection (MOI) of 2. HUVECs were transduced with Luc-UTR/Blank, Luc-UTR/FIH1-FL, Luc-UTR/FIH1-HL, or Luc-UTR/FIH1-HLmut at an MOI of 1. The detailed methods of lentiviral creation, concentration, titration, and transduction were described previously [32, 35].
Cell Transfection
Lipofectamine RNAiMAX (Thermo Fisher Scientific) was used for HUVEC transfection according to the manufacturer’s instructions. Transfection of 10 nM pre-miR-Cont or pre-miR-31 (Thermo Fisher Scientific) was performed in the HUVECs. The transfection medium was changed to regular endothelial cell growth medium 6 hours later. The cells were then incubated to recover for an additional 42 hours for subsequent cell migration assay, tube formation assay, RT-PCR analysis, or incubated overnight for lentiviral transduction and luciferase activity assay.
Aortic Ring Assay
All animal experiments in this study were approved by the Institutional Animal Care and Use Committee of the Atlanta University Center (approval no. 13-09) and complied with the NIH guidelines for the care and use of laboratory animals. An aortic ring assay was performed according to previous reports [36–39]. Male C57BL/6J mice at age 8–10 weeks were obtained from The Jackson Laboratory (Bar Harbor, ME, https://www.jax.org) and were euthanized with CO2 inhalation. Aortae were collected, cleaned, and cut into rings approximately 0.5-mm wide under sterile conditions. Aortic rings were transferred to a 96-well plate at 8 rings per well and incubated with the indicated MVs at 100 µg/ml (protein concentration) in Opti-MEM (Thermo Fisher Scientific) overnight. Each ring was then embedded in 50 μl of Matrigel in an 8-well Chamber Slide (Thermo Fisher Scientific) at 1 ring per well and incubated in 300 μl per well of Opti-MEM supplemented with 2.5% FBS. The medium was carefully changed 3 days after embedding. At day 5, the rings were stained with 10 µM Calcein AM at 37°C for 30 minutes. The chamber was removed and a coverslip was placed to cover the slide. Z-stack images were taken with a fluorescent microscope (Olympus BX41) equipped with a camera (Olympus DP70) and cellSens digital imaging software (Olympus). Outgrowth area was determined with ImagePro Plus software (MediaCybernetics).
Matrigel Plug Assay
Ice-cold Matrigel (300 µl per plug) was mixed with PBS or MVs (100 µg per plug) and then injected subcutaneously into the flanks of male nude mice, 6–8 weeks old (The Jackson Laboratory). The mice were euthanized with CO2 inhalation 2 weeks after implantation. The entire Matrigel plugs were excised, then weighed and frozen for immunohistochemistry.
Immunohistochemistry
Matrigel plugs were embedded in optimal cutting temperature compound (Sakura, The Netherlands, http://www.sakura.eu) and snap-frozen by immersing in liquid nitrogen-cooled 2-methylbutane (Sigma-Aldrich). The frozen blocks were cryosectioned at a thickness of 10 µm with a microtome cryostat (Leica Microsystems, Buffalo Grove, IL, http://www.leica-microsystems.com). The section was mounted onto a glass slide. The slides were dried at room temperature for 2 hours and fixed in cold methanol at −20°C for 15 minutes. The slides were then dried in a fume hood for 30 minutes and washed with PBS twice. Each PBS washing step lasted 5 minutes. After blockade with 3% bovine serum albumin (BSA) in PBS at room temperature for 1 hour, primary anti-CD31 antibody (558736; BD Biosciences) was added to each section at a dilution of 1:200 in blocking buffer and incubated at 4°C overnight. The slides were washed with PBS 3 times and incubated with secondary Alexa Fluor 488 donkey anti-rat IgG (Thermo Fisher Scientific) at a dilution of 1:500 in blocking buffer at room temperature for 1 hour. The slides were then washed with PBS once, counterstained with 4′,6-diamidino-2-phenylindole (DAPI; Thermo Fisher Scientific) at a dilution of 1:5,000 for 3 minutes, and washed with PBS twice. Aqueous mounting medium (Electron Microscopy Sciences, Hatfield, PA, https://www.emsdiasum.com) was added for mounting with a coverslip. The images were acquired using a digital camera-equipped fluorescence microscope. The regions containing the most intense CD31+ areas of neovascularization (hotspots) were chosen for quantification. Five hotspots per section and 3 sections per plug were analyzed at ×200 magnification. ImagePro Plus software (MediaCybernetics) was used to measure CD31+ areas in each hotspot [27].
Luciferase Reporter Assay
HUVECs were transfected with 10 nM pre-miR-Cont or pre-miR-31 and then transduced with lentivirus Luc-UTR/Blank, Luc-UTR/FIH1-FL, Luc-UTR/FIH1-HL, or Luc-UTR/FIH1-HLmut the next day. Two days after the transduction, luciferase activities in HUVECs were determined using a Luciferase Assay Kit (Applied Biological Materials) and a microplate luminometer, Lmax, equipped with SOFTmax PRO software (Molecular Devices), according to manufacturer’s instructions and our previous report [32].
Real-Time PCR Analysis for mRNA
RNA extraction from HUVECs, cDNA reverse transcription, and RT-PCR analysis were performed according to our previous report [32]. The primer pairs used were 5′-taggaccagaagcgagaatg-3′ and 5′-tcctgcatcggatgtaaaat-3′ for human FAT4 [40]; 5′-ttcctgctgaacacggtc-3′ and 5′-cggagcgctgcgtgagct-3′ for human TBXA2R [41]; 5′-gaagaagattttcaacctgatgg-3′ and 5′-gaactcttggctcctttctgc-3′ for human RGS4 [42]; 5′-agtggttggccctggtgatgttct-3′ and 5′-aagcggtgtggacgggatagcagt-3′ for human FIH1; 5′-gatgcggctcgtcagggggtgta-3′ and 5′-ggcgtgtttggggatgctgttgtg-3′ for human TIAM1; and 5′-gctcgtcgtcgacaacggct-3′ and 5′-caaacatgatctgggtcatcttctc-3′ for human β-actin.
Endothelial Cell Differentiation of ASCs
ASCs were incubated in EDM for the indicated days. CD31 and vascular endothelial (VE)-cadherin are endothelial cell markers. The proportion of CD31 or VE-cadherin positive cells was determined using immunocytochemistry and flow cytometry analysis. Briefly, the cells were harvested and stained with anti-CD31 antibody (Abcam) at 1:100 in 3% BSA/PBS, or anti-VE-cadherin antibody (Abcam) at 1:50 in 3% BSA/PBS. The cells were incubated in the primary antibody at room temperature overnight at 4°C in darkness. The cells were washed twice with ice-cold PBS by centrifugation at 400g for 5 minutes and then incubated in AF488-labeled anti-mouse secondary antibody (Thermo Fisher Scientific) diluted in 3% BSA/PBS at room temperature for 30 minutes in darkness. The cells were washed twice with PBS and subjected to flow cytometry analysis. Flow cytometry analysis was performed using a Guava EasyCyte System equipped with CytoSoft software (EMD Millipore, Billerica, MA, http://www.emdmillipore.com). The ExpressPlus program (De Novo Software, Glendale, CA, https://www.denovosoftware.com) was selected to perform the analysis.
Statistical Analysis
Results from the quantitative studies were expressed as mean ± SD in triplicate at a minimum. All experiments were independently repeated at least four times. Statistical comparisons between two groups were performed using a two-tailed Student’s t test. Differences among multiple groups were statistically analyzed using one-way analysis of variance (ANOVA) and a two-way ANOVA test for multiple groups with two main treatments, followed by post hoc Tukey analysis. Differences were considered significant when p < .05.
Results
ASC-Released MVs Promoted Migration and Tube Formation of HUVECs
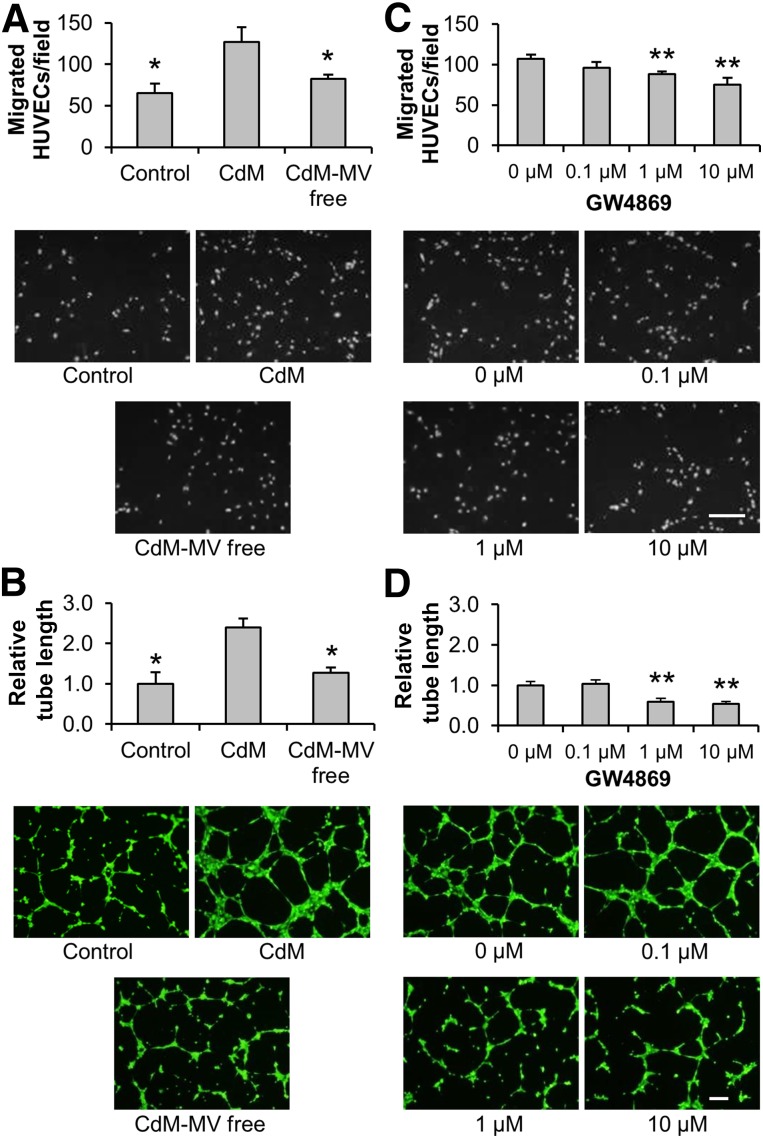
Although ASCs can be efficiently differentiated to adipocytes [32, 35], our observations in this study demonstrated that incubation of ASCs in EDM for 10 days induced very little expression of 2 endothelial markers, CD31 and VE-cadherin (supplemental online Fig. 1). Nevertheless, EDM preconditioning is observed to further enhance the proangiogenic effects of ASCs via the contribution of paracrine mediators [8, 9]. To examine whether ASC-released MVs are an important mediator for angiogenesis, CdM from EDM-preconditioned ASCs was collected and added to HUVECs to detect cell migration and tube formation. Our results demonstrated that HUVEC migration (control: 65.2 ± 11.4 cells; CdM: 126.6 ± 17.9 cells) and tube formation (control: 1.00 ± 0.28; CdM: 2.39 ± 0.22) were induced by CdM. The induction of migration (CdM-MV free: 82.4 ± 5.1 cells) and tube formation (CdM-MV free: 1.28 ± 0.13) was significantly reduced when MVs were removed from CdM (Fig. 1A, 1B). On the other hand, upon inhibiting the generation of MVs by the incubation of ASCs with GW4869 [29], the migration (0 µM: 107.2 ± 5.2 cells; 0.1 µM: 96.2 ± 7.2 cells; 1 µM: 88.0 ± 3.3 cells; 10 µM: 75.0 ± 8.8 cells) and tube formation (0 µM: 1.00 ± 0.09; 0.1 µM: 1.03 ± 0.10; 1µM: 0.60 ± 0.09; 10 µM: 0.54 ± 0.06) effects of CdM were also reduced (Fig. 1C, 1D). These data imply that MVs are a proangiogenic mediator for the ASCs.
Figure 1.
Induction of migration and tube formation of HUVECs by CdM from adipose-derived stem cells (ASCs). ASCs were incubated in endothelial basal medium/1% fetal bovine serum (FBS) for 2 days in the absence (A, B) or presence (C, D) of GW4869, an MV-formation inhibitor. CdM was collected and used to treat HUVECs. Cell migration (A, C) and tube formation (B, D) assays for treated HUVECs were performed. The endothelial basal medium/1% FBS without cells was incubated in parallel and was used as a control. The CdM with removal of MVs via ultracentrifugation was used as CdM-MV free. Representative images of cell migration and tube formation are displayed. Scale bar = 200 µm. (A–D): n = 4. ∗, p < .05 versus CdM; ∗∗, p < .01 versus 0 µM. Abbreviations: CdM, conditioned medium; HUVEC, human umbilical vein endothelial cell; MV, microvesicle.
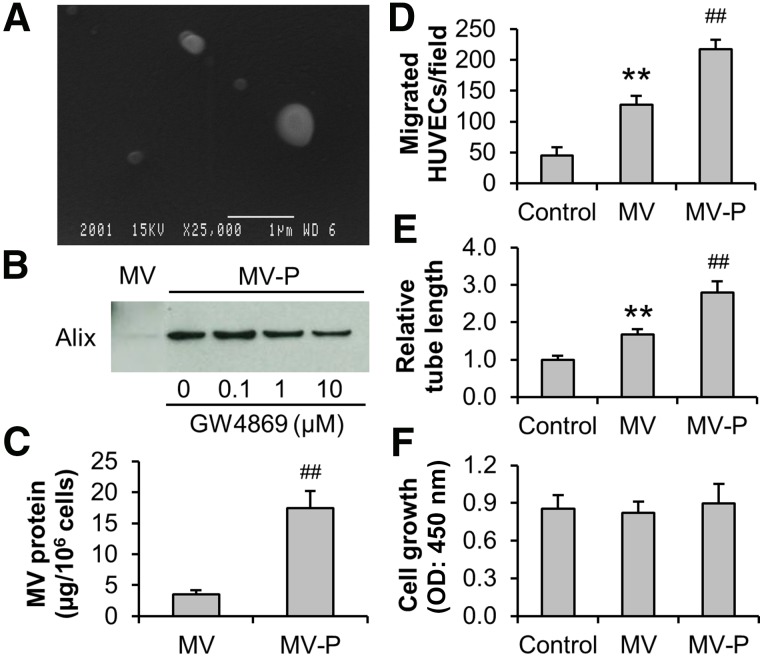
MV isolation was performed with the CdM collected from EDM-preconditioned ASCs. Scanning electron microscopy showed that the particles in the isolated pellet were unevenly sized spheroids of less than 1 µm (Fig. 2A), which agrees with the reported characteristics of MVs [20]. In addition, the presence of Alix, a marker of MV [12, 33], was detected in the isolated pellet. Moreover, our results exhibited that the preconditioning of ASCs with EDM markedly promoted MV release, which was partially inhibited by GW4869 (MV miRNA level set to 1.00. 0 µM: 6.45; 0.1 µM: 7.45; 1 µM: 5.55; 10 µM: 4.37) (Fig. 2B). Quantification of MV protein also demonstrated that EDM enhanced the MV release from ASCs (MV: 3.5 ± 0.6; MV-P: 17.5 ± 2.8) (Fig. 2C). Pretreatment of CdM with detergent, 1%Triton-X100 for 20 minutes at 4°C, resulted in a loss of Alix detection, which suggested that isolated MVs were intact plasma membrane-encapsulated particles that can be broken down by the detergent (data not shown). Equal amounts of MV and MV-P were then used to treat HUVECs. Our data showed that migration (control: 45.3 ± 12.8 cells; MV: 127.0 ± 14.3 cells) and tube formation (control: 1.00 ± 0.10; MV: 1.67 ± 0.15) of HUVECs were significantly elevated upon the treatment of MVs. Interestingly, MV-P exhibited an even stronger migration (217.5 ± 15.0 cells) and tube formation (2.82 ± 0.27) effect (Fig. 2D, 2E), which is similar to the observation in endothelial progenitor cells [20]. These data suggest that MVs from ASCs, particularly those from EDM-preconditioned ASCs, are proangiogenic in vitro. The proliferative effect of MVs on HUVECs was not observed (control: 0.85 ± 0.11; MV: 0.82 ± 0.09; MV-P: 0.90 ± 0.15) (Fig. 2F).
Figure 2.
Induction of migration and tube formation of HUVECs by MVs from adipose-derived stem cells (ASCs). (A): Conditioned medium was subjected to standard serial centrifugation for MV isolation. The isolated pellet was examined with scanning electron microscopy (scale bar = 1 µm). (B): ASCs were maintained in growth medium or preconditioned with endothelial differentiation medium for 4 days. After washing, the ASCs were then incubated for 2 days in endothelial basal medium/1% FBS with or without the presence of GW4869. Western blot analysis of the MVs was performed with Alix, an MV marker. Each lane represented an MV lysate from 3 × 106 cells. (C): The protein content of MVs and MV-P from ASCs were determined. (D–F): HUVECs were treated with 30 µg/ml (protein concentration) MV or MV-P. HUVECs in fresh endothelial basal medium/1% FBS was used as a control. Cell migration (D), tube formation (E), and proliferation (F) assays of the HUVECs were performed as described in Material and Methods. (C–F): n = 4. ##, p < .01 versus MV or control; ∗∗, p < .01 versus control. Abbreviations: HUVEC, human umbilical vein endothelial cell; MV, microvesicle; MV-P, microvesicles from adipose-derived stem cells preconditioned with endothelial differentiation medium; OD, optical density.
MicroRNA Enriched in ASC-Released MVs
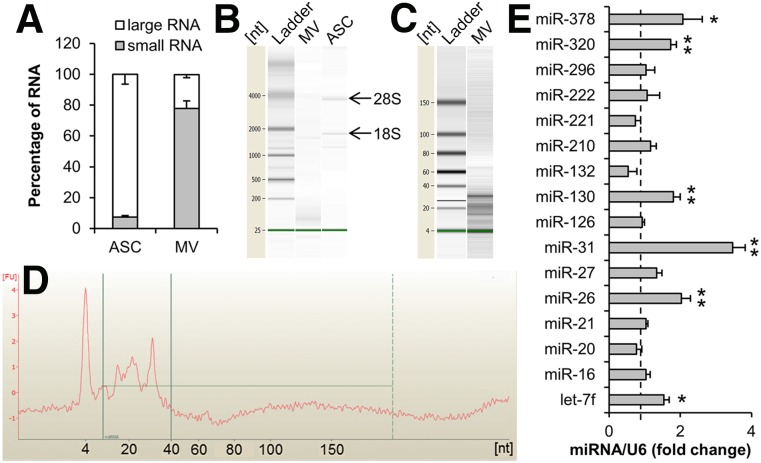
Recent studies have reported that MVs from mesenchymal stem cells shuttle miRNA between cells [23]. To explore the RNA in ASC-released MVs, isolated MV-P was subjected to small RNA (fewer than 200 nucleotides), large RNA, or total RNA extraction and analysis. An average of 3.8 ng of small RNA was harvested per microgram protein of MV. The majority of the RNA in the MVs was small RNA (large RNA: 22.0% ± 2.1%; small RNA: 77.8% ± 4.9%), while majority of the RNA in ASCs was large RNA (large RNA: 92.5% ± 6.4%; small RNA: 7.5% ± 0.8%) (Fig. 3A). Bioanalyzer RNA analysis demonstrated that the majority of total RNA in MVs was fewer than 200 nucleotides. A lack of 18S and 28S ribosomal RNA indicated that the cellular contamination was minimal (Fig. 3B). Analysis of small RNA further revealed that almost all of the small RNA in MVs was in a narrow range of 10–40 nucleotides, suggesting that miRNA was enriched in the MVs (Fig. 3C, 3D). Similar RNA size analysis results were observed in MVs in comparison with MV-P.
Figure 3.
Analysis of RNA extracted from ASC-released MVs. (A): Small RNA (fewer than 200 nucleotides) and large RNA extracted from ASCs and ASC-released MVs were quantified (n = 3– 4). Total RNA was set to 100%. (B): Equal amounts of total RNA from ASCs and MVs were analyzed using Bioanalyzer (Agilent Technologies). 18S and 28S indicated the ribosomal RNA. (C): Small RNA from MVs was analyzed using Bioanalyzer. (D): An electropherogram of the lane MV on (C). (E): Reverse-transcriptase-polymerase chain reaction analysis of miRNA for small RNA extracted from MV-P. The level of miRNA in MV was set to 1 (dashed line). U6 was used as an internal control. n = 4. ∗, p < .05; ∗∗, p < .01. Abbreviations: ASC, adipose-derived stem cell; miRNA, microRNA; MV, microvesicle; nt, nucleotide.
To confirm the presence of miRNA and inspect the pattern of miRNA in MVs, miRNA array analysis of the small RNA extracted from MV-P was performed. A total of 225 miRNAs were detected and the top 15 abundant miRNAs were listed (supplemental online Table 1). Based on the extensive reviews of the reported angiogenesis-regulator miRNAs (AngiomiRs) [43–47], miR-31 is the only proangiogenic miRNA in the ranking list. Moreover, the levels of AngiomiRs in MVs and MV-P were examined using RT-PCR (Fig. 3E). The results demonstrated that miR-31 in MVs was mostly induced by EDM preconditioning in ASCs. These findings imply that miR-31 may be an essential miRNA in MV-P to induce angiogenesis.
MiR-31 Contributed to Proangiogenesis Induced by MVs From EDM-Preconditioned ASCs
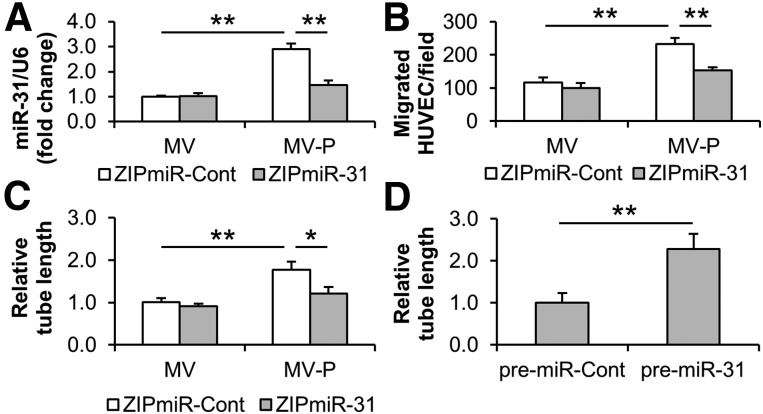
It has been reported that miR-31 is induced by vascular endothelial growth factor (VEGF) in HUVECs [48] and upregulated in regenerative muscle after hindlimb ischemia [49]. Recently, miR-31 has been observed to promote endothelial cell migration and angiogenesis [40, 50]. Our data exhibited that miR-31 levels were elevated in HUVECs upon treatment with miR-31-enriched MV-P (1.00 ± 0.05 vs. 2.90 ± 0.22, MV and MV-P, respectively). The elevation was reduced when the MV-P was isolated from miR-31-silenced donor ASCs (1.47 ± 0.18) (Fig. 4A). Similarly, MV-P-induced cell migration (116.3 ± 15.8 cells vs. 232.0 ± 19.3 cells, MV and MV-P, respectively) and tube formation (1.00 ± 0.10 vs. 1.77 ± 0.19, MV and MV-P, respectively) were significantly decreased for migration (154.0 ± 9.2 cells) and tube formation (1.21 ± 0.16) when the MV-P was isolated from miR-31-silenced ASCs (Fig. 4B, 4C). Direct transfection of pre-miR-31 promoted the tube formation of HUVECs (1.00 ± 0.23 vs. 2.28 ± 0.36) (Fig. 4D). Additionally, the transfection of small RNA from MV-P induced HUVEC migration and tube formation. The induction was decreased when cotransfecting with a commercial anti-miR-31 (supplemental online Fig. 2A, 2B). These findings suggest that miR-31 contributes to the proangiogenesis induced by MV-P in vitro.
Figure 4.
miR-31 contributes to the proangiogenesis induced by MV-P in vitro. (A–C): Adipose-derived stem cells (ASCs) were transduced with lentiviral ZipmiR-31 to silence miR-31. ZipmiR-Cont was used as a control. MVs and MV-P were obtained from the transduced ASCs and were used to treat HUVECs. The miR-31 content in HUVECs (A), and the cell migration (B) and tube formation (C) of HUVECs were determined. (D): HUVECs were transfected with commercial pre-miR-31. A pre-miR-Cont was used as a control. The tube formation of HUVECs was measured 48 hours after transfection. (A–D): n = 4–5. ∗, p < .05; ∗∗, p < .01. Abbreviations: HUVEC, human umbilical vein endothelial cell; miR-31, microRNA-31; MV, microvesicle; MV-P, microvesicles from adipose-derived stem cells preconditioned with endothelial differentiation medium.
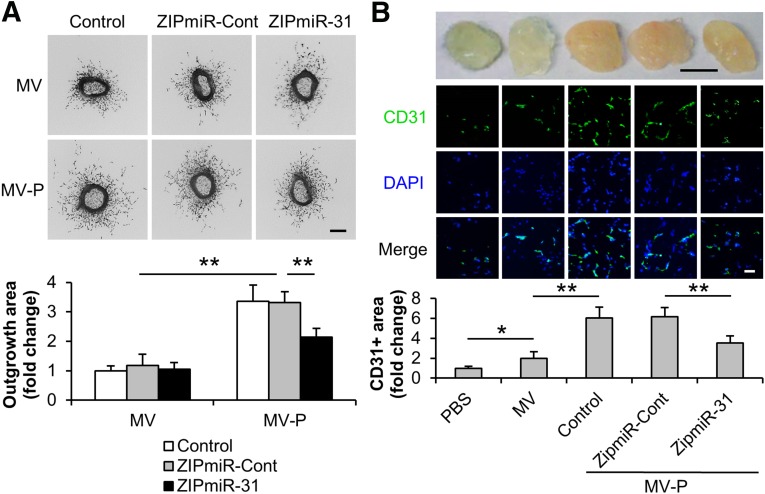
While most in vitro assays are designed to study a particular step in the angiogenic conditions, the aortic ring ex vivo assay recapitulates all of the key steps in angiogenic processes, including matrix degradation, cell migration, and proliferation. Here, our data demonstrated that MV-P enhanced the sprouting outgrowth from aortic rings in comparison with MVs (MV/ZIPmiR-Cont: 1.18 ± 0.38; MV-P/ZIPmiR-Cont: 3.32 ± 0.36) (Fig. 5A). The enhancement was significantly inhibited when MV-P was isolated from miR-31 depleted ASCs (MV-P/ZIPmiR-31: 2.14 ± 0.30). A mouse Matrigel plug assay was also used to inspect the effect of ASC-released MVs on angiogenesis. Our results revealed that MVs induced functional vasculature formation, compared with PBS, as indicated by the red plug appearance and CD31-positive area on the plug section (1.00 ± 0.19 vs. 2.01 ± 0.66, PBS and MV, respectively), and that MV-P exhibited even more proangiogenic activity than MV (6.04 ± 1.11). The effect of MV-P was attenuated if the MV-P were from miR-31 knockdown cells (MV-P/ZIPmiR-Cont: 6.18 ± 0.93; MV-P/ZIPmiR-31: 3.54 ± 0.70) (Fig. 5B). Taken together, we present the evidence that miR-31 mediates MV-P induced proangiogenesis ex vivo and in vivo.
Figure 5.
miR-31 contributes to the proangiogenesis induced by MV-P ex vivo and in vivo. Adipose-derived stem cells (ASCs) were transduced with lentiviral ZipmiR-31 to silence miR-31. ASCs untransduced (control) or transduced with a ZipmiR-Cont were used as controls. MV and MV-P were obtained from these ASCs. (A): Mouse aortic rings were collected and treated with various MVs, as indicated, for 5 days (n = 8). Representative images (upper panel) and a statistical analysis of the outgrowth area of aortic rings (lower panel) for each treatment condition are displayed. The outgrowth area of aortic rings treated with MV from untransduced cells was set to 1. Scale bar = 100 µm. (B): PBS, MV, or MV-P was mixed with Matrigel and injected subcutaneously into the flanks of the nude mice. The Matrigel plugs were harvested 2 weeks postimplantation (n = 6). Upper panel: Representative pictures of the plugs are exhibited. Scale bar = 5 mm. Middle panel: The sections of the plugs were subject to immunohistochemistry analysis for CD31, an endothelial cell marker, and counterstained with DAPI. Scale bar = 100 µm. Lower panel: Quantification of the CD31-positive area was performed. The positive area in the slide from the plugs containing PBS was set to 1. ∗, p < .05; ∗∗, p < .01. Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; miR-31, microRNA-31; MV, microvesicle; MV-P, microvesicles from adipose-derived stem cells preconditioned with endothelial differentiation medium; PBS, phosphate-buffered saline.
FIH1 Is a Target of MV-P-Delivered miR-31 in HUVECs
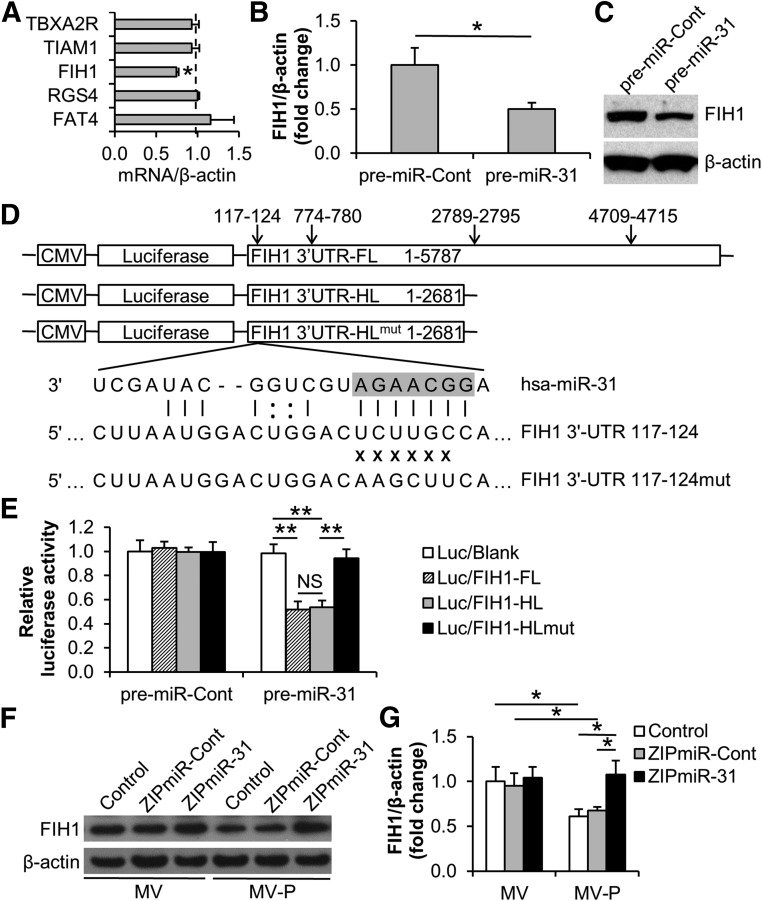
To further investigate the molecular mechanism of MV-delivered miR-31 in HUVECs, the target genes of miR-31 were investigated. The genes that have been reported as both miR-31 targets and angiogenesis regulators, so far as we know, include fih1 [34, 51], fat4 [40], tbxa2r [50, 52], tiam1 [53, 54], and rgs4 [55, 56]. Among these genes, only fih1 was downregulated by MV-P in HUVECs in comparison with that by MVs, while the others presented no significant difference (Fig. 6A). Indeed, the FIH1 mRNA levels (1.00 ± 0.19 vs. 0.50 ± 0.07) and protein levels (1.00 vs. 0.65) were reduced in HUVECs upon the transfection of miR-31 precursor (Fig. 6B, 6C). A conserved miR-31 target site residing at nucleotides 117–124 and 3 poorly conserved sites at 774–780, 2,789–2,795, and 4,709–4,715 in the FIH1 3′-UTR were predicted by miRNA target prediction websites TargetScan Human 6.0 (http://www.targetscan.org/vert_60) and PicTar (http://pictar.mdc-berlin.de) (Fig. 6D; supplemental online Fig. 3A–3C). To experimentally validate the targeting activity, luciferase reporter constructs, a Luc/FIH1-FL (full-length) containing all of the 4 sites and a Luc/FIH1-HL (half-length) containing only first 2 sites at nucleotides 117–124 and 774–780, were created (Fig. 6D). Our data showed that the luciferase activity in Luc/FIH1-FL or Luc/FIH1-HL-transduced HUVECs was similarly suppressed by miR-31, which suggested that FIH1 was a direct target of miR-31 in HUVECs and that the actual target sites might be at nucleotides 117–124 and/or 774–780 in FIH1 3′-UTR. Furthermore, a mutation at the site 117–124 in FIH1 3′-UTR nearly completely restored the luciferase activity, which suggests that miR-31 targets FIH1 in HUVECs at the conserved nucleoside site 117–124 of 3′-UTR (in the pre-miR-31 transduced group, Luc/blank: 0.98 ± 0.08; Luc/FIH1-FL: 0.52 ± 0.07; Luc/FIH1-HL: 0.54 ± 0.06; Luc/FIH1-HLmut: 0.94 ± 0.08) (Fig. 6E). As expected, the downregulation of FIH1 expression in HUVECs by the treatment of MV-P was restored when MV-P originated from miR-31 depleted ASCs (MV/control: 1.00 ± 0.16; MV/ZIPmiR-Cont: 0.95 ± 0.14; MV/ZIPmiR-31: 1.04 ± 0.12; MV-P/control: 0.61 ± 0.08; MV-P/ZIPmiR-Cont: 0.68 ± 0.04; MV-P/ZIPmiR-31: 1.08 ± 0.16) (Fig. 6F, 6G). These findings suggest that FIH1 may be a direct target of MV-P-delivered miR-31 in HUVECs.
Figure 6.
MV-P suppresses FIH1 expression in human umbilical vein endothelial cells (HUVECs) via the delivery of miR-31. (A): The levels of predicted miR-31 target mRNA in MV or MV-P-treated HUVECs were examined with reverse-transcriptase polymerase chain reaction (RT-PCR). β-actin was used as an internal control. The mRNA levels in MV-treated HUVECs were set to 1.0 (dashed line) (n = 5). (B, C): HUVECs were transfected with pre-miR-Cont (control) or pre-miR-31. The mRNA and protein levels of FIH1 in transfected HUVECs were determined with RT-PCR (n = 3) (B) and Western blot (C), respectively. (D): Schematic representation of the luciferase-reporter lentivirus containing insert of FL or HL of FIH1 3′-UTR. Four predictive target sites by miR-31 are shown. A mutation at the site of 117–124 of FIH1 3′-UTR is illustrated. The seed site of mature miR-31 is highlighted. (E): HUVECs were transfected with pre-miR-Cont or pre-miR-31 followed by transduction of indicated lentiviral reporters the next day. The luciferase activity in HUVECs was determined using luminometry 2 days posttransduction (n = 5). (F): MV and MV-P from ZIPmiR-31-transduced adipose-derived stem cells (ASCs) were used to treat HUVECs. MV and MV-P from untransduced (control) or ZIPmiR-Cont-transduced ASCs were used as controls. The protein levels of FIH1 in treated HUVECs were determined using Western blot. β-actin was used as an internal control. (G): A statistical analysis of the density of the Western blot bands in (F) using Image J software. ∗, p < .05; ∗∗, p < .01. Abbreviations: FL, full length; HL, half length; miR-31, microRNA-31; MV, microvesicle; MV-P, microvesicles from adipose-derived stem cells preconditioned with endothelial differentiation medium; NS, not statistically significant.
Discussion
As a cell-based angiogenic therapy, implantation of ASCs is efficacious in the ischemic hindlimb/heart animal models mainly through secretory effects [8, 9]. MVs secreted from stem cells recently have been recognized as a potential alternative therapeutic approach to ischemic diseases [14]. Here, we demonstrate that ASC-released MVs enhance angiogenesis, and that miR-31 in the MVs contributes to the induction of angiogenesis by targeting FIH1 in vascular endothelial cells.
The characterization and proangiogenic effect of ASCs on cardiovascular repair have been recently reviewed [4]. The underlying mechanism of the proangiogenic effects of ASCs remains unclear. Although the endothelial differentiation of ASCs is not efficient and is not considered a the major proangiogenic mechanism, preconditioning ASCs with EDM is still observed to promote angiogenesis [8, 9]. There is a growing acknowledgment that the proangiogenic effect of the preconditioned ASCs is mainly a result of cell secretions [9]. Apart from soluble growth factors, cell-released MVs have been identified as a new mechanism of intercellular communication [22]. MVs are plasma membrane-derived vesicles [21] released into the microenvironment by various cell types, including stem cells and progenitors [19, 20]. Emerging evidence in MV-based therapy indicates that MVs released from bone marrow mesenchymal stem cells can promote angiogenesis in animal myocardial infarction models [26, 28]. Our data reveal that the amount of ASC-released MVs is dramatically increased by EDM-preconditioning. In addition, MVs from EDM-preconditioned ASCs demonstrate a greater proangiogenic effect in vitro than the same amount of MVs from regular cultured ASCs. These findings suggest that EDM preconditioning of ASCs enhances both the quantity and the proangiogenic effect of the MVs. In contrast, MV release is reduced by pretreatment of ASCs with GW4869, an inhibitor of neutral sphingomyelinase that catalyzes the production of plasma membrane ceramide. Ceramide promotes the sorting and formation of secretory vesicles from multivesicular bodies in cells [29]. Although we have not observed an MV-induced cell proliferation, which may be induced by paracrine growth factors from ASCs, EDM preconditioning is still beneficial for MVs in cell migration and tube formation. Other preconditioning methods, such as hypoxia, are also worthwhile for future evaluation.
An interesting publication has demonstrated that stem cell-released MVs/exosomes from healthy people inhibit the growth of tumor cells, whereas stem cell-released MVs/exosomes from patients with tumors promote tumor growth [57]. This suggests the healthy donors as the source of ASCs. Genetic materials are known to be transferred vertically from parent cell to daughter cells in the process of mitosis. However, recent findings have revealed that functional mRNA and miRNA can be transferred horizontally between cells via the MVs [19, 20, 22]. The concept of genetic material delivery from MVs to target cells is similar to the concept of gene delivery from sperm to ovum, but is ubiquitous in physiology, pathology, and pharmacology. Our results have demonstrated that miRNA is enriched in ASC-released MVs. There is increasing evidence that miRNA is involved in various biological processes, including angiogenesis [43–47]. MiRNA in MVs has been reported to protect the kidney from ischemia-reperfusion injury [58].
Based on our miRNA profiling analysis and AngiomiR analysis, we have demonstrated that the level of miR-31 in MVs is markedly upregulated when the donor ASCs are EDM preconditioned. MiR-31 is a pleiotropically acting miRNA and is reported to decrease or increase cell motility in different cancer cells [34, 54, 55, 59]. This may be because the miRNA-mRNA target relationship differs among tissues, cells, and conditions [60]. We and other investigators have shown that miR-31 consistently augments cell migration in vascular endothelial cells or endothelial progenitor cells [40, 50, 61]. In addition, miR-31 is elevated in VEGF-treated HUVECs and ischemic hindlimbs [48, 49]. In patients suffering from coronary artery disease, overexpressing miR-31 in endothelial progenitor cells rescues their angiogenic abilities both in vitro and in vivo [50]. Our current study demonstrates that miR-31 contributes to MV-P-induced angiogenesis. These findings not only uncover an underlying molecular mechanism for MV-based angiogenic therapy but also suggest a potential strategy to elevate the angiogenic effect of MVs through miRNA-engineering of MV-donor cells.
miRNAs are a group of small (21–24 nucleotides), noncoding RNAs that regulate gene expression by binding to the 3′-UTR of mRNA to repress gene expression [62]. Computational predictive analysis with TargetScan illustrates that miR-31 has 368 conserved targets, with a total of 394 broadly conserved sites and 153 poorly conserved sites. Certain genes have been experimentally validated to be the direct targets of miR-31, including fih1 in cancer cells and keratinocytes [34, 63], fat4 and tbxa2r in 293T cells [40, 50], and tiam1 and rgs4 in cancer cells [54, 55]. Additionally, all of these genes are reported to be antiangiogenic [40, 51–53, 56]. Among these five genes, only fih1, rgs4, and tbxa2r are on the predicted list, in which fih1 has the highest aggregate probability of conserved targeting (PCT). In fact, the PCT score of fih1 ranks second among those 368 predicted target genes. In this study, we have validated that FIH1 mRNA is a target of miR-31 in HUVECs and is repressed by MVs isolated from EDM-preconditioned ASCs. Moreover, we have identified that the target site is at a broadly conserved domain located at nucleotides 117–124 in the FIH1 3′-UTR. FIH1 is an asparaginyl hydroxylase enzyme that inhibits the transcriptional activity of HIF-1, a well-known proangiogenic factor [64, 65]. Taken together, miR-31 may mediate the proangiogenic effect of MVs by targeting FIH1 in vascular endothelial cells.
Small RNAs, including antisense, small interfering RNA, and microRNA, have emerged as promising therapeutic agents against a wide array of diseases [66]. Effective delivery of these molecules is crucial to their successful clinical application. MVs are naturally produced membrane vesicles that circulate in the bloodstream and have entered the limelight in delivering therapeutic nucleic acids, because of their ability to transfer exogenous miRNAs into target cells. Based on these aspects, MVs might be an attractive alternative in therapeutic angiogenesis with regard to the durability, stability, and safety of in vivo delivery.
Conclusion
This study demonstrates that release of MVs is an important mechanistic effect of ASCs on angiogenesis. Direct administration of MVs from ASCs, particularly from EDM-preconditioned ASCs, promotes angiogenesis. The majority of RNA in MVs is small RNA, in which miR-31 contributes to MV-triggered angiogenesis by targeting FIH1 in vascular endothelial cells. These advances may provide a framework for implantation of engineered MVs as an alternative approach for the therapy of ischemic diseases. However, extensive investigation in animal models is still required to proceed with clinical trials.
Supplementary Material
Acknowledgments
We thank Dr. Ming Bo Huang and Dr. Yan Xiao at the Morehouse School of Medicine for their technical assistance with microvesicle isolation and immunohistochemistry, respectively. This work was supported, in whole or in part, by NIH Grants SC2GM099629 and G12RR003034 to D.L.
Author Contributions
T.K.: collection and assembly of data, data analysis and interpretation, and manuscript writing; T.M.J. and C.N.: collection and assembly of data; M.B., J.W.C., and W.E.T.: data analysis and interpretation; V.C.B. and Y.E.C.: conception and design; D.L.: conception and design, financial support, collection and assembly of data, data analysis and interpretation, manuscript writing, and final approval of the manuscript.
Disclosure of Potential Conflicts of Interest
The authors indicated no potential conflicts of interest.
References
- 1.Gupta R, Tongers J, Losordo DW. Human studies of angiogenic gene therapy. Circ Res. 2009;105:724–736. doi: 10.1161/CIRCRESAHA.109.200386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beohar N, Rapp J, Pandya S, et al. Rebuilding the damaged heart: The potential of cytokines and growth factors in the treatment of ischemic heart disease. J Am Coll Cardiol. 2010;56:1287–1297. doi: 10.1016/j.jacc.2010.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tongers J, Losordo DW, Landmesser U. Stem and progenitor cell-based therapy in ischaemic heart disease: Promise, uncertainties, and challenges. Eur Heart J. 2011;32:1197–1206. doi: 10.1093/eurheartj/ehr018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Madonna R, Geng YJ, De Caterina R. Adipose tissue-derived stem cells: Characterization and potential for cardiovascular repair. Arterioscler Thromb Vasc Biol. 2009;29:1723–1729. doi: 10.1161/ATVBAHA.109.187179. [DOI] [PubMed] [Google Scholar]
- 5.Cai X, Lin Y, Hauschka PV, et al. Adipose stem cells originate from perivascular cells. Biol Cell. 2011;103:435–447. doi: 10.1042/BC20110033. [DOI] [PubMed] [Google Scholar]
- 6.Szöke K, Brinchmann JE. Concise review: Therapeutic potential of adipose tissue-derived angiogenic cells. Stem Cells Translational Medicine. 2012;1:658–667. doi: 10.5966/sctm.2012-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cronk SM, Kelly-Goss MR, Ray HC, et al. Adipose-derived stem cells from diabetic mice show impaired vascular stabilization in a murine model of diabetic retinopathy. Stem Cells Translational Medicine. 2015;4:459–467. doi: 10.5966/sctm.2014-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miranville A, Heeschen C, Sengenès C, et al. Improvement of postnatal neovascularization by human adipose tissue-derived stem cells. Circulation. 2004;110:349–355. doi: 10.1161/01.CIR.0000135466.16823.D0. [DOI] [PubMed] [Google Scholar]
- 9.Nakagami H, Maeda K, Morishita R, et al. Novel autologous cell therapy in ischemic limb disease through growth factor secretion by cultured adipose tissue-derived stromal cells. Arterioscler Thromb Vasc Biol. 2005;25:2542–2547. doi: 10.1161/01.ATV.0000190701.92007.6d. [DOI] [PubMed] [Google Scholar]
- 10.Valina C, Pinkernell K, Song YH, et al. Intracoronary administration of autologous adipose tissue-derived stem cells improves left ventricular function, perfusion, and remodelling after acute myocardial infarction. Eur Heart J. 2007;28:2667–2677. doi: 10.1093/eurheartj/ehm426. [DOI] [PubMed] [Google Scholar]
- 11.Camussi G, Deregibus MC, Bruno S, et al. Exosomes/microvesicles as a mechanism of cell-to-cell communication. Kidney Int. 2010;78:838–848. doi: 10.1038/ki.2010.278. [DOI] [PubMed] [Google Scholar]
- 12.Raposo G, Stoorvogel W. Extracellular vesicles: Exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez MC, Andriantsitohaina R. Microparticles in angiogenesis: Therapeutic potential. Circ Res. 2011;109:110–119. doi: 10.1161/CIRCRESAHA.110.233049. [DOI] [PubMed] [Google Scholar]
- 14.Ratajczak MZ, Kucia M, Jadczyk T, et al. Pivotal role of paracrine effects in stem cell therapies in regenerative medicine: Can we translate stem cell-secreted paracrine factors and microvesicles into better therapeutic strategies? Leukemia. 2012;26:1166–1173. doi: 10.1038/leu.2011.389. [DOI] [PubMed] [Google Scholar]
- 15.Lopatina T, Bruno S, Tetta C, et al. Platelet-derived growth factor regulates the secretion of extracellular vesicles by adipose mesenchymal stem cells and enhances their angiogenic potential. Cell Commun Signal. 2014;12:26. doi: 10.1186/1478-811X-12-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marędziak M, Marycz K, Lewandowski D, et al. Static magnetic field enhances synthesis and secretion of membrane-derived microvesicles (MVs) rich in VEGF and BMP-2 in equine adipose-derived stromal cells (EqASCs)-a new approach in veterinary regenerative medicine. In Vitro Cell Dev Biol Anim. 2015;51:230–240. doi: 10.1007/s11626-014-9828-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farinazzo A, Turano E, Marconi S, et al. Murine adipose-derived mesenchymal stromal cell vesicles: In vitro clues for neuroprotective and neuroregenerative approaches. Cytotherapy. 2015;17:571–578. doi: 10.1016/j.jcyt.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 18.Lai RC, Chen TS, Lim SK. Mesenchymal stem cell exosome: A novel stem cell-based therapy for cardiovascular disease. Regen Med. 2011;6:481–492. doi: 10.2217/rme.11.35. [DOI] [PubMed] [Google Scholar]
- 19.Ratajczak J, Miekus K, Kucia M, et al. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: Evidence for horizontal transfer of mRNA and protein delivery. Leukemia. 2006;20:847–856. doi: 10.1038/sj.leu.2404132. [DOI] [PubMed] [Google Scholar]
- 20.Deregibus MC, Cantaluppi V, Calogero R, et al. Endothelial progenitor cell derived microvesicles activate an angiogenic program in endothelial cells by a horizontal transfer of mRNA. Blood. 2007;110:2440–2448. doi: 10.1182/blood-2007-03-078709. [DOI] [PubMed] [Google Scholar]
- 21.Skog J, Würdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valadi H, Ekström K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 23.Chen TS, Lai RC, Lee MM, et al. Mesenchymal stem cell secretes microparticles enriched in pre-microRNAs. Nucleic Acids Res. 2010;38:215–224. doi: 10.1093/nar/gkp857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eirin A, Riester SM, Zhu XY, et al. MicroRNA and mRNA cargo of extracellular vesicles from porcine adipose tissue-derived mesenchymal stem cells. Gene. 2014;551:55–64. doi: 10.1016/j.gene.2014.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park JE, Tan HS, Datta A, et al. Hypoxic tumor cell modulates its microenvironment to enhance angiogenic and metastatic potential by secretion of proteins and exosomes. Mol Cell Proteomics. 2010;9:1085–1099. doi: 10.1074/mcp.M900381-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lai RC, Arslan F, Lee MM, et al. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res (Amst) 2010;4:214–222. doi: 10.1016/j.scr.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 27.Sahoo S, Klychko E, Thorne T, et al. Exosomes from human CD34(+) stem cells mediate their proangiogenic paracrine activity. Circ Res. 2011;109:724–728. doi: 10.1161/CIRCRESAHA.111.253286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arslan F, Lai RC, Smeets MB, et al. Mesenchymal stem cell-derived exosomes increase ATP levels, decrease oxidative stress and activate PI3K/Akt pathway to enhance myocardial viability and prevent adverse remodeling after myocardial ischemia/reperfusion injury. Stem Cell Res (Amst) 2013;10:301–312. doi: 10.1016/j.scr.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 29.Trajkovic K, Hsu C, Chiantia S, et al. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 30.Ali SA, Huang MB, Campbell PE, et al. Genetic characterization of HIV type 1 Nef-induced vesicle secretion. AIDS Res Hum Retroviruses. 2010;26:173–192. doi: 10.1089/aid.2009.0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu D, Hou J, Hu X, et al. Neuronal chemorepellent Slit2 inhibits vascular smooth muscle cell migration by suppressing small GTPase Rac1 activation. Circ Res. 2006;98:480–489. doi: 10.1161/01.RES.0000205764.85931.4b. [DOI] [PubMed] [Google Scholar]
- 32.Kang T, Lu W, Xu W, et al. MicroRNA-27 (miR-27) targets prohibitin and impairs adipocyte differentiation and mitochondrial function in human adipose-derived stem cells. J Biol Chem. 2013;288:34394–34402. doi: 10.1074/jbc.M113.514372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soo CY, Song Y, Zheng Y, et al. Nanoparticle tracking analysis monitors microvesicle and exosome secretion from immune cells. Immunology. 2012;136:192–197. doi: 10.1111/j.1365-2567.2012.03569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu CJ, Tsai MM, Hung PS, et al. miR-31 ablates expression of the HIF regulatory factor FIH to activate the HIF pathway in head and neck carcinoma. Cancer Res. 2010;70:1635–1644. doi: 10.1158/0008-5472.CAN-09-2291. [DOI] [PubMed] [Google Scholar]
- 35.Liu D, Lin Y, Kang T, et al. Mitochondrial dysfunction and adipogenic reduction by prohibitin silencing in 3T3-L1 cells. PLoS One. 2012;7:e34315. doi: 10.1371/journal.pone.0034315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei Y, Gong J, Thimmulappa RK, et al. Nrf2 acts cell-autonomously in endothelium to regulate tip cell formation and vascular branching. Proc Natl Acad Sci USA. 2013;110:E3910–E3918. doi: 10.1073/pnas.1309276110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jain S, Gabunia K, Kelemen SE, et al. The anti-inflammatory cytokine interleukin 19 is expressed by and angiogenic for human endothelial cells. Arterioscler Thromb Vasc Biol. 2011;31:167–175. doi: 10.1161/ATVBAHA.110.214916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pang X, Yi T, Yi Z, et al. Morelloflavone, a biflavonoid, inhibits tumor angiogenesis by targeting rho GTPases and extracellular signal-regulated kinase signaling pathways. Cancer Res. 2009;69:518–525. doi: 10.1158/0008-5472.CAN-08-2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brill A, Dashevsky O, Rivo J, et al. Platelet-derived microparticles induce angiogenesis and stimulate post-ischemic revascularization. Cardiovasc Res. 2005;67:30–38. doi: 10.1016/j.cardiores.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 40.Wu YH, Hu TF, Chen YC, et al. The manipulation of miRNA-gene regulatory networks by KSHV induces endothelial cell motility. Blood. 2011;118:2896–2905. doi: 10.1182/blood-2011-01-330589. [DOI] [PubMed] [Google Scholar]
- 41.Benndorf RA, Schwedhelm E, Gnann A, et al. Isoprostanes inhibit vascular endothelial growth factor-induced endothelial cell migration, tube formation, and cardiac vessel sprouting in vitro, as well as angiogenesis in vivo via activation of the thromboxane A(2) receptor: A potential link between oxidative stress and impaired angiogenesis. Circ Res. 2008;103:1037–1046. doi: 10.1161/CIRCRESAHA.108.184036. [DOI] [PubMed] [Google Scholar]
- 42.Albig AR, Schiemann WP. Identification and characterization of regulator of G protein signaling 4 (RGS4) as a novel inhibitor of tubulogenesis: RGS4 inhibits mitogen-activated protein kinases and vascular endothelial growth factor signaling. Mol Biol Cell. 2005;16:609–625. doi: 10.1091/mbc.E04-06-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suárez Y, Sessa WC. MicroRNAs as novel regulators of angiogenesis. Circ Res. 2009;104:442–454. doi: 10.1161/CIRCRESAHA.108.191270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang S, Olson EN. AngiomiRs--key regulators of angiogenesis. Curr Opin Genet Dev. 2009;19:205–211. doi: 10.1016/j.gde.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang SH, Hla T. Gene regulation by RNA binding proteins and microRNAs in angiogenesis. Trends Mol Med. 2011;17:650–658. doi: 10.1016/j.molmed.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patella F, Rainaldi G. MicroRNAs mediate metabolic stresses and angiogenesis. Cell Mol Life Sci. 2012;69:1049–1065. doi: 10.1007/s00018-011-0775-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thum T. MicroRNA therapeutics in cardiovascular medicine. EMBO Mol Med. 2012;4:3–14. doi: 10.1002/emmm.201100191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suárez Y, Fernández-Hernando C, Yu J, et al. Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc Natl Acad Sci USA. 2008;105:14082–14087. doi: 10.1073/pnas.0804597105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Greco S, De Simone M, Colussi C, et al. Common micro-RNA signature in skeletal muscle damage and regeneration induced by Duchenne muscular dystrophy and acute ischemia. FASEB J. 2009;23:3335–3346. doi: 10.1096/fj.08-128579. [DOI] [PubMed] [Google Scholar]
- 50.Wang HW, Huang TS, Lo HH, et al. Deficiency of the microRNA-31-microRNA-720 pathway in the plasma and endothelial progenitor cells from patients with coronary artery disease. Arterioscler Thromb Vasc Biol. 2014;34:857–869. doi: 10.1161/ATVBAHA.113.303001. [DOI] [PubMed] [Google Scholar]
- 51.Huang M, Nguyen P, Jia F, et al. Double knockdown of prolyl hydroxylase and factor-inhibiting hypoxia-inducible factor with nonviral minicircle gene therapy enhances stem cell mobilization and angiogenesis after myocardial infarction. Circulation. 2011;124(suppl):S46–S54. doi: 10.1161/CIRCULATIONAHA.110.014019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ashton AW, Cheng Y, Helisch A, et al. Thromboxane A2 receptor agonists antagonize the proangiogenic effects of fibroblast growth factor-2: Role of receptor internalization, thrombospondin-1, and alpha(v)beta3. Circ Res. 2004;94:735–742. doi: 10.1161/01.RES.0000122043.11286.57. [DOI] [PubMed] [Google Scholar]
- 53.Lee SH, Kunz J, Lin SH, et al. 16-kDa prolactin inhibits endothelial cell migration by down-regulating the Ras-Tiam1-Rac1-Pak1 signaling pathway. Cancer Res. 2007;67:11045–11053. doi: 10.1158/0008-5472.CAN-07-0986. [DOI] [PubMed] [Google Scholar]
- 54.Cottonham CL, Kaneko S, Xu L. miR-21 and miR-31 converge on TIAM1 to regulate migration and invasion of colon carcinoma cells. J Biol Chem. 2010;285:35293–35302. doi: 10.1074/jbc.M110.160069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang T, Wang Q, Zhao D, et al. The oncogenetic role of microRNA-31 as a potential biomarker in oesophageal squamous cell carcinoma. Clin Sci (Lond) 2011;121:437–447. doi: 10.1042/CS20110207. [DOI] [PubMed] [Google Scholar]
- 56.Jaba IM, Zhuang ZW, Li N, et al. NO triggers RGS4 degradation to coordinate angiogenesis and cardiomyocyte growth. J Clin Invest. 2013;123:1718–1731. doi: 10.1172/JCI65112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roccaro AM, Sacco A, Maiso P, et al. BM mesenchymal stromal cell-derived exosomes facilitate multiple myeloma progression. J Clin Invest. 2013;123:1542–1555. doi: 10.1172/JCI66517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cantaluppi V, Gatti S, Medica D, et al. Microvesicles derived from endothelial progenitor cells protect the kidney from ischemia-reperfusion injury by microRNA-dependent reprogramming of resident renal cells. Kidney Int. 2012;82:412–427. doi: 10.1038/ki.2012.105. [DOI] [PubMed] [Google Scholar]
- 59.Valastyan S, Reinhardt F, Benaich N, et al. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis [retracted Cell. 2015 Apr 9;161(2):417] Cell. 2009;137:1032–1046. doi: 10.1016/j.cell.2015.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Krek A, Grün D, Poy MN, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 61.Tsai YH, Wu MF, Wu YH, et al. The M type K15 protein of Kaposi’s sarcoma-associated herpesvirus regulates microRNA expression via its SH2-binding motif to induce cell migration and invasion. J Virol. 2009;83:622–632. doi: 10.1128/JVI.00869-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 63.Peng H, Kaplan N, Hamanaka RB, et al. microRNA-31/factor-inhibiting hypoxia-inducible factor 1 nexus regulates keratinocyte differentiation. Proc Natl Acad Sci USA. 2012;109:14030–14034. doi: 10.1073/pnas.1111292109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lando D, Peet DJ, Gorman JJ, et al. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 2002;16:1466–1471. doi: 10.1101/gad.991402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rey S, Semenza GL. Hypoxia-inducible factor-1-dependent mechanisms of vascularization and vascular remodelling. Cardiovasc Res. 2010;86:236–242. doi: 10.1093/cvr/cvq045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Castanotto D, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457:426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.