Colorectal cancer stem cell (CSC)-enriched cultures were obtained efficiently from primary tumor specimens by optimizing a CSC-isolation protocol. Based on in vitro and in vivo validation, genetic characterization, and drug sensitivity analysis, panels of CSC lines were generated with defined patterns of genetic mutations and therapy sensitivity. An analysis of CSC response to EGFR-targeted therapy in vivo and an overview of factors implicated in therapy response/resistance are presented.
Keywords: Cancer stem cells, Colorectal cancer, Anti-EGFR therapy, Cetuximab, Proteomic arrays
Abstract
Colorectal cancer (CRC) therapy mainly relies on the use of conventional chemotherapeutic drugs combined, in a subset of patients, with epidermal growth factor receptor [EGFR]-targeting agents. Although CRC is considered a prototype of a cancer stem cell (CSC)-driven tumor, the effects of both conventional and targeted therapies on the CSC compartment are largely unknown. We have optimized a protocol for colorectal CSC isolation that allowed us to obtain CSC-enriched cultures from primary tumor specimens, with high efficiency. CSC isolation was followed by in vitro and in vivo validation, genetic characterization, and drug sensitivity analysis, thus generating panels of CSC lines with defined patterns of genetic mutations and therapy sensitivity. Colorectal CSC lines were polyclonal and maintained intratumor heterogeneity in terms of somatically acquired mutations and differentiation state. Such CSC-enriched cultures were used to investigate the effects of both conventional and targeted therapies on the CSC compartment in vivo and to generate a proteomic picture of signaling pathways implicated in sensitivity/resistance to anti-EGFR agents. We propose CSC lines as a sound preclinical framework to test the effects of therapies in vitro and in vivo and to identify novel determinants of therapy resistance.
Significance
Colorectal cancer stem cells (CSCs) have been shown to be responsible for tumor propagation, metastatic dissemination, and relapse. However, molecular pathways present in CSCs, as well as mechanisms of therapy resistance, are mostly unknown. Taking advantage of genetically characterized CSC lines derived from colorectal tumors, this study provides an extensive analysis of CSC response to EGFR-targeted therapy in vivo and an overview of factors implicated in therapy response or resistance. Furthermore, the implementation of a biobank of molecularly annotated CSC lines provides an innovative resource for future investigations in colorectal cancer.
Introduction
Colorectal cancer (CRC) represents one of the major causes of cancer-related deaths worldwide. Although the use of epidermal growth factor receptor (EGFR)-targeted agents has improved survival in a subset of patients with CRC, even sensitive tumors invariably develop therapy resistance, mainly because of the emergence of mutations that confer epidermal growth factor (EGF)-independent proliferation [1]. The hierarchical structure of CRC is dominated by cancer stem cells (CSCs), which have been shown to be responsible for tumor progression, metastasis, chemotherapy resistance, and tumor relapse [2]. Recent studies provided significant advancements in identifying CSC derivation, phenotype, and function, indicating a tight link between CSC features and patient prognosis [3–5]. However, the effect of anticancer drugs on the CSC compartment in vivo and the molecular mechanisms responsible for CSC resistance remain largely unexplored. We have implemented a protocol for CSC isolation as multicellular spheroids, followed by genomic, proteomic, and functional analyses of drug sensitivity. This method allowed us to generate genomic and proteomic profiles of CSCs sensitive and resistant to anti-EGFR agents, thus identifying differentially regulated factors that can contribute to therapy response. CRC tumor spheroids were used to investigate the in vivo effect of conventional and targeted therapies on the CSC compartment, providing evidence that anti-EGFR therapy can affect CSC number and can counteract CSC expansion induced by chemotherapy. Finally, we analyzed the sensitivity of KRASwt and KRASmut CSC lines to a panel of targeted pathway inhibitors as single agents and in combination with anti-EGFR. Altogether, the generation of a molecularly annotated bank of CSC lines represents a resource for future investigations of the molecular basis of CRC and, in particular, for studies on the role of CSC in therapy resistance.
Material and Methods
CSC Isolation and Culture
Fresh human colorectal cancer tissues were obtained in accordance with the standards of the ethics committee on human experimentation of the Istituto Superiore di Sanità (authorization no. CE5ISS 09/282). Tissue samples were collected by a surgeon or a pathologist immediately after each patient’s surgery, quickly washed 2–3 times in cold saline, then transferred in Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fisher Scientific, Carlsbad, CA, https://www.thermofisher.com) containing 3% penicillin-streptomycin-amphotericin B solution (Lonza Group, Walkersville, MD, http://www.lonza.com), and kept in this medium at 4°C until processing within 24–48 hours. For tissue dissociation, CRC samples were first washed 3–4 times in phosphate-buffered saline (PBS), then cut by forceps and/or scalpel in pieces of approximately 0.5 mm or smaller. Fragments were further washed twice by centrifugation at 150g for 3 minutes, then incubated in DMEM (Thermo Fisher Scientific) with 1.5 mg/ml collagenase type II (Thermo Fisher Scientific) and 20 μg/ml DNAse (Roche Diagnostics, Indianapolis, IN, https://usdiagnostics.roche.com) for 1 hour at 37°C under shaking. The cell suspension was then filtered through a 100-μm nylon mesh, and washed by 2 further centrifugation steps in DMEM. Finally, pellets containing cells, cell clusters, and tissue fragments were resuspended in CSC medium [6] supplemented with 10 mM nicotinamide, 1 µM Y-27632 (both from Sigma-Aldrich, St. Louis, MO, http://www.sigmaaldrich.com), 20 ng/ml human EGF and 10 ng/ml human basic fibroblast growth factor (both from PeproTech, London, U.K., https://www.peprotech.com). The resulting suspension was plated in ultra-low attachment tissue culture flasks (Corning Costar, Cambridge, MA, https://www.corning.com), and cultured in humidified atmosphere at 37°C, 5% CO2. Every 2 to 3 days, half of the culture medium was refreshed. In the first weeks of culture, cells were periodically centrifuged at 150g for 5 minutes, and the pellet was delicately passed 3 to 5 times through a 200 μl Gilson pipette tip in a small volume of medium; then the final medium volume was added and cells were replated. Clusters of proliferating cells became evident after a variable length of time, ranging from 5 to 7 days to 3 weeks. Bacterial contamination usually developed in approximately 20% of specimens within 3–4 days of culture. Cultures in which no proliferating clusters were detected after 4 weeks were discarded. Regular culture splitting (1:2) was usually needed after 3–6 weeks from isolation. Spheroids were then passaged weekly by mechanical dissociation or by incubation for 3–5 minutes at 37°C with TrypLE Express (Thermo Fisher). Cultures were usually used to prepare frozen stocks around the fifth passage and used for in vitro and in vivo experiments within the 12th passage.
Animal Procedures
All animal procedures were performed according to the Italian national animal experimentation guidelines (D.L.116/92) upon approval of the experimental protocol by the Italian Ministry of Health's Animal Experimentation Committee. We used 4- to 6-week-old female NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (The Jackson Laboratory, Bar Harbor, ME, https://www.jax.org) were used for all the validations and experiments. For CSC validation, 5 × 105 cells were injected subcutaneously in the flank of 3 replicate mice, in 100 μl 1:1 PBS/Matrigel (BD, Franklin Lakes, NJ, http://www.bd.com). In all the CSCs validated, xenografts were detectable within 3–5 weeks in at least 2 out of 3 mice. Palpable xenografts were extracted, formalin-fixed, and paraffin-embedded. Hematoxylin and eosin-stained sections were subsequently evaluated by a pathologist in comparison with the human tumor of origin. For drug testing, 5 × 105 CSCs were inoculated subcutaneously as described above. Tumors were measured twice weekly by an external digital caliper, and volumes were calculated using the following formula: π/6 × d2 × D, where d and D represent shorter and longer tumor measurements, respectively. Drug treatments were started after tumor establishment (100–150 mm3). Mice were treated with 10 mg/kg cetuximab (Merck KGaA, Darmstad, Germany, http://www.emdgroup.com) intravenously twice weekly. Irinotecan (Pfizer, New York, NY, http://www.pfizer.com) was administered intraperitoneally weekly at the dose of 15 mg/kg. Control animals were treated with vehicle only. For secondary transplantation experiments, tumors (six tumors per treatment group) were harvested and dissociated into single cells. For each individual tumor, cells were injected into secondary mice at serial doses ranging from 10 to 103. Mice were recorded as negative when no graft was observed after 24 weeks from inoculation.
Flow Cytometry
Single cells dissociated from spheroids or xenografts were washed with PBS/bovine serum albumin (BSA) 1% and incubated with the appropriate dilution of control or specific antibody in the same buffer for 45 minutes at 4°C. Fluorescence intensity of labeled cells was evaluated by a FACSCanto instrument (BD). Antibodies and controls used for routine analysis and experiments reported in the study were the following: anti-EpCAM-APC, anti-EpCAM-PE, and isotype controls mouse IgG2b-APC, mouse IgG1-APC (all from BD); mouse IgG1-PE, anti-Lgr5-PE, and anti-CD133-PE (Miltenyi Biotec, Bergisch Gladbach, Germany, http://www.miltenyibiotec.com); anti CD44v6-APC (R&D Systems, Minneapolis, MN, https://www.rndsystems.com). We always added 7-aminoactinomycin D (10 μg/ml; BD) to exclude dead cells.
Clonogenicity Assay
The clonogenic units in xenograft-derived cells were assessed by plating 500 cells/ml per well in triplicate in 24-well plates containing a soft agar bilayer (0.3% top and 0.4% bottom layer; SeaPlaque Agarose; Cambrex, East Rutherford, NJ, http://www.cambrex.com). Cultures were incubated in humidified atmosphere at 37°C and 5% CO2 for 21 days. Colonies were stained with crystal violet (0.01% in 10:1 methanol to water), and counted under a light microscope. Data represent the percentage of colonies normalized to the number of cells plated.
Reverse-Phase Protein Array Analysis
CSC pellets were lysed in buffer containing T-PER reagent (Thermo Fisher Scientific), 300 mM NaCl, 2 mM Pefabloc (Roche), 1 mM orthovanadate, 5 µg/ml aprotinin, 5 µg/ml pepstatin A, and 5 µg/ml leupeptin (all from Sigma-Aldrich), and were incubated on ice for 20 minutes. Samples were centrifuged at 10,000g for 5 minutes, supernatants were transferred to fresh tubes and total protein concentration was measured by Bradford reagent method (Coomassie protein assay; Thermo Fisher Scientific). Lysates were then diluted for printing in extraction buffer containing 50% T-PER, 47.5% 2× sodium dodecyl sulfate, and 2.5% β-mercaptoethanol (all from Thermo Fisher Scientific) to final concentrations of 0.5 mg/ml.
All samples were printed in technical triplicate spots and in four-point twofold dilution curve format onto nitrocellulose-coated glass slides (GRACE Bio-Laboratories, Bend, OR, http://www.gracebio.com). Reference standard lysates (i.e., HeLa plus pervanadate (BD), A431 plus EGF (BD), Jurkat plus etoposide (Cell Signaling Technology, Danvers, MA, http://www.cellsignal.com), and Jurkat plus calyculin A (Cell Signaling) were printed in 10-point decreasing mixtures of treated to untreated sample as procedural controls and as positive controls for antibody staining. Each reference standard curve was printed in technical triplicate at a final concentration of 0.5 mg/ml. An Aushon 2470 arrayer equipped with 185-µm pins (Aushon Biosystems, Billerica, MA, https://www.aushon.com) was used to print samples/slides according to the manufacturer’s instructions.
A selected subset of the printed microarray slides was stained with Sypro Ruby protein blot stain (Thermo Fisher) to estimate sample total protein concentration, and the remaining slides were stored under desiccated conditions at −20°C. Immediately before antibody staining, printed slides were treated with 1× Re-Blot Mild Solution (EMP Millipore, Temecula, CA, http://www.emdmillipore.com) for 15 minutes, washed for 5 minutes twice with PBS and incubated for 1 hour in blocking solution containing 2% I-Block (Applied Biosystems, Foster City, CA, http://www.appliedbiosystems.com) and 0.1% Tween-20 in PBS. Immunostaining was carried out using a tyramide-biotin signal amplification kit (Dako, Carpinteria, CA, http://www.dako.com). Arrays were probed with a library of 40 antibodies against antigens to either total, cleaved, or phosphorylated protein targets. Primary antibody binding was detected using a biotinylated goat anti-rabbit immunoglobulin G (IgG) H+L (1:7,500; Vector Laboratories, Burlingame, CA, https://www.vectorlabs.com) or rabbit anti-mouse Ig (1:10; Dako) followed by streptavidin-conjugated IRDye680LT fluorophore (LI-COR Biosciences, Lincoln, NE, https://www.licor.com). Primary antibodies had undergone pre- and post-reverse-phase protein array (RPPA) validation for single-band specificity by Western blot analysis using complex cellular lysates. Negative control slides were incubated with secondary antibody only and included in each staining run.
All Sypro- and immunostained slides were scanned using a Tecan Power Scanner (Tecan Group, Mannedorf, Switzerland, http://www.tecan.com) at 5-μm resolution. Acquired images were analyzed with MicroVigene version 5.2 (VigeneTech, Carlisle, MA, http://www.vigenetech.com) for spot detection, local background subtraction, negative control subtraction, replicate averaging, and total protein normalization. The packages ggplot2, Bioconductor, coin, and gplots of the “R” software for statistical computing (https://www.r-project.org) [7] were used to carry out quality control, internal standardization, two-way hierarchical clustering (Spearman correlation distance and Ward method), and two-sample Wilcoxon rank-sum tests (significance cutoff set at .05).
Western Blotting
Cell lysates were obtained from approximately 5 × 105 spheroid cells by incubation of cell pellets in 1% NP40 lysis buffer (20 mM TrisHCl pH 7.2; 200 mM NaCl; 1% NP40) supplemented with protease inhibitor cocktail and phosphatase inhibitor cocktails I and II (all from Sigma-Aldrich). Lysate concentrations were determined by Bradford assay (Bio-Rad Laboratories, Hercules, CA, http://www.bio-rad.com) and equal amounts of proteins were loaded on a gradient precast gel (4%–12% or 3%–8%; Thermo Fisher), then transferred to nitrocellulose membranes (GE Healthcare, Piscataway, NJ, http://www.gelifesciences.com). Blots were blocked with TBST 5% nonfat dry milk and incubated overnight at 4°C with primary antibodies, then incubated for 45 minutes with secondary horseradish peroxidase-conjugated antibodies dissolved in TBST 1% BSA. Chemiluminescent signals were detected with Super Signal West Pico (Thermo Fisher Scientific). Monoclonal anti-α-tubulin and monoclonal anti-β-actin were from Sigma-Aldrich; other antibodies were from Cell Signaling Technology.
Viability Assay and Drug Screening
CSC viability upon treatment with cetuximab and/or kinase inhibitors was determined by CellTiter-Glo luminescent cell viability assay (Promega, Madison, WI, http://www.promega.ca) according to the manufacturer’s directions. Briefly, CSCs (2.5–3 × 103 per well) were dissociated with TrypLE Express, seeded in 96-well plates (six replicates per experimental point) in CSC medium, and incubated in a humidified atmosphere at 37°C, 5% CO2. Luminescence was detected with a DTX880 multimode microplate reader (Beckman Coulter, Brea, CA, https://www.beckmancoulter.com). Cetuximab was always used at a concentration of 100 μg/ml and kinase inhibitors (Selleck Chemicals, Houston, TX, http://www.selleckchem.com) at 100 nM.
Statistical Analysis
Statistical significance of data was evaluated by analysis of variance (ANOVA) and Bonferroni posttests or by unpaired Student’s t test with Welch’s correction, depending on the experiment. Statistical significance up to .05 was accepted. Statistical analyses were performed using GraphPad Prism version 4.0 for Windows (GraphPad Software, San Diego, CA, http://www.graphpad.com). Serial transplantation/limiting dilution assays were analyzed by extreme limiting dilution analysis (ELDA) software [8].
DNA Extraction and Short Tandem Repeats Analysis
Genomic DNA was obtained by CSCs and patient-matched nontumoral tissues (Dnasy mini kit; Qiagen, Limburg, The Netherlands, https://www.qiagen.com). Short tandem repeats (STR) analysis was performed by using the AmpFlSTRIdentifiler Plus kit (Applied Biosystems). A unique STR profile was generated for each CSC line, which was used to monitor purity of the line over time and to confirm its matching with the biological material available for each patient included in the study.
Whole Exome Sequencing, Data Analysis, and Validation
Target enrichment was performed using in-solution technology (NimbleGen SeqCap EZ Library version 3.0; Roche) and the resulting target libraries were sequenced by Illumina sequencing technology (HiSeq2000; Illumina, San Diego, CA, http://www.illumina.com). Raw image files were processed by Illumina base-calling software (CASAVA 1.7) using default parameters. Paired-end reads were aligned to the human genome (UCSC GRCh37/hg19; University of California, Santa Cruz, Bioinformatics, Santa Cruz, CA, https://genome.ucsc.edu) with the Burrows-Wheeler aligner (version 0.7.10) [9]. Presumed polymerase chain reaction duplicates were removed using Picard’s MarkDuplicates (Broad Institute, Cambridge, MA, http://broadinstitute.github.io/picard). The Genome Analysis Toolkit (GATK 3.3; Broad Institute) [10] was used for realignment of sequences encompassing insertions and deletions (indels) and for base quality recalibration. Somatic single-nucleotide variants were detected using Mutect software version 1.1.6 (Broad Institute) [11], and small indels were identified through a comparison between indels called in individual CSC lines and their matched nontumoral samples by means of the GATK Haplotype Caller algorithm [12], applying the following quality filters: quality score greater than 100 and quality-by-depth score greater than 1.5. Indels below these thresholds or resulting from 4 or more reads having ambiguous mapping (this number being greater than 10% of all aligned reads) were discarded. The resulting single-nucleotide variants and small indels were annotated by SnpEff version 3.6 (http://snpeff.sourceforge.net) [13] and dbNSFP2.8 [14] in terms of functional impact of variants (i.e., missense or nonsense; coding or noncoding; location with respect to exon-intron junction, depth, reference/variant reads ratio, single nucleotide polymorphism database identification, amino acid change, and position; cancer association data; and structural/functional impact). Variant validation and genotyping were performed by Sanger sequencing. Amplicons were directly sequenced using the ABI BigDye Terminator Sequencing kit (Applied Biosystems) and an automated capillary sequencer (ABI 3500; Applied Biosystems). Sequence electropherograms were analyzed using Sequencing Analysis Software version 5.4 (Applied Biosystems).
Results
A Protocol for the Isolation, Profiling, and Functional Analysis of Colorectal CSC
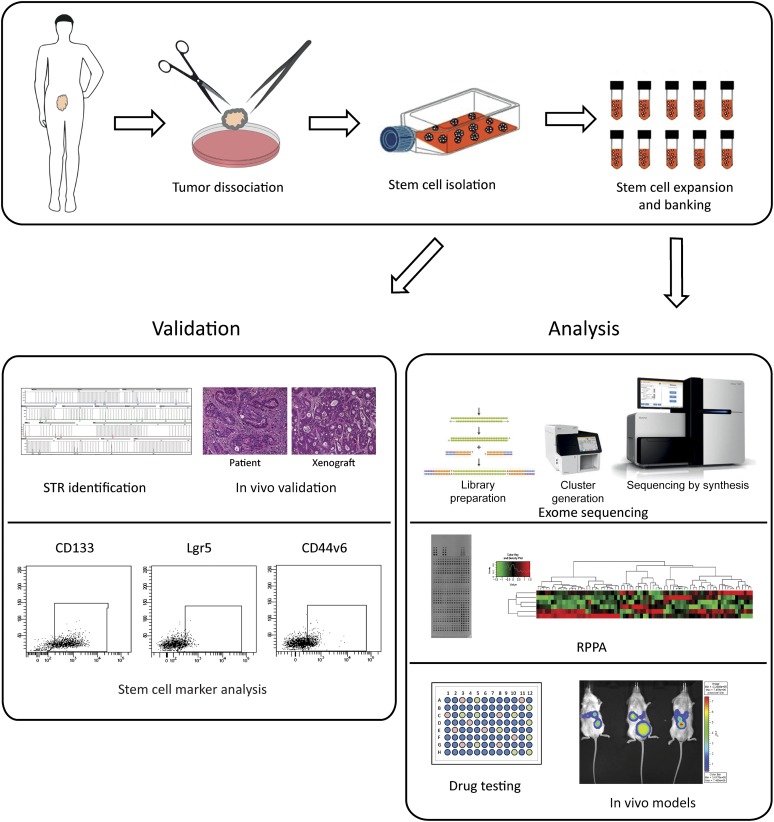
Previous studies by our and other laboratories reported that multicellular spheroid suspensions enriched in CSCs could be obtained by the selective culture of cells extracted from colorectal tumor specimens in serum-free media [6, 15–18]. We improved previous culture methods through supplementation with factors identified by Sato et al. (nicotinamide and Y-27632), which are also essential for the in vitro growth of normal intestinal stem cells [19], reaching approximately 40% of success in the generation of CSC cultures from tumor specimens (supplemental online Table 1). It is of note that our method for the generation of CSC-enriched cultures (called “CSC lines” hereafter) avoids previous in vivo passages that could exert a selective pressure on CRC cells by allowing the exclusive growth of clones compatible with the murine microenvironment. The resulting spheroid cultures are routinely validated for the expression of the stem cell markers CD133, Lgr5, and CD44v6 (which are usually present respectively at greater than 70%, approximately 10%, and approximately 30% positivity) and for their capability to reproduce a histological copy of the original patient tumor when inoculated in immunocompromised mice (Fig. 1; supplemental online Fig. 1), which, together, provide evidence for an enrichment in stem/progenitor cells. The high content of stem/progenitor cells in CSC lines is further corroborated by the elevated percentage of cells expressing GPF under the TCF/LEF promoter (TOP-GFP), which is an indicator of high WNT activity and is a functional marker of CSCs (supplemental online Fig. 2) [20]. Having fulfilled the in vitro and in vivo validation steps, CSC lines undergo identity verification through STR analysis and are subsequently frozen in a continuously expanding CSC bank comprising more than 60 CSC lines at the time of writing. Cells are then used for molecular analyses such as whole exome sequencing (WES), RPPA, and drug sensitivity tests (Fig. 1). Moreover, banked CSC lines can be used to produce subcutaneous tumor xenografts and models of tumor metastasis. The latter can be generated by injecting CSC lines transduced with a luciferase-encoding lentiviral vector either in the colon wall, where they give rise to a colon tumor that, in turn, produces spontaneous liver metastases, or in the spleen, where they generate a local tumor and subsequent liver metastases [21] (supplemental online Fig. 3).
Figure 1.
Workflow for cancer stem cell (CSC) isolation and banking. Newly isolated CSCs (top panel) are validated by STR identification and matching with normal patient’s tissue, by tumorigenicity assessment in immunodeficient mice and xenograft comparison with primary patient’s tumor, and, finally, by stem cell marker analysis (lower left panel). Following banking, CSCs are exome sequenced, analyzed by RPPA analysis, screened for drug sensitivity in vitro and used to produce xenograft models to allow preclinical drug validation (lower right panel). Abbreviations: RPPA, reverse phase proteomic array; STR, short tandem repeats analysis.
Mutational Layout and Genomic Profiling of Colorectal CSC Lines
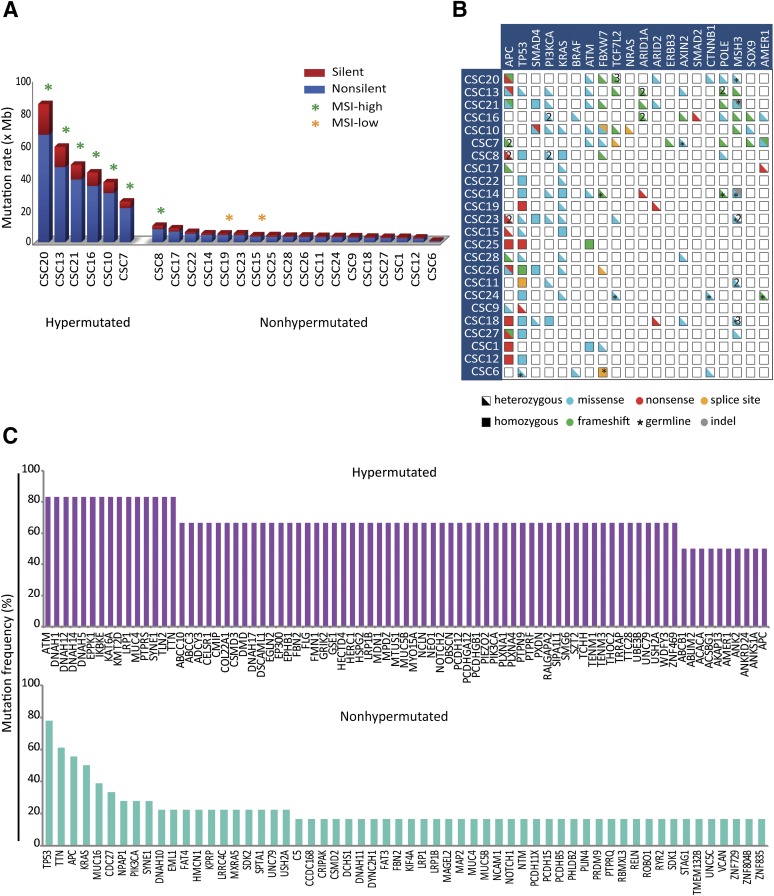
To provide an exhaustive profiling of somatically acquired mutations present in CSC lines, WES was systematically carried out on genomic DNA extracted from CSCs and patient-matched nontumoral tissues, as described in Materials and Methods. Approximately 90 million high-quality paired end reads, corresponding to 8 Gb, were obtained for each sample. The median depth across all samples was 74×. On average, 99% of all targeted bases were covered by at least 1 read, and 91% were covered by at least 20 reads (supplemental online Table 2). WES data analysis was directed to identify and annotate somatic variants within the coding sequence and flanking intronic regions, as well as copy-number variations and loss of heterozygosity regions. In a representative sample of 24 CSC lines, the average number of identified functionally relevant somatic variants (i.e., nonsynonymous and splice site changes) was 808, displaying marked differences among individual CSCs (supplemental online Table 2). Based on mutation rate per Mb, CSCs could be distinguished in hypermutated (more than 10 mutations/Mb) and nonhypermutated (fewer than 10 mutations/Mb) (Fig. 2A). The presence of both hypermutated and nonhypermutated CSC lines and their relative frequency (25%, slightly higher than 16% reported by the Cancer Genome Atlas Network [22]) implies a balanced generation of CSC cultures with respect to mutational rates.
Figure 2.
Whole exome sequencing (WES)-based mutation profiling of cancer stem cells (CSCs). (A): WES analysis allows estimation of CSC mutation rates, distinguishing hypermutated (more than 10 mutations per Mb) and nonhypermutated (fewer than 10 mutations per Mb) lines. High- and low-grade MSI is indicated by green and orange asterisks, respectively. Both silent (blue) and functionally relevant (red) somatic variants are reported. (B): An OncoPrint (cBioPortal, New York, NY, http://www.cbioportal.org) showing functionally relevant intragenic lesions in recurrently mutated genes for individual CSC lines. Half boxes and full boxes represent heterozygous and homozygous variants, respectively; colors are used to specify the type of mutation. Asterisks indicate germline mutations; multiple hits affecting the same gene are indicated by numbers. (C): Graphs showing the most frequently mutated genes in hypermutated (upper graph) and nonhypermutated (lower graph) CSC lines. Abbreviation: MSI, microsatellite instability.
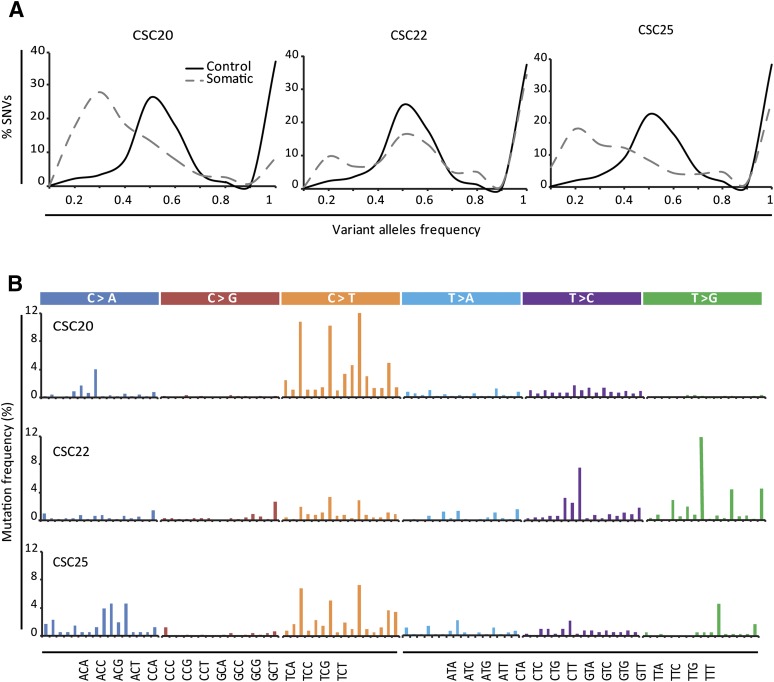
Analysis of the most common CRC mutations (Fig. 2B; supplemental online Table 3) revealed an occurrence of KRAS, BRAF, PIK3CA, APC, TP53, and SMAD4 mutations roughly corresponding to those reported in the COSMIC database, thus indicating the absence of gross mutational biases in the method of CSC generation. Also, a differential prevalence of gene mutations was documented in the hypermutated and nonhypermutated CSC lines (Fig. 2C); this was consistent with previously reported mutational profiles in CRC [22]. For most lines, the observed difference in the frequency distribution profiles of individual germline-inherited and somatically acquired variants supported their polyclonality (Fig. 3A). CSCs also differed in their mutational signatures, including the presence of distinctive signatures associated with colon cancer and defective mismatch repair [23] (Fig. 3B). Overall, these observations indicate that CSC lines faithfully reproduce the genomic complexity of colorectal tumors, thus representing a valuable tool to discover novel correlations between genetic mutations and therapy outcomes.
Figure 3.
Distribution of variant alleles in cancer stem cells (CSCs). (A): Frequency distribution patterns for the annotated germline inherited (solid line) and somatically acquired (dashed line) variants in a representative subset of CSC lines. (B): Mutational signatures of the same CSC lines as in (A). Each signature is displayed according to the 96 substitution classification defined by the substitution class and sequence context immediately 3′ and 5′ to the mutated base. Bars for the six types of substitutions are displayed in different colors. Abbreviation: SNV, single-nucleotide variant.
In Vitro and In Vivo Sensitivity of Colorectal CSC Lines to EGFR Targeting
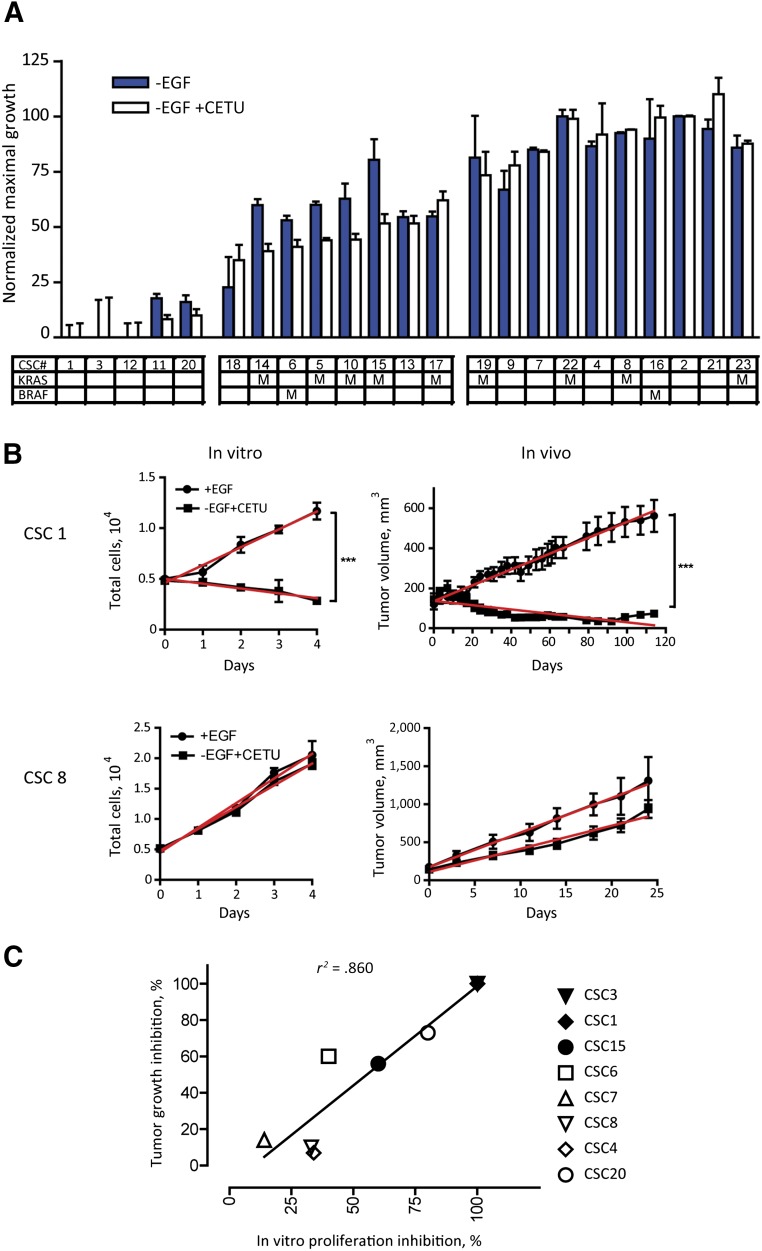
Monoclonal antibodies targeting the EGF receptor (EGFR), such as cetuximab and panitumumab, achieve clinically significant responses in a portion of patients with CRC with KRASwt. The molecular pathways active in resistant tumors that drive EGFR-independent proliferation have only been partly elucidated [24]. We hypothesized that colorectal CSC lines would represent a valuable model to investigate both the molecular determinants associated with anti-EGFR response and the in vivo effects of cetuximab on the CSC compartment. To validate this hypothesis, we first investigated cetuximab sensitivity in 23 CSC lines in relation to KRAS and BRAF mutational status. We performed this test in the absence of EGF, because we (supplemental online Fig. 4) and others [25] previously verified that the effect of cetuximab is masked by EGF normally present in CSC media, probably due to competition for the occupancy of the EGFR extracellular domain.
As shown in Figure 4A, individual CSC lines displayed a variable response to cetuximab treatment that was partly related to their mutational status. CSC lines with mutated KRAS or BRAF were all found to display partial or high resistance to anti-EGFR treatment. By contrast, all the lines with high sensitivity to cetuximab had wild-type KRAS and BRAF. As expected, KRASwt/BRAFwt CSCs were also found among cetuximab-resistant lines, confirming that other mechanisms are responsible for anti-EGFR resistance. Then, we investigated the therapeutic response of CSC-derived xenografts to cetuximab treatment. Figure 4B shows that a KRASwt CSC line sensitive to cetuximab-mediated growth inhibition in vitro gives rise to cetuximab-sensitive tumors in mice. Likewise, a KRASmut CSC line resistant to cetuximab in vitro generates resistant tumors in vivo. The correlation between CSC sensitivity to cetuximab in vitro and tumor response in vivo (r2 = 0.86) (Fig. 4C) was confirmed by analyzing 6 additional CSC lines (supplemental online Fig. 5) and indicates the presence of parallel sensitivity patterns in multicellular spheroids and in CSC-derived xenografts.
Figure 4.
Effect of epidermal growth factor receptor inhibition on CSC spheroids and matched xenografts. (A): CSC sensitivity to cetuximab in EGF-deprived cultures. Cell growth is expressed as the percentage of maximal growth occurring in complete CSC medium (i.e., in the presence of 20 ng/ml exogenous EGF). Mutational status for KRAS and BRAF is indicated for each CSC line. Spheroid culture viability was assessed at 72 hours by the CellTiter-Glo luminescent cell viability assay (Promega), as described in Materials and Methods. Data represent the average of three independent experiments ± SEM. (B): Time course of cetuximab inhibition on the growth of spheroid cultures (left panels) and of tumor xenografts (right panels) in representative CSC 1 (KRASwt) and CSC 8 (KRASmut). Spheroid viability was assessed as in (A); data represent the average of three independent experiments ± SEM. CSC subcutaneous xenografts were generated as described in Materials and Methods. After tumor establishment, mice were treated with 10 mg/kg cetuximab intravenously twice weekly. Data represent the average of 6–10 tumors per group. For all the tests, statistical significance was calculated by two-tailed Student’s t test. Linear regression is shown in red. For each test, inhibition can be expressed as the ratio of slopes (treated to untreated). (C): Plot of cetuximab-induced growth inhibition in spheroids versus xenografts calculated for eight CSC lines (panel B; supplemental online Fig. 5). Cetuximab inhibition of CSC spheroids strongly correlates with xenograft reduction (r2 = .860). All calculations were done with the GraphPad Prism software. Abbreviations: CETU, cetuximab; CSC, cancer stem cell; EGF, epidermal growth factor.
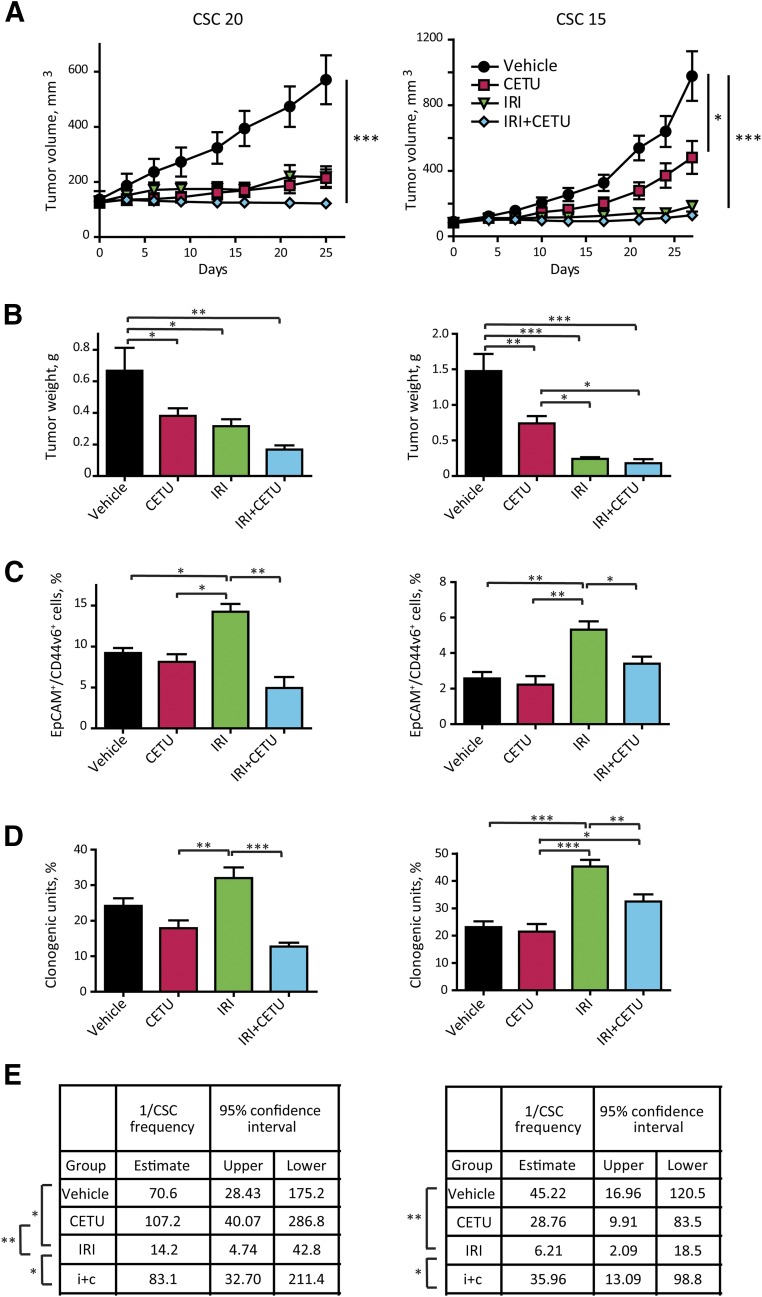
Starting from the assumption that the effect of EGFR inhibition on the CSC compartment in vivo is poorly understood, we took advantage of xenografts derived from two cetuximab-sensitive CSC lines (CSC 20, KRASwt/highly sensitive and CSC 15, KRASmut/medium sensitive) to investigate the effect of cetuximab on CSCs. Mice bearing subcutaneous CSC-derived tumors were treated with cetuximab, irinotecan, or with the cetuximab-irinotecan combination and, at the end of the treatment, the CSC compartment was evaluated through multiple methods. In xenografts derived from both CSC lines, cetuximab treatment induced a reduction of tumor growth that was proportional to the cells’ sensitivity to growth inhibition observed in vitro (Fig. 5A, 5B; supplemental online Fig. 5). In xenografts derived from highly sensitive CSCs, the effect of irinotecan was comparable to that of cetuximab, whereas in medium-sensitive tumors, irinotecan (and the irinotecan-cetuximab combination) was more effective than cetuximab alone in reducing tumor growth.
Figure 5.
Effect of epidermal growth factor receptor inhibition on the CSC compartment in vivo. (A): Effect of cetuximab and irinotecan on the growth of xenografts derived from highly cetuximab-responsive CSC 20 and medium-responsive CSC 15. After tumor establishment, mice were treated with either cetuximab (10 mg/kg intravenously twice weekly) or irinotecan (15 mg/kg intraperitoneally weekly) or their combination; control animals were treated with vehicle only. Data represent the average of 8–10 tumors ± SEM (B): Tumor weight at the time of excision. (C): Flow cytometry analysis for EpCAM/CD44v6 on cells dissociated from xenografts in (A). Data represent the average of six xenografts for group ± SD (D): Clonogenicity assay performed ex vivo on cells isolated from xenografts as in (A). Cells were plated in triplicate, at 500 cells/ml per dish as described in Materials and Methods. Colonies were scored at day 21. Data represent the average of six to eight xenografts per group ± SEM. Statistical significance in (A–D) was calculated with one-way analysis of variance. (E): Second transplantation of cells dissociated from xenografts of in (A). Cells dissociated from individual tumors (six tumors per treatment group) were retransplanted subcutaneously into secondary recipient mice. For each individual tumor, different cell doses were tested as explained in Materials and Methods. Mice were recorded as negative when no graft was observed after 24 weeks from the inoculation. CSC frequency was calculated with the extreme limiting dilution analysis software, as described in the Materials and Methods. Statistical significance of the results was calculated with chi-square analysis. ∗, p < .05, ∗∗, p < .01, ∗∗∗, p < .001. Abbreviations: CETU, cetuximab; CSC, cancer stem cell; IRI, irinotecan.
Then, we assessed the size of the CSC compartment in control and treated tumors through three different methods: frequency of CD44v6+ cells (Fig. 5C), ability of tumor cells to form colonies ex vivo in semisolid culture (Fig. 5D), and functional ability to form tumors upon serial transplantation (where CSC frequency in control and treated tumors was calculated with the ELDA software [8]) (Fig. 5E). Interestingly, the three stem-cell assays all showed that irinotecan increased the proportion of CSCs as compared with both vehicle-treated and cetuximab-treated tumors, while cetuximab did not significantly change the percentage of CSCs as compared with controls, indicating that anti-EGFR treatment affects stem and nonstem tumor cells to the same extent. Notably, the combination of cetuximab and irinotecan resulted in a significantly lower percentage of CSCs as compared with irinotecan alone, indicating that cetuximab efficiently counteracts chemotherapy-induced CSC selection.
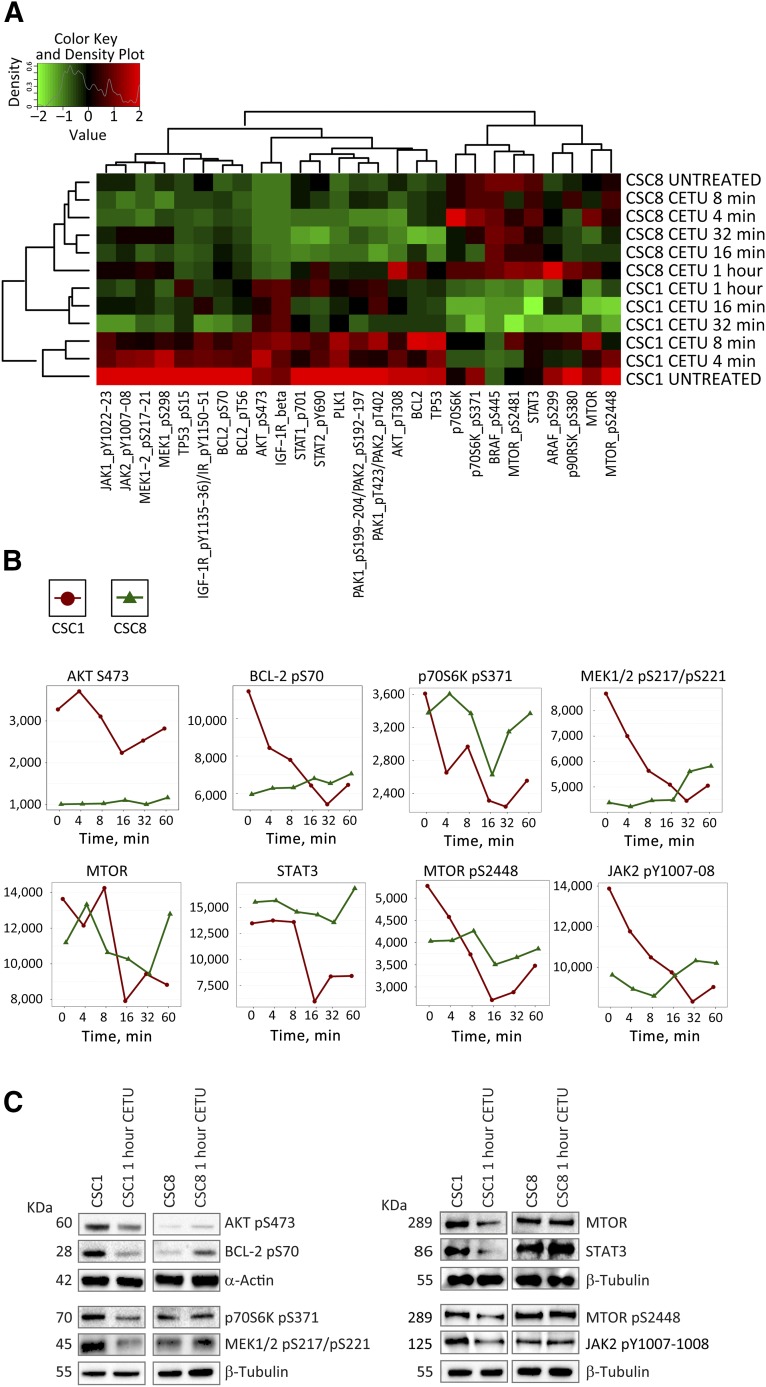
Proteomic-Based Identification of Molecular Pathways Involved in Anti-EGFR Sensitivity/Resistance
RPPA allows the simultaneous evaluation of phosphorylated, cleaved, or unmodified proteins generating comprehensive profiles of pathway activation in untreated and drug-treated cells [26]. To define the signaling pathways affected by anti-EGFR treatment in KRASwt/anti-EGFR-sensitive and KRASmut/resistant CSCs, we performed a RPPA analysis with 40 antibodies (supplemental online Table 4) of 2 CSC lines with opposite cetuximab sensitivity at different times of treatment (Fig. 6A). Selected significantly different endpoints (Fig. 6B) were validated by immunoblotting (Fig. 6C). Sensitive CSCs showed a decrease in phosphorylated AKT and its downstream target p70S6K upon cetuximab treatment, in parallel with a decrease in MEK1/2, BCL2, JAK2, and MTOR phosphorylation and in total STAT3. By contrast, resistant CSC showed variable levels of phospho-AKT, phospho-p70S6K, phospho-MTOR, and total MTOR, which, however, returned to the initial levels after 1 hour of treatment. Moreover, in resistant CSC, anti-EGFR treatment increased the levels of phospho-BCL2, phospho-JAK2, phospho-MEK1/2, and total STAT3. Overall, these observations suggest that cetuximab-sensitive cells undergo a coordinated downregulation of multiple survival pathways (specifically, the PI3K/AKT/MTOR, RAS/MEK/ERK, and JAK/STAT pathways) upon anti-EGFR treatment. By contrast, cetuximab-resistant cells may show a transitory decrease in survival factor activation but shortly recover baseline levels and may even show a reactive increase in the expression/activation of some factors at longer times of treatment upon anti-EGFR blockade.
Figure 6.
Reverse phase proteomic array (RPPA) for the identification of pathways differently activated in anti-epidermal growth factor receptor-sensitive and -resistant CSC. (A): Highly responsive (CSC1) and resistant (CSC8) CSC lines were treated with cetuximab and analyzed at different time points by RPPA. Statistical analysis of unsupervised clustering identified 27 endpoints differentially modulated in CSC1 and CSC8. Time-course plots (B) and Western blot validation (C) of eight relevant pathway effectors. Abbreviations: CETU, cetuximab; CSC, cancer stem cell.
Effects of Targeted Pathway Inhibition in Anti-EGFR Sensitive and Resistant CSCs
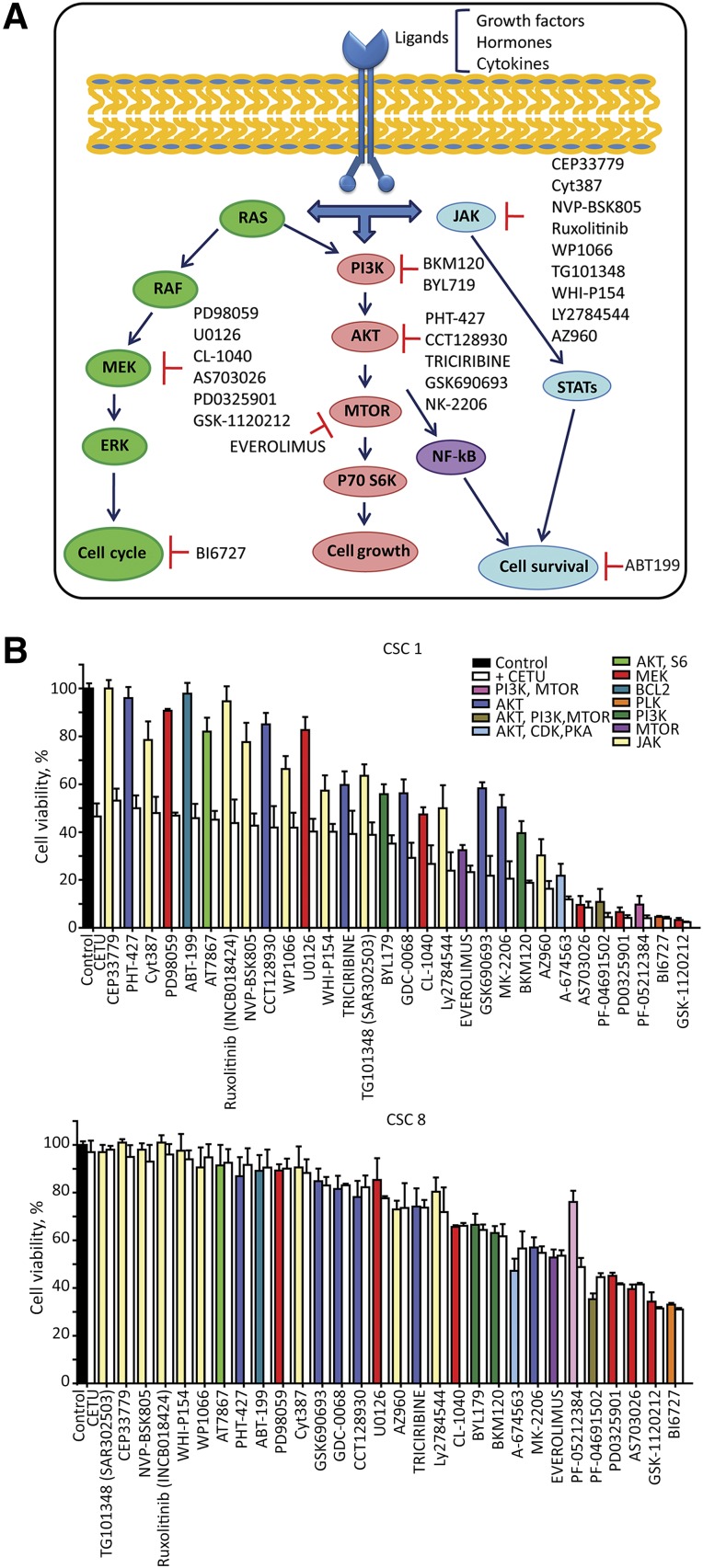
To further investigate the involvement of pathways identified by RPPA analysis in CSC survival, we compared the sensitivity of two CSC lines, one KRASwt (sensitive to anti-EGFR) and one KRASmut, with that of a panel of inhibitors directed against factors belonging to the three main survival pathways identified through RPPA analysis (Fig. 7A; supplemental online Table 5). Spheroids were dissociated 24 hours before treatment, seeded in 96-well plates, incubated with targeted inhibitors, and viability was assessed after 72 hours. In KRASwt cells, cetuximab as a single agent induced approximately a 50% loss of vitality. The combination of cetuximab with pathway inhibitors was invariably more effective than inhibitors alone in inducing CSC death, and, in some cases (particularly for multiple pathway inhibitors), had a strongly increased effect as compared with the single drugs (Fig. 7B, top panel). Cetuximab-resistant CSCs showed a completely different pattern of sensitivity to targeted inhibitors, being highly resistant to the majority of drugs tested, either alone or in combination with anti-EGFR (Fig. 7B, bottom panel). However, in cetuximab-resistant CSCs, we observed a synergy between anti-EGFR and the double PI3K-MTOR inhibitor PF-05212384 in decreasing cell survival, indicating a specific vulnerability to the combined inhibition of EGFR, PI3K, and MTOR pathways (Fig. 7B, bottom panel).
Figure 7.
Targeted pathway inhibition in anti-epidermal growth factor receptor (EGFR)-sensitive and -resistant CSCs. (A): Schematic representation of EGFR downstream pathway effectors. Small molecule inhibitors directed to relevant pathway effectors identified by reverse phase proteomic array are further specified in supplemental online Table 5. (B): Drug screening performed on dissociated spheroid cells highly responsive (CSC 1) or resistant (CSC 8) to anti-EGFR inhibition. Cells were treated as described in the Materials and Methods, viability was assessed with the CellTiter-Glo assay (Promega) and expressed as the percentage of dimethyl sulfoxide-treated control cells (black bar). Bars relative to inhibitor-treated cells were assigned different colors according to the pathway inhibited. Each compound was tested as a single agent at a 100 nM concentration (colored bars) or in combination with 100 μg/ml cetuximab (white bars). Samples were ordered on the basis of their sensitivity to the combination of cetuximab plus targeted inhibitor. Data represent the average of three independent experiments ± SEM. Abbreviations: CETU, cetuximab; CSC, cancer stem cell.
Discussion
The development of faithful in vitro and in vivo models to investigate the molecular basis of tumor development, progression, and relapse is a central issue in CRC research [2]. Advancements made in recent years include patient-derived spheroid cultures, organoid cultures, xenospheres, patient-derived xenografts, and short-term cultures of stem cells [15, 25, 27–29]. Each method of CRC cell isolation, expansion, and culture is associated with a series of drawbacks encompassing, from time to time, the emergence of culture-associated changes, genetic biases due to selective pressures acting during xenograft or culture establishment, low rates of success in cell isolation, inadequate cell expansion, and scarce manageability of cultures for molecular studies. Several reports have described methods for the isolation of CRC cells from primary tumors [30–35]. However, the ultimate value of such methods depends on multiple factors, including cell purity, presence and quantity of tumor-initiating cells, reproducibility, and the ability of cultured cells to phenocopy the original tumor in mice.
We have set up a protocol for CRC cell isolation, expansion, and characterization that allowed us to create a biobank of molecularly annotated patient-derived cells enriched in CSCs. Such cells represent a resource available for in vitro studies and in vivo evaluations of anticancer drug efficacy in subcutaneous or metastatic tumors. In comparison with previous methods used for the establishment of tumor-derived multicellular spheroid cultures, we have increased the yield of CSC lines from surgical specimens through an optimization of culture media and conditions that permits the development of CSC-enriched lines highly representative of patient tumors in terms of genetic landscape and drug sensitivity. Although the derivation of spheroid cultures from surgical specimens has a lower rate of success than organoid cultures, the resulting populations have highly increased cell yields and are suitable for studies that require considerable amounts of cellular materials, such as proteomic or metabolomic arrays. Moreover, tumor spheroids are easily amenable to genetic modification and can be used to modulate the expression of putative stem cell genes and to assess their effects on population dynamics [15]. Importantly, we show that spheroid cultures retain, at least in part, the genetic heterogeneity of tumors of origin, as demonstrated by the polyclonality and the complex pattern of mutations present within single CSC lines. Spheroid lines display heterogeneity also in terms of differentiated state, as shown by the expression of surface markers and nuclear β-catenin (revealed by the TOP-GFP system). The expression of “stem” cell markers (CD133, CD44v6, and Lgr5) in spheroid cultures suggests that they are prevalently composed by early progenitors (positive for CD133) with a significant presence of CSCs (positive for Lgr5 and/or CD44v6) and a fraction of terminally differentiated (TOP-GFPnegative) cells.
We used spheroid cultures to investigate the impact of anti-EGFR treatment on the CSC compartment in vivo, to identify the main pathways affected by anti-EGFR and to perform a proof-of-principle test of sensitivity to targeted pathway inhibitors in combination with cetuximab. In this context, we found that anti-EGFR treatment in KRASwt/cetuximab-sensitive xenografts affects both stem and nonstem cells, as shown by the smaller volume but unvaried percentage of CSCs found in cetuximab-treated tumors as compared with controls. This is in contrast with the increased CSC content of chemotherapy-treated tumors, which we found to be efficiently counteracted by the combination with anti-EGFR. These observations provide a strong rationale for the combined use of cetuximab-chemotherapy combinations, because surviving tumor cells should be less aggressive than those treated with chemotherapy alone.
Proteomic analysis of pathways affected by cetuximab treatment revealed coordinated changes in key factors belonging to the PI3K/AKT/MTOR, RAS/MEK/ERK, and JAK/STAT pathways that result in the death of KRASwt/cetuximab-sensitive cells. The same pathways present time-dependent variations in KRASmut cells but invariably return to a baseline state and, in some cases (as for BCL2), to a phosphorylation level higher than untreated cells, suggesting the possible occurrence of a reactive antiapoptotic state. Treatment of KRASwt/cetuximab-sensitive cells with a panel of inhibitors directed against the PI3K/AKT/MTOR, RAS/MEK/ERK, and JAK/STAT pathways showed an overall sensitivity to targeted drugs, which, in several cases, was increased by combination with cetuximab. By contrast, KRASmut cells were highly resistant to the majority of targeted drugs and, as predicted by the presence of constitutively activated KRAS, were insensitive to cetuximab as a single agent or in combination with pathway inhibitors. Notably, however, the dual PI3K/MTOR inhibitor PF-05212384 displayed a synergistic activity with cetuximab in KRASmut cells, suggesting a potential efficacy in inhibiting PI3K activity (both downstream of EGFR and from other sources) simultaneously with MTOR inhibition. This observation corroborates the clinical development of PF-05212384 (which recently showed a manageable safety profile and antitumor activity in phase I clinical trials [36]) and its potential use in combination with anti-EGFR in therapy-resistant colorectal cancer.
Conclusion
In aggregate, our results show that CSC-enriched spheroid cultures faithfully capture important features of primary colorectal tumors. Even more importantly, we show that CSC lines provide an efficient experimental system to perform panomic analyses and preclinical drug testing, thus leading to a deeper understanding of the molecular determinants of therapy resistance and of the tumor sensitivity to combination therapies.
Supplementary Material
Acknowledgments
We thank Dr. Marcella Mottolese for collection and useful comments on clinical information. We thank Paola Di Matteo, Roberto Ricci, Stefano Guida, and Paola Simeone for excellent technical assistance, and Enrico Duranti for immunohistochemical assistance. This work was financially supported by the Italian Association for Cancer Research (AIRC) with a 5 per Mille 9979 grant to Ruggero De Maria, Marco Tartaglia, and Giorgio Stassi, and an IG grant 15749 to Ann Zeuner.
Author Contributions
M.L.D.A.: experimental data generation, interpretation and assembly; A.Z.: data assembly, manuscript writing; E. Policicchio, A. Bruselles, and A.P.: genetic data collection and analysis; G.R., S.V., G.S., and L.R.-V.: CSC banking contribution; M.S.: proteomic data generation and analysis; G.D.L.: animal experiment assistance; E. Pilozzi and C.A.A.: tissue collection, histological analyses; A. Boe: flow cytometry; F.F.: experimental data generation; M.T.: genetic data supervision and assembly; R.D.M.: conception and design, financial support; M.B.: conception, design, data assembly and interpretation, manuscript writing.
Disclosure of Potential Conflicts of Interest
The authors indicated no potential conflicts of interest.
References
- 1.Chong CR, Jänne PA. The quest to overcome resistance to EGFR-targeted therapies in cancer. Nat Med. 2013;19:1389–1400. doi: 10.1038/nm.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zeuner A, Todaro M, Stassi G, et al. Colorectal cancer stem cells: From the crypt to the clinic. Cell Stem Cell. 2014;15:692–705. doi: 10.1016/j.stem.2014.11.012. [DOI] [PubMed] [Google Scholar]
- 3.De Sousa E Melo F, Wang X, Jansen M, et al. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat Med. 2013;19:614–618. doi: 10.1038/nm.3174. [DOI] [PubMed] [Google Scholar]
- 4.Merlos-Suárez A, Barriga FM, Jung P, et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell. 2011;8:511–524. doi: 10.1016/j.stem.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 5.Todaro M, Gaggianesi M, Catalano V, et al. CD44v6 is a marker of constitutive and reprogrammed cancer stem cells driving colon cancer metastasis. Cell Stem Cell. 2014;14:342–356. doi: 10.1016/j.stem.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 6.Francescangeli F, Patrizii M, Signore M, et al. Proliferation state and polo-like kinase1 dependence of tumorigenic colon cancer cells. Stem Cells. 2012;30:1819–1830. doi: 10.1002/stem.1163. [DOI] [PubMed] [Google Scholar]
- 7.R Foundation. The R Project for statistical computing. Available at http://www.R-project.org.
- 8.Hu Y, Smyth GK. ELDA: Extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 9.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cingolani P, Platts A, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu X, Jian X, Boerwinkle E. dbNSFP v2.0: A database of human non-synonymous SNVs and their functional predictions and annotations. Hum Mutat. 2013;34:E2393–E2402. doi: 10.1002/humu.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Francescangeli F, Contavalli P, De Angelis ML, et al. Dynamic regulation of the cancer stem cell compartment by Cripto-1 in colorectal cancer. Cell Death Differ. 2015;22:1700–1713. doi: 10.1038/cdd.2015.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vermeulen L, Todaro M, de Sousa Mello F, et al. Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity [published correction appears in Proc Natl Acad Sci USA 2009;106] Proc Natl Acad Sci USA. 2008;105:13427–13432. doi: 10.1073/pnas.0805706105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dieter SM, Ball CR, Hoffmann CM, et al. Distinct types of tumor-initiating cells form human colon cancer tumors and metastases. Cell Stem Cell. 2011;9:357–365. doi: 10.1016/j.stem.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 18.Cammareri P, Scopelliti A, Todaro M, et al. Aurora-a is essential for the tumorigenic capacity and chemoresistance of colorectal cancer stem cells. Cancer Res. 2010;70:4655–4665. doi: 10.1158/0008-5472.CAN-09-3953. [DOI] [PubMed] [Google Scholar]
- 19.Sato T, Stange DE, Ferrante M, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 20.Horst D, Chen J, Morikawa T, et al. Differential WNT activity in colorectal cancer confers limited tumorigenic potential and is regulated by MAPK signaling. Cancer Res. 2012;72:1547–1556. doi: 10.1158/0008-5472.CAN-11-3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baiocchi M, Biffoni M, Ricci-Vitiani L, et al. New models for cancer research: Human cancer stem cell xenografts. Curr Opin Pharmacol. 2010;10:380–384. doi: 10.1016/j.coph.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 22.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Emburgh BO, Sartore-Bianchi A, Di Nicolantonio F, et al. Acquired resistance to EGFR-targeted therapies in colorectal cancer. Mol Oncol. 2014;8:1084–1094. doi: 10.1016/j.molonc.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luraghi P, Reato G, Cipriano E, et al. MET signaling in colon cancer stem-like cells blunts the therapeutic response to EGFR inhibitors. Cancer Res. 2014;74:1857–1869. doi: 10.1158/0008-5472.CAN-13-2340-T. [DOI] [PubMed] [Google Scholar]
- 26.Gallagher RI, Espina V. Reverse phase protein arrays: Mapping the path towards personalized medicine. Mol Diagn Ther. 2014;18:619–630. doi: 10.1007/s40291-014-0122-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van de Wetering M, Francies HE, Francis JM, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161:933–945. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bertotti A, Migliardi G, Galimi F, et al. A molecularly annotated platform of patient-derived xenografts (“xenopatients”) identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer Discov. 2011;1:508–523. doi: 10.1158/2159-8290.CD-11-0109. [DOI] [PubMed] [Google Scholar]
- 29.D'Arcangelo M, Todaro M, Salvini J, et al. Cancer stem cells sensitivity assay (STELLA) in patients with advanced lung and colorectal cancer: A feasibility study. PLOS One. 2015;10:e0125037. doi: 10.1371/journal.pone.0125037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fang DD, Kim YJ, Lee CN, et al. Expansion of CD133(+) colon cancer cultures retaining stem cell properties to enable cancer stem cell target discovery. Br J Cancer. 2010;102:1265–1275. doi: 10.1038/sj.bjc.6605610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kondo J, Endo H, Okuyama H, et al. Retaining cell-cell contact enables preparation and culture of spheroids composed of pure primary cancer cells from colorectal cancer. Proc Natl Acad Sci USA. 2011;108:6235–6240. doi: 10.1073/pnas.1015938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohata H, Ishiguro T, Aihara Y, et al. Induction of the stem-like cell regulator CD44 by Rho kinase inhibition contributes to the maintenance of colon cancer-initiating cells. Cancer Res. 2012;72:5101–5110. doi: 10.1158/0008-5472.CAN-11-3812. [DOI] [PubMed] [Google Scholar]
- 33.Rajcevic U, Knol JC, Piersma S, et al. Colorectal cancer derived organotypic spheroids maintain essential tissue characteristics but adapt their metabolism in culture. Proteome Sci. 2014;12:39. doi: 10.1186/1477-5956-12-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karlsson H, Fryknäs M, Larsson R, et al. Loss of cancer drug activity in colon cancer HCT-116 cells during spheroid formation in a new 3-D spheroid cell culture system. Exp Cell Res. 2012;318:1577–1585. doi: 10.1016/j.yexcr.2012.03.026. [DOI] [PubMed] [Google Scholar]
- 35.Lee SH, Hong JH, Park HK, et al. Colorectal cancer-derived tumor spheroids retain the characteristics of original tumors. Cancer Lett. 2015;367:34–42. doi: 10.1016/j.canlet.2015.06.024. [DOI] [PubMed] [Google Scholar]
- 36.Shapiro GI, Bell-McGuinn KM, Molina JR, et al. First-in-human study of PF-05212384 (PKI-587), a small-molecule, intravenous, dual inhibitor of PI3K and mTOR in patients with advanced cancer. Clin Cancer Res. 2015;21:1888–1895. doi: 10.1158/1078-0432.CCR-14-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.