Highlights
-
•
We examine TP53 and lacZ mutagenesis induced by 3-nitrobenzanthrone (3-NBA).
-
•
We utilize Hupki (Human TP53 knock-in) fibroblasts (HUFs) wild-type and null for Xpa.
-
•
Xpa-Null HUFs are more sensitive to 3-NBA than Xpa-WT HUFs.
-
•
3-NBA induces a similar TP53 and lacZ mutant frequency in Xpa-WT and Xpa-Null HUFs.
-
•
3-NBA induces mutations at TP53 codons that are hotspots in smokers’ lung cancer.
Keywords: TP53, Mutation, Nucleotide excision repair, 3-Nitrobenzanthrone, DNA adduct, Environmental carcinogen
Abstract
3-Nitrobenzanthrone (3-NBA) is a highly mutagenic compound and possible human carcinogen found in diesel exhaust. 3-NBA forms bulky DNA adducts following metabolic activation and induces predominantly G:C > T:A transversions in a variety of experimental systems. Here we investigated the influence of nucleotide excision repair (NER) on 3-NBA-induced mutagenesis of the human tumour suppressor gene TP53 and the reporter gene lacZ. To this end we utilised Xpa -knockout (Xpa-Null) human TP53 knock-in (Hupki) embryo fibroblasts (HUFs). As Xpa is essential for NER of bulky DNA adducts, we hypothesized that DNA adducts induced by 3-NBA would persist in the genomes of Xpa-Null cells and lead to an increased frequency of mutation. The HUF immortalisation assay was used to select for cells harbouring TP53 mutations following mutagen exposure. We found that Xpa-Null Hupki mice and HUFs were more sensitive to 3-NBA treatment than their wild-type (Xpa-WT) counterparts. However, following 3-NBA treatment and immortalisation, a similar frequency of TP53-mutant clones arose from Xpa-WT and Xpa-Null HUF cultures. In cells from both Xpa genotypes G:C > T:A transversion was the predominant TP53 mutation type and mutations exhibited bias towards the non-transcribed strand. Thirty-two percent of 3-NBA-induced TP53 mutations occurred at CpG sites, all of which are hotspots for mutation in smokers’ lung cancer (codons 157, 158, 175, 245, 248, 273, 282). We also examined 3-NBA-induced mutagenesis of an integrated lacZ reporter gene in HUFs, where we again observed a similar mutant frequency in Xpa-WT and Xpa-Null cells. Our findings suggest that 3-NBA-DNA adducts may evade removal by global genomic NER; the persistence of 3-NBA adducts in DNA may be an important factor in its mutagenicity.
1. Introduction
Lung cancer is the most common malignant disease worldwide. While tobacco smoking is the predominant cause of lung cancer, vehicular exhaust and ambient air pollution are also implicated [1], [2]. In Great Britain diesel engine exhaust is the sixth most important occupational carcinogen [3]. Moreover, two recent epidemiological studies of miners provide strong evidence for a link between diesel exposure and lung cancer risk, finding a 3-fold increased risk for lung cancer overall and a 5-fold increased risk for miners most heavily exposed to diesel exhaust [4], [5]. Subsequently, the International Agency for Research on Cancer (IARC) has classified diesel engine exhaust as Group 1 human carcinogen [2].
The extremely potent mutagen 3-nitrobenzanthrone (3-nitro-7H-benz[de]anthracen-7-one, 3-NBA) can be found on the surface of ambient air particulate matter and diesel exhaust particles [6] and has been classified as a possible human carcinogen (Group 2B) by IARC [2]. Human exposure to 3-NBA has been demonstrated in workers occupationally exposed to diesel emissions [7] and intratracheal instillation of 3-NBA induces squamous cell carcinoma in rat lung [8]. Premutagenic bulky DNA adducts generated by 3-NBA are critical for its high mutagenic potency [9], [10]. In order to form DNA adducts, 3-NBA requires metabolic activation via reduction of its nitro group [11], [12]. The predominant DNA adducts detected in vitro and in vivo after treatment with 3-NBA are at guanine residues, i.e. 2-(2′-deoxyguanosin-N2-yl)-3-aminobenzanthrone (dG-N2-3-ABA) and N-(2′-deoxyguanosin-8-yl)-3-aminobenzanthrone (dG-C8-N-3-ABA) (see Supplementary Fig. 1) [12], [13]. The mutagenic potency of 3-NBA has been shown in a variety of experimental systems [14], [15]. 3-NBA-DNA adducts preferentially induce G:C > T:A transversions in vitro and in vivo in reporter genes [9], [10], [16].
3-NBA was also shown to induce G:C > T:A transversions in the tumour suppressor gene TP53 in immortalized embryonic fibroblasts derived from the human TP53 knock-in (Hupki) mouse, which harbours exons 4–9 of human TP53 in place of the corresponding mouse exons [17] [18]. TP53, which encodes the protein p53, is a key cancer gene and is somatically mutated in approximately half of human cancers [19], [20]. Over 28,000 TP53 mutations detected in human tumours have been catalogued in the IARC TP53 mutation database [21]. Some of these mutations in TP53 bear hallmarks of carcinogen exposure and can offer clues to tumour aetiology [22]. Carcinogen-induced TP53 mutagenesis can be studied in vitro using the Hupki mouse embryo fibroblast (HUF) immortalization assay (HIMA) [23], [24]. HUFs that acquire TP53 mutations following mutagen treatment can bypass culture-induced senescence and become immortalised clones, in which mutations can be identified by DNA sequencing. The HIMA allows a direct comparison between mutations induced in vitro in a human cancer gene (i.e. TP53) and mutations that occur in tumours of populations exposed to a particular carcinogen.
Bulky DNA adducts, such as those formed by polycyclic aromatic hydrocarbons (PAHs), can be removed from the genome by the nucleotide excision repair (NER) pathway [25], [26]. We developed a Hupki mouse strain harbouring a knockout allele for a critical NER component, Xpa (xeroderma pigmentosum complementation group A) (Hupki; Xpa+/−), with the aim of increasing TP53 mutation frequency in the HIMA [27]. Xpa is critical for both global genomic NER (GG-NER) and transcription-coupled NER (TC-NER), thus Xpa−/− (Xpa-Null) HUFs derived from Hupki; Xpa+/− mice are completely deficient in NER and incapable of removing bulky DNA adducts from their genome [25]. We recently showed that Xpa-Null HUFs, compared with Xpa+/+ (Xpa-WT) HUFs, are extremely sensitive to the PAH benzo[a]pyrene (BaP) and exhibit increased TP53 mutagenesis on the transcribed strand when treated with the reactive intermediate of BaP, benzo[a]pyrene-7,8-diol-9,10-epoxide (BPDE) [27].
In the present study, we examined the impact of NER-deficiency on 3-NBA-induced adduct formation (in vitro and in vivo) and TP53 mutagenesis in HUFs. We hypothesized that 3-NBA-induced DNA adducts would persist in the genomes of Xpa-Null Hupki mice and HUFs, and that these persistent adducts would lead to increased mutation induction during replication of adducted DNA [28], [29]. In addition to TP53 mutagenesis, we examined 3-NBA-induced mutations in a lacZ reporter gene in Xpa-WT and Xpa-Null HUFs derived from Hupki; Xpa +/− mice, as the pUR288 plasmid locus has also been integrated into this strain [30]. The pUR288 plasmid can be extracted from the genomic DNA of mutagen-treated cells following 1–3 days of proliferation and lacZ mutations identified by selection in Escherichia coli host cells [31]. In this way, the frequency of mutations induced by carcinogen treatment may be studied in a short-term reporter gene assay (in a manner that is not dependent on selection within HUFs) in parallel to TP53. As the HIMA takes several months to complete, we aimed to evaluate whether the mutagenic activity of a carcinogen (i.e. 3-NBA) could be rapidly assessed in HUFs using the lacZ system. This would potentially aid in the optimisation of experimental treatment conditions prior to initiation of the HIMA.
2. Materials and methods
2.1. Carcinogen information
3-NBA was synthesised as described previously [32]. For in vivo treatments 3-NBA was dissolved in tricaprylin at a concentration of 0.5 mg/mL. For treatment of cells in culture, 3-NBA was dissolved in DMSO to a stock concentration of 4 mM and stored at −20 °C.
2.2. Description of mouse strain and details of genotyping
A mouse strain homozygous for the Hupki allele (a knock-in allele harbouring exons 4–9 of the human TP53 gene) and heterozygous for an Xpa-knockout allele was generated as described previously [27]. This strain is on a mixed 129/Sv and C57Bl/6 background and also harbours the pUR288 plasmid (containing the bacterial lacZ mutation marker gene) [30], [33]. The pUR288 plasmid is chromosomally integrated in ∼20 tandem copies per haploid genome. More information can be found at the European Mouse Mutant Archive (EMMA; www.infrafrontier.eu) where this strain has been deposited (EMMA ID EM:08137). Hupki+/+; Xpa+/−; lacZ + mice were bred to produce Xpa+/+ (Xpa-WT) and Xpa−/− (Xpa-Null) mice and embryos for experiments. Genotyping for Hupki, Xpa and lacZ was performed as described previously [27] using the primers and PCR conditions detailed in Supplementary Table 1. Note that genotyping for lacZ cannot differentiate between genomes that are haploid or diploid for pUR288 but this does affect the outcome of the lacZ mutation assay.
2.3. In-vivo 3-NBA treatment
All animal experiments were carried out under license according to protocols approved by the Home Office under the Animals (Scientific Procedures) Act (1986) after approval by the institutional ethics committee. Groups of female Xpa-WT and Xpa-Null Hupki mice (∼3 months old; 20–25 g; n = 3/group) were treated with 3-NBA following a treatment protocol published previously [34]. In Groups 1–3 mice were treated with 2 mg/kg bw 3-NBA by intraperitoneal (i.p.) injection. Groups 1 and 2 received a single dose and were sacrificed 1 day and 5 days after the last administration, respectively. Group 3 was dosed once daily for 5 days and sacrificed 24 h after the last injection. Groups 4 and 5 were treated i.p. with 0.2 mg/kg bw 3-NBA. Whereas Group 4 was treated with a single dose and sacrificed 24 h later, Group 5 was dosed once daily for 5 days and sacrificed 24 h after the last administration. Matched control mice (n = 3) to Groups 3 and 5 received tricaprylin only. Liver, lung, small intestine, spleen, colon and kidney tissues were removed, snap frozen in liquid N2, and stored at −80 °C until analysis.
2.4. DNA adduct analysis by 32P-postlabelling
Genomic DNA was isolated from cells or tissue by a standard phenol/chloroform extraction method and stored at −20 °C. DNA adducts were measured in each DNA sample using the butanol enrichment version of the 32P-postlabelling method as described previously [32], [35] with minor modifications. Briefly, DNA samples (4 μg) were digested with micrococcal nuclease (120 mU; Sigma, #N3755) and calf spleen phosphodiesterase (40 mU; Calbiochem, #524711), enriched and labelled with 50 μCi [γ-32P]ATP (Hartmann Analytic #HP601ND) as reported. 3-NBA-derived DNA adducts were identified as reported [11], [12], [13].
2.5. Isolation and culture of primary HUFs
Xpa-WT and Xpa-Null Hupki mouse embryonic fibroblasts (HUFs) were isolated from day 13.5 embryos of intercrosses of Hupki+/+; Xpa+/−; lacZ+ mice according to a standard procedure, as described previously [27]. Isolated HUFs were cultured to 80–90% confluence for 2–3 days in growth medium (Dulbecco’s modified medium (Invitrogen #31966-021) supplemented with 10% fetal bovine serum (Invitrogen #26140-079) and 100 U/mL penicillin and streptomycin (Invitrogen #15140-130) at 37 °C/5% CO2/3% O2 before preparing frozen stocks (passage 0) and genotyping.
Cells were cultured in growth medium at 37 °C/5% CO2 in either 20% or 3% O2, adjusted using an incubator fitted with an oxygen sensor and a nitrogen source. Manipulations conducted outside the incubator were performed at 20% O2. For passaging cells were detached with 0.05% trypsin-EDTA (Invitrogen #25300-062), suspended in growth media and reseeded at the desired dilution or cell number.
2.6. Cell survival assay
Cell survival was assessed using a crystal violet staining assay as described previously [27]. Xpa-WT or Xpa-Null HUFs were seeded into 96-well plates at 2.5–5.0 × 103 cells/well and treated the following day with 3-NBA diluted in growth medium to a highest final concentration of 4 μM (0.1% DMSO). Cells were treated at 37 °C/5% CO2/3% O2 in 5 replicate wells per concentration of 3-NBA or DMSO (control). Following 24 or 48 h of treatment, cells were rinsed with PBS, stained for 15 min with 0.1% (w/v) crystal violet (Sigma #C3886) in 10% ethanol, washed again with PBS to remove excess dye and then allowed to air-dry. To quantify the crystal violet retained by surviving cells, 100 μL of 50% ethanol was added per well to solubilise the dye and the absorbance at 595 nm was determined using a plate reader. Data are presented as the percentage of A595nm in 3-NBA treated cells relative to that of control cells and are representative of at least three independent experiments.
2.7. 3-NBA treatment of HUFs for DNA adduct analysis
Xpa-WT and Xpa-Null HUFs were cultured and treated at 37 °C/5% CO2 in either 3% or 20% O2. HUFs (passage 1, ≤ 7 days in culture) were seeded at 1.0–1.5 × 106 cells/75-cm2 flask and treated the following day for 48 h. 3-NBA was diluted in growth medium to a final concentration of 0.1, 0.5, 1.0 or 2.0 μM (0.1% DMSO) and cells were treated in duplicate flasks per condition. Following treatment cells were trypsinised, pelleted and stored at −20 °C until further analysis.
2.8. Western blotting
Treated cells were lysed with 62.5 mM Tris–HCl pH 6.8, 500 mM EDTA pH 8.0, 2% sodium dodecyl sulphate (SDS) and 10% glycerol, supplemented with fresh protease inhibitors (78425; Thermo Fisher, UK). The expression of p53, phospho-p53 (S15), p21 and Gapdh in the lysates was assessed by SDS-PAGE and Western blotting as reported previously [36] using the following primary antibodies: anti-p53 (NCL-p53-CM5, Leica Microsystems, UK; 1:1,000), anti-phospho-p53 (#9284, Cell Signaling Technology, UK; 1:1,000); anti-p21 (#556431, BD Bioscience, UK; 1:2,000); and Gapdh (#MAB374, Millipore, UK; 1:25,000).
2.9. Hupki mouse embryo fibroblast immortalisation assay (HIMA)
Primary HUFs treated with 3-NBA were immortalised essentially as described previously [27], [37]. Frozen stocks (passage 0) of Xpa-WT and Xpa-Null primary HUFs were thawed and expanded in 175-cm2 flasks at 37 °C/5% CO2/3% O2 for 72 h until 80–90% confluent. Cells were trypsinised, counted and seeded at 2.0 × 105 cells/well into 6-well Corning CellBind® plates. Cells were treated the following day with 1 μM 3-NBA for 48 h (36 cultures, each Xpa genotype) or 2 × 48 h (24 cultures, each Xpa genotype). For cultures receiving 2 treatments, ∼300,000 cells from each culture (Xpa-WT and Xpa-Null) were reseeded following the first 48 h treatment with 3-NBA and then retreated the following day for a further 48 h. Additionally, a set of HUF cultures (54 cultures, each Xpa genotype) were treated with 0.1% DMSO in parallel [27].
Following treatment, as cultures approached confluence, HUFs were subcultured on 6-well Corning CellBind® plates at a dilution of 1:2–1:4. At Day 11 (post-thaw), all cultures were transferred to 20% O2 to select for senescence bypass. Cultures were serially passaged at dilutions of 1:2–1:4 until senescence/crisis (ceasing of cell division and enlarged morphology); during crisis cells received fresh media every 3 days. Serial passaging was resumed once regions of dividing cells emerged (immortalised clones). Cultures containing clones were split at dilutions of at least 1:2–1:4 for several passages, followed by further passaging at dilutions up to 1:20. Cultures were expanded to larger flasks (25-, 75-cm2) once they appeared homogenous and achieved a doubling rate of ≤ 48 h; frozen stocks were prepared and a portion of cells was pelleted for DNA extraction (≥ passage 12; 8–16 weeks).
2.10. TP53 mutation analysis
DNA for TP53 mutations analysis was prepared as described recently [27]. Human TP53-specific primers (exon 4 to exon 9, including introns, and cycling conditions are described in Supplementary Table 2). PCR amplification products were submitted to Beckman Coulter Genomics (Takeley, UK) for Sanger dideoxy fluorescent sequencing using the sequencing primers indicated in Supplementary Table 2. Sequences were assessed for the presence of TP53 mutations as described [27]. Variations (e.g. single base substitutions, deletions) were assessed using the mutation validation tool available at the IARC TP53 mutation database (http://www-p53.iarc.fr/MutationValidationCriteria.asp) and could be classified as either homo-/hemi-zygous or heterozygous. Mutations were confirmed by sequencing DNA from an independent sample of cells from the same clone.
2.11. lacZ mutation assay
Xpa-WT and Xpa-Null primary HUFs (passage 0) carrying the lacZ reporter gene were thawed and expanded at 37 °C/5% CO2/3% O2 for 72 h. Cells (1.0–1.5 × 106) were seeded into 75-cm2 flasks (5 flasks per treatment). The following day cells were treated with 3-NBA (0.1, 0.5, 1.0 or 2.0 μM; cytotoxic and sub-cytotoxic concentrations) or solvent control (0.1% DMSO) for 48 h. After treatment, cells from each flask were trypsinised, counted and reseeded at 1.75 × 106 cells into 175-cm2 flasks to allow cells to proliferate and fix mutations. After 96 h, when control cultures were ∼90% confluent, cells were trypsinised, pelleted and stored at −20 °C until DNA isolation. DNA was isolated from cells by standard phenol/chloroform extraction. pUR288 (lacZ) plasmid rescue and mutant frequency determination were performed as described previously [38], [39], [40]. Briefly, 10–30 μg of DNA were digested with HindIII and then incubated with magnetic beads coated with lacI fusion protein. Plasmids were eluted from the beads using isopropyl-β-D-thiogalactopyranoside (IPTG), circularised at the cohesive HindIII sites using T4 DNA ligase and then electroporated into E. coli deficient in β-galactosidase (lacZ−) and galactose epimerase (galE−). One thousandth of the transformed bacteria was plated on nonselective, titre plates (containing X-Gal (5- bromo-4-chloro-3-indolyl-β-D-galactopyranoside)) and the remainder was plated onto selective plates (containing the lactose analog P-Gal (phenyl-β-D galactosidase)). Mutant frequencies were calculated as the ratio between the number of mutant colonies on the selective plate to the total number of colonies on the titre plate (times the dilution factor of 1,000).
3. Results
3.1. DNA adduct formation by 3-NBA in Xpa-WT and Xpa-Null Hupki mice
The effect of NER deficiency on 3-NBA-induced DNA adduct formation in vivo was assessed by 32P-postlabelling (Fig. 1). The DNA adduct pattern induced by 3-NBA (see Supplementary Fig. 2) consisted of up to four adduct spots. These were identified previously [11], [12], [13] as deoxyadenosine (adduct spots 1 and 2) and deoxyguanosine adducts (adduct spots 3 and 4), of which three have been structurally characterised as 2-(2′-deoxyadenosin-N6 aminobenzanthrone (dA-N6-3-ABA; spot 1), 2-(2′-deoxyguanosin-N2-yl)-3-aminobenzanthrone (dG-N2-3-ABA; spot 3) and N-(2′-deoxyguanosin-8-yl)-3-aminobenzanthrone (dG-C8-N-3-ABA; spot 4). The structures of these adducts are shown in Supplementary Fig. 1.
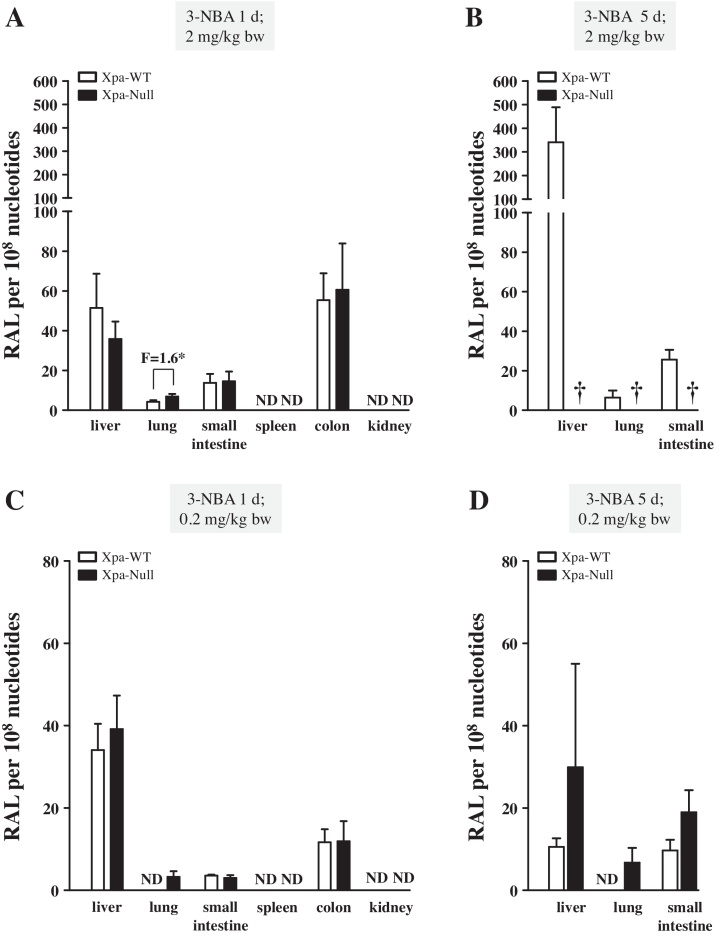
Fig. 1.
DNA adduct formation in Xpa-WT and Xpa-Null Hupki mice treated with 3-NBA. Mice were treated with 3-NBA (2 or 0.2 mg/kg bw) either once (A) and (C) or once a day for 5 days (B) and (D). DNA adduct levels (RAL, relative adduct labelling) in the indicated tissues were assessed by 32P-postlabelling. See Supplementary Fig. 2 for representative autoradiograms showing the adduct profiles in examined tissues. Values represent means ± SD from 3 animals, and each DNA sample was measured by two independent 32P-postlabelling analyses. ND = not detected; † = not determined due to death of Xpa-Null animals. Statistical analysis, comparing adduct levels in tissues from Xpa-WT and Xpa-Null mice, was performed using the Student’s t -test; * P < 0.05.
One day after a single administration of 2 mg/kg bw 3-NBA, Xpa-Null mice had a 1.6-fold higher level of DNA adducts in the lung compared to Xpa-WT animals but similar DNA adduct levels in the liver, small intestine and colon (Fig. 1A). No adducts were detected in spleen or kidney. The highest levels of adducts were detected in liver (51 ± 17 [Xpa-WT] versus 36 ± 9 adducts per 108 nt [Xpa-Null]) and colon (55 ± 13 [Xpa-WT] versus 61 ± 23 adducts per 108 nt [Xpa-Null]). While 3-NBA was well tolerated by Xpa-WT mice, all Xpa-Null mice died within 2–3 days of 3-NBA treatment (2 mg/kg bw) (Fig. 1B and Supplementary Fig. 3). 3-NBA-DNA adduct levels persisted in the tissues of Xpa-WT mice 5 days after a single treatment (Supplementary Fig. 3). Further, 3-NBA-DNA adduct levels increased nearly 6-fold in the liver and ∼2-fold in the small intestine of mice that were treated daily for 5 consecutive days (Fig. 1B).
As Xpa-Null mice were highly sensitive to treatment with 3-NBA at 2 mg/kg bw, animals were subsequently treated with a 10-fold lower dose (0.2 mg/kg bw) in a follow-up experiment (Fig. 1C and D). In mice treated once with 0.2 mg/kg bw 3-NBA, 3-NBA-DNA adducts were detectable in the lungs of Xpa-Null mice but not Xpa-WT mice; no significant difference between DNA adduct formation in Xpa-WT and Xpa-Null animals was found in other organs with detectable levels of 3-NBA-DNA adducts (i.e. liver, small intestine, and colon). Following a single treatment with 0.2 mg/kg bw 3-NBA, DNA adduct levels in the lung, small intestine and colon were at least 2-fold lower, or undetectable, (Fig. 1C) compared with tissues from mice that received 2 mg/kg bw (Fig. 1A). In contrast, adduct levels in the liver were similar after low or high dose treatment. Both Xpa-WT and Xpa-Null mice survived 5 daily treatments of 0.2 mg/kg bw 3-NBA. Again 3-NBA-DNA adducts were detectable in the lungs of Xpa-Null mice but not in Xpa-WT mice, indicating that 3-NBA-DNA adducts were repaired by NER. In the other two tissues (liver and small intestine) assessed from Group 5 animals, a trend for higher DNA adduct levels in Xpa-Null mice than in Xpa-WT mice was found (Fig. 1D), but the observed difference did not reach statistical significance. Interestingly, adduct levels in the livers of both genotypes from Group 5 mice were lower than those detected for Group 4 animals (Fig. 1C).
3.2. 3-NBA-induced DNA adduct formation in Xpa-WT HUFs cultured in 3% or 20% O2
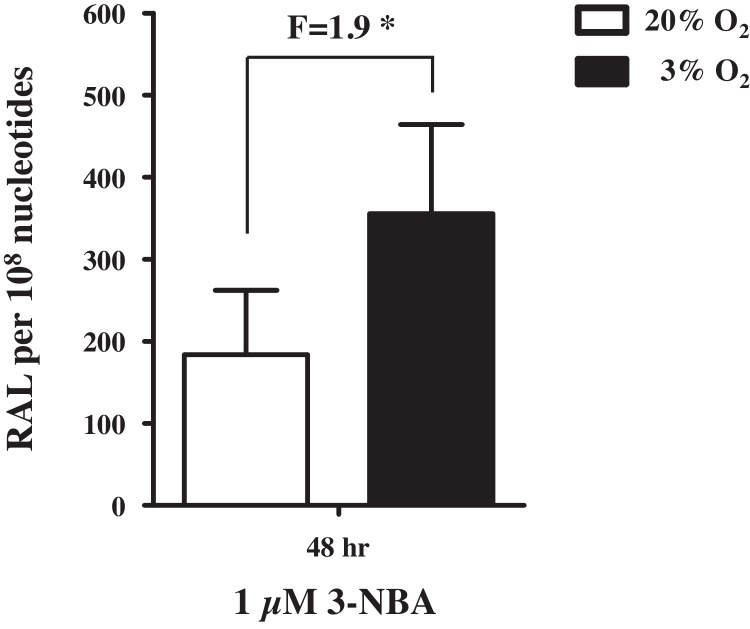
We then went on to examine DNA adduct formation in HUFs treated with 3-NBA. Previously, we showed that when HUFs were cultured in physiological O2 (3%) their replicative potential was enhanced [27]. This suggested that cells could be cultured in 3% O2 before and during mutagen treatment to increase the number of cells available for experiments. Cells would then be transferred to 20% O2 to select for mutated cells capable of senescence bypass. The impact of oxygen on 3-NBA metabolism and 3-NBA-induced DNA damage is unknown. Thus, we treated primary Xpa-WT HUFs grown in 3% or 20% O2 with 1 μM 3-NBA for 48 h to assess DNA adduct formation. We found that DNA adduct levels were 2-fold higher in HUFs treated with 3-NBA in 3% O2 compared with cells treated in 20% O2 (Fig. 2). Therefore, all subsequent 3-NBA treatments of HUFs in this study were performed in 3% O2.
Fig. 2.
3-NBA-induced DNA adduct formation in HUFs cultured at 20% or 3% O2. Xpa-WT HUFs cultured in 20% or 3% O2 were treated for 48 h with 1 μM 3-NBA. DNA adduct formation (RAL, relative adduct labelling) was assessed by 32P-postlabelling. Values represent means ± SD of two biological replicates where each DNA sample was measured by two independent 32P-postlabelling analyses. Statistical analysis, comparing DNA adduct formation at 20% and 3% O2, was performed using the Student’s t -test; * P < 0.05.
3.3. DNA adduct formation and survival of 3-NBA-treated Xpa-WT and Xpa-Null HUFs
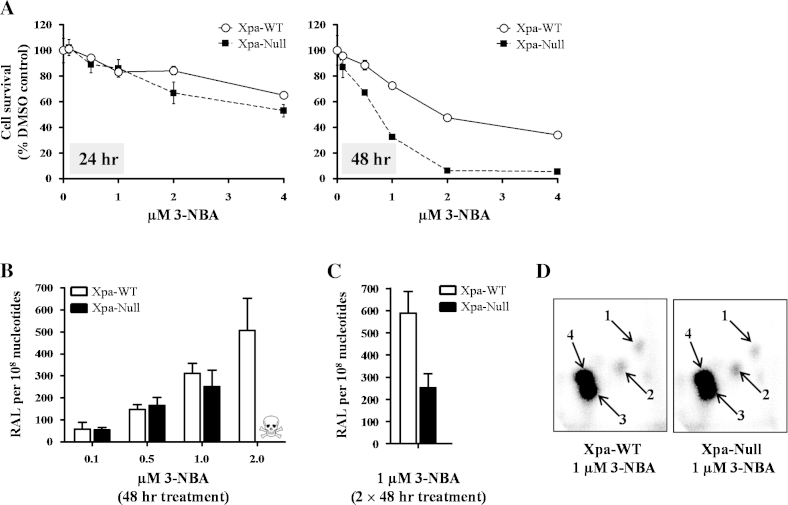
Xpa-WT and Xpa-Null HUFs were compared for their sensitivity (i.e. cell survival) to 3-NBA after 24 or 48 h treatment (Fig. 3A). Xpa-Null HUFs were more sensitive to 3-NBA treatment; however, the decrease in cell survival was more pronounced after 48 h. After 24-h treatment with 1 μM 3-NBA, 80% of Xpa-Null cells had survived (similar for Xpa-WT), whereas after 48-h treatment, the surviving fraction was ∼30% of control (∼70% for Xpa-WT). At higher doses of 3-NBA (2–4 μM), most Xpa-Null cells were dead after 48 h, whereas at least 40% of Xpa-WT cells had survived.
Fig. 3.
Survival and DNA adduct formation in Xpa-WT and Xpa-Null HUFs treated with 3-NBA. (A) Cells were treated for 24 or 48 h with the indicated doses of 3-NBA and survival was measured using a crystal violet staining assay. Cells treated with 0.1% DMSO only served as control. Mean values are presented as% of control ± SD of 5 replicate wells and are representative of at least three independent experiments (variation ≤ 15%). (B) and (C) Cells were treated with 3-NBA (as indicated) for 48 h or 2 × 48 h and DNA adduct formation (RAL, relative adduct labelling) was assessed by 32P-postlabelling. Values represent means ± SD of two biological replicates where each DNA sample was measured by two independent 32P-postlabelling analyses. Due to severe loss of viability, DNA adduct formation was not assessed in Xpa-Null cells treated with 2 μM 3-NBA (indicated by skull and crossbones). (D) Autoradiographic profiles of DNA adducts obtained in 3-NBA-treated HUFs; arrows indicate spot 1: 2-(2′-deoxyadenosin-N6-yl)-3-aminobenzanthrone (dA-N6-3-ABA), spot 3: 2-(2′-deoxyguanosin-N2-yl)-3-aminobenzanthrone (dG-N2-3-ABA) and spot 4: N-(2′-deoxyguanosin-8-yl)-3-aminobenzanthrone (dG-C8-N-3-ABA). Spot 2 is a deoxyadenosine adduct that has not yet been structurally characterised. The origins, in the bottom left-hand corners, were cut off before exposure.
Next, DNA adduct formation induced by 3-NBA was assessed in Xpa-WT and Xpa-Null HUFs. Cells were treated with 0.1–2.0 μM of 3-NBA for 48 h (Fig. 3B). Although proficient in NER, Xpa-WT HUFs accumulated similar levels of DNA adducts (up to ∼300 adducts per 108 nt) to Xpa-Null HUFs after treatment with 0.1–1.0 μM 3-NBA. However, while Xpa-WT HUFs accrued 507 ± 144 adducts per 108 nt after 2 μM 3-NBA treatment, Xpa-Null HUFs did not survive treatment at that concentration. We concluded from these data that a 48-h treatment with 1 μM 3-NBA induced substantial DNA damage in both Xpa-WT and Xpa-Null HUFs (Xpa-WT: ∼300 adducts per 108 nt; Xpa-Null: ∼250 adducts per 108 nt) while preserving a population of viable cells that could proliferate and accumulate mutations (Xpa-WT: ∼85% survival; Xpa-Null: ∼30% survival). Further, as previous HIMAs have often employed repeated treatments with a mutagen [18], [41], we also assessed DNA adduct formation in HUFs following 2 × 48-h treatments with 1 μM 3-NBA (Fig. 3C). In Xpa-WT HUFs, two treatments with 1 μM 3-NBA increased the DNA adduct level ∼2-fold over the level induced by a single treatment, whereas in Xpa-Null HUFs DNA adduct levels were nearly the same after one or two treatments. Cell survival of both Xpa-WT and Xpa-Null HUFs was ∼2-fold lower following repeated treatment with 3-NBA than after a single treatment (data not shown).
The adduct patterns induced by 3-NBA (Fig. 3D) consisted of four adduct spots that were identical to those observed in several organs of Xpa-WT and Xpa-Null Hupki mice treated with 3-NBA in vivo (see Supplementary Fig. 2). No DNA adducts were detected in HUFs treated with solvent (DMSO) only (data not shown).
3.4. Induction of the p53 pathway in Xpa-WT and Xpa-Null HUFs after 3-NBA treatment
Induction of p53 in 3-NBA-treated Xpa-WT and Xpa-Null HUFs was assessed by immunoblotting (Supplementary Fig. 4). Expression of p53, and phosphorylation on serine 15, was highly induced after treatment of Xpa-Null HUFs with 0.5 and 1.0 μM 3-NBA, coupled with a moderate induction of p21, a p53 transcriptional target. Similar treatment of Xpa-WT cells only slightly induced expression and phosphorylation of p53, and resulted in a very modest increase in p21 expression. The highest dose of 3-NBA (2 μM) tested in Xpa-WT HUFs strongly induced phosphorylation of p53 and increased p53 expression to a level similar to that observed in Xpa-Null HUFs treated with 0.5 μM 3-NBA. HUFs were also treated with 0.25 μM BaP for comparison. BaP treatment strongly induced p53 expression and phosphorylation in Xpa-Null cells, whereas the p53 response was much weaker in the Xpa-WT cells.
3.5. TP53 mutations induced by 3-NBA in Xpa-WT and Xpa-Null HUFs
3.5.1. Mutation frequency
We next examined the effect of NER-deficiency on 3-NBA-induced TP53 mutagenesis using the HIMA. Xpa-WT and Xpa-Null HUFs were treated either 1 × 48 h or 2 × 48 h with 1 μM 3-NBA and then serially passaged for ≥12 passages (8–16 weeks) until established into immortalised clones. Untreated cells were immortalised in parallel [27]. Mutations in exons 4–9 and adjacent introns of the human TP53 sequence in immortal HUF clones are shown in Table 1, Table 2.
Table 1.
Summary of TP53 mutation frequency and pattern detected in immortalised clones of Xpa-WT and Xpa-Null HUFs treated with 3-NBA.
| 3-NBA | Xpa-WT | Xpa-Null | control | Xpa-WT | Xpa-Null |
|---|---|---|---|---|---|
| Total 3-NBA-treated HUF cultures (#) | 60 | 60 | Total 0.1% DMSO-treated HUF cultures (#) | 54 | 54 |
| TP53-mutant immortalised clones (#) | 11 | 11 | TP53-mutant immortalised clones (#) | 2 | 2 |
| Total TP53 mutations detected (#) | 14 | 12 | Total TP53 mutations detected (#) | 2 | 2 |
| Frequency of TP53-mutant clones | 18.3% (11/60) | 18.3% (11/60) | Frequency of TP53-mutant clones | 3.7% (2/54) | 3.7% (2/54) |
| Mutations on the transcribed strand | 42.8% (6/14) | 8.3% (1/12) | Mutations on the transcribed strand | 50% (1/2) | 100% (2/2) |
| TP53 mutation pattern | TP53 mutation pattern | ||||
| G to A | 21.4% (3/14) | 16.7% (2/12) | G to A | 0% (0/2) | 0% (0/2) |
| G to C | 21.4% (3/14) | 25% (3/12) | G to C | 50% (1/2) | 0% (0/2) |
| G to T | 28.6% (4/14) | 50% (6/12) | G to T | 0% (0/2) | 0% (0/2) |
| A to C | 14.3% (2/14) | 0% (0/12) | A to C | 50% (1/2) | 100% (2/2) |
| A to G | 7.1% (1/14) | 0% (0/12) | A to G | 0% (0/2) | 0% (0/2) |
| A to T | 7.1% (1/14) | 0% (0/12) | A to T | 0% (0/2) | 0% (0/2) |
| Ins. G | 0% (0/14) | 8.3% (1/12) | Ins. G | 0% (0/2) | 0% (0/2) |
| TP53 mutation pattern at CpG sites | TP53 mutation pattern at CpG sites | ||||
| Total G mutations at CpG | 40% (4/10) | 41.7% (5/12) | Total G mutations at CpG | 0% (0/1) | 0% (0/0) |
| G to A at CpG | 66.7% (2/3) | 0% (0/2) | G to A at CpG | 0% (0/0) | 0% (0/0) |
| G to C at CpG | 0% (0/3) | 33.3% (1/3) | G to C at CpG | 0% (0/1) | 0% (0/0) |
| G to T at CpG | 50% (2/4) | 66.7% (4/6) | G to T at CpG | 0% (0/0) | 0% (0/0) |
| Ins. G at CpG | 0% (0/0) | 0% (0/1) | Ins. G at CpG | 0% (0/0) | 0% (0/0) |
Table 2.
TP53 mutations induced by 3-NBA in immortalised Xpa-WT and Xpa-Null HUFs.
| Xpa status | Treatmenta | Clone Idb | Codon # | Exon | Mutation type | Strand | WT codon | MUT codon | Coding change | CpG | Zygosity | Contact, structure, zinc | Activityc |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TP53-mutated immortalised clones from HUFs treated with 1 μM 3-NBA | |||||||||||||
| WT | 2 | XW-3N-9 | 120 | 4 | A:T > T:A | NTS | AAG | ATG | K120M | Homo-/hemi- | C | NF | |
| WT | 1 | XW-3N-59 | 158 | 5 | G:C > T:A | NTS | CGC | CTC | R158L | CpG | Homo-/hemi- | NF | |
| WT | 2 | XW-3N-14 | 161 | 5 | G:C > C:G | NTS | GCC | CCC | A161P | Hetero- | PF | ||
| WT | 2 | XW-3N-15 | 161 | 5 | G:C > C:G | TS | GCC | GGC | A161G | Hetero- | PF | ||
| WT | 1 | XW-3N-55 | 175 | 5 | G:C > T:A | NTS | CGC | CTC | R175L | CpG | Homo-/hemi- | S | PF |
| WT | 1 | XW-3N-54 | 178 | 5 | G:C > T:A | TS | CAC | CAA | H178Q | Hetero- | NF | ||
| WT | 2 | XW-3N-8 | 179 | 5 | G:C > T:A | TS | CAT | AAT | H179N | Homo-/hemi- | Z | PF | |
| WT | 1 | XW-3N-54 | 179 | 5 | A:T > C:G | NTS | CAT | CCT | H179P | Hetero- | Z | PF | |
| WT | 2 | XW-3N-15 | 194 | 6 | A:T > C:G | TS | CTT | CGT | L194R | Hetero- | NF | ||
| WT | 2 | XW-3N-7 | 236 | 7 | A:T > G:C | NTS | TAC | TGC | Y236C | Homo-/hemi- | NF | ||
| WT | 1 | XW-3N-43 | 248 | 7 | G:C > A:T | TS | CGG | TGG | R248W | CpG | Hetero- | C | NF |
| WT | 2 | XW-3N-14 | 249 | 7 | G:C > C:G | NTS | AGG | AGC | R249S | Hetero- | S | NF | |
| WT | 1 | XW-3N-37 | 282 | 8 | G:C > A:T | TS | CGG | TGG | R282W | CpG | Homo-/hemi- | S | NF |
| WT | 1 | XW-3N-29 | in. 3 (SA) | G:C > A:T | NTS | Homo-/hemi- | N/A | ||||||
| Null | 1 | XN-3N-151 | 124 | 4 | insertion, G | NTS | TGC | TG-G | Frameshift | Homo-/hemi- | N/A | ||
| Null | 2 | XN-3N-117 | 157 | 5 | G:C > T:A | NTS | GTC | TTC | V157F | CpG | Hetero- | NF | |
| Null | 2 | XN-3N-120 | 157 | 5 | G:C > T:A | NTS | GTC | TTC | V157F | CpG | Hetero- | NF | |
| Null | 1 | XN-3N-137 | 244 | 7 | G:C > C:G | NTS | GGC | GCC | G244A | Hetero- | NF | ||
| Null | 1 | XN-3N-136 | 245 | 7 | G:C > C:G | NTS | GGC | CGC | G245R | CpG | Homo-/hemi- | S | NF |
| Null | 1 | XN-3N-137 | 249 | 7 | G:C > C:G | NTS | AGG | ACG | R249T | Hetero- | S | NF | |
| Null | 1 | XN-3N-156 | 273 | 8 | G:C > T:A | TS | CGT | AGT | R273S | CpG | Hetero- | C | NF |
| Null | 2 | XN-3N-105 | 273 | 8 | G:C > T:A | NTS | CGT | CTT | R273L | CpG | Hetero- | C | NF |
| Null | 1 | XN-3N-140 | 277 | 8 | G:C > T:A | NTS | TGT | TTT | C277F | Hetero- | C | NF | |
| Null | 1 | XN-3N-141 | 277 | 8 | G:C > A:T | NTS | TGT | TAT | C277Y | Hetero- | C | NF | |
| Null | 2 | XN-3N-110 | in. 5 (SA) | G:C > A:T | NTS | splice | Homo-/hemi- | N/A | |||||
| Null | 1 | XN-3N-157 | in. 8 (SA) | G:C > T:A | NTS | splice | Homo-/hemi- | N/A | |||||
| TP53-mutated spontaneously immortalised clones from control HUFs (treated with 0.1% DMSO)d | |||||||||||||
| WT | XW-C-115 | 138 | 5 | G:C > C:G | NTS | GCC | CCC | A138P | Homo-/hemi- | NF | |||
| WT | XW-C-137 | 173 | 5 | A:T > C:G | TS | GTG | GGG | V173G | Homo-/hemi- | NF | |||
| Null | XN-C-325 | 113 | 4 | A:T > C:G | TS | TTC | GTC | F113V | Homo-/hemi- | NF | |||
| Null | XN-C-338 | 113 | 4 | A:T > C:G | TS | TTC | GTC | F113V | Hetero- | NF | |||
Treatment refers to 1 or 2 × 48 h.
XW = Xpa-WT; XN = Xpa-Null; 3N = 3-NBA- treated; C = control.
The overall transactivation activity of the mutant in a yeast functional assay published by Kato et al. [46]. (NF = non-functional, PF = partially functional, F = functional, NA = not assessed).
As published previously [27].
As described previously [27], TP53 mutations were only detected in two spontaneously immortalised HUF clones from untreated cultures of either genotype (3.7%) (Table 1). Following 3-NBA treatment the frequency of TP53-mutated clones increased to 18.3% in both Xpa-WT and Xpa- Null cultures. A similar frequency of TP53-mutated clones was generated following a single or double treatment with 3-NBA. A total of 14 TP53 mutations were detected in 11 immortal clones derived from 3-NBA-treated Xpa-WT cultures (three clones contained two mutations each) and 12 TP53 mutations were detected in 11 immortal clones derived from 3-NBA-treated Xpa-Null cultures (1 clone harboured 2 mutations). Treatment with 3-NBA clearly increased the frequency of TP53 mutations over that of untreated cultures, but, interestingly, Xpa-deficiency did not further increase the mutation frequency induced by 3-NBA.
3.5.2. Mutation pattern and sequence context
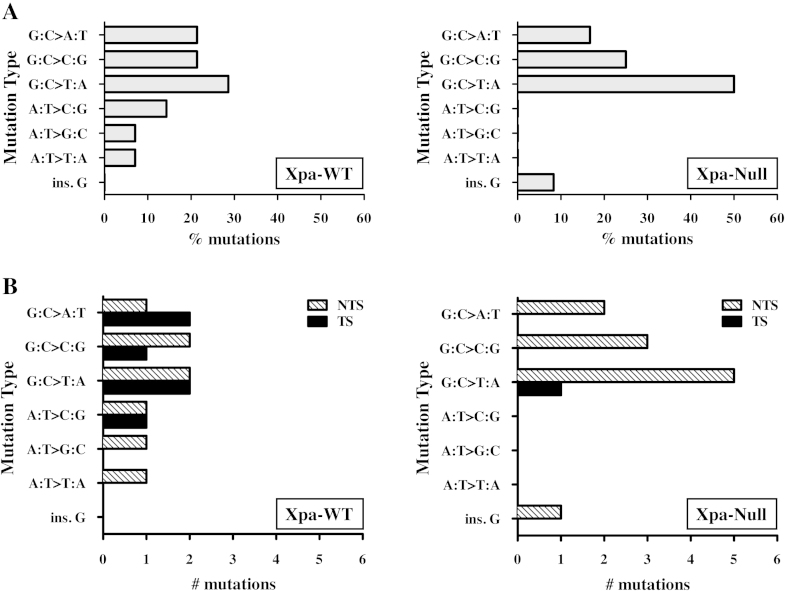
All but one of the TP53 mutations (an insertion) were single base substitutions and occurred at splice sites or in coding sequences, resulting in an amino acid substitution. No silent or nonsense mutations were found. The predominant mutation type identified in clones from 3-NBA-treated cultures was G:C > T:A transversion, representing 29% of the mutations in Xpa-WT clones and 50% of the mutations in Xpa-Null clones (Table 1 and Fig. 4). With the exception of one clone that contained an insertion (XN-3N-151), all of the remaining mutations found in the 3-NBA-treated Xpa-Null clones also occurred at G:C base pairs; 25% were G:C > C:G transversions and 17% were G:C > A:T transitions. G:C > C:G and G:C > A:T base changes were similarly found in 3-NBA-treated Xpa-WT clones (∼20% for both mutation types). Guanines at CpG dinucleotides were often mutational targets in both Xpa-WT and Xpa-Null clones. Overall, 40% of G:C > A:T transitions and 60% of G:C > T:A transversions occurred at CpG sites (Supplementary Table 3). G:C > G:C transversions, on the other hand, occurred most frequently in a sequence context of 5′G-G-C3′.
Fig. 4.
TP53 mutation pattern and strand bias in 3-NBA-treated Xpa-WT and Xpa-Null HUFs. (A) The % of each type of single base substitution or insertion is shown. (B) The number of each type of single base substitution or insertion occurring on the non-transcribed strand (NTS) or transcribed strand (TS) is shown.
In contrast to 3-NBA-treated Xpa-Null clones, ∼30% of the mutations in Xpa-WT clones occurred at A:T base pairs. This mutation type was not found in any 3-NBA-treated Xpa-Null clones. In untreated control cultures, three of the four TP53 mutations identified were A:T > C:G base substitutions. This mutation type was not found in 3-NBA-treated Xpa-Null cultures, but was found in 2 clones from 3-NBA-treated Xpa-WT HUFs (XW-3N-15 and -54; Table 2); notably, these clones also harboured a second TP53 mutation.
3.5.3. Mutational strand bias
As the non-transcribed strand of TP53 is repaired more slowly than the transcribed strand in cells with normal NER capacity, mutations induced by carcinogens such as BaP or UV are preferentially found on the non-transcribed strand [42], [43]. In cells deficient in TC-NER, however, an increase in mutations on the transcribed strand may occur [44]. Indeed, we previously showed that BPDE-induced TP53 mutations were more frequently induced on the transcribed strand in Xpa-Null HUFs compared with Xpa-WT HUFs [27]. Interestingly, here we did not find an excess of 3-NBA-induced TP53 mutations on the transcribed strand in Xpa-Null HUFs (Table 2, Fig. 4). In fact, only one Xpa-Null clone harboured a mutated guanine on the transcribed strand (G:C > T:A at codon 273) compared with five such mutations in Xpa-WT clones (G:C > T:A at codons 178 and 179; G:C > C:G at codon 161; G:C > A:T at codons 248 and 282).
3.5.4. Codon distribution and effect of mutations on p53 function
3-NBA-induced TP53 mutations occurred in all sequenced exons (4–9). Only one codon, 249, was mutated in both Xpa-WT and Xpa-Null HUFs. In both cases the mutation type at codon 249 was G:C > C:G, although a different guanine was targeted in the Xpa-WT and Xpa-Null clones (AGG to ACG vs . AGG to AGC, respectively). Of the seven residues in the p53 core DNA binding domain that make direct contact with DNA bases (K120, S241, R248, R273, A276, C277, R280), two were mutated in Xpa-Null clones (R273 and C277) and two were mutated in Xpa-WT clones (K120 and R248) [45]. Additionally, several mutations were induced at residues that support the structure of the DNA binding surface of p53: G245 and R249 in Xpa-Null clones; R175, R249 and R282 in Xpa-WT clones. H179, a residue important for coordinating zinc-binding, was mutated in two Xpa-WT clones.
The majority of the 3-NBA-induced TP53 mutations were classified as non-functional, based on a comprehensive study of mutant transactivation function in yeast [46]; five mutations were classified as partially functional. Three of the mutations with partial functionality (A161P, A161G, H179P) were detected in clones harbouring two mutations (XW-3N-14, -15, 54), where the second mutation was non-functional (R249S, L194R, and H178Q, respectively). This suggests, as discussed previously [27], that the HIMA selects for clones harbouring non-functional p53 mutants and that p53 mutants retaining partial functionality may not be capable of senescence bypass.
3.5.5. Comparison of 3-NBA-induced TP53 mutations with mutations found in human cancer
The TP53 mutations induced by 3-NBA were compared with TP53 mutations found in human cancer, as catalogued in the IARC TP53 mutation database, version R17 (Fig. 5). The frequency of each mutation was assessed in lung cancer of smokers and non-smokers, as well as cancer overall (Fig. 5B–D). Each mutation induced by 3-NBA in this study has been found in at least one human tumour. Further, all codons and splice sites mutated by 3-NBA have been targeted in lung tumours, including 16/25 identical mutations.
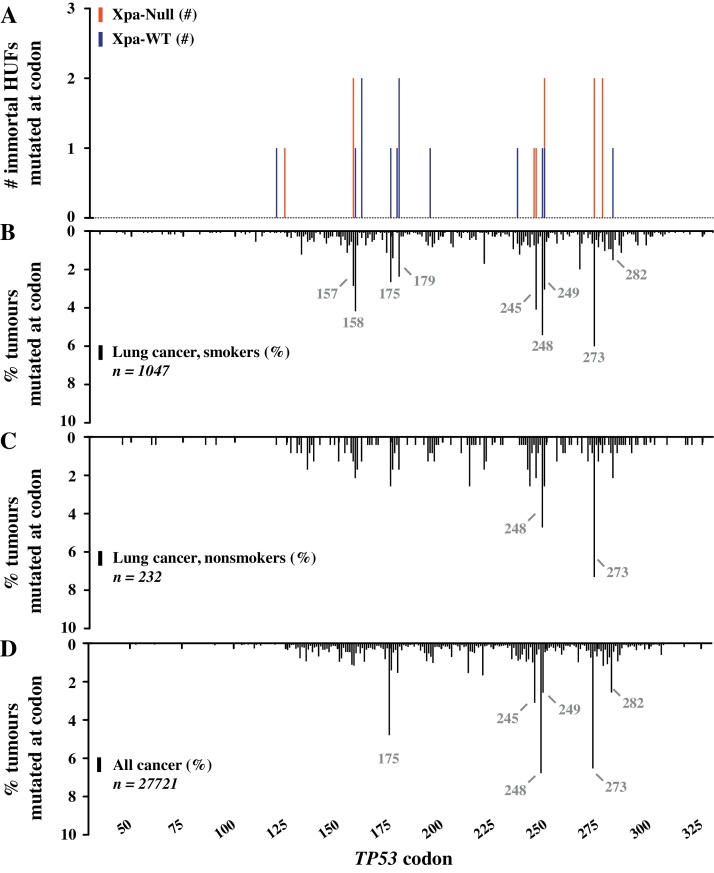
Fig. 5.
The spectrum of TP53 mutations induced by 3-NBA in Xpa-WT and Xpa-Null HUFs compared with mutations found in human tumours. Shown are (A) the number of mutations at each codon (within exons 4–9) in the TP53 gene induced by 3-NBA in HUFs compared with the frequency of mutation at each codon in (B) lung cancer of smokers (exclusions: radon, asbestos, mustard gas, and coal [65], [66], (C) lung cancer of non- and passive-smokers (exclusions: radon, asbestos, mustard gas, and coal [65], [66] and (D) cancer overall. Reference for the codon distribution of TP53 mutations in human cancer: IARC TP53 Mutation Database, R17 (November 2013). Mutation hotspots are indicated in grey.
Interestingly, all of the most frequently mutated hotspot codons in smokers’ lung cancer (157, 158, 175, 179, 245, 248, 249, 273, 282) or cancer overall (175, 245, 248, 249, 273, 282) were mutated following 3-NBA treatment in the HIMA (Fig. 5A). Six hotspots were mutated in Xpa-WT clones (158, 175, 179, 248, 249, 282) and four hotspots were mutated in Xpa-Null clones (157, 245, 249, 273). In fact, the mutation spectrum induced by 3-NBA in the current HIMA closely mirrors that of smokers' lung cancer (Fig. 5B).
Specific mutations induced by 3-NBA in this HIMA that are most frequently detected in human cancer include GTC to TTC at codon 157, CGC to CTC at codon 158, and CGG to TGG at codons 248 and 282 (Supplementary Table 4). Interestingly, those four hotspot codons have never been reported as mutated in spontaneously immortalised HUFs, suggesting that they may represent specific targets for carcinogen-induced mutations. Further, G:C > A:T transversions located at codons 157, 158, 175, 273 and 277, all of which were induced by 3-NBA in this study, are more commonly found in lung cancer than in cancer overall. Therefore, one could speculate that these mutations in lung tumours may indicate exposure to compounds such as nitro-PAHs or PAHs found in air pollution or tobacco smoke.
3.5.6. Comparison to mutations from previous HIMAs
The TP53 mutations induced by 3-NBA in the current study were compared to the mutations generated in previous HIMAs (Supplementary Table 5). Four codons (236, 244, 245, 273) mutated by 3-NBA in the current assay were also targeted by 3-NBA in a previous assay [18]. Identical mutations at three codons occurred in both 3-NBA studies (236: TAC to TGC; 244: GGC to GCC; 273: CGT to CTT). Further, identical mutations to those induced by 3-NBA at five codons in the current study were also induced by treatment with BaP/BPDE (157: GTC to TTC; 158: CGC to CTC; 245: GGC to CGC; 249: AGG to AGC; 273: CGT to AGT or CTT). While aristolochic acid I (AAI) and UV also targeted several similar hotspot codons to 3-NBA (AAI: 158, 179, 245, 249, 273; UV: 179, 248, 249) the mutations induced at these sites by AAI and UV were not identical to those generated by 3-NBA.
3.6. 3-NBA-induced mutation of lacZ reporter gene in Xpa-WT and Xpa-Null HUFs
The impact of NER deficiency on 3-NBA-induced mutagenesis in HUFs was further assessed at the lacZ reporter gene. The Xpa-WT and Xpa-Null HUFs utilised in this study harbour a pUR288 plasmid locus containing lacZ. Within a few days of treating HUFs with a carcinogen, the pUR288 plasmid can be recovered from HUF DNA and amplified in E. coli to determine lacZ mutant frequencies.
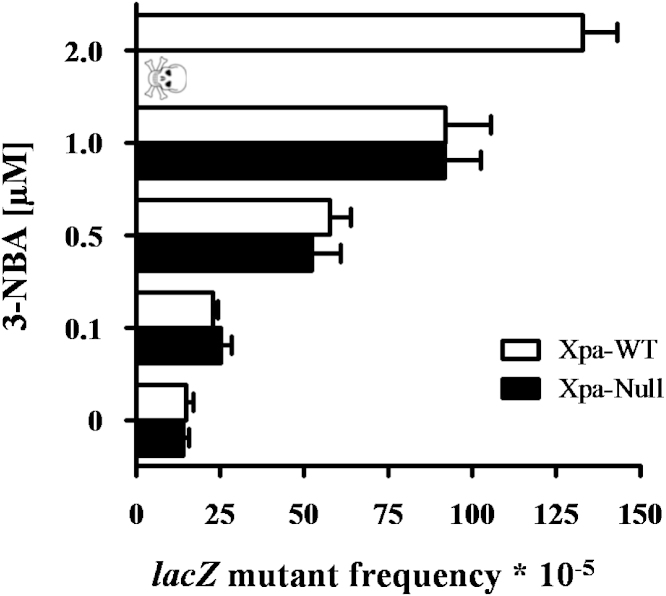
Xpa-WT and Xpa-Null HUFs were treated with 0.1–2.0 μM 3-NBA for 48 h, and then assessed for frequency of lacZ mutations after 4 days of mutation fixation (Fig. 6). A background mutant frequency of 14–15 × 10−5 was detected in both Xpa-WT and Xpa-Null HUFs. Following treatment with increasing concentrations of 3-NBA, a dose-dependent increase in lacZ mutant frequency was observed. Interestingly, as found for TP53 in the HIMA, the mutant frequency of lacZ was very similar between Xpa-WT and Xpa-Null HUFs at all concentrations, with the exception of cells treated with 2 μM 3-NBA, where the lack of viable cells precluded the assessment of mutants in Xpa-Null HUFs. Treatment with 1 μM 3-NBA increased the mutant frequency ∼6-fold over the background to 92 × 10−5 in Xpa-WT and Xpa-Null HUFs. The mutant frequency in Xpa-WT HUFs rose to ∼9-fold over background (133 × 10−5) following treatment with 2 μM 3-NBA.
Fig. 6.
Mutant frequency at the lacZ locus in Xpa-WT and Xpa-Null HUFs treated with 3-NBA. HUFs were treated for 48 h with the indicated concentrations of 3-NBA (or 0.1% DMSO solvent control) and then allowed to proliferate for 4 days to fix DNA damage into mutations. LacZ mutant frequencies were calculated as the number of mutant colonies per number of recovered transformants.
4. Discussion
In this study, TP53 and lacZ mutagenesis by the carcinogenic air pollutant 3-NBA was assessed in Xpa-WT and Xpa-Null HUFs. As Xpa is required for the removal of bulky adducts from the DNA by NER, it was hypothesised that greater persistence of 3-NBA-DNA adducts in Xpa-Null HUFs would lead to increased mutation frequency. However, a similar frequency of 3-NBA-induced TP53 and lacZ mutations was detected in Xpa-WT and Xpa-Null HUFs.
Xpa-Null HUFs and Hupki mice were much more sensitive to 3-NBA than their Xpa-WT counterparts. A pronounced decrease in survival was observed for Xpa-Null HUFs treated with the highest concentrations of 3-NBA and Xpa-Null Hupki mice died within 2–3 days of receiving 2 mg/kg bw 3-NBA. A similar response was found previously for Xpa-Null HUFs and Hupki mice treated with BaP [27]. Further, this sensitivity to genotoxicants was also shown for Xpa-Null mice with wild-type Trp53 after exposure to UV or 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) [33], [47]. While Xpa-Null and Csb (Cockayne syndrome B)-Null cells, both deficient in TC-NER, undergo apoptosis after DNA damage induced by UV or BaP, Xpc-Null cells or mice, deficient only in GG-NER, do not [48], [49]. The inability of TC-NER-deficient cells to remove DNA lesions from the transcribed strand of active genes is a strong trigger for the induction of apoptosis [50]. Lesions that persist on the transcribed strand can block the progression of RNAPII transcription complexes, which may subsequently collide with DNA replication forks during S-phase [51]. Therefore, the sensitivity of Xpa-Null HUFs and Hupki mice to 3-NBA could be due to the blocking of RNAPII progression by 3-NBA-DNA adducts. In this context it is noteworthy that a recent study found that 3-NBA-DNA adducts markedly block DNA replication [52]. The replication frequency of dG-C8-N-3-ABA was ∼17% compared with ∼33% for dG-N2-3-ABA and ∼43% for dA-N6-3-ABA [52].
Despite increased sensitivity to 3-NBA, Xpa-WT and Xpa-Null HUFs accumulated similar 3-NBA-DNA adduct levels. Further, 24 h after a single administration of 3-NBA, a difference in 3-NBA-DNA adduct formation between Xpa-WT and Xpa-Null Hupki mice was only observed in the lung (see Fig. 1). Previously, DNA adduct levels induced by BaP were found to be higher in several tissues (spleen, colon, kidney) in Xpa-Null relative to Xpa-WT Hupki mice [27]. These results indicate that repair of bulky DNA adducts may be tissue- and lesion-specific.
The fact that Xpa-Null HUFs did not exhibit an increase in 3-NBA-induced mutation frequency at the TP53 and lacZ loci nor an increase in DNA adduct levels, compared to Xpa-WT HUFs, suggests that, contrary to expectation, 3-NBA-derived DNA adducts may not be recognised and repaired by GG-NER. GG-NER is initiated following damage recognition by XPC/hHR23B and may occur with varying efficiency, depending on the structure and sequence context of the lesion [53]. Recognition of bulky adducts by XPC/hHR23B is correlated with the degree of thermodynamic destabilisation induced by the lesion [54], [55]. Lesions that distort and destabilise the DNA helix locally (e.g. pyrimidine (6–4) pyrimidone photoproducts [6-4PPs] or 10-(deoxyguanosin-N2-yl)-7,8,9-trihydroxy-7,8,9,10-tetrahydrobenzo[a]pyrene [dG-N2-BPDE]) are repaired efficiently by GG-NER [56], [57], whereas lesions that distort DNA to a lesser degree (e.g. cyclobutane pyrimidine dimers [CPDs]) or that stabilise DNA (e.g. 3-(deoxyguanosin-N2-yl)-2-acetylaminofluorene [dG-N2-AAF] or 8,9-dihydro-8-(2,6-diamino-4-oxo-3,4-dihydropyrimid5-yl-formamido)-9-hydroxyaflatoxin B1 [AFB1-FAPY]) are repaired less efficiently [58], [59].
It was recently shown that the major DNA adduct formed after metabolic activation of 3-NBA, dG-N2-3-ABA, had a stabilising effect when incorporated into a synthetic oligonucleotide [60]. The dG-N2-3-ABA adduct-containing duplex formed a regular double helical structure, with the adduct residing in the minor groove. Therefore, it is possible that the thermodynamic stability of dG- N2-3-ABA, which is the most abundant and persistent 3-NBA-DNA adduct formed in vivo [61], may indeed prevent its recognition and removal by GG-NER. It is important to consider, however, that metabolic activation of 3-NBA generates 4 major adducts, including dG-N2-3-ABA (see Supplementary Fig. 1) with ∼80% of adducts occurring at deoxyguanosine and ∼20% at deoxyadenosine [9], [11]; the thermodynamic stability of the other moieties is unknown. A recent study evaluating the formation and removal of dA-N6-3-ABA, dG-N2-3-ABA and dG-C8-N-3-ABA in human cells found that dA-N6-3-ABA and dG-C8-N-3-ABA were repaired more efficiently than dG-N2-3-ABA [52]. After a 24-hour period ∼85% of the dG-N2-3-ABA adducts remained, whereas only ∼50% of the dG-C8-N-3-ABA adducts and ∼40% of dA-N6-3-ABA adducts were present in the cells [52].
In contrast to our findings, however, are the results of an earlier study where a supF shuttle vector plasmid was treated with N-Aco-3-ABA, a reactive metabolite of 3-NBA, and then propagated in NER-proficient and -deficient human fibroblasts (WI-38-VA13 and XP2OS(SV), respectively) [16]. Nearly 2-fold more supF mutations were induced in plasmids recovered from the XP2OS(SV) cells compared with the WI-38-VA13 cells. Using a polymerase stop assay, the authors determined that 92% of the N-Aco-3-ABA-induced DNA adducts were located at G:C base pairs in supF. DNA adducts were not measured in this study, however, therefore their exact composition, such as the proportion of dG-N2-3-ABA adducts, is unknown. Interestingly, a higher frequency of supF mutations induced in the XP2OS(SV) cells occurred at A:T base pairs, compared with the NER-proficient cell line (34% versus 17%, respectively) [16]. The authors concluded that dA-3-ABA adducts may be more readily recognised by NER than dG-3-ABA adducts, leading to an increase in mutations at A:T sites in NER-deficient cells. However, this was not the case for TP53 mutations induced by 3-NBA in Xpa-Null HUFs. While ∼30% of the 3-NBA-induced TP53 mutations in Xpa-WT HUFs occurred at A:T base pairs, all of the mutations in Xpa-Null HUFs occurred at G:C base pairs.
There are several possible explanations for the differences between our findings and those of Nishida et al. [16]. Firstly, the supF gene is not transcribed in human cells, therefore dA-3-ABA adducts in supF would not cause RNAPII blockage or apoptosis in XP2OS(SV) cells, but could instead induce mutations. Further, the relative proportion of individual 3-NBA-derived DNA adducts in our systems may be different. The adducts detected in the present study were generated following enzymatic activation of 3-NBA in HUFs, whereas the supF shuttle vector was directly reacted with N-Aco-3-ABA in situ prior to cell transfection. As discussed above, DNA repair efficiency of the four major 3-NBA-derived DNA adducts varies [52], ultimately influencing mutagenesis. Additionally, it could be that 3-NBA-derived DNA adducts are recognised and repaired differently in human cells and mouse cells, or that GG-NER of adducts contained on plasmids operates differently to GG-NER of adducted genomic DNA.
Most 3-NBA-induced mutations occurred at G:C base pairs, with G:C > T:A transversions the predominant mutation type in both Xpa-WT and Xpa-Null HUFs. This is in agreement with the results of an earlier HIMA using 3-NBA performed by vom Brocke et al. [18]. Of note, the dG-N2-3-ABA and dG-C8-N-3-ABA adducts are the predominant 3-NBA-DNA adducts formed in HUFs and both lesions have recently been shown to induce G:C > T:A transversions in site-specific mutagenesis assays in human cells [52]. Four codons were targeted by 3-NBA in both our study and that of vom Brocke et al. (236, 244, 245, 273) [18]. We found that 32% (8/25) of unique TP53 mutations occurred at CpG sites, compared with only 14% (3/22) in the previous HIMA using 3-NBA. Interestingly, we also found that all of the CpG sites targeted by 3-NBA are hotspots for mutation in human cancer, including smokers' lung cancer (i.e. codons 157, 158, 175, 245, 248, 273, 282). Carcinogens such as BPDE show enhanced adduct formation at many of these sites (codons 157, 158, 245, 248, 273) [62], [63]. Our results suggest that metabolites of 3-NBA may also exhibit preferential binding at such sites, although this would have to be confirmed in future studies. The existence of mutation hotspots in cancer can also be influenced by other factors besides enhanced carcinogen binding. These sites may be more resistant to repair or may be more readily bypassed by error-prone translesion polymerases. Additionally, it is possible that these mutations are highly selected for due to their deleterious impact on the function of p53 or their gain-of-function properties [20]. Nevertheless, the spectrum of mutations generated in this study indicates that similar factors influence TP53 mutagenesis in the HIMA and human cancer.
For the first time, we were able to assess mutagenesis at a lacZ reporter gene in HUFs in addition to TP53 mutagenesis. This allowed us to compare the frequency of 3-NBA-induced TP53 mutations, detected following a several-month selection process in mammalian cells, to the frequency of lacZ mutations generated, but not subjected to selection, in the same cells. In future studies having lacZ integrated into HUFs could aid in the optimisation of carcinogen treatment conditions, where the mutagenic activity of an agent could be determined at the lacZ locus prior to initiating the more laborious HIMA. The data generated in the present study, comparing 3-NBA-induced mutagenesis of TP53 and lacZ in HUFs, may serve as an initial point of reference for identifying a level of lacZ mutation frequency in primary HUFs commensurate with TP53 mutation frequency in immortalised clones.
While a lack of recognition of 3-NBA-DNA adducts by GG-NER could explain why TP53 mutant frequency was not increased in Xpa-Null HUFs, as discussed above, it is unclear why these cells did not at least harbour more mutations on the transcribed strand compared with Xpa-WT HUFs, as was observed previously for BPDE [27]. It will be important to examine additional mutagens in future studies, particularly those with firm evidence for removal by NER (e.g. UV). One drawback of using Xpa-Null HUFs in the HIMA is their extreme sensitivity to genotoxicants that block RNAPII. This sensitivity necessitates treating Xpa-Null HUFs with a much lower concentration of carcinogen than Xpa-WT cells can tolerate, and therefore lowers the level of premutagenic DNA damage. Xpa-Null HUFs may be particularly useful for improving HIMA mutation frequencies when assessing mutagens that do not generate high levels of DNA damage in HUFs, if such damage is recognised by GG-NER. Furthermore, Xpa-Null HUFs may be useful in determining the role NER plays in shaping TP53 mutagenesis, rather than simply increasing mutation frequencies.
Although NER-deficient HUFs did not increase the frequency of 3-NBA-induced mutations in the HIMA, data from this study contribute to our understanding of the TP53 mutation spectrum induced by 3-NBA and further indicate that 3-NBA-derived DNA adducts may evade removal by GG-NER. The persistence of 3-NBA adducts in DNA may be an important factor in its mutagenicity [9], [52], [61]. 3-NBA is an air pollutant, predominantly originating from diesel emissions [2], [6]. Occupational diesel exhaust exposure has been shown to increase the risk of developing lung cancer [2], [64], and compounds such as 3-NBA may contribute to the process. The current evidence from experimental models indicates that 3-NBA is a potent mutagen and carcinogen, but it will be important to assess TP53 mutations in tumour samples from diesel exhaust-exposed individuals in order to assess more fully what impact this compound has on human carcinogenesis.
Acknowledgements
The study was funded, in part, by Cancer Research UK (grant C313/A14329) and the Wellcome Trust (grants 101126/Z/13/Z and 101126/B/13/Z). Jill E. Kucab was supported by a PhD studentship (2008–2012) from the Institute of Cancer Research, London, United Kingdom. Jill E. Kucab, David H. Phillips and Volker M. Arlt are members of the Wellcome Trust funded COMSIG (Causes of Mutational SIGnatures) consortium. We thank Allan Thornhill and Carla Foley at the Biological Services Unit at the Institute of Cancer Research, Sutton, Surrey, for their excellent assistance with animal breeding and in-vivo experiments.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.dnarep.2015.11.004.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.IARC . vol. 46. IARC; Lyon: 1989. IARC monograph on the evaluation of carcinogenic risk to humans. (Diesel and Gasoline Engine Exhausts and Some Nitroarenes). [PMC free article] [PubMed] [Google Scholar]
- 2.IARC . IARC Science Publication; 2013. Diesel and Gasoline Engine Exhaust and Some Nitroarenes. [Google Scholar]
- 3.Rushton L., Bagga S., Bevan R., Brown T.P., Cherrie J.W., Holmes P., Fortunato L., Slack R., Van Tongeren M., Young C., Hutchings S.J. Occupation and cancer in Britain. Br. J. Cancer. 2010;102:1428–1437. doi: 10.1038/sj.bjc.6605637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Attfield M.D., Schleiff P.L., Lubin J.H., Blair A., Stewart P.A., Vermeulen R., Coble J.B., Silverman D.T. The diesel exhaust in miners study: a cohort mortality study with emphasis on lung cancer. J. Natl. Cancer Inst. 2012 doi: 10.1093/jnci/djs035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Silverman D.T., Samanic C.M., Lubin J.H., Blair A.E., Stewart P.A., Vermeulen R., Coble J.B., Rothman N., Schleiff P.L., Travis W.D., Ziegler R.G., Wacholder S., Attfield M.D. The diesel exhaust in miners study: a nested case-control study of lung cancer and diesel exhaust. J. Natl. Cancer Inst. 2012 doi: 10.1093/jnci/djs034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arlt V.M. 3-Nitrobenzanthrone, a potential human cancer hazard in diesel exhaust and urban air pollution: a review of the evidence. Mutagenesis. 2005;20:399–410. doi: 10.1093/mutage/gei057. [DOI] [PubMed] [Google Scholar]
- 7.Seidel A., Dahmann D., Krekeler H., Jacob J. Biomonitoring of polycyclic aromatic compounds in the urine of mining workers occupationally exposed to diesel exhaust. Int. J. Hyg. Environ. Health. 2002;204:333–338. doi: 10.1078/1438-4639-00116. [DOI] [PubMed] [Google Scholar]
- 8.Nagy E., Zeisig M., Kawamura K., Hisamatsu Y., Sugeta A., Adachi S., Moller L. DNA adduct and tumor formations in rats after intratracheal administration of the urban air pollutant 3-nitrobenzanthrone. Carcinogenesis. 2005;26:1821–1828. doi: 10.1093/carcin/bgi141. [DOI] [PubMed] [Google Scholar]
- 9.Arlt V.M., Zhan L., Schmeiser H.H., Honma M., Hayashi M., Phillips D.H., Suzuki T. DNA adducts and mutagenic specificity of the ubiquitous environmental pollutant 3-nitrobenzanthrone in muta mouse. Environ. Mol. Mutagen. 2004;43:186–195. doi: 10.1002/em.20014. [DOI] [PubMed] [Google Scholar]
- 10.Arlt V.M., Gingerich J., Schmeiser H.H., Phillips D.H., Douglas G.R., White P.A. Genotoxicity of 3- nitrobenzanthrone and 3-aminobenzanthrone in MutaMouse and lung epithelial cells derived from muta mouse. Mutagenesis. 2008;23:483–490. doi: 10.1093/mutage/gen037. [DOI] [PubMed] [Google Scholar]
- 11.Arlt V.M., Bieler C.A., Mier W., Wiessler M., Schmeiser H.H. DNA adduct formation by the ubiquitous environmental contaminant 3-nitrobenzanthrone in rats determined by (32) P-postlabeling. Int. J. Cancer. 2001;93:450–454. doi: 10.1002/ijc.1346. [DOI] [PubMed] [Google Scholar]
- 12.Arlt V.M., Schmeiser H.H., Osborne M.R., Kawanishi M., Kanno T., Yagi T., Phillips D.H., Takamura- Enya T. Identification of three major DNA adducts formed by the carcinogenic air pollutant 3-nitrobenzanthrone in rat lung at the C8 and N2 position of guanine and at the N6 position of adenine. Int. J. Cancer. 2006;118:2139–2146. doi: 10.1002/ijc.21622. [DOI] [PubMed] [Google Scholar]
- 13.Gamboa da Costa G., Singh R., Arlt V.M., Mirza A., Richards M., Takamura-Enya T., Schmeiser H.H., Farmer P.B., Phillips D.H. Quantification of 3-nitrobenzanthrone-DNA adducts using online column-switching HPLC-electrospray tandem mass spectrometry. Chem. Res. Toxicol. 2009;22:1860–1868. doi: 10.1021/tx900264v. [DOI] [PubMed] [Google Scholar]
- 14.Enya T., Suzuki H., Watanabe T., Hirayama T., Hisamatsu Y. 3- Nitrobenzanthrone, a powerful bacterial mutagen and suspected human carcinogen found in diesel exhausts and airborne particulates. Environ. Sci. Technol. 1997;31:2772–2776. [Google Scholar]
- 15.Phousongphouang P.T., Grosovsky A.J., Eastmond D.A., Covarrubias M., Arey J. The genotoxicity of 3 -nitrobenzanthrone and the nitropyrene lactones in human lymphoblasts. Mutat. Res. 2000;472:93–103. doi: 10.1016/s1383-5718(00)00135-2. [DOI] [PubMed] [Google Scholar]
- 16.Nishida H., Kawanishi M., Takamura-Enya T., Yagi T. Mutagenic specificity of N-acetoxy-3-aminobenzanthrone, a major metabolically activated form of 3-nitrobenzanthrone, in shuttle vector plasmids propagated in human cells. Mutat. Res. 2008;654:82–87. doi: 10.1016/j.mrgentox.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 17.Luo J.L., Yang Q., Tong W.M., Hergenhahn M., Wang Z.Q., Hollstein M. Knock-in mice with a chimeric human/murine p53 gene develop normally and show wild-type p53 responses to DNA damaging agents: a new biomedical research tool. Oncogene. 2001;20:320–328. doi: 10.1038/sj.onc.1204080. [DOI] [PubMed] [Google Scholar]
- 18.vom Brocke J., Krais A., Whibley C., Hollstein M.C., Schmeiser H.H. The carcinogenic air pollutant 3-nitrobenzanthrone induces GC to TA transversion mutations in human p53 sequences. Mutagenesis. 2009;24:17–23. doi: 10.1093/mutage/gen049. [DOI] [PubMed] [Google Scholar]
- 19.Lane D.P. Cancer p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 20.Olivier M., Hollstein M., Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harbor Perspect. Biol. 2010;2:a001008. doi: 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petitjean A., Mathe E., Kato S., Ishioka C., Tavtigian S.V., Hainaut P., Olivier M. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum. Mutat. 2007;28:622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 22.Schetter A.J., Harris C.C. Tumor suppressor p53 (TP53) at the crossroads of the exposome and the cancer genome. Proc. Natl. Acad. Sci. U. S. A. 2012;109:7955–7956. doi: 10.1073/pnas.1205457109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kucab J.E., Phillips D.H., Arlt V.M. Linking environmental carcinogen exposure to TP53 mutations in human tumours using the human TP53 knock-in (Hupki) mouse model. FEBS J. 2010;277:2567–2583. doi: 10.1111/j.1742-464X.2010.07676.x. [DOI] [PubMed] [Google Scholar]
- 24.Besaratinia A., Pfeifer G.P. Applications of the human p53 knock-in (Hupki) mouse model for human carcinogen testing. FASEB J. 2010;24:2612–2619. doi: 10.1096/fj.10-157263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedberg E.C. How nucleotide excision repair protects against cancer. Nat. Rev. Cancer. 2001;1:22–33. doi: 10.1038/35094000. [DOI] [PubMed] [Google Scholar]
- 26.Naegeli H., Geacintov N.E. Imperial College Press; London: 2005. Mechanisms of Repair of Polycyclic Aormatic Hydrocarbon-Induced DNA Damage. [Google Scholar]
- 27.Kucab J.E., van Steeg H., Luijten M., Schmeiser H.H., White P.A., Phillips D.H., Arlt V.M. TP53 mutations induced by BPDE in Xpa-WT and Xpa-Null human TP53 knock-in (Hupki) mouse embryo fibroblasts. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2015;773:48–62. doi: 10.1016/j.mrfmmm.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rebel H., Mosnier L.O., Berg R.J., Westerman-de Vries A., van Steeg H., van Kranen H.J., de Gruijl F.R. Early p53-positive foci as indicators of tumor risk in ultraviolet-exposed hairless mice: kinetics of induction, effects of DNA repair deficiency, and p53 heterozygosity. Cancer Res. 2001;61:977–983. [PubMed] [Google Scholar]
- 29.Tanaka K., Kamiuchi S., Ren Y., Yonemasu R., Ichikawa M., Murai H., Yoshino M., Takeuchi S., Saijo M., Nakatsu Y., Miyauchi-Hashimoto H., Horio T. UV-induced skin carcinogenesis in xeroderma pigmentosum group A (XPA) gene-knockout mice with nucleotide excision repair- deficiency. Mutat. Res. 2001;477:31–40. doi: 10.1016/s0027-5107(01)00093-8. [DOI] [PubMed] [Google Scholar]
- 30.de Vries A., Dolle M.E., Broekhof J.L., Muller J.J., Kroese E.D., van Kreijl C.F., Capel P.J., Vijg J., van Steeg H. Induction of DNA adducts and mutations in spleen, liver and lung of XPA-deficient/lacZ transgenic mice after oral treatment with benzo[a]pyrene: correlation with tumour development. Carcinogenesis. 1997;18:2327–2332. doi: 10.1093/carcin/18.12.2327. [DOI] [PubMed] [Google Scholar]
- 31.Busuttil R.A., Rubio M., Dolle M.E., Campisi J., Vijg J. Mutant frequencies and spectra depend on growth state and passage number in cells cultured from transgenic lacZ-plasmid reporter mice. DNA Rep. (Amst.) 2006;5:52–60. doi: 10.1016/j.dnarep.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 32.Arlt V.M., Glatt H., Muckel E., Pabel U., Sorg B.L., Schmeiser H.H., Phillips D.H. Metabolic activation of the environmental contaminant 3- nitrobenzanthrone by human acetyltransferases and sulfotransferase. Carcinogenesis. 2002;23:1937–1945. doi: 10.1093/carcin/23.11.1937. [DOI] [PubMed] [Google Scholar]
- 33.de Vries A., van Oostrom C.T., Hofhuis F.M., Dortant P.M., Berg R.J., de Gruijl F.R., Wester P.W., van Kreijl C.F., Capel P.J., van Steeg H. Increased susceptibility to ultraviolet-B and carcinogens of mice lacking the DNA excision repair gene XPA. Nature. 1995;377:169–173. doi: 10.1038/377169a0. [DOI] [PubMed] [Google Scholar]
- 34.Arlt V.M., Stiborova M., Henderson C.J., Osborne M.R., Bieler C.A., Frei E., Martinek V., Sopko B., Wolf C.R., Schmeiser H.H., Phillips D.H. Environmental pollutant and potent mutagen 3-nitrobenzanthrone forms DNA adducts after reduction by NAD(P)H: quinone oxidoreductase and conjugation by acetyltransferases and sulfotransferases in human hepatic cytosols. Cancer Res. 2005;65:2644–2652. doi: 10.1158/0008-5472.CAN-04-3544. [DOI] [PubMed] [Google Scholar]
- 35.Phillips D.H., Arlt V.M. The 32P-postlabeling assay for DNA adducts. Nat. Protoc. 2007;2:2772–2781. doi: 10.1038/nprot.2007.394. [DOI] [PubMed] [Google Scholar]
- 36.Wohak L.E., Krais A.M., Kucab J.E., Stertmann J., Ovrebo S., Seidel A., Phillips D.H., Arlt V.M. Carcinogenic polycyclic aromatic hydrocarbons induce CYP1A1 in human cells via a p53-dependent mechanism. Arch. Toxicol. 2014 doi: 10.1007/s00204-014-1409-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Z., Belharazem D., Muehlbauer K.R., Nedelko T., Knyazev Y., Hollstein M. Mutagenesis of human p53 tumor suppressor gene sequences in embryonic fibroblasts of genetically-engineered mice. Genet. Eng. (N. Y.) 2007;28:45–54. doi: 10.1007/978-0-387-34504-8_3. [DOI] [PubMed] [Google Scholar]
- 38.Boerrigter M.E., Dolle M.E., Martus H.J., Gossen J.A., Vijg J. Plasmid-based transgenic mouse model for studying in vivo mutations. Nature. 1995;377:657–659. doi: 10.1038/377657a0. [DOI] [PubMed] [Google Scholar]
- 39.Dolle M.E., Martus H.J., Gossen J.A., Boerrigter M.E., Vijg J. Evaluation of a plasmid-based transgenic mouse model for detecting in vivo mutations. Mutagenesis. 1996;11:111–118. doi: 10.1093/mutage/11.1.111. [DOI] [PubMed] [Google Scholar]
- 40.Mahabir A.G., Zwart E., Schaap M., van Benthem J., de Vries A., Hernandez L.G., Hendriksen C.F., van Steeg H. lacZ mouse embryonic fibroblasts detect both clastogens and mutagens. Mutat. Res. 2009;666:50–56. doi: 10.1016/j.mrfmmm.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 41.Reinbold M., Luo J.L., Nedelko T., Jerchow B., Murphy M.E., Whibley C., Wei Q., Hollstein M. Common tumour p53 mutations in immortalized cells from Hupki mice heterozygous at codon 72. Oncogene. 2008;27:2788–2794. doi: 10.1038/sj.onc.1210932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Denissenko M.F., Pao A., Pfeifer G.P., Tang M. Slow repair of bulky DNA adducts along the nontranscribed strand of the human p53 gene may explain the strand bias of transversion mutations in cancers. Oncogene. 1998;16:1241–1247. doi: 10.1038/sj.onc.1201647. [DOI] [PubMed] [Google Scholar]
- 43.Ford J.M., Lommel L., Hanawalt P.C. Preferential repair of ultraviolet light-induced DNA damage in the transcribed strand of the human p53 gene. Mol. Carcinog. 1994;10:105–109. doi: 10.1002/mc.2940100208. [DOI] [PubMed] [Google Scholar]
- 44.Takeuchi S., Nakatsu Y., Nakane H., Murai H., Hirota S., Kitamura Y., Okuyama A., Tanaka K. Strand specificity and absence of hot spots for p53 mutations in ultraviolet B-induced skin tumors of XPA-deficient mice. Cancer Res. 1998;58:641–646. [PubMed] [Google Scholar]
- 45.Joerger A.C., Fersht A.R. Structural biology of the tumor suppressor p53. Annu. Rev. Biochem. 2008;77:557–582. doi: 10.1146/annurev.biochem.77.060806.091238. [DOI] [PubMed] [Google Scholar]
- 46.Kato S., Han S.Y., Liu W., Otsuka K., Shibata H., Kanamaru R., Ishioka C. Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc. Natl. Acad. Sci. U. S. A. 2003;100:8424–8429. doi: 10.1073/pnas.1431692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Imaida K., Ogawa K., Takahashi S., Ito T., Yamaguchi T., Totsuka Y., Wakabayashi K., Tanaka K., Ito N., Shirai T. Delay of DNA- adduct repair and severe toxicity in xeroderma pigmentosum group A gene (XPA) deficient mice treated with 2-amino-1-methyl-6-phenyl-imidazo [4,5-b] pyridine (PhIP) Cancer Lett. 2000;150:63–69. doi: 10.1016/s0304-3835(99)00377-8. [DOI] [PubMed] [Google Scholar]
- 48.de Waard H., Sonneveld E., de Wit J., Esveldt-van Lange R., Hoeijmakers J.H., Vrieling H., van der Horst G.T. Cell-type-specific consequences of nucleotide excision repair deficiencies: embryonic stem cells versus fibroblasts. DNA Rep. (Amst.) 2008;7:1659–1669. doi: 10.1016/j.dnarep.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 49.Berg R.J., Ruven H.J., Sands A.T., de Gruijl F.R., Mullenders L.H. Defective global genome repair in XPC mice is associated with skin cancer susceptibility but not with sensitivity to UVB induced erythema and edema. J. Invest. Dermatol. 1998;110:405–409. doi: 10.1111/j.1523-1747.1998.00173.x. [DOI] [PubMed] [Google Scholar]
- 50.Ljungman M., Zhang F. Blockage of RNA polymerase as a possible trigger for UV light-induced apoptosis. Oncogene. 1996;13:823–831. [PubMed] [Google Scholar]
- 51.Ljungman M., Lane D.P. Transcription—guarding the genome by sensing DNA damage. Nat. Rev. Cancer. 2004;4:727–737. doi: 10.1038/nrc1435. [DOI] [PubMed] [Google Scholar]
- 52.Kawanishi M., Fujikawa Y., Ishii H., Nishida H., Higashigaki Y., Kanno T., Matsuda T., Takamura-Enya T., Yagi T. Adduct formation and repair, and translesion DNA synthesis across the adducts in human cells exposed to 3- nitrobenzanthrone. Mutat. Res. 2013;753:93–100. doi: 10.1016/j.mrgentox.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 53.Sidorenko V.S., Yeo J.E., Bonala R.R., Johnson F., Scharer O.D., Grollman A.P. Lack of recognition by global-genome nucleotide excision repair accounts for the high mutagenicity and persistence of aristolactam-DNA adducts. Nucleic Acids Res. 2012;40:2494–2505. doi: 10.1093/nar/gkr1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dip R., Camenisch U., Naegeli H. Mechanisms of DNA damage recognition and strand discrimination in human nucleotide excision repair. DNA Rep. (Amst.) 2004;3:1409–1423. doi: 10.1016/j.dnarep.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 55.Geacintov N.E., Broyde S., Buterin T., Naegeli H., Wu M., Yan S., Patel D.J. Thermodynamic and structural factors in the removal of bulky DNA adducts by the nucleotide excision repair machinery. Biopolymers. 2002;65:202–210. doi: 10.1002/bip.10239. [DOI] [PubMed] [Google Scholar]
- 56.Gunz D., Hess M.T., Naegeli H. Recognition of DNA adducts by human nucleotide excision repair. Evidence for a thermodynamic probing mechanism. J. Biol. Chem. 1996;271:25089–25098. doi: 10.1074/jbc.271.41.25089. [DOI] [PubMed] [Google Scholar]
- 57.Szymkowski D.E., Lawrence C.W., Wood R.D. Repair by human cell extracts of single (6–4) and cyclobutane thymine–thymine photoproducts in DNA. Proc. Natl. Acad. Sci. U. S. A. 1993;90:9823–9827. doi: 10.1073/pnas.90.21.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mao H., Deng Z., Wang F., Harris T.M., Stone M.P. An intercalated and thermally stable FAPY adduct of aflatoxin B1 in a DNA duplex: structural refinement from 1H NMR. Biochemistry. 1998;37:4374–4387. doi: 10.1021/bi9718292. [DOI] [PubMed] [Google Scholar]
- 59.Zaliznyak T., Bonala R., Johnson F., de Los Santos C. Structure and stability of duplex DNA containing the 3- (deoxyguanosin-N2-yl)-2-acetylaminofluorene (dG (N2)-AAF) lesion: a bulky adduct that persists in cellular DNA. Chem. Res. Toxicol. 2006;19:745–752. doi: 10.1021/tx060002i. [DOI] [PubMed] [Google Scholar]
- 60.Lukin M., Zaliznyak T., Johnson F., de Los Santos C.R. Incorporation of 3-aminobenzanthrone into 2′-deoxyoligonucleotides and its impact on duplex stability. J. Nucleic Acids. 2011;2011:521035. doi: 10.4061/2011/521035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bieler C.A., Cornelius M.G., Stiborova M., Arlt V.M., Wiessler M., Phillips D.H., Schmeiser H.H. Formation and persistence of DNA adducts formed by the carcinogenic air pollutant 3-nitrobenzanthrone in target and non-target organs after intratracheal instillation in rats. Carcinogenesis. 2007;28:1117–1121. doi: 10.1093/carcin/bgl219. [DOI] [PubMed] [Google Scholar]
- 62.Denissenko M.F., Chen J.X., Tang M.S., Pfeifer G.P. Cytosine methylation determines hot spots of DNA damage in the human P53 gene. Proc. Natl. Acad. Sci. U. S. A. 1997;94:3893–3898. doi: 10.1073/pnas.94.8.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pfeifer G.P. p53 mutational spectra and the role of methylated CpG sequences. Mutat. Res. 2000;450:155–166. doi: 10.1016/s0027-5107(00)00022-1. [DOI] [PubMed] [Google Scholar]
- 64.Benbrahim-Tallaa L., Baan R.A., Grosse Y., Lauby-Secretan B., El Ghissassi F., Bouvard V., Guha N., Loomis D., Straif K. Carcinogenicity of diesel-engine and gasoline-engine exhausts and some nitroarenes. Lancet Oncol. 2012;13:663–664. doi: 10.1016/s1470-2045(12)70280-2. [DOI] [PubMed] [Google Scholar]
- 65.DeMarini D.M., Landi S., Tian D., Hanley N.M., Li X., Hu F., Roop B.C., Mass M.J., Keohavong P., Gao W., Olivier M., Hainaut P., Mumford J.L. Lung tumor KRAS and TP53 mutations in nonsmokers reflect exposure to PAH-rich coal combustion emissions. Cancer Res. 2001;61:6679–6681. [PubMed] [Google Scholar]
- 66.Sarkar F.H., Li Y., Vallyathan V. Molecular analysis of p53 and K-ras in lung carcinomas of coal miners. Int. J. Mol. Med. 2001;8:453–459. doi: 10.3892/ijmm.8.4.453. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.