Abstract
Covalent modifications of both DNA histones act in concert to define the landscape of our epigenome. In this review, we explore the interconnections between histone and DNA modifications by focusing on a conserved chromatin-binding regulatory domain, the ATRX-DNMT3-DNMT3L (ADD) domain. New studies show that the ADD domain is capable of sensing, and therefore integrating, the status of multiple histone modifications. This in turn dictates the in vivo localization or allosteric regulation of the full-length ADD-containing protein and its ability to function in downstream chromatin remodeling events. Strategies to re-engineer the ADD “reader pocket” in the de novo DNA methyltransferase DNMT3A such that it redirects this “writer” to new genomic loci proved useful in understanding important biological downstream consequences of mis-targeting of DNA methylation via altered reading of histone marks. Combined with genome-editing tools, this approach stands as a poof-of-principle and will be broadly applicable to the elucidation of epigenetic networks that have been altered by “reader” mutations, either artificially or as naturally occurs in some human diseases.
Graphical Abstract
In the postgenomic era, rapidly expanding literature defines the exciting field of “epigenetics,” wherein DNA sequence, the cornerstone of classical “genetics,” is further interpreted in cellular and developmental contexts giving rise to heritable phenotypes that cannot be explained by alterations in DNA sequence alone.1 Covalent modifications including DNA methylation and the acetylation, methylation, phosphorylation, or ubiquitination of histones, the principal proteins that package the DNA, all contribute to the “epigenome.” Pioneering studies carried out in fungal and plant models suggest that there is a close interplay between histone and DNA modifications, particularly with regard to methylation.2–4 In the mammalian system, however, the precise details of this interplay remain poorly understood. DNA methylation and histone modifications are often considered separately, in part due to the remarkable complexity of each modification and their known intermediates that require separate assays and approaches by experts in the respective camp. One goal of this review is to reinforce the general idea that histone and DNA methylation are closely linked to one another and lead to a final epigenetic state that is dependent on the interplay between both classes of modification (see refs 5 and 6). Equally, genetics and epigenetics are closely intertwined supported by the explosion of examples where driver mutations, even in genes encoding histone proteins, influence cancer epigenomes with exciting new therapeutic potential.7–9
The “histone/epigenetic code” hypothesis10,11 posits that distinct combinations of histone post-translational modifications (PTMs) on one or more histone protein provide an additional layer of regulatory complexity to the well-accepted genetic (DNA) code. Mechanistically, this hypothesis suggests that chromatin-associated effector complexes “read” distinct patterns of histone PTMs to bring about distinct biological outcomes. A wealth of evidence has accumulated on the existence of such “readers” of covalent histone modifications, including X-ray structures detailing the exquisite specificity of protein-ligand interactions at atomic resolution.12–14 Notably, genetic mutations that result in improper histone reading have been linked to human disease,15,16 and in some cases, drugs targeting histone-binding domains (“pockets”) have led to successful therapeutic outcomes.17–19 This general concept of “reading” covalent modifications articulated in the histone code hypothesis can also be readily applied to DNA. Methylated DNA readers, such as MeCP2, are well documented, and mutations in these proteins have equally been linked with human disease (e.g., Rett Syndrome, see ref 20).
In this review, we will focus on the histone-binding ATRX-DNMT3-DNMT3L (ADD) domain, an integration of GATA-like and plant homeodomain (PHD) zinc fingers that is present in the chromatin-associated alpha thalassemia/mental retardation syndrome X-linked (ATRX) protein and the de novo DNA methyltransferases DNMT3A/B/L (DNMT3; Figure 1A–C). This domain has a role in establishing and maintaining patterns of DNA methylation, and we will use this binding module as a paradigm to illustrate concepts that are likely to apply to other histone-binding domains. This includes (1) cross-talk relationships between nearby histone modifications that affect domain binding, (2) structure-guided mutagenesis to not only define crucial residues for domain binding but also to allow the re-engineering of histone pockets to accommodate new histone-binding properties, (3) the clear interplay between histone methylation/phosphorylation and DNA methylation, and (4) the importance of histone-binding pockets in human biology and disease. Within the scope of this review, we cannot provide a comprehensive survey of other histone-binding domains such as bromodomains, chromodomains, PHD fingers, etc., and for these we refer the reader to other excellent reviews.12–14
Figure 1.
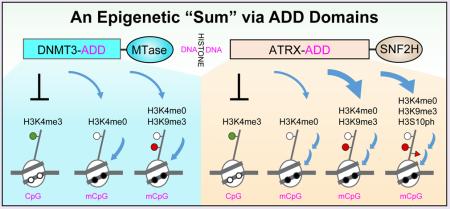
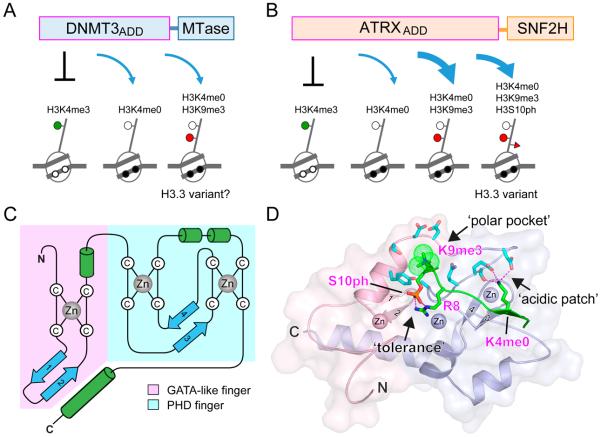
Linking histone modification, DNA methylation, and chromatin remodeling through ADD domain. (A) DNMT3 family ADD domain binds to unmodified H3K4 N-tails regardless of H3K9 methylation. DNMT3 proteins function broadly throughout the genome although their link to H3.3 variant is unclear. MTase, DNA methyltransferase domain. (B) ATRX ADD domain binds to H3K9 methylation in conjunction with unmodified H3K4 which limits its location largely to the repeat elements. ATRX deposits H3.3 variant in the repeat elements. ATRX ADD also tolerates H3S10 phosphorylation. SNF2H, SWI/SNF homology ATPase domain that has chromatin remodeling activity. (C) Topology of the ADD fold. ADD domain consists of a GATA-like zinc finger, a PHD finger and a C-terminal α-helix. Blue arrow, β-strand; green cylinder, α-helix. Tight association of the GATA-like finger and the PHD finger is notably stabilized by an extended C-terminal α-helical segment. (D) A structural illustration of the ATRX ADD fold. The GATA-like finger is colored light pink, and the PHD finger is colored light blue. Three zinc ions are depicted as spheres. A semitransparent surface of ATRX ADD is presented to highlight tight integration of the GATA-like and PHD fingers as well as the reader pocket. ATRX ADD contains an acidic surface patch and a polar pocket to recognize unmodified H3K4 (H3K4me0) and trimethylated H3K9 (H3K9me3, dotted spheres), respectively. Histone H3 peptide (green) forms an antiparallel β-sheet within the PHD finger subdomain around H3K4. Recognition of H3K4me0 is conserved among all ADDs. However, H3K9me3 readout and H3S10ph tolerance is unique to ATRX due to lack of functional pocket in DNMT3 ADDs. S10ph tolerance is partly contributed by intramolecular hydrogen bonding with H3R8. PDB entry 4W5A was used for figure preparation.
LINKING THE ADD DOMAIN TO HISTONE BINDING: INTERPRETING “OFF” AND “ON” HISTONE METHYLATION
A central question in epigenetic research is how patterns of DNA methylation that mediate heritable transcriptional silencing of affected sequences (e.g., retrotransposons or imprinted genes) are interpreted and targeted to the appropriate genomic regions. The methylation of lysine 4 on histone 3 (H3K4me) is an “active” mark that switches a gene “on” and is associated with gene promoters. By contrast, the methylation of lysine 9 on histone H3 (H3K9me) is a heterochromatin-associated mark that switches a gene “off.” Pioneering studies have shown that the H3K9 mark is required for DNA methylation in Neurospora crassa3 and for DNA methylation of non-CpG sequences such as CHG and CHH (H corresponds to A, T, or C) in Arabidopsis thaliana.2 Furthermore, mouse embryonic stem cells (ESCs) that lack the H3 lysine 9 methyltransferases Suv39h1 and Suv39h2 show demethylation of pericentromeric satellite DNA.21 Taken together, these studies suggest that a chromatin-based signal for DNA methylation occurs in the presence of H3K9 methylation and the absence of the H3K4 methylation. How these opposing “off” versus “on” histone methylation marks are recognized and translated into patterns of DNA methylation leads us toward structural and functional studies of de novo DNA methyltransferases themselves.
The de novo establishment of DNA methylation patterns in early mammalian development involves the DNMT3 family members DNMT3A and DNMT3B and the DNMT3-like nonenzymatic regulatory factor DNMT3L. DNMT3L is required for the de novo methylation of imprinting control regions in female germ cells and of dispersed repeated sequences in male germ cells through its recruitment or activation of active DNMT3 family members.22,23 Although catalytically inactive, DNMT3L has provided some important insights into how histone methylation marks are interpreted within epigenomes. Peptide-binding studies, using collections of biotinylated amino (N)-terminal tail peptides from each of the four core histones, have shown that DNMT3L binds to the extreme N-terminus of the H3 tail using a cysteine-rich domain. This domain shares some similarities with a PHD finger-like domain of BHC80, which is a subunit of the H3K4 demethylase complex LSD1.24 Importantly, and in keeping with a complex associated with transcriptional silencing, the binding of DNMT3L to the H3 N-tail was abolished by mono-, di-, or trimethylation of the “active” H3K4 mark but was insensitive to modifications at other positions such as “silencing” H3K9 methylation. This study provided an early indication that the cysteine-rich domain of DNMT3L—later defined as the conserved ADD domain—recognizes histone H3 tails that are either unmethylated or lack “active” (H3K4) methyl marks, whereas it tolerates “inactive” (H3K9) marks. Together, these properties suggest an attractive mechanism for targeting de novo DNA methylation to genomic regions that are to be silenced based upon the histone modification “signature.” Similarly, the ADD domain of de novo DNMT3A and DNMT3B binds to unmodified H3 N-tails regardless of H3K9 methylation but is disrupted by H3K4 methylation (Figure 1A).25,26 It is noteworthy that the ADD domain of DNMT3A/B/L (DNMT3ADD) does not contain a specific reading pocket for H3K9 methylation and so DNMT3 proteins probably function broadly throughout the genome (see below ATRXADD).
The other ADD domain-containing protein, ATRX, is an ATP-dependent chromatin remodeler and has been shown to interpret histone methylation marks in a similar manner to DNMT3ADD: ATRXADD exhibits a binding for H3K9 methylation, whereas binding is inhibited by H3K4 methylation. Interestingly, unlike DNMT3ADD, ATRXADD recognizes unmodified H3K4 and methylated H3K9 in a combinatorial manner; co-structural studies revealed an atypical H3K9me3-binding polar pocket at the GATA-like-PHD interface, which is distinct from the conventional trimethyllysine-binding aromatic cage (Figure 1B–D).27,28 Thus, ATRXADD represents a new reader module that senses the methylation states of both H3K4 and H3K9, which limits the location of ATRX mostly to the repeat elements enriched in these marks. In support, H3K9me3-pocket mutants and ATRX syndrome mutants are defective in both H3K9me3 binding and localization at pericentromeric heterochromatin.27,28 In line with genetic mutations mapping to other classes of histone-binding domains,29,30 this study supports the general view that mistakes in histone recognition have important human disease implications. In addition, while ATRX does not contain a DNA methyltransferase domain, ATRX syndrome mutations cause abnormal patterns of DNA methylation.31 Such a consequence is likely assisted by additional ATRX interacting partners, such as the H3K9me3 reader HP1 and the methyl-CpG-binding protein MeCP2, which act in concert with ATRX to establish connections among histone recognition, chromatin remodeling, DNA methylation, and heterochromatic silencing.32–34
LINKING HISTONE AND DNA METHYLATION THROUGH INTERACTIONS OF HISTONE VARIANTS, CHROMATIN REMODELERS, AND DE NOVO DNA METHYLTRANSFERASES
Histone variants often differ from their canonical counterparts by only a small number of amino-acid replacements (see ref 35 for review and references). A number of studies, all designed to investigate the biology of a histone variant in mammalian cells, independently revealed an unexpected association of a histone H3 family member, H3.3, with the chromatin remodeler ATRX, Daxx, and DNMT3 in locations closely associated with pericentromeric heterochromatin and telomeres.36–38 Recently, ATRX-dependent deposition of H3.3 has also been shown to occur in other distinct heterochromatic regions across the genome, notably at imprinted alleles.39 Preferential ATRX-driven H3.3 uptake in these regions localizes to the DNA-methylated allele, reinforcing the link between histones and DNA methylation. How histone variants such as H3.3 participate in the “epigenetic memory” of silenced heterochromatin in some genomic regions is poorly understood. However, despite these uncertainties, the importance of the ATRX/Daxx/DNMT3/H3.3 pathway is underscored by clear links to certain cancers that, in some cases, display striking alternative lengthening of telomere (ALT) phenotypes and genomic instability.40–42
Adding complexity to H3.3 biology is the fact that this histone variant can be incorporated into the genome by a distinct and nonoverlapping set of deposition machinery in a pathway much more closely associated with transcriptional activation. This pathway involves the H3.3-specific chaperone HIRA and other less well-characterized components that deposit H3.3 carrying “active” covalent modifications at genomic loci such as promoters, gene bodies, and enhancers.43–45 This “split personality” of H3.3 is both interesting and confusing. How does the ADD domain in ATRX or the DNMTs respond to activating H3.3 marks if indeed it evolved to be a heterochromatin-based chromatin reader? If, for example, active histone PTMs like H3K4 methylation repel the ADD domain as described above, can H3.3 discard these marks by some active mechanism, such as PTM “erasers” or rapid H3.3 turnover to allow for heterochromatin formation and gene silencing? What, if any, are the “rules” for ADD binding with respect to the wealth of PTMs other than H3K4 and H3K9 methylation, and can these rules be deciphered to allow its chromatin association to be altered in predictable ways in either active or inactive regions of the epigenome? Future studies addressing these questions will be highly informative as there are currently no comprehensive data regarding the effect of the H3.3 variant in DNA methylation.
ALLOSTERIC REGULATION AND AUTOINHIBITION OF DNA METHYLTRANSFERASES
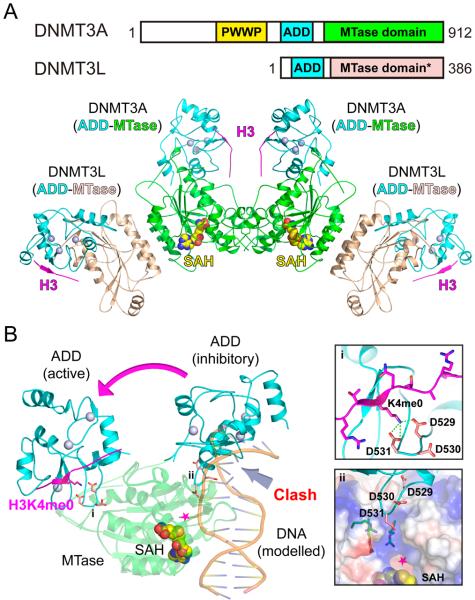
Initial biochemical studies showed that binding of DNMT3AADD to unmodified histone H3 stimulates the catalytic activity of DNMT3A, and such an allosteric regulation is required for de novo DNA methylation by DNMT3A in ESCs.26,46 The molecular basis underlying this activation was recently uncovered by elegant structural studies of a minimal DNMT3A-DNMT3L complex in its free state and bound to a H3K4me0 peptide (Figure 2A).47 Without the bound histone peptide, DNMT3AADD interacts with the C-terminal catalytic domain intramolecularly through electrostatic contacts between an acidic loop of DNMT3AADD and a basic patch of the catalytic domain (Figure 2B). This interaction impedes DNA binding and thus inhibits methyltransferase activity. When the ADD domain binds to an unmodified H3 tail, DNMT3A undergoes a large conformational change that allows DNA to access the catalytic site, suggesting an allosteric regulatory role of histone H3 in DNA methylation (Figure 2B). It is worth noting that the current allosteric model was proposed based on modeled DNA. Additional new insights could emerge from structural studies of the DNMT3A-H3 complex in the presence of bound DNA. Nevertheless, such an allosteric regulation by the histone H3 tail inversely correlates with the methylation state at H3K4, consistent with the genome-wide counter correlation between DNA methylation and histone H3K4 trimethylation. Collectively, these studies reveal that in addition to a role in recruitment, the ADD domain of DNMT3A also serves as an autoinhibitory switch that safeguards its catalytic activity in direct response to the presence of an unmethylated histone H3K4 tail.
Figure 2.
Complex structure, allosteric regulation, and autoinhibition of de novo DNA methyltransferases. (A) Top, domain architecture of mammalian de novo DNA methyltransferases DNMT3A and its cofactor DNMT3L. “*” denotes that the methyltransferase (MTase) domain of DNMT3L is catalytically inactive. Bottom, structural model of DNMT3L–DNMT3A–DNMT3A–DNMT3L tetramer bound to histone H3 peptides and S-adenosyl-l-homocysteine (SAH). The model was built by combining experimental structures of a minimal DNMT3A (ADD-MTase)-DNMT3L (MTase) tetramer bound to histone H3 peptide (PDB code: 4U7T) and full length DNMT3L (ADD-MTase) bound to histone H3 (PDB code: 2PVC). The protein structure is shown in ribbon representation, and domains of DNMT3A and 3L are color-coded as defined in domain architecture. The cofactor SAH is shown in space-filling spheres. Three zinc ions are depicted as light blue spheres. (B) Superimposed structures of H3K4me0-bound (PDB code: 4U7T) and peptide free (PDB code: 4U7P) DNMT3A ADD-MTase domains. MTase domain (green) was superimposed for structural alignment. Note the large domain movement of ADD from an inhibitory state to an active state upon H3 peptide (magenta) binding. Without histone H3, DNMT3A ADD adopts an autoinhibitory conformation and interacts with the MTase domain to block DNA substrate (light orange) binding. The modeled DNA was introduced by structural alignment with a bacterial CpG-specific DNA methyltransferase M. MpeI (PDB code: 4DKJ). Magenta star denotes the active site where the methyl transfer reaction occurs. Acidic residues that participate in autoinhibition and H3K4me0 recognition are shown as salmon sticks, with close-up views in (i) for H3K4me0 recognition and (ii) for autoinhibition near the active pocket. The surface of the active pocket is colored by its electrostatic potential with blue for positive charge and red for negative charge.
A similar mechanism of allosteric regulation and autoinhibition has been demonstrated for the maintenance DNA methyltransferase DNMT1. Crystal structure studies of DNMT1 have shown that an N-terminal replication foci domain (RFD) overlaps with the DNA substrate captured in an active state, suggesting that DNMT1 RFD has a cis-inhibitory role by blocking DNA substrate binding.48,49 Unlike DNMT3A, there is no direct evidence supporting an interaction between histones and DNMT1. However, the cross-talk between histone methylation and DNMT1-mediated methylation can be compensated by an essential DNMT1 binding partner, the ubiquitin-like PHD finger/RING domain-containing protein 1 (UHRF1) that contains paired reader modules including tandem Tudor, PHD finger, and SET and RING-associated (SRA). Structure and binding studies have established that the tandem Tudor and PHD finger domains act combinatorially to recognize a modification pattern of unmodified histone H3R2 and H3K9me3.50–52 The UHRF1 SRA domain preferentially binds to hemimethylated DNA.53–56 Subsequent biochemical studies have further revealed that UHRF1 stimulates the activity of DNMT1 mainly via a physical interaction between its SRA and the DNMT1 RFD domains.57 Collectively, the above epigenetic reader modules within UHRF1 provide a DNMT3AADD-like function to form molecular connections between repressive histone H3K9me3 readout, hemimethylated CG recognition, and allosteric stimulation during maintenance DNA methylation by DNMT1 (see ref 58 for review).
ENGINEERING CHROMATIN-BINDING DOMAINS TO ALTER EPIGENETIC LANDSCAPES
Given the complexities in histone-reader interactions noted above, re-engineering histone-binding modules to tolerate new PTM signatures is clearly challenging. This approach would allow the controlled altering of epigenetic states, but few such engineered chromatin-binding domains have been created to date. That said, numerous histone-reader structures have become available, partly due to the growing interest in epigenetics, particularly when combined with disease implications. Taking clues from the lower methylation state properties of the malignant brain tumor (MBT) repeats of L3MBT—which was shown to prefer monomethyl over dimethyl lysines—an early, proof-of-principle example has been provided by a single amino-acid substitution in the aromatic cage of the PHD finger-containing protein BPTF. This substitution (Y to E) altered the binding preference of BPTF from H3K4me3 to H3K4me2.59 Although this study did not probe the function of these changes, it provided insights into how histone “pockets” can be “decoded” to change methylation-state preference.
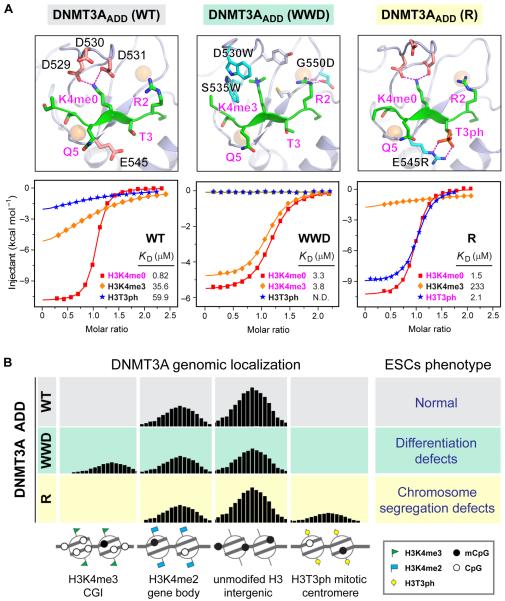
Following this example, and building on the growing literature of ADD domain structure-function analyses, the ADD domain of DNMT3A was recently re-engineered to expand its binding preference to allow all methylation states of “active” H3K4 (me1/me2/me3), which are not normally accommodated in wild-type DNMT3A.60 Systematic mutagenesis of key DNMT3AADD pocket residues created mutant DNMT3 alleles with gain-of-function properties with respect to its ADD domain; specifically, a “WWD” mutant contained two novel residues W–W in its aromatic cage to interact with H3K4me and a critical D to interact with H3R2 (Figure 3A). To avoid complications from wild-type copies of DNMT3, mutant expression studies were performed in triple knockout mouse ESCs lacking all copies of active de novo DNMT3A/B and the maintenance DNMT1. Targeting of Dnmt3a to H3K4me3 promoters decreased gene expression in a subset of developmental genes and affected ESC differentiation. Another mutant allowed aberrant binding to phosphorylated H3T3 during mitosis, which promoted genomic instability (Figure 3B).
Figure 3.
Structure-based reader engineering of DNMT3A ADD and its functional outcomes. (A) Structures (top) and calorimetric titration fitting curves (bottom) of wild type (WT) (PDB code: 4QBQ), WWD mutant (modeled based on a G550D structure, PDB code: 4QBR), and R mutant DNMT3A ADDs (PDB code: 4QBS). Histone peptides are shown as green ribbons. Mutant residues are shown as cyan sticks. Binding KD's are listed below the fitting curve with strong binding highlighted in magenta. (B) Diagram of genome-wide distribution of wild type and re-engineered DNMT3A and its phenotypic outcomes in embryonic stem cells (ESCs).
Although these pioneering studies of histone-binding pockets have yet to engineer a “black-and-white” switch in histone reading, they support the general notion that binding pockets can be altered to mis-read histone/epigenetic codes. Importantly, the structure-function studies of DNMT3AADD make a compelling argument in support of the interplay between histone methylation and DNA methylation. We look forward to more experimental studies aimed at perturbing histone-reading function in systems in which phenotypes can be examined. As mentioned above, the development of small molecules that can disrupt chromatin reading and the uncovering of mutations in histone-binding pockets underscore the need to continue to determine the “rules” of these PTM-binding interactions, not only between chromatin-associated proteins and histones but equally nonhistone targets that use what has been termed “histone mimicry.”18,61 The concept that de novo DNA methylation writers have evolved to exploit these interactions to bind to and enrich epigenetic landscapes is a central point that the ADD domain serves to illustrate.
NEIGHBORING HISTONE MODIFICATIONS: DOES EVERY AMINO ACID IN HISTONES MATTER, AND IF SO, HOW?
With respect to other chromatin-binding domains, phosphorylation nearby or adjacent to the principal site of modification that is being “read” has been shown to influence the overall binding reaction both in vitro and in vivo. One of the better-known examples of this cross-talk comes from the chromodomain-containing, heterochromatin-associated protein HP1. HP1 is anchored, at least in part, to H3K9 methylated chromatin through its chromodomain. Phosphorylation of an adjacent serine 10 (H3S10ph) has been shown to disrupt HP1 binding in a mechanism termed “phospho-methyl switching.”62,63 “Acetyl-methyl switching” has also been reported to regulate crucial protein-protein interactions of nonhistone proteins with their binding partners, suggesting that this may be a broadly used mechanism.64
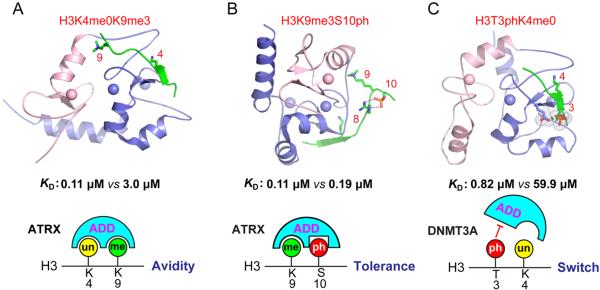
As with every rule, exceptions exist, but these provide important insights into the process under examination. Recently, several examples have been described for which phospho-methyl switching may not apply to every chromatin-binding module. For example, unlike HP1, UHRF1 binds to methylated H3K9 but is insensitive to mitotic H3S10 phosphorylation.52 As UHRF1 association with the maintenance DNA methyltransferase, DNMT1, is required for maintaining DNA methylation through mitosis, this exception to phospho-methyl switching has important biological consequences. Similarly, the ADD domain of ATRX is insensitive to H3S10 phosphorylation (Figure 4A, B), giving it the unique property of binding to “dual” H3K9meS10ph marks in postmitotic neurons.27,65 Upon neuronal stimulation, the dual marks mainly localize to centromeric and pericentromeric regions within the nucleus (e.g., heterochromatic structures that surround the nucleolus and nuclear membrane). The transient nature of activity-dependent histone phosphorylation enables a rapid transcriptional response while maintaining cellular identity. It is suggested that this H3K9meS10ph binding property allows ATRX to maintain repeat element silencing during periods of activity-dependent, H3S10-mediated phosphorylation.
Figure 4.
Neighboring histone modification cross-talk centered on ADD domains. (A) ATRX ADD domain recognizes a combinatorial methylation pattern of unmodified H3K4 (H3K4me0; KD = 3.0 μM) and trimethylated H3K9 (H3K9me3; KD = 0.11 μM) with enhanced binding affinity. (B) ATRX ADD domain recognition of H3K9me3 (KD = 0.11 μM) tolerates H3S10 phosphorylation (KD = 0.19 μM) with minimally affected binding affinity. (C) The binding of DNMT3A ADD domain to H3K4me0 (KD = 0.82 μM) can be disrupted by phosphorylation at H3T3 (H3T3ph; KD = 59.9 μM) through a binary switch mechanism. Histone peptides are depicted as green ribbons. The ADD domains are in ribbon representation with pink for the GATA-like finger and blue for the PHD finger. Zinc ions are shown as spheres. Binding KD's are taken from refs 60 and 65.
Increasing this complexity, multivalent modifications (e.g., methylation, acetylation, and phosphorylation) in neighboring residues on the H3 N-tail add a further layer of cooperative, antagonistic, or independent relationships between modified histones and bound protein.66,67 It has been shown that modification of H3R2, H3K4, H3T3, H3T6, and H3S10, but not H3R8 and H3K9, disrupts high-affinity binding of the chromodomain- and PHD finger-containing protein CHD5.68 Given that CHD5 is a chromatin-remodeling enzyme, this multivalent recognition can enhance the specificity of its target genomic regions. For the ATRX and DNMT3 ADD domains, modification of H3K4, H3T3, and H3T6 disrupts binding to the H3 N-terminus (Figure 4C); however, modification of H3R2, H3R8, H3K9, and H3S10 is compatible with binding.60,65 In relation to phosphorylation, H3T3 appears exclusively during mitosis enriched at centromere regions69,70 and H3T6 occurs during interphase at the gene promoters.71,72 Considering that phosphorylation at both H3T3 and H3S10 appear during mitosis, selective ADD disruption by H3T3 is probably linked to a specific function (see Figure 3B). Taken together, these findings from the H3 N-tail alone demonstrate the extent to which complex cross-talk interactions can disrupt crucial histone and reader interactions.
FUTURE DIRECTIONS AND CHALLENGES
In this review, we have highlighted the ADD domain as an informative example of how histone “reading” not only wprobably—if not certainly—drives interactions between a wealth of other epigenetic regulators, including protein, DNA and RNA components. A staggering array of histone PTMs are being identified by new approaches73,74 as well as novel chromatin-reading domains (e.g., see75). Considerable structural information is revealing exquisite specificity that characterizes the binding of histone-binding domain and ligand interactions.13 Although most published cocrystal structures utilize PTM-modified (or unmodified) histone peptides with cognate readers, several studies have extended these concepts and have examined modified nucleosomes with multiple histone-binding domains, illustrating the concept of multivalency in histone/chromatin reading.66,76,77 High-throughput studies using immobilized histone peptide arrays78 or bar-coded “designer nucleosomes”79,80 will enable the rapid screening of combinatorial histone PTMs, which is an explicit prediction of the histone code hypothesis.
Despite these advances, significant challenges remain as to how well-defined, in vitro histone-protein interactions can be translated into in vivo functional insights. Structural-guided protein engineering via mutagenesis of key histone-binding residues will undoubtedly allow detailed abinding “rules” to be uncovered. Combining this information with novel genome-editing approaches, such as the CRISPR-Cas system, will permit more elaborate knockout/add-back studies than the DNMT3AADD structure-function studies described above60 and could be extended to the interplay of other key epigenetic regulators.81 Mutagenesis of essential residues in histone proteins is complicated by the multicopy nature of histone-encoding genes. That said, novel synthetic biology methods have been developed to engineer histones that bear site-specific modifications on cellular chromatin using protein trans-splicing with fast-acting inteins.82 Meanwhile, with the rise of functional genomic and proteomic technologies, numerous signature chromatin modifications have been identified genome-wide to dictate critical regulatory functions in vivo.83,84 One special case is the discovery of bivalent modifications that demarcate promoters (H3K4me3-H3K27me3)85 or enhancers (e.g., H3K4me1-H3K27me3, H3K4me1-H3K27ac)86,87 of developmentally regulated genes. Thus, defining the readers of these bivalent marks among other hallmark histone modification(s) represents an outstanding challenge in the field with important implications in development and stem cell biology.
Finally, the identification of monoallelic, single-copy mutations at or nearby key sites of histone PTMs in various cancers (referred to as “oncohistones”42,88), as well as the better-documented mutations in genes that encode histone-binding domains (discussed above for the ADD domain), underscore the importance of human genetics in dissecting histone reading by associated epigenetic regulators. Mistakes made on either “side” of these protein-protein interactions have profound disease implications and pave the way for new and exciting therapeutic strategies to treat them as exemplified by the effective drugging of bromodomain/acetyl-histone interactions.17,18 Learning how to smartly “pick pockets” promises to lead to a lucrative (and legal) scientific career for members of the epigenetics community.
KEYWORDS.
ADD domain: A histone-binding zinc finger module named after “ATRX-DNMT3-DNMT3L”. Topologically, ADD is an integration of a GATA-like finger and a PHD finger.
DNA methylation: A process or modification state by which methyl groups are added to DNA. Methylation of cytosine at the 5 position within “CpG” dinucleotide is often, but not always, linked to gene silencing.
Histone modification: A covalent post-translational modification to histone proteins which includes the addition of chemical groups such as methylation, phosphorylation, acetylation, ubiquitination, and sumoylation.
Histone variant: Noncanonical and nonallelic variants of histones—representing one or a few amino acid differences—that are expressed at very low levels compared with their conventional counterparts.
Reader: Proteins and domains capable of binding to a specific epigenetic mark to recruit certain proteins to the target epigenetic mark.
Writer: Enzymes that generate a specific epigenetic mark to establish a modification state of chromatin.
Combinatorial readout: Cooperative recognition of multiple histone or DNA modifications by reader module(s) with significantly enhanced binding as compared to a single mark readout.
Bivalent tolerance: Introduction of an adjacent modification has a minimal effect on preexisting “mark-reader” interaction.
Binary switch: Introduction of an adjacent or nearby modification acts to disrupt (switch off), or in some cases, facilitate the binding (switch on) of a particular “mark-reader” pair.
Epigenetic code: A second system “above” or “in addition to” the genetic code that give rises to heritable phenotypes brought about by alterations in chromatin states without changes in DNA sequences. Epigenetic modifications, which define these states, may include histone modifications and DNA methylation
ACKNOWLEDGMENTS
We apologize to all authors whose important contributions could not be acknowledged because of space constraints. We thank D. Zhao, B. Xiang, and H. Wang from the Li laboratory for their structural studies of ATRXADD and DNMT3AADD. This work is supported by grants from the General Program of National Natural Science Foundation of China (31270763 to H.L.), the Major State Basic Research Development Program in China (2015CB910503 to H.L.), the Women & Science Postdoctoral Fellowship (to K.-M.N.), a NIMH grant (PHS MH 094698 to C.D.A.), and a Leukemia Lymphoma Society Program Project Award (NORTHWESTERN-LLS 7006-13 to C.D.A.).
Footnotes
The authors declare no competing financial interest.
REFERENCES
- (1).Allis CD, Caparros M-L, Jenuwein T, Reinberg D. Epigenetics. 2nd ed. Chapter 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor; New York: 2015. Overview and Concepts. [Google Scholar]
- (2).Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- (3).Tamaru H, Selker EU. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- (4).Tamaru H, Zhang X, McMillen D, Singh PB, Nakayama J, Grewal SI, Allis CD, Cheng X, Selker EU. Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nat. Genet. 2003;34:75–79. doi: 10.1038/ng1143. [DOI] [PubMed] [Google Scholar]
- (5).Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- (6).Lay FD, Liu Y, Kelly TK, Witt H, Farnham PJ, Jones PA, Berman BP. The role of DNA methylation in directing the functional organization of the cancer epigenome. Genome Res. 2015;25:467–477. doi: 10.1101/gr.183368.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Morgan MA, Shilatifard A. Chromatin signatures of cancer. Genes Dev. 2015;29:238–249. doi: 10.1101/gad.255182.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Roy DM, Walsh LA, Chan TA. Driver mutations of cancer epigenomes. Protein Cell. 2014;5:265–296. doi: 10.1007/s13238-014-0031-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Shi J, Vakoc CR. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol. Cell. 2014;54:728–736. doi: 10.1016/j.molcel.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- (11).Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- (12).Musselman CA, Lalonde ME, Cote J, Kutateladze TG. Perceiving the epigenetic landscape through histone readers. Nat. Struct. Mol. Biol. 2012;19:1218–1227. doi: 10.1038/nsmb.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Patel DJ, Wang Z. Readout of epigenetic modifications. Annu. Rev. Biochem. 2013;82:81–118. doi: 10.1146/annurev-biochem-072711-165700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Baker LA, Allis CD, Wang GG. PHD fingers in human diseases: disorders arising from misinterpreting epigenetic marks. Mutat. Res., Fundam. Mol. Mech. Mutagen. 2008;647:3–12. doi: 10.1016/j.mrfmmm.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Wang GG, Allis CD, Chi P. Chromatin remodeling and cancer, Part I: Covalent histone modifications. Trends Mol. Med. 2007;13:363–372. doi: 10.1016/j.molmed.2007.07.003. [DOI] [PubMed] [Google Scholar]
- (17).Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, Philpott M, Munro S, McKeown MR, Wang Y, Christie AL, West N, Cameron MJ, Schwartz B, Heightman TD, La Thangue N, French CA, Wiest O, Kung AL, Knapp S, Bradner JE. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, Chandwani R, Marazzi I, Wilson P, Coste H, White J, Kirilovsky J, Rice CM, Lora JM, Prinjha RK, Lee K, Tarakhovsky A. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Wang CY, Filippakopoulos P. Beating the odds: BETs in disease. Trends Biochem. Sci. 2015;40:468–479. doi: 10.1016/j.tibs.2015.06.002. [DOI] [PubMed] [Google Scholar]
- (20).Lyst MJ, Bird A. Rett syndrome: a complex disorder with simple roots. Nat. Rev. Genet. 2015;16:261–275. doi: 10.1038/nrg3897. [DOI] [PubMed] [Google Scholar]
- (21).Lehnertz B, Ueda Y, Derijck AAHA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen TP, Li E, Jenuwein T, Peters AHFM. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- (22).Bourc'his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- (23).Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129:1983–1993. doi: 10.1242/dev.129.8.1983. [DOI] [PubMed] [Google Scholar]
- (24).Ooi SKT, Qiu C, Bernstein E, Li KQ, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, Cheng XD, Bestor TH. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–U713. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Otani J, Nankumo T, Arita K, Inamoto S, Ariyoshi M, Shirakawa M. Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain. EMBO Rep. 2009;10:1235–1241. doi: 10.1038/embor.2009.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Zhang Y, Jurkowska R, Soeroes S, Rajavelu A, Dhayalan A, Bock I, Rathert P, Brandt O, Reinhardt R, Fischle W, Jeltsch A. Chromatin methylation activity of Dnmt3a and Dnmt3a/3L is guided by interaction of the ADD domain with the histone H3 tail. Nucleic Acids Res. 2010;38:4246–4253. doi: 10.1093/nar/gkq147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Eustermann S, Yang JC, Law MJ, Amos R, Chapman LM, Jelinska C, Garrick D, Clynes D, Gibbons RJ, Rhodes D, Higgs DR, Neuhaus D. Combinatorial readout of histone H3 modifications specifies localization of ATRX to heterochromatin. Nat. Struct. Mol. Biol. 2011;18:777–782. doi: 10.1038/nsmb.2070. [DOI] [PubMed] [Google Scholar]
- (28).Iwase S, Xiang B, Ghosh S, Ren T, Lewis PW, Cochrane JC, Allis CD, Picketts DJ, Patel DJ, Li H, Shi Y. ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome. Nat. Struct. Mol. Biol. 2011;18:769–776. doi: 10.1038/nsmb.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, Kou Y, Liu L, Fromer M, Walker S, Singh T, Klei L, Kosmicki J, Shih-Chen F, Aleksic B, Biscaldi M, Bolton PF, Brownfeld JM, Cai J, Campbell NG, Carracedo A, Chahrour MH, Chiocchetti AG, Coon H, Crawford EL, Curran SR, Dawson G, Duketis E, Fernandez BA, Gallagher L, Geller E, Guter SJ, Hill RS, Ionita-Laza J, Jimenz Gonzalez P, Kilpinen H, Klauck SM, Kolevzon A, Lee I, Lei I, Lei J, Lehtimaki T, Lin CF, Ma'ayan A, Marshall CR, McInnes AL, Neale B, Owen MJ, Ozaki N, Parellada M, Parr JR, Purcell S, Puura K, Rajagopalan D, Rehnstrom K, Reichenberg A, Sabo A, Sachse M, Sanders SJ, Schafer C, Schulte-Ruther M, Skuse D, Stevens C, Szatmari P, Tammimies K, Valladares O, Voran A, Li-San W, Weiss LA, Willsey AJ, Yu TW, Yuen RK, Study DDD, Homozygosity Mapping Collaborative for, A. Consortium UK, Cook EH, Freitag CM, Gill M, Hultman CM, Lehner T, Palotie A, Schellenberg GD, Sklar P, State MW, Sutcliffe JS, Walsh CA, Scherer SW, Zwick ME, Barett JC, Cutler DJ, Roeder K, Devlin B, Daly MJ, Buxbaum JD. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Iossifov I, O'Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, Stessman HA, Witherspoon KT, Vives L, Patterson KE, Smith JD, Paeper B, Nickerson DA, Dea J, Dong S, Gonzalez LE, Mandell JD, Mane SM, Murtha MT, Sullivan CA, Walker MF, Waqar Z, Wei L, Willsey AJ, Yamrom B, Lee YH, Grabowska E, Dalkic E, Wang Z, Marks S, Andrews P, Leotta A, Kendall J, Hakker I, Rosenbaum J, Ma B, Rodgers L, Troge J, Narzisi G, Yoon S, Schatz MC, Ye K, McCombie WR, Shendure J, Eichler EE, State MW, Wigler M. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Gibbons RJ, McDowell TL, Raman S, O'Rourke DM, Garrick D, Ayyub H, Higgs DR. Mutations in ATRX, encoding a SWI/SNF-like protein, cause diverse changes in the pattern of DNA methylation. Nat. Genet. 2000;24:368–371. doi: 10.1038/74191. [DOI] [PubMed] [Google Scholar]
- (32).Lechner MS, Schultz DC, Negorev D, Maul GG, Rauscher FJ., 3rd The mammalian heterochromatin protein 1 binds diverse nuclear proteins through a common motif that targets the chromoshadow domain. Biochem. Biophys. Res. Commun. 2005;331:929–937. doi: 10.1016/j.bbrc.2005.04.016. [DOI] [PubMed] [Google Scholar]
- (33).Nan X, Hou J, Maclean A, Nasir J, Lafuente MJ, Shu X, Kriaucionis S, Bird A. Interaction between chromatin proteins MECP2 and ATRX is disrupted by mutations that cause inherited mental retardation. Proc. Natl. Acad. Sci. U. S. A. 2007;104:2709–2714. doi: 10.1073/pnas.0608056104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Ratnakumar K, Bernstein E. ATRX: The case of a peculiar chromatin remodeler. Epigenetics. 2013;8:3–9. doi: 10.4161/epi.23271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Maze I, Noh KM, Soshnev AA, Allis CD. Every amino acid matters: essential contributions of histone variants to mammalian development and disease. Nat. Rev. Genet. 2014;15:259–271. doi: 10.1038/nrg3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Drane P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010;24:1253–1265. doi: 10.1101/gad.566910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, Wen D, Chapgier A, DeKelver RC, Miller JC, Lee YL, Boydston EA, Holmes MC, Gregory PD, Greally JM, Rafii S, Yang C, Scambler PJ, Garrick D, Gibbons RJ, Higgs DR, Cristea IM, Urnov FD, Zheng D, Allis CD. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Wong LH, McGhie JD, Sim M, Anderson MA, Ahn S, Hannan RD, George AJ, Morgan KA, Mann JR, Choo KH. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 2010;20:351–360. doi: 10.1101/gr.101477.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Voon HP, Hughes JR, Rode C, De La Rosa-Velazquez IA, Jenuwein T, Feil R, Higgs DR, Gibbons RJ. ATRX Plays a Key Role in Maintaining Silencing at Interstitial Heterochromatic Loci and Imprinted Genes. Cell Rep. 2015;11:405–418. doi: 10.1016/j.celrep.2015.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, Schulick RD, Tang LH, Wolfgang CL, Choti MA, Velculescu VE, Diaz LA, Jr., Vogelstein B, Kinzler KW, Hruban RH, Papadopoulos N. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331:1199–1203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Lovejoy CA, Li W, Reisenweber S, Thongthip S, Bruno J, de Lange T, De S, Petrini JH, Sung PA, Jasin M, Rosenbluh J, Zwang Y, Weir BA, Hatton C, Ivanova E, Macconaill L, Hanna M, Hahn WC, Lue NF, Reddel RR, Jiao Y, Kinzler K, Vogelstein B, Papadopoulos N, Meeker AK, Consortium ALTSC. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet. 2012;8:e1002772. doi: 10.1371/journal.pgen.1002772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tonjes M, Hovestadt V, Albrecht S, Kool M, Nantel A, Konermann C, Lindroth A, Jager N, Rausch T, Ryzhova M, Korbel JO, Hielscher T, Hauser P, Garami M, Klekner A, Bognar L, Ebinger M, Schuhmann MU, Scheurlen W, Pekrun A, Fruhwald MC, Roggendorf W, Kramm C, Durken M, Atkinson J, Lepage P, Montpetit A, Zakrzewska M, Zakrzewski K, Liberski PP, Dong Z, Siegel P, Kulozik AE, Zapatka M, Guha A, Malkin D, Felsberg J, Reifenberger G, von Deimling A, Ichimura K, Collins VP, Witt H, Milde T, Witt O, Zhang C, Castelo-Branco P, Lichter P, Faury D, Tabori U, Plass C, Majewski J, Pfister SM, Jabado N. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- (43).Banaszynski LA, Wen D, Dewell S, Whitcomb SJ, Lin M, Diaz N, Elsasser SJ, Chapgier A, Goldberg AD, Canaani E, Rafii S, Zheng D, Allis CD. Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell. 2013;155:107–120. doi: 10.1016/j.cell.2013.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Elsaesser SJ, Goldberg AD, Allis CD. New functions for an old variant: no substitute for histone H3.3. Curr. Opin. Genet. Dev. 2010;20:110–117. doi: 10.1016/j.gde.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Ray-Gallet D, Woolfe A, Vassias I, Pellentz C, Lacoste N, Puri A, Schultz DC, Pchelintsev NLA, Adams PD, Jansen LET, Almouzni G. Dynamics of Histone H3 Deposition In Vivo Reveal a Nucleosome Gap-Filling Mechanism for H3.3 to Maintain Chromatin Integrity. Mol. Cell. 2011;44:928–941. doi: 10.1016/j.molcel.2011.12.006. [DOI] [PubMed] [Google Scholar]
- (46).Li BZ, Huang Z, Cui QY, Song XH, Du L, Jeltsch A, Chen P, Li GH, Li E, Xu GL. Histone tails regulate DNA methylation by allosterically activating de novo methyltransferase. Cell Res. 2011;21:1172–1181. doi: 10.1038/cr.2011.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Guo X, Wang L, Li J, Ding Z, Xiao J, Yin X, He S, Shi P, Dong L, Li G, Tian C, Wang J, Cong Y, Xu Y. Structural insight into autoinhibition and histone H3-induced activation of DNMT3A. Nature. 2015;517:640–644. doi: 10.1038/nature13899. [DOI] [PubMed] [Google Scholar]
- (48).Song J, Teplova M, Ishibe-Murakami S, Patel DJ. Structure-based mechanistic insights into DNMT1-mediated maintenance DNA methylation. Science. 2012;335:709–712. doi: 10.1126/science.1214453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Syeda F, Fagan RL, Wean M, Avvakumov GV, Walker JR, Xue S, Dhe-Paganon S, Brenner C. The Replication Focus Targeting Sequence (RFTS) Domain Is a DNA-competitive Inhibitor of Dnmt1. J. Biol. Chem. 2011;286:15344–15351. doi: 10.1074/jbc.M110.209882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Arita K, Isogai S, Oda T, Unoki M, Sugita K, Sekiyama N, Kuwata K, Hamamoto R, Tochio H, Sato M, Ariyoshi M, Shirakawa M. Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1. Proc. Natl. Acad. Sci. U. S. A. 2012;109:12950–12955. doi: 10.1073/pnas.1203701109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Rothbart SB, Dickson BM, Ong MS, Krajewski K, Houliston S, Kireev DB, Arrowsmith CH, Strahl BD. Multivalent histone engagement by the linked tandem Tudor and PHD domains of UHRF1 is required for the epigenetic inheritance of DNA methylation. Genes Dev. 2013;27:1288–1298. doi: 10.1101/gad.220467.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Rothbart SB, Krajewski K, Nady N, Tempel W, Xue S, Badeaux AI, Barsyte-Lovejoy D, Martinez JY, Bedford MT, Fuchs SM, Arrowsmith CH, Strahl BD. Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat. Struct. Mol. Biol. 2012;19:1155–1160. doi: 10.1038/nsmb.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008;455:818–821. doi: 10.1038/nature07249. [DOI] [PubMed] [Google Scholar]
- (54).Avvakumov GV, Walker JR, Xue S, Li Y, Duan S, Bronner C, Arrowsmith CH, Dhe-Paganon S. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature. 2008;455:822–825. doi: 10.1038/nature07273. [DOI] [PubMed] [Google Scholar]
- (55).Hashimoto H, Horton JR, Zhang X, Bostick M, Jacobsen SE, Cheng X. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature. 2008;455:826–829. doi: 10.1038/nature07280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Qian C, Li S, Jakoncic J, Zeng L, Walsh MJ, Zhou MM. Structure and hemimethylated CpG binding of the SRA domain from human UHRF1. J. Biol. Chem. 2008;283:34490–34494. doi: 10.1074/jbc.C800169200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Bashtrykov P, Jankevicius G, Jurkowska RZ, Ragozin S, Jeltsch A. The UHRF1 protein stimulates the activity and specificity of the maintenance DNA methyltransferase DNMT1 by an allosteric mechanism. J. Biol. Chem. 2014;289:4106–4115. doi: 10.1074/jbc.M113.528893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Du J, Johnson LM, Jacobsen SE, Patel DJ. DNA methylation pathways and their crosstalk with histone methylation. Nat. Rev. Mol. Cell Biol. 2015;16:519–532. doi: 10.1038/nrm4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Li H, Fischle W, Wang W, Duncan EM, Liang L, Murakami-Ishibe S, Allis CD, Patel DJ. Structural basis for lower lysine methylation state-specific readout by MBT repeats of L3MBTL1 and an engineered PHD finger. Mol. Cell. 2007;28:677–691. doi: 10.1016/j.molcel.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Noh KM, Wang HB, Kim HR, Wenderski W, Fang F, Li CH, Dewell S, Hughes SH, Melnick AM, Patel DJ, Li HT, Allis CD. Engineering of a Histone-Recognition Domain in Dnmt3a Alters the Epigenetic Landscape and Phenotypic Features of Mouse ESCs. Mol. Cell. 2015;59:89–103. doi: 10.1016/j.molcel.2015.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Marazzi I, Ho JSY, Kim J, Manicassamy B, Dewell S, Albrecht RA, Seibert CW, Schaefer U, Jeffrey KL, Prinjha RK, Lee K, Garcia-Sastre A, Roeder RG, Tarakhovsky A. Suppression of the antiviral response by an influenza histone mimic. Nature. 2012;483:428–433. doi: 10.1038/nature10892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- (63).Fischle W, Wang YM, Allis CD. Binary switches and modification cassettes in histone biology and beyond. Nature. 2003;425:475–479. doi: 10.1038/nature02017. [DOI] [PubMed] [Google Scholar]
- (64).Tong Q, Mazur SJ, Rincon-Arano H, Rothbart SB, Kuznetsov DM, Cui GF, Liu WH, Gete Y, Klein BJ, Jenkins L, Mer G, Kutateladze AG, Strahl BD, Groudine M, Appella E, Kutateladze TG. An Acetyl-Methyl Switch Drives a Conformational Change in p53. Structure. 2015;23:322–331. doi: 10.1016/j.str.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Noh KM, Maze I, Zhao D, Xiang B, Wenderski W, Lewis PW, Shen L, Li H, Allis CD. ATRX tolerates activity-dependent histone H3 methyl/phos switching to maintain repetitive element silencing in neurons. Proc. Natl. Acad. Sci. U. S. A. 2015;112:6820–6827. doi: 10.1073/pnas.1411258112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Li H, Zhao S, Patel D. Histone Recognition by Tandem Modules and Modulation by Multiple PTMs. In: Zhou M-M, editor. Histone Recognition. Springer International Publishing; 2015. pp. 149–172. [Google Scholar]
- (67).Oliver SS, Denu JM. Dynamic Interplay between Histone H3Modifications and Protein Interpreters: Emerging Evidence for a “Histone Language”. ChemBioChem. 2011;12:299–307. doi: 10.1002/cbic.201000474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Oliver SS, Musselman CA, Srinivasan R, Svaren JP, Kutateladze TG, Denu JM. Multivalent recognition of histone tails by the PHD fingers of CHD5. Biochemistry. 2012;51:6534–6544. doi: 10.1021/bi3006972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Dai J, Sultan S, Taylor SS, Higgins JM. The kinase haspin is required for mitotic histone H3 Thr 3 phosphorylation and normal metaphase chromosome alignment. Genes Dev. 2005;19:472–488. doi: 10.1101/gad.1267105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Markaki Y, Christogianni A, Politou AS, Georgatos SD. Phosphorylation of histone H3 at Thr3 is part of a combinatorial pattern that marks and configures mitotic chromatin. J. Cell Sci. 2009;122:2809–2819. doi: 10.1242/jcs.043810. [DOI] [PubMed] [Google Scholar]
- (71).Garske AL, Oliver SS, Wagner EK, Musselman CA, LeRoy G, Garcia BA, Kutateladze TG, Denu JM. Combinatorial profiling of chromatin binding modules reveals multisite discrimination. Nat. Chem. Biol. 2010;6:283–290. doi: 10.1038/nchembio.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Metzger E, Imhof A, Patel D, Kahl P, Hoffmeyer K, Friedrichs N, Muller JM, Greschik H, Kirfel J, Ji S, Kunowska N, Beisenherz-Huss C, Gunther T, Buettner R, Schule R. Phosphorylation of histone H3T6 by PKCbeta(I) controls demethylation at histone H3K4. Nature. 2010;464:792–796. doi: 10.1038/nature08839. [DOI] [PubMed] [Google Scholar]
- (73).Huang H, Sabari BR, Garcia BA, Allis CD, Zhao Y. SnapShot: histone modifications. Cell. 2014;159:458–458.e1. doi: 10.1016/j.cell.2014.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Zhao Y, Garcia BA. Comprehensive Catalog of Currently Documented Histone Modifications. Cold Spring Harbor Perspect. Biol. 2015;7:a025064. doi: 10.1101/cshperspect.a025064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Li Y, Wen H, Xi Y, Tanaka K, Wang H, Peng D, Ren Y, Jin Q, Dent SY, Li W, Li H, Shi X. AF9 YEATS domain links histone acetylation to DOT1L-mediated H3K79 methylation. Cell. 2014;159:558–571. doi: 10.1016/j.cell.2014.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Ruthenburg AJ, Li H, Milne TA, Dewell S, McGinty RK, Yuen M, Ueberheide B, Dou Y, Muir TW, Patel DJ, Allis CD. Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell. 2011;145:692–706. doi: 10.1016/j.cell.2011.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Rothbart SB, Dickson BM, Raab JR, Grzybowski AT, Krajewski K, Guo AH, Shanle EK, Josefowicz SZ, Fuchs SM, Allis CD, Magnuson TR, Ruthenburg AJ, Strahl BD. An Interactive Database for the Assessment of Histone Antibody Specificity. Mol. Cell. 2015;59:502–511. doi: 10.1016/j.molcel.2015.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Muller MM, Muir TW. Histones: at the crossroads of peptide and protein chemistry. Chem. Rev. 2015;115:2296–2349. doi: 10.1021/cr5003529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Nguyen UT, Bittova L, Muller MM, Fierz B, David Y, Houck-Loomis B, Feng V, Dann GP, Muir TW. Accelerated chromatin biochemistry using DNA-barcoded nucleosome libraries. Nat. Methods. 2014;11:834–840. doi: 10.1038/nmeth.3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Hilton IB, D'Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, Gersbach CA. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 2015;33:510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).David Y, Vila-Perello M, Verma S, Muir TW. Chemical tagging and customizing of cellular chromatin states using ultrafast trans-splicing inteins. Nat. Chem. 2015;7:394–402. doi: 10.1038/nchem.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Han Y, Garcia BA. Combining genomic and proteomic approaches for epigenetics research. Epigenomics. 2013;5:439–452. doi: 10.2217/epi.13.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- (86).Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Zentner GE, Tesar PJ, Scacheri PC. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 2011;21:1273–1283. doi: 10.1101/gr.122382.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, Qu C, Ding L, Huether R, Parker M, Zhang J, Gajjar A, Dyer MA, Mullighan CG, Gilbertson RJ, Mardis ER, Wilson RK, Downing JR, Ellison DW, Zhang J, Baker SJ, St. Jude Children's Research Hospital-Washington University Pediatric Cancer Genome, P Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat. Genet. 2012;44:251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]