Abstract
F-box proteins, subunits of SKP1-cullin 1-F-box protein (SCF) type of E3 ubiquitin ligase complexes, have been validated to play a crucial role in governing various cellular processes such as cell cycle, cell proliferation, apoptosis, migration, invasion and metastasis. Recently, a wealth of evidence has emerged that F-box proteins is critically involved in tumorigenesis in part through governing the ubiquitination and subsequent degradation of cell cycle proteins, and dysregulation of this process leads to aberrant cell cycle progression and ultimately, tumorigenesis. Therefore, in this review, we describe the critical role of F-box proteins in the timely regulation of cell cycle. Moreover, we discuss how F-box proteins involve in tumorigenesis via targeting cell cycle-related proteins using biochemistry studies, engineered mouse models, and pathological gene alternations. We conclude that inhibitors of F-box proteins could have promising therapeutic potentials in part through controlling of aberrant cell cycle progression for cancer therapies.
Keywords: Cell cycle, F-box protein, Ubiquitination, UPS, Cancer, Degradation
1. Introduction
Ubiquitin proteasome system (UPS) governs the process of cellular protein degradation, known as ubiquitination, and subsequently controls various cellular processes such as cell proliferation, cell cycle progression, transcription and apoptosis (Eldridge and O'Brien, 2010, Hershko and Ciechanover, 1998, Hoeller and Dikic, 2009, Komander and Rape, 2012, Varshavsky, 2012). It has been known that ubiquitinated proteins are degraded by the 26S proteasome complex after the ubiquitin molecules are conjugated to the targeted substrate proteins. These processes include a three-step enzymatic reaction, which is catalyzed by the ubiquitin-activating enzyme (E1), the ubiquitin-conjugating enzyme (E2), and the ubiquitin ligase (E3). Specifically, ubiquitin molecule is activated by the E1 using an ATP, and transfers to the E2. Then, ubiquitin ligase E3 binds to, and facilitates the transfer of the ubiquitin moiety to its substrates, leading to degradation by the 26S proteasome in an ATP-dependent manner (Nakayama and Nakayama, 2006, Nalepa et al., 2006, Pickart, 2001) (Figure 1). It is noteworthy that the E3 ubiquitin ligase is most important to specifically determine the substrates for ubiquitination and further degradation.
Figure 1.
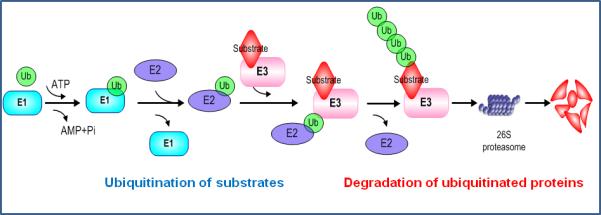
A schematic illustration of the E1-E2-E3 cascade-mediated ubiquitin transfer process to a given ubiquitin substrate.
Up to date, more than 600 E3 ubiquitin ligases have been discovered in the human genome (Li et al., 2008). According to their protein sequence homology, the major types of E3 ubiquitin ligases consist of the HECT (homologous to the E6-AP carboxyl terminus) type, the RING (really interesting new gene) finger type, and the RBR (ring between ring fingers) domain-containing ubiquitin ligases (Bedford et al., 2011, Deshaies and Joazeiro, 2009, Petroski and Deshaies, 2005). The Cullin-Ring Ligases (CRL-type of E3s) are the most well-studied RING type of E3 ubiquitin ligases (Hua and Vierstra, 2011, Sarikas et al., 2011), which contain CRL-1, CRL-2, CRL-3, CRL-4A, CRL-4B, CRL-5, CRL-7 and CRL-9 subfamilies (Duda et al., 2011, Metzger et al., 2012). CRL-1, also known as the SKP1-cullin 1-F-box protein (SCF) type of E3 ubiquitin ligase complex, has been most well characterized (Frescas and Pagano, 2008, Skaar et al., 2013). The SCF complex consists of cullin1 as the scaffold protein, the RING finger protein Rbx1 to recruit the E2 enzyme, and Skp1 (S phase kinase associated protein 1) as the adaptor protein to bridge F-box proteins (Peters, 2006, Zheng et al., 2002). It has been validated that the human genome encodes 69 F-box proteins, which contain several functional domains such as various carboxy-terminal domain for binding specific substrates and the F-box motif (Bai et al., 1996). Depending on the different binding domains, F-box proteins are classified into three major sub-families: the FBXW sub-family (contains WD40 substrate binding domains), the FBXL sub-family (contains leucine-rich repeats substrate binding domains), and the FBXO sub-family (contains other motifs such as kelch repeats or proline-rich motifs to bind substrates) (Frescas and Pagano, 2008, Welcker and Clurman, 2008).
The SCF complex has been reported to regulate multiply cell processes including cell cycle (Eldridge and O'Brien, 2010, Hershko and Ciechanover, 1998, Hoeller and Dikic, 2009, Komander and Rape, 2012, Varshavsky, 2012). Dysregulation of cell cycle progression is a key event in cancer development since it causes uncontrolled cell growth. It is clear that the cell cycle is mainly governed by cyclin-dependent kinases (Cdks) (Besson et al., 2008, Malumbres and Barbacid, 2009). Specifically, Cdks are activated by Cyclins and inhibited by Cdk inhibitors (CdkIs). Multiple mechanisms are involved in strictly controlling cell cycle progression such as regulatory factors (Cyclins and CdkIs), posttranslational modifications (phosphorylation, acetylation, or methylation), and degradation of cell cycle related proteins (Dai and Grant, 2003). A wealth of evidence has emerged that F-box proteins targets certain regulatory proteins as ubiquitin substrates that are critically involved in cell cycle regulation. Therefore, in this review, we describe the function of F-box proteins in regulation of cell cycle. Moreover, we elucidate how F-box proteins involve in tumorigenesis via targeting cell cycle regulatory proteins. We conclude that targeting F-box proteins could be a novel strategy for the control of cell cycle and cancer therapies.
2. Roles of FBXW sub-family in the regulation of cell cycle
The FBXW sub-family comprises 10 proteins including FBXW-1 (also known as β-TRCP1), FBXW-2, FBXW-4, FBXW-5, FBXW-7, FBXW-8, FBXW-9, FBXW-10, FBXW-1 1 (also known as β-TRCP2), and FBXW-12 (Wang et al., 2014b). The FBXW sub-family has been revealed to be critically involved in the regulation of cell cycle. In the following paragraphs, we will discuss how the FBXW sub-family governs the cell cycle progression through targeting cell cycle regulatory proteins for ubiquitination and destruction (Table 1).
Table 1.
A list of reported representative substrates of FBXW sub-family proteins in cell cycle regulation.
| Substrates | F-box | Functions | References |
|---|---|---|---|
| Emi1 | β-TRCP | Inhibitor of APC/C, Cell cycle | (Guardavaccaro, Kudo, 2003, Margottin-Goguet, Hsu, 2003) |
| Cdc25A | β-TRCP | Protein phosphatase, Cell cycle | (Busino, Donzelli, 2003, Jin, Shirogane, 2003) |
| Cdc25B | β-TRCP | Protein phosphatase, Cell cycle | (Kanemori, Uto, 2005, Uchida, Watanabe, 2011) |
| Wee1A | β-TRCP | Protein kinase, Cell cycle | (Watanabe, Arai, 2004) |
| Cyclin D1 | β-TRCP | Cyclin, Cell cycle | (Seki, Coppinger, 2008, Wei, Yang, 2008) |
| BTG | β-TRCP | Transcription factor, Cell cycle | (Sasajima, Nakagawa, 2012) |
| Plk4 | β-TRCP | Protein kinase, Centrosome duplication | (Cunha-Ferreira, Rodrigues-Martins, 2009, Guderian, Westendorf, 2010) |
| CEP68 | β-TRCP | Centrosome separation | (Pagan, Marzio, 2015) |
| Bora | β-TRCP | Aurora A kinase activator, Cell cycle | (Seki, Coppinger, 2008) |
| Securin | β-TRCP | Regulatory protein, Cell cycle | (Limon-Mortes, Mora-Santos, 2008) |
| REST | β-TRCP | Spindle assembly checkpoint | (Guardavaccaro, Frescas, 2008) |
| Claspin | β-TRCP | DNA replication checkpoint | (Peschiaroli, Dorrello, 2006) |
| hGCMa | FBXW2 | Transcription factor, Cell cycle | (Yang, Yu, 2005) (Zhuang, Li, 2014) |
| COP9 | FBXW4 | Cell growth, Cell cycle | (Lockwood, Chandel, 2013) |
| Eps8 | FBXW5 | Mitosis, Cell cycle | (Werner, Disanza, 2013) |
| DLC1 | FBXW5 | Tumor suppressor, Cell growth | (Jiang, Li, 2015, Kim, Jackson, 2013) |
| HsSAS-6 | FBXW5 | Centrosome duplication, Cell cycle | (Hu, Zacharek, 2008, Pagan and Pagano, 2011, Puklowski, Homsi, 2011) |
| Cyclin E | FBXW7 | Protein kinase, Cell cycle | (Perciavalle and Opferman, 2013, Siu, Rosner, 2012, Spruck, Won, 1999) |
| Aurora A/B | FBXW7 | Mitosis, Cell cycle | (Kwon, Kim, 2012, Slattery, Moore, 2008, Teng, Hsieh, 2012) |
| Cyclin D1 | FBXW8 | Cell growth, Cell cycle | (Okabe, Lee, 2006) |
| HPK1 | FBXW8 | Cell growth, Cell cycle | (Wang, Chen, 2014a) |
2.1 FBXW1 (β-TRCP1) and FBXW11 (β-TRCP2)
It has been well documented that β-TRCP recognizes the consensus sequence D-pS-G-X-X-pS (X represents any amino acid) degron and phosphorylation of both serine residues by specific kinases (Wertz et al., 2011). It has been known that β-TRCP includes two homologues, namely β-TRCP1 and β-TRCP2, which have similar structure with an F-box domain and seven WD-40 repeats. These two isoforms have noticeable sequence differences in their N-terminal regions (Yaron et al., 1998). In addition, the functions of β-TRCP1 and β-TRCP2 are non-distinguishable as least using in vitro biochemical assays (Nakayama et al., 2003). Some studies have demonstrated that β-TRCP plays a vital regulatory role by recognizing specific substrates in cell cycle process. For instance, certain cell cycle regulators including Emi1, Cdc25A, Cdc25B, Wee1A, Cyclin D1, BTG, Plk4, Bora, and Securin have been identified as the substrates of β-TRCP. Specifically, the dysregulation of Emi1 by β-TRCP led to mitotic catastrophe (Margottin-Goguet et al., 2003). Similarly, it has been previously shown that the dysregulation of Emi1 by β-TRCP caused delayed progression through mitosis (Guardavaccaro et al., 2003). Another independent study showed that cdc25A is required for the S-phase entry and its phosphorylation can be targeted by β-TRCP and thus slow down cell-cycle progression to response to DNA damage (Busino et al., 2003, Jin et al., 2003). Besides Cdc25A, Cdc25B can also be recognized by β-TRCP and subsequently degraded to mediate the stress-activated MAPK signaling pathway (Kanemori et al., 2005, Uchida et al., 2011). Similarly, β-TRCP targets the Wee1, a Cdc2 inhibitory kinase, to ensure normal mitosis (Watanabe et al., 2004). Moreover, Cyclin D1 has been recently found to be degraded by SCF/β-TRCP, leading to regulating the cell cycle and cell proliferation (Wei et al., 2008).
Additionally, both β-TRCP1 and β-TRCP2 can recognize and degrade BTG (B-cell translocation gene), thereby regulating the cell cycle and cell proliferation (Sasajima et al., 2012). The centriolar protein Plk4 (Polo-like kinase 4) plays a role in cell cycle via regulation of centriole biogenesis and maintaining constant centriole number. β-TRCP was found to induce Plk4 degradation after Plk4 autophosphorylates itself and bound to β-TRCP (Cunha-Ferreira et al., 2009, Guderian et al., 2010). One study also showed that Plk1 (Polo-like kinase 1) can phosphorylate Bora and promote its degradation by β-TRCP, resulting in subsequent regulation of mitotic progression (Seki et al., 2008). Similarly, β-TRCP was reported to govern centrosome separation in part by targeting CEP68 for proteasomal degradation (Pagan et al., 2015). Moreover, Securin is a complicated protein with its bifunctional properties in cell cycle. In the expose of UV irradiation, Securin is found to be degraded by β-TRCP and leads to a sharp decrease both in nucleus and cytoplasm, resulting in cell cycle arrest (Limon-Mortes et al., 2008). Moreover, it has been reported that REST (repressor-element-1-silencing transcription factor) is degraded by β-TRCP during the G2 phase of the cell cycle, leading to derepression of Mad2, which is a key component of the spindle assembly checkpoint (Guardavaccaro et al., 2008). Therefore, the overexpression of REST found in certain human cancers could be partly due to dysregulation of β-TRCP, which contributed to cellular transformation through enhancing genomic instability (Guardavaccaro, Frescas, 2008). In keeping with this notion, another study showed that β-TRCP-dependent degradation of Claspin is important for the efficient and timely termination of the DNA replication checkpoint (Peschiaroli et al., 2006). Specifically, inhibition of Claspin proteolysis by β-TRCP led to subsequent activation of Chk1 and attenuating the recovery from the DNA replication stress response, thereby delaying entry into mitosis (Peschiaroli, Dorrello, 2006). Recently, we reported that β-TRCP earmarks Set8 for ubiquitination and degradation in a casein kinase I-dependent manner, leading to control of cell cycle progression, and governing the onset of DNA damage-induced checkpoints (Wang et al., 2015). However, further in-depth studies are required to fully understand the critical role for both β-TRCP1 and β-TRCP2 and their potential redundancy in governing cell cycle progression.
2.2 FBXW2 and FBXW4
FBXW2 has been reported to regulate cell cycle in part via targeting hGCMa (human glial cell missing homolog 1) protein for degradation (Yang et al., 2005). It has been known that hGCMa, a zinc-containing transcription factor, regulates the syncytin-1 gene, which takes part in cell cycle progression (Zhuang et al., 2014). FBXW2 was found to promote the degradation of hGCMa by ubiquitination (Yang, Yu, 2005). Mechanistically, FBXW2 interacts with hGCMa in a phosphorylation-dependent manner and promotes hGCMa ubiquitination. Moreover, depletion of FBXW2 using its siRNA led to lower level ubiquitination of hGCMa and increased hGCMa protein stability (Yang, Yu, 2005). These findings suggest that hGCMa is a substrate of FBXW2. Due to the fact that hGCMa plays a key role in regulation of cell cycle, FBXW2 could be involved in cell cycle regulation through targeting hGCMa. However, further investigation is required to discover the direct evidence of FBXW2-regulated cell cycle.
FBXW4, encoded by gene dactylin, plays a critical role in SHFM (split hand/foot malformation) diseases. The level of FBXW4 is decreased in the mouse dactylaplasia mutant, suggesting that FBXW4 could have an important function in SHFM (Basel et al., 2003, Ianakiev et al., 1999, Sidow et al., 1999). Recently, it has been reported that FBXW4 interacts with COP9 signalosome and mediates the specific protein degradation and maintains the normal cell growth. Furthermore, mutation, lost and low expression of FBXW4 were found in multiple cancer cell lines, indicating that FBXW4 may be a tumor suppressor by controlling cell cycle progression (Lockwood et al., 2013). Without a doubt, in-depth exploration is necessary to determine the mechanism how FBXW4 regulates cell cycle in human cancer cells.
2.3 FBXW5
Several studies have highlighted the important role of FBXW5 in regulation of cell cycle. For example, Eps8, a regulator in cell proliferation, is stable in G1 and S phase, but its expression drops in G2 by ubiquitination-mediated degradation (Werner et al., 2013). To this end, FBXW5 targets and degrades Eps8 to maintain the proper mitotic progression. Moreover, in NSCLC (non-small cell lung cancer), tumor suppressor DLC1 is degraded by the FBXW5-CRL4A E3 ubiquitin ligase, leading to promoting cell growth (Jiang et al., 2015, Kim et al., 2013). Consistently, inhibition of FBXW5 causes cell proliferation (Kim, Jackson, 2013). Since HsSAS-6 is a centriolar protein and plays important role in centrosome duplication, FBXW5 targets and ubiquitylates HsSAS-6, resulting in controlling centrosome number (Pagan and Pagano, 2011, Puklowski et al., 2011). Consistently, depletion or mutation of FBXW5 causes centrosome overduplication and formation of multipolar spindles (Puklowski, Homsi, 2011). Another independent study identified that FBXW5 regulates TSC2 (tuberous sclerosis 2) protein stability, indicating that FBXW5 might regulate the cell cycle and growth partly via targeting TSC2 (Hu et al., 2008). Furthermore, it was found that FBXW5 itself is controlled by PLK4 and APC/C (anaphase-promoting) complex for degradation during mitosis and G1 (Puklowski, Homsi, 2011). Taken together, FBXW5 plays a critical role in cell cycle regulation, but in-depth studies are required in the future to fully dissect the downstream targets that may mediate the critical function of FBXW5 in cell cycle progression.
2.4 FBXW7
Some FBXW7 downstream substrates are vital regulatory effectors involved in cell cycle. For instance, Cyclin E is a critical regulator of cell cycle procession (Siu et al., 2012). It binds to and activates Cdk2 and thus promotes the entry from the G1 phase to the S phase of the cell cycle, while the mount of the Cyclin E is tightly controlled by ubiquitin- mediated proteolysis (Perciavalle and Opferman, 2013, Siu, Rosner, 2012, Spruck et al., 1999). FBW7 interacts specifically with phosphorylated Cyclin E, leading to its ubiquitination and subsequent degradation by the 26S proteasome (Minella et al., 2007). Deregulation of Cyclin E has been frequently found in cancer, and overexpression of Cyclin E leads to genomic instability and tumorigenesis (Spruck, Won, 1999). Importantly, deletion of FBW7 caused accumulation and stabilization of Cyclin E in various types of human malignances (Koepp et al., 2001). Aurora kinases play important roles in mitosis (Carmena and Earnshaw, 2003, Ducat and Zheng, 2004, Fu et al., 2007, Katayama et al., 2003, Lens et al., 2010, Lindqvist et al., 2009, Ruchaud et al., 2007). Notably, there are three Aurora kinase family members, namely Aurora A, Aurora B, and Aurora C, which are highly expressed during mitosis. Aurora A is initially located at the centrosomes during prophase and later moved to the spindle poles during prometaphase and metaphase. Aurora B and Aurora C are chromosomal passenger proteins. Aurora B is located on the chromosome arms during the prophase and at the centromeres during prometaphase and metaphase (Carmena and Earnshaw, 2003, Sampath et al., 2004). Both Aurora A and Aurora B play pivotal roles in mitosis by ensuring correct chromosome segregation and normal progression through mitosis (Kwon et al., 2012, Teng et al., 2012). Interestingly, the function of Aurora C is similar as Aurora B (Slattery et al., 2008). Hence, the abundance of Aurora kinases is strictly controlled during each stage of mitosis, primarily through ubiquitination-mediated degradation (Teng, Hsieh, 2012). It has been found that FBXW7 interacts with and negatively regulates Aurora A and Aurora B via the ubiquitination pathway (Kwon, Kim, 2012, Teng, Hsieh, 2012). Notably, loss of FBXW7 in various cancers leads to an abnormal elevation of Aurora A and B and results in deregulated mitosis, which misleads cell cycle and accelerates cancer cell growth (Kwon, Kim, 2012, Rajagopalan et al., 2004). Collectively, FBW7 is considered as a key regulator in cell cycle, but further study is warranted to understand the physiological downstream substrate through which Fbw7 suppresses cell cycle progress to inhibit tumorigenesis.
2.5 Other FBXW proteins
FBXW8 (also known as FBW6, FBW8, FBX29, FBXW6, or FBXO29) plays a critical role in cancer cell proliferation by increasing the degradation of Cyclin D1 (Okabe et al., 2006). Interestingly, FBXW8 did not regulate the proteolysis of Cyclin D in normal cell cycle (Kanie et al., 2012). Furthermore, depletion of FBXW8 results in pre- and postnatal growth retardation in mice, suggesting that FBXW8 is important in regulating cell growth (Tsutsumi et al., 2008). Lin et al. reported that FBXW8 regulated the G2/M phase transition to control the proliferation of human choriocarcinoma cells, which is associated with several cell cycle regulators such as CDK1, CDK2, Cyclin A, Cyclin B1 and p27 expression (Lin et al., 2011). In pancreatic cancer cells, FBXW8 increases the degradation of HPK1 (hematopoietic progenitor kinase 1) and promotes the cell growth (Wang et al., 2014a). Moreover, miR-218 targets the FBXW8 and inhibits the proliferation of human choriocarcinoma cells (Shi et al., 2014).
It has been recently reported that the protein level and function of FBXW10 depend on the protein O-GlcNAcylation as the levels of FBXW10 mRNA and protein were reduced in GlcN-treated cell (Feng et al., 2013). Furthermore, O-GlcNA protein modification exerts function in many cellular processes such as cell cycle, insulin signaling, calcium handling as well as the cellular stress response (Zachara and Hart, 2006). This indicates that FBXW10 could be associated with regulation of cell cycle. On the other hand, FBXW12 is reported to be deleted or methylated in epithelial ovarian cancer (Chesnaye Ede et al., 2015). Interestingly, knockdown of FBXW12 increases human epithelial cell growth and cell cycle progression (Franz et al., 2015), suggesting that FBXW12 is an epithelial growth suppressor probably by inhibiting cell cycle progression.
3. Roles of FBXL sub-family in cell cycle
The FBXL sub-family composes 22 members including FBXL1 (also known as Skp2) and FBXL2 to FBXL21. All of FBXL proteins contain an F-box motif and a C-terminal Leu-rich repeat (LRR) domain. The FBXL proteins have been characterized as cell cycle regulators in the control of cell cycle. In this section, we will describe the roles of FBXL proteins in governing cell cycle (Table 2).
Table 2.
A list of reported representative substrates of FBXL sub-family proteins in cell cycle regulation.
| Substrates | F-box | Functions | References |
|---|---|---|---|
| P27 | FBXL1 | Cdk inhibitor, Cell cycle | (Nakayama, Hatakeyama, 2001, Suzuki, Fukasawa, 2012) |
| P21 | FBXL1 | Cdk inhibitor, Cell cycle | (Bornstein, Bloom, 2003, Nakayama, Nagahama, 2004, Yu, Gervais, 1998) |
| Cyclin D1 | FBXL1 | Cyclin, Cell cycle | (Nakayama, Nagahama, 2004, Yu, Gervais, 1998) |
| P57 | FBXL1 | Cdk inhibitor, Cell cycle | (Pateras, Apostolopoulou, 2006, Yang, Nan, 2015) |
| P130 | FBXL1 | Rb protein family, Cell cycle | (Bhattacharya, Garriga, 2003, Tedesco, Lukas, 2002) |
| Cyclin A | FBXL1 | Cyclin, Cell cycle | (Ji, Goldin, 2006, Michel and Xiong, 1998, Yam, Ng, 1999) |
| Cyclin E | FBXL1 | Cyclin, Cell cycle | (Li, Li, 2004, Ungermannova, Gao, 2005) |
| Cyclin G2 | FBXL1 | Cyclin, Cell cycle | (Xu, Bernaudo, 2008) |
| Cdh1 | FBXL1 | DNA replication factor, Cell cycle | (Kurland and Tansey, 2004) |
| Cyclin D2 | FBXL2 | Cyclin, Cell cycle | (Chen, Glasser, 2012b) |
| Cyclin D3 | FBXL2 | Cyclin, Cell cycle | (Chen, Glasser, 2011a, 2012a) |
| P85beta | FBXL2 | Cell cycle | (Kuchay, Duan, 2013) |
| CRY | FBXL3 FBXL21 |
Circadian clock, Cell cycle | (Busino, Bassermann, 2007, Godinho, Maywood, 2007, Siepka, Yoo, 2007) (Hirano, Yumimoto, 2013, Yoo, Mohawk, 2013) |
| JMJD2A | FBXL4 | Cell cycle | (Das, Chai, 2014) |
| CITED2 | FBXL5 | Cell cycle | (Machado-Oliveira, Guerreiro, 2015) |
| Aurora A | FBXL7 | Mitosis, Cell cycle | (Coon, Glasser, 2012) |
| EZH2 | FBXL10 FBXL11 |
Cell cycle | (Kawakami, Tokunaga, 2015, Tzatsos, Paskaleva, 2011) |
| CaMK1 | FBXL12 | Cell cycle | (Mallampalli, Kaercher, 2013) |
| Smurf1 | FBXL15 | Cell cycle | (Fei, He, 2014) |
3.1 FBXL1 (Skp2)
FBXL1, also called Skp2 (S-phase kinase-associated protein 2), is one of the well-characteristic F-box proteins (Chan et al., 2010). Some studies have demonstrated that Skp2 is a key cell cycle regulator through targeting multiple cell cycle related proteins such as p27, p21, p57, p130, Cyclin A, Cyclin E, Cyclin D1 and Cyclin G2. It has been reported that p27 is a primary target of Skp2 (Nakayama et al., 2001, Suzuki et al., 2012), and it is inversely related to Skp2 expression during the differentiation of human embryonic stem cells and in many human tumors (Dombrowski et al., 2013, Egozi et al., 2007, Kitagawa et al., 2009). The higher expression of Skp2 induces the degradation of p27 and promotes the entry of S phase from G phase, leading to induction of the immortalized cell proliferation (Carrano and Pagano, 2001). In mammalian cells, silencing Skp2 induced the accumulation of Cyclin D and p21 (Bornstein et al., 2003, Nakayama et al., 2004, Yu et al., 1998). In some cancer cells, p57 (also name as KIP2) plays important roles in controlling cell cycle and is reported to be negatively correlated with Skp2 (Pateras et al., 2006, Yang et al., 2015). Moreover, p130, a member of the retinoblastoma family of pocket proteins, decreases sharply in the mid-G1 phase. This is due to its hyperphosphorylation on Serine 672 and degradation by Skp2, and subsequently promoting the cell into S phase (Bhattacharya et al., 2003, Tedesco et al., 2002).
Notably, Cyclin A can promote G1 entering into the S phase. Previous studies have shown that the kinase activity of Cdk2/Cyclin A was blocked by the p27, while Skp2 binds with Cyclin A and conceals the site targeted by p27, suggesting that Skp2 has a crucial role in cell cycle via regulation of Cyclin A (Ji et al., 2006, Michel and Xiong, 1998, Yam et al., 1999). In addition, Skp2 combines with the Cyclin E and mediates its degradation (Yeh et al., 2001). Interestingly, Cyclin E also involves in the degradation of p27 (Li et al., 2004, Ungermannova et al., 2005). Overexpression of Cyclin G2 inhibits cell proliferation and the normal amount of Cyclin G2 is controlled by Skp2 through promoting its degradation (Xu et al., 2008). Our previous study also demonstrated Skp2 expression reaches its summit during the entry to S phase from G1 when the protein levels of Cdh1 are low (Wei et al., 2004). Notably, Cdh1 can bind the N-terminal D-box motif of Skp2 and the deficiency or mutation of the motif leads to resistance of Skp2 to ubiquitination and degradation mediated by Cdh1 (Kurland and Tansey, 2004). Altogether, Skp2 is an essential regulator to control cell cycle progression and its own expression is also subjected to cell cycle dependent regulation to achieve timely entry into the S phase at optimized growth conditions.
3.2 FBXL2 and FBXL3
FBXL2 has a CAAX motif that targets it to cell membranes. Interestingly, one of identified substrates of FBXL2, p85-beta, is localized to cell membrane. FBXL2-mediated degradation of p85-beta is essential for the efficient response of quiescent cells to mitogens and their reentry into the cell cycle (Kuchay et al., 2013). Additionally, FBXL2 targets and degrades Cyclin D2 in leukemic and B-lymphoblastoid cell lines, leading to G0 phase arrest and apoptosis (Chen et al., 2012b). Depletion of endogenous FBXL2 stabilizes Cyclin D2 levels, while overexpression of FBXL2 promotes Cyclin D2 degradation (Chen, Glasser, 2012b). Additionally, Cyclin D3 was identified as a substrate of FBXL2 as well (Chen et al., 2011a, 2012a). Moreover, degradation of Cyclin D3 inhibits lung cancer proliferation and cell cycle arrest. This could be due to that ectopically expressed FBXL2 elicits G2/M-phase arrest and thus suppressed tumorigenesis (Chen, Glasser, 2011a, 2012a). Overexpression of FBXL2 also hinders tumor formation in athymic nude mice, implicating that FBXL2 could serve as a tumor suppressor in part via governing cell cycle in human cancer cells (Chen, Glasser, 2012a).
FBXL3 has been reported to regulate circadian clock by directly degrading CRY (cryptochrome) protein (Busino et al., 2007, Godinho et al., 2007, Siepka et al., 2007). In normal conditions, mammalian CRY interacts with Per (PERIOD) and together depresses their own genes expression, while FBXL3 negatively controls this loop by inducing the ubiquitination and degradation of CRY (Anand et al., 2013, Busino, Bassermann, 2007, Xing et al., 2013). Furthermore, FBXL3 interacts with the cofactor pocket, which is formed by both FAD (flavin adenine dinucleotide) and CRY, to insure the timely degradation process (Xing, Busino, 2013). Deletion of FBXL3 results in the stabilization of CRY and thus inhibits the expression of CRY and Per proteins, and subsequently interferes the normal circadian clock (Siepka, Yoo, 2007). Furthermore, CRY1 is critical in formation of functional E3 complex (Yumimoto et al., 2013). However, it is required to investigate whether FBXL3 could regulate cell cycle via targeting CRY and Per proteins, which will reveal an intrinsic connection between circadian and cell cycle machineries.
3.3 FBXL4 and FBXL5
FBXL4 targets and degrades JMJD2A (Jumonji domain-containing 2A), which plays an essential role in cell cycle (Van Rechem et al., 2011). The overexpression of JMJD2A promotes the S phase and cancer cell proliferation, while depletion of JMJD2A suppresses cell growth in lung and bladder cancer cells (Kogure et al., 2013, Van Rechem, Black, 2011). JMJD2A attenuation affects cell cycle and tumourigenic inflammatory gene regulation in lipopolysaccharide syimulated neurodermal stem cells (Das et al., 2014), suggesting that FBXL4 could be involved in cell cycle through targeting JMJD2A.
Notably, FBXL5 is reported as a substrate of miR-290-295 (Lichner et al., 2011). Specially, miR-290-295 expresses specifically in mouse ESC (embryonic stem cells) and ECC (embryonic carcinoma cells). Overexpression of miR-290-295 promotes the entry of S phase from G1 phase, indicating that this cell cycle process may be controlled by the substrate of miR-290-295, FBXL5 (Lichner, Pall, 2011). Another independent study revealed that FBXL5 targets CITED2 (with Glu/Asp-Rich Carboxy-Terminal Domain, 2) for degradation to regulate the HIF-1α (hypoxia-inducible factor-1α) (Machado-Oliveira et al., 2015). Moreover, Snail 1 is validated as a substrate of FBXL5 and the degradation of Snail 1 leads to inhibition of metastasis in gastric cancer cells (Vinas-Castells et al., 2014, Wu et al., 2015). Additionally, the ubiquitination and degradation of cortactin by FBXL5 suppresses the migration and invasion of gastric cancer cells (Cen et al., 2014). One group reported that FBXL5 regulates the DNA damage response by targeting the hSSB1 (Human single-strand DNA binding proteins 1) (Chen et al., 2014). It is required to define mechanistically whether FBXL5 directly targets cell cycle regulatory proteins in the near future.
3.4 FBXL7 and FBXL10
FBXL7 exerts a critical role in mitotic process through targeting Aurora A and thus disturbing spindle formation and cell proliferation (Coon et al., 2012). Consistently, the deregulation of FBXL7 results in G2/M arrest, and subsequently leads to mitotic arrest (Coon, Glasser, 2012). FBXL10, also named as JHDM1B or Kdm2b, which is an H3K36 demethylase, regulates cell proliferation and senescence by regulating p15 (He et al., 2008). Depletion of FBXL10 induces the expression of let-7 and miR-101 and inhibits their target EZH2 (zester homolog 2). On the other hands, overexpression of FBXL10 inhibits the let-7 and miR-101 and subsequently promotes the level of EZH2, leading to increased immortal cells, suggesting that the FBXL10-let-7-EZH2 pathway plays an important role in cell cycle and cell progression (Tzatsos et al., 2011). In NPC (nasopharyngeal carcinoma), FBXL10 promotes cell progression in part by controlling the PI3K/mTOR pathway (Ren et al., 2015). Moreover, loss of FBXL10 induces the genomic DNA hyper-methylation (Boulard et al., 2015). As a nucleolar protein, FBXL10 represses transcription of ribosomal RNA genes and involves in cancer development (Frescas et al., 2007). Furthermore, FBXL10 captures PRC1 (polycomb repressive complex 1) to CpG islands and regulates H2A ubiquitination (Wu et al., 2013). In pancreatic cancer, FBXL10 promotes tumorigenesis by Polycomb-dependent and independent transcriptional programs (Tzatsos et al., 2013). Taken together, FBXL10 plays a central role in cell progression and involves in cancer development, but further studies are warranted to fully understand its physiological contribution to this process.
3.5 FBXL11 and FBXL12
FBXL11 as a histone demethylase, also known as Kdm2a, plays a role in cell proliferation, apoptosis and senescence (Ishimura et al., 2012, Kawakami et al., 2015). FBXL11 KO mice exhibit embryonic lethality with growth defects. Cells without FBXL11 decrease proliferation and promote apoptosis. Moreover, knockout of FBXL11 depresses the level of EZH2, the ubiquitination of H2A and upregulates of p21 (Kawakami, Tokunaga, 2015). Thus, FBXL11 is important in cell cycle regulation. Additionally, overexpression of FBXL11 inhibits activity of NF-κB, while knockdown of FBXL11 promotes NF-κB binding DNA and activates its gene expression (Lu et al., 2009).
The mechanism of regulating NF-κB may be due to the reversible lysine methylation of p65 (Lu et al., 2010). In NSCLC, FBXL11 promotes lung tumorigenesis by epigenetically enhancing ERK1/2 signaling (Wagner et al., 2013). Additionally, transcriptional repression of histone deacetylase 3 by FBXL11 is coupled to tumorigenicity of lung cancer cells (Dhar et al., 2014). On the other hand, FBXL12 targets and degrades CaMK1 (calmodulin kinase 1) to induce G1 arrest (Mallampalli et al., 2013). Interestingly, one group independently showed that FBXL12 has a novel function in regulating the DNA damage by inducing Ku80 ubiquitination (Postow and Funabiki, 2013). Taken together, FBXL12 may be considered as a cell cycle regulator but its role in tumorigenesis warrants further investigation.
3.6 other FBXL proteins
FBXL15 interacts with the Smurf1 (Smad ubiquitination regulatory factor 1) E3 ubiquitin ligase and directly degrades Smurfe (Cui et al., 2011, Fei et al., 2014). Notably, the C2 domain of Smurf1 performs a critical role in targeting Axin for ubiquitination. The interaction between Smurf1 and Axin locates Smurf1 to membrane and decreases the process of G2/M phase of cell cycle, and this state promotes the response to Wnt stimulation (Fei, He, 2014). The degradation of Smurf1 mediated by FBXL15 also involves the BMP (bone morphogenetic protein) signaling pathway during embryonic development and adult bone formation (Cui, He, 2011).
Notably, FBXL18 could bind Lys 109 site of FBXL7 and then ubiquitinate and degrade FBXL7, indicating that FBXL18 regulates cell cycle possibly by controlling the level of FBXL7 (Liu et al., 2015). On the other hand, FBXL20, known as SCRAPPER, has been reported to involve in tumorigenesis (Yao et al., 2007). Depletion of FBXL20 causes inhibition of cell proliferation, G1 cell cycle arrest and induction of apoptosis in colorectal adenocarcinoma cells (Zhu et al., 2012). Furthermore, overexpression of FBXL20 increases cell viability and invasion capacity in colon cancer cells, accompanied by the upregulation of β-catenin and c-Myc, and downregulation of E-cadherin (Zhu et al., 2014). Therefore, FBXL20 plays a critical role in colon cancer development and cell cycle. Notably, FBXL21 interacts with CRY and to promote its degradation (Hirano et al., 2013, Yoo et al., 2013). Since FBXL3 could lengthen and FBXL21 shorten circadian period (Yoo, Mohawk, 2013), FBXL21 could antagonize the function of FBXL3 and their combined actions ensure the stable oscillation of the circadian clock (Hirano, Yumimoto, 2013), but additional studies are required to understand the physiological role and their functional interplay between FBXL3 and FBXL21 in both cell cycle regulation and tumorigenesis.
4. Roles of FBXO sub-family in cell cycle
Except for FBXW sub-family and the FBXL sub-family, the last 36 F-box proteins are defined as F-box only (FBXO) proteins. FBXO sub-family proteins contain the F-box motif in its N-terminus and multiple types of functional domains in its C-terminus. Different from the FBXW with the WD40 motif and FBXL proteins with the LRR motif, the FBXO sub-family contains 21 functional homology domains. The function of FBXO subfamily proteins is not fully characterized and we will focus on the role of FBXO subfamily in cell cycle in the following paragraphs (Table 3).
Table 3.
A list of reported representative substrates of FBXO sub-family prorteins in cell cycle.
| Substrates | F-box | Functions | References |
|---|---|---|---|
| Cyclin B | FBXO1 | Cyclin, Cell cycle | (Fung, Siu, 2002, Kong, Barnes, 2000) |
| CP110 | FBXO1 | Centrosome duplication, Cell cycle | (D'Angiolella, Donato, 2010) |
| RRM2 | FBXO1 | DNA repair, Cell cycle | (D'Angiolella, Donato, 2012) |
| NUSAP1 | FBXO1 | Microtubule, Cell cycle | (Emanuele, Elia, 2011) |
| Smurf1 | FBXO3 | Cell cycle | (Li, Xie, 2015) |
| Cyclin D1 | FBXO4 | Cyclin, Cell cycle | (Barbash, Zamfirova, 2008, Lin, Barbash, 2006, Santra, Wajapeyee, 2009) |
| P53 | FBXO5, FBXO11 | Tumor suppressor, Cell cycle | (Lehman, Verschuren, 2006) |
| Chk1 | FBXO6 | Replication checkpoint, Cell cycle | (Zhang, Brognard, 2009) |
| Cyclin D/Cdk6/p27 | FBXO7 | Cyclin, Cell cycle | (Laman, Funes, 2005) |
| HURP | FBXO7 | Oncogene, Cell cycle | (Hsu, Lee, 2004) |
| Cdt2 | FBXO11 | Cell cycle | (Abbas, Keaton, 2013a) |
| Bcl6 | FBXO11 | Cell cycle | (Duan, Cermak, 2012) |
| EID1 | FBXO21 | Cell cycle | (Zhang, Li, 2015) |
| KDM4A | FBXO22 | Cell cycle | (Tan, Lim, 2011) |
| Cdt1 | FBXO11, FBXO31 | Cell cycle | (Johansson, Jeffery, 2014) |
| MDM2 | FBXO31 | Cell cycle | (Malonia, Dutta, 2015) |
| BRCA1 | FBXO44 | DNA repair, Cell cycle | (Lu, Li, 2012) |
| Par-4 | FBXO45 | Apoptosis, Cell cycle | (Wang and Wei, 2014) |
| P73 | FBXO45 | Tumor suppressor, Cell cycle | (Peschiaroli, Scialpi, 2009) |
4.1 FBXO1
FBXO1 (also known as FBX1 or Cyclin F), which localizes to both the centrosome and nucleus, is believed as a novel mammalian cyclin because its amino acid sequences are much similar to Cyclin A. The amount of FBXO1 accumulates in the S phase and summits in G2 and finally disappears in mitosis (Bai et al., 1994, Fung et al., 2002). FBXO1 has a critical function in cell cycle while the mechanism is still unclear. It has been believed that FBXO1 binds Cyclin B and transports it into the nucleus, leading to governing Cyclin B/Cdk1 localization and functions during mitosis (Fung, Siu, 2002, Kong et al., 2000). Moreover, FBXO1 targets CP110 protein for degradation, which is essential in centrosome duplication, subsequently regulates the fidelity of mitosis and genome integrity (D'Angiolella et al., 2010). RRM2 is identified as a degraded substrate of FBXO1. The decrease of RRM2 controls the amount of dNTP pools and genome stability, and thus provides the DNA repair from genotoxic stress (D'Angiolella et al., 2012). Furthermore, FBXO1 targets NUSAP1 during the S and G2 phases in cell cycle, leading to sensitizing cells to microtubule-based chemotherapeutics (Emanuele et al., 2011). Notably, deletion of FBXO1 results in cell cycle doubling time and delays cell cycle reentry from quiescence (Tetzlaff et al., 2004). In support of this, MEFs without FBXO1 displays cell cycle defects, suggesting that FBXO1 is critical in cell cycle progression (Tetzlaff, Bai, 2004).
4.2 FBXO3 and FBXO4
FBXO3 targets and degrades Smurf1 to regulate cell cycle (Li et al., 2015). Unlike FBXL15, FBXO3 targets all the Nedd4 family and controls their stability (Li, Xie, 2015). On the other hand, FBXO4 (also known as FBX4) binds chaperone αβ-crystallin to form an active SCF E3 ubiquitin ligase. The SCF FBXO4-aβ-crystallin complex mediates the ubiquitination of Cyclin D1 in cytoplasm. Mutation or deletion of FBXO4 attenuates degradation of Cyclin D1 and leads to Cyclin D1 accumulation and promotion of cell cycle progression (Barbash et al., 2008, Lin et al., 2006). Moreover, control of FBXO4 activity attenuates the degradation of Cyclin D1 and oncogenic transformation. Mutation of FBXO4 inhibits dimerization of SCF (FBXO4) ligase and leads to tumorigenesis (Barbash, Zamfirova, 2008). However, there are no changes in genetic analysis or protein level of Cyclin D1 in Fbxo4−/− mice, suggesting that the regulatory of FBXO4 may be compensated by others proteins (Kanie, Onoyama, 2012).
4.3 FBXO5 and FBXO6
FBXO5, also named as EMI1 and FBX5, is an endogenous inhibitor of APC/C. FBXO5 could play an oncogenic role in human cancers. For instance, the high expression of FBXO5 contributes to increased proliferation, tetraploidy and instability of p53-deficient cells, indicating that loss of p53 may contribute to tumorigenesis together with FBXO5 (Lehman et al., 2006). Additionally, FBXO5 has been reported to promote the entry of S phase and mitosis by controlling the Cyclin A, Cyclin B or securin (Hsu et al., 2002). Interestingly, the stability of FBXO5 is promoted by BCR-ABL fusion oncoprotein, and inhibits SKP2 degradation, subsequently increases cell proliferation in chronic myeloid leukaemia cells (Chen et al., 2011b). Notably, FBXO6 binds the carboxyl terminus of Chk1, which plays a key role in replication checkpoint with ATR, and then degrades Chk1 (Zhang et al., 2009). The protein levels of FBXO6 and Chk1 are negatively correlated in both cultured cancer cells and human breast tumor tissues. Conclusively, FBXO6-dependent Chk1 degradation leads to S phase checkpoint arrest and cell cycle defection (Zhang, Brognard, 2009), but their physiological role in cell cycle regulation warrants further in-depth studies.
4.4 FBXO7
FBXO7 have a C-terminal specific proline-rich region (PRR) that binds various substrates (Chang et al., 2006, Hsu et al., 2004). Multiple reports showed FBXO7 function in a tissue-specific manner (Hsu, Lee, 2004, Laman et al., 2005, Lomonosov et al., 2011, Meziane el et al., 2011). FBXO7 was defined as a putative proto-oncogene and directly bound to the Cyclin D/Cdk6/p27 complex in immortalized fibroblasts. The binding activates Cdk6 and promotes the cell cycle progression and tumorigenesis (Laman, Funes, 2005). Conversely, reduction of FBXO7 promotes cell progression, decreases cell size and shortens G1 phase due to enhanced Cyclins in S phase and CDK2 activity (Meziane el, Randle, 2011). Notably, FBXO7 targets HURP (hepatoma upregulated protein), a cell cycle regulated oncogene, and controls cell growth in human liver cancer (Hsu, Lee, 2004). Moreover, one independent study revealed that depletion of FBXO7 promotes cell proliferation by shortening G1 phase (Meziane el, Randle, 2011). Altogether, FBXO7 may be a tumor suppressor through regulation of cell cycle in human cancer, but its physiological contribution to this process awaits further investigation.
4.5 FBXO11, FBXO18, FBXO21 and FBXO22
The CRL4 (Cdt2) E3 ubiquitin ligase controls the cell cycle progression by regulating Cdt1, p21 and Set8 during S phase (Abbas et al., 2013a, Abbas et al., 2013b, Rossi et al., 2013). Importantly, Cdt2 is polyubiquitylated and degraded by FBXO11, and decrease of Cdt2 stabilizes cell cycle regulators including Cdt1, p21 and Set8. The stability of Set8 regulates cell response to TGF-β exiting from the cell cycle and cellular migration (Abbas, Keaton, 2013a). This cross-regulation between specific Cullin 4 and Cullin 1 E3 ubiquitin ligase may play a critical role in cell cycle regulation (Abbas, Mueller, 2013b). Notably, FBXO11 targets BCL6, a repressor of the cell cycle regulator p53, for degradation (Duan et al., 2012). FBXL11 is inactivated in diffuse large B-cell lymphomas, indicating that FBXO11 is a haplo-insufficient tumor suppressor gene (Duan, Cermak, 2012). Furthermore, FBXO11 promotes the neddylation of p53 both in vivo and in vitro and thus inhibits its function of transcription (Abida et al., 2007).
In addition, upon DNA replication stress, FBXO18 (FBHI) promotes double-strand breakage, leading to activation of the DNA-PK and ATM signaling cascades and apoptosis (Jeong et al., 2013). On the other hand, FBXO21 targets EID1 (EP300-interacting inhibitor of differentiation 1) for degradation in G0 phase cells. The peptidic degron of EID1 is a binding site of FBXO21 and the polyubiquitylation is required in both cycling and quiescent cells (Zhang et al., 2015). FBXO21 may regulate cell cycle in part by mediating the protein level of EID1. Moreover, KDM4A is regulated by FBXO22 since overexpression or knockdown of FBXO22 decreases or increases the protein level of KDM4A, which oscillates the cell cycle progression (Tan et al., 2011).
4.6 FBXO31
FBXO31 is a senescence-related gene that located in the chromosome 16q24.3 (Kumar et al., 2005). FBXO31 targets and degrades Cyclin D1 that plays a critical role in G1/S entry. Overexpression of FBXO31 leads to low level of Cyclin D1 and subsequently causes G1 arrest in cell cycle (Santra et al., 2009). This happens specifically after DNA damage, but FBXO31 may not degrade Cyclin D1 in normal cell cycle progression (Kanie, Onoyama, 2012). Moreover, FBXO31 is identified as a tumor suppressor in breast, ovarian, hepatocellular and prostate cancers (Johansson et al., 2014, Kumar, Neilsen, 2005). Consistently, FBXO31 targets Cdt1 for degradation in G2 phase of cell cycle to prevent re-replication and tumorigenesis (Johansson, Jeffery, 2014). Notably, MDM2, a negative regulator of p53, has been identified as a substrate of FBXO31. In this context, FBXO31 promotes cell growth arrest by degrades MDM2 in genotoxic stress (Malonia et al., 2015). In gastric cancer cells, FBXO31 is negatively regulated by miR-17 and miR-20a (Zhang et al., 2014). The depression of FBXO31 promotes the expression of Cyclin D1 and decreases G1 phase (Zhang, Kong, 2014). Taken together, FBXO31 may act as a tumor suppressor by regulating cell cycle progression, but its downstream effect pathway as well as its own regulation in cell cycle progress and tumorigenesis awaits further investigation.
4.7 FBXO44 and FBXO45
BRCA1 is associated in sporadic cancer cases and plays a role in cell cycle checkpoint and DNA repair. The N terminus of BRCA1 has a binding site of FBXO44, and FBXO44 targets BRCA1 for degradation and regulates cell cycle in sporadic breast cancer (Lu et al., 2012). On the other hand, FBXO45 has been found to play a role in neural development and tumorigenesis (Peschiaroli et al., 2009, Saiga et al., 2009, Wang and Wei, 2014). To this end, previous studies have shown that FBXO45 is an estrogen-induced gene that contains estrogen receptor-binding sequences (Han et al., 2016, Yoshida, 2005). Further study suggests that Era could be a substrate of FBXO45 (Han, Begum, 2016). Par-4 (prostate apoptosis response protein 4), a novel specific substrate of FBXO45, is a tumor suppressor by inducing cancer cells apoptosis (Wang and Wei, 2014). Moreover, FBXO45 ubiquitylates and degrades p73, which is a member of p53 family that responses to DNA damage by mediating cell cycle arrest and apoptosis.(Peschiaroli, Scialpi, 2009). Taken together, FBXO45 may control cell cycle by regulating its substrates such as p73 and Par-4, but its physiological role in cell cycle and tumorigenesis warrant further in-depth studies.
5. Conclusions
In conclusion, F-box proteins exert their function in cell cycle regulation mainly via targeting the various cell cycle regulatory substrates. Since most studies focus on the role of Fbw7, Skp2, and β-TRCP among the identified 69 F-box proteins in cell cycle and tumorigenesis, the remaining members are required to elucidate their biological functions by genetic, biochemical, and cell biological approaches. Due to the fact that dysregulation of cell cycle contributes to tumorigenesis, comprehensively understanding of cellular functions for all 69 F-box proteins could be helpful for finding a novel strategy via targeting F-box proteins for the treatment of human cancers. It is known that F-box proteins play an oncogenic or tumor suppressive role in context-dependent manner. Therefore, we need develop the inhibitors of oncogenic F-box proteins to suppress their expression. As many F-box proteins such as FBXW7 act as tumor suppressors, discovering inhibitors for upstream regulatory proteins to activate them, or inhibiting their downstream oncoprotein targets would be viable therapeutic approaches (Skaar et al., 2014). Without a doubt, it is essential to design personalized medicine targeting the individual dysregulated F-box protein. To this end, in-depth investigation of functions of F-box proteins will elucidate their role in cell cycle and develop novel strategies for the treatment and prevention of human cancers.
ACKNOWLEDGEMENTS
This work was also supported by the National Natural Science Foundation of China (81172087, 81572936), and a projected funded by the priority academic program development of Jiangsu higher education institutions and by the NIH grants to W.W. (GM094777 and CA177910).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
References
- Abbas T, Keaton M, Dutta A. Regulation of TGF-beta signaling, exit from the cell cycle, and cellular migration through cullin cross-regulation: SCF-FBXO11 turns off CRL4-Cdt2. Cell Cycle. 2013a;12:2175–82. doi: 10.4161/cc.25314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbas T, Mueller AC, Shibata E, Keaton M, Rossi M, Dutta A. CRL1-FBXO11 promotes Cdt2 ubiquitylation and degradation and regulates Pr-Set7/Set8-mediated cellular migration. Mol Cell. 2013b;49:1147–58. doi: 10.1016/j.molcel.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abida WM, Nikolaev A, Zhao W, Zhang W, Gu W. FBXO11 promotes the Neddylation of p53 and inhibits its transcriptional activity. J Biol Chem. 2007;282:1797–804. doi: 10.1074/jbc.M609001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand SN, Maywood ES, Chesham JE, Joynson G, Banks GT, Hastings MH, et al. Distinct and separable roles for endogenous CRY1 and CRY2 within the circadian molecular clockwork of the suprachiasmatic nucleus, as revealed by the Fbxl3(Afh) mutation. J Neurosci. 2013;33:7145–53. doi: 10.1523/JNEUROSCI.4950-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai C, Richman R, Elledge SJ. Human cyclin F. EMBO J. 1994;13:6087–98. doi: 10.1002/j.1460-2075.1994.tb06955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, et al. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–74. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- Barbash O, Zamfirova P, Lin DI, Chen X, Yang K, Nakagawa H, et al. Mutations in Fbx4 inhibit dimerization of the SCF(Fbx4) ligase and contribute to cyclin D1 overexpression in human cancer. Cancer Cell. 2008;14:68–78. doi: 10.1016/j.ccr.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basel D, DePaepe A, Kilpatrick MW, Tsipouras P. Split hand foot malformation is associated with a reduced level of Dactylin gene expression. Clin Genet. 2003;64:350–4. doi: 10.1034/j.1399-0004.2003.00153.x. [DOI] [PubMed] [Google Scholar]
- Bedford L, Lowe J, Dick LR, Mayer RJ, Brownell JE. Ubiquitin-like protein conjugation and the ubiquitin-proteasome system as drug targets. Nat Rev Drug Discov. 2011;10:29–46. doi: 10.1038/nrd3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besson A, Dowdy SF, Roberts JM. CDK inhibitors: cell cycle regulators and beyond. Dev Cell. 2008;14:159–69. doi: 10.1016/j.devcel.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Bhattacharya S, Garriga J, Calbo J, Yong T, Haines DS, Grana X. SKP2 associates with p130 and accelerates p130 ubiquitylation and degradation in human cells. Oncogene. 2003;22:2443–51. doi: 10.1038/sj.onc.1206339. [DOI] [PubMed] [Google Scholar]
- Bornstein G, Bloom J, Sitry-Shevah D, Nakayama K, Pagano M, Hershko A. Role of the SCFSkp2 ubiquitin ligase in the degradation of p21Cip1 in S phase. J Biol Chem. 2003;278:25752–7. doi: 10.1074/jbc.M301774200. [DOI] [PubMed] [Google Scholar]
- Boulard M, Edwards JR, Bestor TH. FBXL10 protects Polycomb-bound genes from hypermethylation. Nat Genet. 2015;47:479–85. doi: 10.1038/ng.3272. [DOI] [PubMed] [Google Scholar]
- Busino L, Bassermann F, Maiolica A, Lee C, Nolan PM, Godinho SI, et al. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science. 2007;316:900–4. doi: 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- Busino L, Donzelli M, Chiesa M, Guardavaccaro D, Ganoth D, Dorrello NV, et al. Degradation of Cdc25A by beta-TrCP during S phase and in response to DNA damage. Nature. 2003;426:87–91. doi: 10.1038/nature02082. [DOI] [PubMed] [Google Scholar]
- Carmena M, Earnshaw WC. The cellular geography of aurora kinases. Nat Rev Mol Cell Biol. 2003;4:842–54. doi: 10.1038/nrm1245. [DOI] [PubMed] [Google Scholar]
- Carrano AC, Pagano M. Role of the F-box protein Skp2 in adhesion-dependent cell cycle progression. J Cell Biol. 2001;153:1381–90. doi: 10.1083/jcb.153.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cen G, Ding HH, Liu B, Wu WD. FBXL5 targets cortactin for ubiquitination-mediated destruction to regulate gastric cancer cell migration. Tumour Biol. 2014;35:8633–8. doi: 10.1007/s13277-014-2104-9. [DOI] [PubMed] [Google Scholar]
- Chan CH, Lee SW, Wang J, Lin HK. Regulation of Skp2 expression and activity and its role in cancer progression. ScientificWorldJournal. 2010;10:1001–15. doi: 10.1100/tsw.2010.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YF, Cheng CM, Chang LK, Jong YJ, Yuo CY. The F-box protein Fbxo7 interacts with human inhibitor of apoptosis protein cIAP1 and promotes cIAP1 ubiquitination. Biochem Biophys Res Commun. 2006;342:1022–6. doi: 10.1016/j.bbrc.2006.02.061. [DOI] [PubMed] [Google Scholar]
- Chen BB, Glasser JR, Coon TA, Mallampalli RK. FBXL2 is a ubiquitin E3 ligase subunit that triggers mitotic arrest. Cell Cycle. 2011a;10:3487–94. doi: 10.4161/cc.10.20.17742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BB, Glasser JR, Coon TA, Mallampalli RK. F-box protein FBXL2 exerts human lung tumor suppressor-like activity by ubiquitin-mediated degradation of cyclin D3 resulting in cell cycle arrest. Oncogene. 2012a;31:2566–79. doi: 10.1038/onc.2011.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BB, Glasser JR, Coon TA, Zou C, Miller HL, Fenton M, et al. F-box protein FBXL2 targets cyclin D2 for ubiquitination and degradation to inhibit leukemic cell proliferation. Blood. 2012b;119:3132–41. doi: 10.1182/blood-2011-06-358911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JY, Wang MC, Hung WC. Bcr-Abl-induced tyrosine phosphorylation of Emi1 to stabilize Skp2 protein via inhibition of ubiquitination in chronic myeloid leukemia cells. J Cell Physiol. 2011b;226:407–13. doi: 10.1002/jcp.22346. [DOI] [PubMed] [Google Scholar]
- Chen ZW, Liu B, Tang NW, Xu YH, Ye XY, Li ZM, et al. FBXL5-mediated degradation of single-stranded DNA-binding protein hSSB1 controls DNA damage response. Nucleic Acids Res. 2014;42:11560–9. doi: 10.1093/nar/gku876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesnaye Ede L, Mendez JP, Lopez-Romero R, Romero-Tlalolini Mde L, Vergara MD, Salcedo M, et al. FBXW12, a novel F box protein-encoding gene, is deleted or methylated in some cases of epithelial ovarian cancer. Int J Clin Exp Pathol. 2015;8:10192–203. [PMC free article] [PubMed] [Google Scholar]
- Coon TA, Glasser JR, Mallampalli RK, Chen BB. Novel E3 ligase component FBXL7 ubiquitinates and degrades Aurora A, causing mitotic arrest. Cell Cycle. 2012;11:721–9. doi: 10.4161/cc.11.4.19171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, He S, Xing C, Lu K, Wang J, Xing G, et al. SCFFBXL(1)(5) regulates BMP signalling by directing the degradation of HECT-type ubiquitin ligase Smurf1. EMBO J. 2011;30:2675–89. doi: 10.1038/emboj.2011.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha-Ferreira I, Rodrigues-Martins A, Bento I, Riparbelli M, Zhang W, Laue E, et al. The SCF/Slimb ubiquitin ligase limits centrosome amplification through degradation of SAK/PLK4. Curr Biol. 2009;19:43–9. doi: 10.1016/j.cub.2008.11.037. [DOI] [PubMed] [Google Scholar]
- D'Angiolella V, Donato V, Forrester FM, Jeong YT, Pellacani C, Kudo Y, et al. Cyclin F-mediated degradation of ribonucleotide reductase M2 controls genome integrity and DNA repair. Cell. 2012;149:1023–34. doi: 10.1016/j.cell.2012.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angiolella V, Donato V, Vijayakumar S, Saraf A, Florens L, Washburn MP, et al. SCF(Cyclin F) controls centrosome homeostasis and mitotic fidelity through CP110 degradation. Nature. 2010;466:138–42. doi: 10.1038/nature09140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y, Grant S. Cyclin-dependent kinase inhibitors. Curr Opin Pharmacol. 2003;3:362–70. doi: 10.1016/s1471-4892(03)00079-1. [DOI] [PubMed] [Google Scholar]
- Das A, Chai JC, Jung KH, Das ND, Kang SC, Lee YS, et al. JMJD2A attenuation affects cell cycle and tumourigenic inflammatory gene regulation in lipopolysaccharide stimulated neuroectodermal stem cells. Exp Cell Res. 2014;328:361–78. doi: 10.1016/j.yexcr.2014.08.029. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Dhar SS, Alam H, Li N, Wagner KW, Chung J, Ahn YW, et al. Transcriptional repression of histone deacetylase 3 by the histone demethylase KDM2A is coupled to tumorigenicity of lung cancer cells. J Biol Chem. 2014;289:7483–96. doi: 10.1074/jbc.M113.521625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrowski C, Helledie T, Ling L, Grunert M, Canning CA, Jones CM, et al. FGFR1 signaling stimulates proliferation of human mesenchymal stem cells by inhibiting the cyclin-dependent kinase inhibitors p21(Waf1) and p27(Kip1). Stem Cells. 2013;31:2724–36. doi: 10.1002/stem.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan S, Cermak L, Pagan JK, Rossi M, Martinengo C, di Celle PF, et al. FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas. Nature. 2012;481:90–3. doi: 10.1038/nature10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducat D, Zheng Y. Aurora kinases in spindle assembly and chromosome segregation. Exp Cell Res. 2004;301:60–7. doi: 10.1016/j.yexcr.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Duda DM, Scott DC, Calabrese MF, Zimmerman ES, Zheng N, Schulman BA. Structural regulation of cullin-RING ubiquitin ligase complexes. Curr Opin Struct Biol. 2011;21:257–64. doi: 10.1016/j.sbi.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egozi D, Shapira M, Paor G, Ben-Izhak O, Skorecki K, Hershko DD. Regulation of the cell cycle inhibitor p27 and its ubiquitin ligase Skp2 in differentiation of human embryonic stem cells. FASEB J. 2007;21:2807–17. doi: 10.1096/fj.06-7758com. [DOI] [PubMed] [Google Scholar]
- Eldridge AG, O'Brien T. Therapeutic strategies within the ubiquitin proteasome system. Cell Death Differ. 2010;17:4–13. doi: 10.1038/cdd.2009.82. [DOI] [PubMed] [Google Scholar]
- Emanuele MJ, Elia AE, Xu Q, Thoma CR, Izhar L, Leng Y, et al. Global identification of modular cullin-RING ligase substrates. Cell. 2011;147:459–74. doi: 10.1016/j.cell.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei C, He X, Xie S, Miao H, Zhou Z, Li L. Smurf1-mediated axin ubiquitination requires Smurf1 C2 domain and is cell cycle-dependent. J Biol Chem. 2014;289:14170–7. doi: 10.1074/jbc.M113.536714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Hui Y, Ling L, Xiaoyan L, Yuqiu W, Peng W, et al. FBXW10 is negatively regulated in transcription and expression level by protein O-GlcNAcylation. Biochem Biophys Res Commun. 2013;438:427–32. doi: 10.1016/j.bbrc.2013.07.091. [DOI] [PubMed] [Google Scholar]
- Franz J, Jerome J, Lear T, Gong Q, Weathington NM. The Human IL-22 Receptor Is Regulated through the Action of the Novel E3 Ligase Subunit FBXW12, Which Functions as an Epithelial Growth Suppressor. J Immunol Res. 2015;2015:912713. doi: 10.1155/2015/912713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frescas D, Guardavaccaro D, Bassermann F, Koyama-Nasu R, Pagano M. JHDM1B/FBXL10 is a nucleolar protein that represses transcription of ribosomal RNA genes. Nature. 2007;450:309–13. doi: 10.1038/nature06255. [DOI] [PubMed] [Google Scholar]
- Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008;8:438–49. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Bian M, Jiang Q, Zhang C. Roles of Aurora kinases in mitosis and tumorigenesis. Mol Cancer Res. 2007;5:1–10. doi: 10.1158/1541-7786.MCR-06-0208. [DOI] [PubMed] [Google Scholar]
- Fung TK, Siu WY, Yam CH, Lau A, Poon RY. Cyclin F is degraded during G2-M by mechanisms fundamentally different from other cyclins. J Biol Chem. 2002;277:35140–9. doi: 10.1074/jbc.M205503200. [DOI] [PubMed] [Google Scholar]
- Godinho SI, Maywood ES, Shaw L, Tucci V, Barnard AR, Busino L, et al. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science. 2007;316:897–900. doi: 10.1126/science.1141138. [DOI] [PubMed] [Google Scholar]
- Guardavaccaro D, Frescas D, Dorrello NV, Peschiaroli A, Multani AS, Cardozo T, et al. Control of chromosome stability by the beta-TrCP-REST-Mad2 axis. Nature. 2008;452:365–9. doi: 10.1038/nature06641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guardavaccaro D, Kudo Y, Boulaire J, Barchi M, Busino L, Donzelli M, et al. Control of meiotic and mitotic progression by the F box protein beta-Trcp1 in vivo. Dev Cell. 2003;4:799–812. doi: 10.1016/s1534-5807(03)00154-0. [DOI] [PubMed] [Google Scholar]
- Guderian G, Westendorf J, Uldschmid A, Nigg EA. Plk4 trans-autophosphorylation regulates centriole number by controlling betaTrCP-mediated degradation. J Cell Sci. 2010;123:2163–9. doi: 10.1242/jcs.068502. [DOI] [PubMed] [Google Scholar]
- Han SJ, Begum K, Foulds CE, Hamilton RA, Bailey S, Malovannaya A, et al. The Dual Estrogen Receptor alpha Inhibitory Effects of the Tissue-Selective Estrogen Complex for Endometrial and Breast Safety. Mol Pharmacol. 2016;89:14–26. doi: 10.1124/mol.115.100925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Kallin EM, Tsukada Y, Zhang Y. The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b). Nat Struct Mol Biol. 2008;15:1169–75. doi: 10.1038/nsmb.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–79. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hirano A, Yumimoto K, Tsunematsu R, Matsumoto M, Oyama M, Kozuka-Hata H, et al. FBXL21 regulates oscillation of the circadian clock through ubiquitination and stabilization of cryptochromes. Cell. 2013;152:1106–18. doi: 10.1016/j.cell.2013.01.054. [DOI] [PubMed] [Google Scholar]
- Hoeller D, Dikic I. Targeting the ubiquitin system in cancer therapy. Nature. 2009;458:438–44. doi: 10.1038/nature07960. [DOI] [PubMed] [Google Scholar]
- Hsu JM, Lee YC, Yu CT, Huang CY. Fbx7 functions in the SCF complex regulating Cdk1-cyclin B-phosphorylated hepatoma up-regulated protein (HURP) proteolysis by a proline-rich region. J Biol Chem. 2004;279:32592–602. doi: 10.1074/jbc.M404950200. [DOI] [PubMed] [Google Scholar]
- Hsu JY, Reimann JD, Sorensen CS, Lukas J, Jackson PK. E2F-dependent accumulation of hEmi1 regulates S phase entry by inhibiting APC(Cdh1). Nat Cell Biol. 2002;4:358–66. doi: 10.1038/ncb785. [DOI] [PubMed] [Google Scholar]
- Hu J, Zacharek S, He YJ, Lee H, Shumway S, Duronio RJ, et al. WD40 protein FBW5 promotes ubiquitination of tumor suppressor TSC2 by DDB1-CUL4-ROC1 ligase. Genes Dev. 2008;22:866–71. doi: 10.1101/gad.1624008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z, Vierstra RD. The cullin-RING ubiquitin-protein ligases. Annu Rev Plant Biol. 2011;62:299–334. doi: 10.1146/annurev-arplant-042809-112256. [DOI] [PubMed] [Google Scholar]
- Ianakiev P, Kilpatrick MW, Dealy C, Kosher R, Korenberg JR, Chen XN, et al. A novel human gene encoding an F-box/WD40 containing protein maps in the SHFM3 critical region on 10q24. Biochem Biophys Res Commun. 1999;261:64–70. doi: 10.1006/bbrc.1999.0963. [DOI] [PubMed] [Google Scholar]
- Ishimura A, Minehata K, Terashima M, Kondoh G, Hara T, Suzuki T. Jmjd5, an H3K36me2 histone demethylase, modulates embryonic cell proliferation through the regulation of Cdkn1a expression. Development. 2012;139:749–59. doi: 10.1242/dev.074138. [DOI] [PubMed] [Google Scholar]
- Jeong YT, Rossi M, Cermak L, Saraf A, Florens L, Washburn MP, et al. FBH1 promotes DNA double-strand breakage and apoptosis in response to DNA replication stress. J Cell Biol. 2013;200:141–9. doi: 10.1083/jcb.201209002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P, Goldin L, Ren H, Sun D, Guardavaccaro D, Pagano M, et al. Skp2 contains a novel cyclin A binding domain that directly protects cyclin A from inhibition by p27Kip1. J Biol Chem. 2006;281:24058–69. doi: 10.1074/jbc.M603105200. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Li JM, Luo HQ. Clinicopathological Significance of DLC-1 Expression in Cancer: a Meta-Analysis. Asian Pac J Cancer Prev. 2015;16:7255–60. doi: 10.7314/apjcp.2015.16.16.7255. [DOI] [PubMed] [Google Scholar]
- Jin J, Shirogane T, Xu L, Nalepa G, Qin J, Elledge SJ, et al. SCFbeta-TRCP links Chk1 signaling to degradation of the Cdc25A protein phosphatase. Genes Dev. 2003;17:3062–74. doi: 10.1101/gad.1157503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson P, Jeffery J, Al-Ejeh F, Schulz RB, Callen DF, Kumar R, et al. SCF FBXO31 E3 ligase targets DNA replication factor Cdt1 for proteolysis in the G2 phase of cell cycle to prevent re-replication. J Biol Chem. 2014;289:18514–25. doi: 10.1074/jbc.M114.559930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanemori Y, Uto K, Sagata N. Beta-TrCP recognizes a previously undescribed nonphosphorylated destruction motif in Cdc25A and Cdc25B phosphatases. Proc Natl Acad Sci U S A. 2005;102:6279–84. doi: 10.1073/pnas.0501873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanie T, Onoyama I, Matsumoto A, Yamada M, Nakatsumi H, Tateishi Y, et al. Genetic reevaluation of the role of F-box proteins in cyclin D1 degradation. Mol Cell Biol. 2012;32:590–605. doi: 10.1128/MCB.06570-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama H, Brinkley WR, Sen S. The Aurora kinases: role in cell transformation and tumorigenesis. Cancer Metastasis Rev. 2003;22:451–64. doi: 10.1023/a:1023789416385. [DOI] [PubMed] [Google Scholar]
- Kawakami E, Tokunaga A, Ozawa M, Sakamoto R, Yoshida N. The histone demethylase Fbxl11/Kdm2a plays an essential role in embryonic development by repressing cell-cycle regulators. Mech Dev. 2015;135:31–42. doi: 10.1016/j.mod.2014.10.001. [DOI] [PubMed] [Google Scholar]
- Kim TY, Jackson S, Xiong Y, Whitsett TG, Lobello JR, Weiss GJ, et al. CRL4A FBXW5-mediated degradation of DLC1 Rho GTPase-activating protein tumor suppressor promotes non-small cell lung cancer cell growth. Proc Natl Acad Sci U S A. 2013;110:16868–73. doi: 10.1073/pnas.1306358110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa K, Kotake Y, Kitagawa M. Ubiquitin-mediated control of oncogene and tumor suppressor gene products. Cancer Sci. 2009;100:1374–81. doi: 10.1111/j.1349-7006.2009.01196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepp DM, Schaefer LK, Ye X, Keyomarsi K, Chu C, Harper JW, et al. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 2001;294:173–7. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- Kogure M, Takawa M, Cho HS, Toyokawa G, Hayashi K, Tsunoda T, et al. Deregulation of the histone demethylase JMJD2A is involved in human carcinogenesis through regulation of the G(1)/S transition. Cancer Lett. 2013;336:76–84. doi: 10.1016/j.canlet.2013.04.009. [DOI] [PubMed] [Google Scholar]
- Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–29. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- Kong M, Barnes EA, Ollendorff V, Donoghue DJ. Cyclin F regulates the nuclear localization of cyclin B1 through a cyclin-cyclin interaction. EMBO J. 2000;19:1378–88. doi: 10.1093/emboj/19.6.1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchay S, Duan S, Schenkein E, Peschiaroli A, Saraf A, Florens L, et al. FBXL2- and PTPL1-mediated degradation of p110-free p85beta regulatory subunit controls the PI(3)K signalling cascade. Nat Cell Biol. 2013;15:472–80. doi: 10.1038/ncb2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Neilsen PM, Crawford J, McKirdy R, Lee J, Powell JA, et al. FBXO31 is the chromosome 16q24.3 senescence gene, a candidate breast tumor suppressor, and a component of an SCF complex. Cancer Res. 2005;65:11304–13. doi: 10.1158/0008-5472.CAN-05-0936. [DOI] [PubMed] [Google Scholar]
- Kurland JF, Tansey WP. Crashing waves of destruction: the cell cycle and APC(Cdh1) regulation of SCF(Skp2). Cancer Cell. 2004;5:305–6. doi: 10.1016/s1535-6108(04)00091-1. [DOI] [PubMed] [Google Scholar]
- Kwon YW, Kim IJ, Wu D, Lu J, Stock WA, Jr., Liu Y, et al. Pten regulates Aurora-A and cooperates with Fbxw7 in modulating radiation-induced tumor development. Mol Cancer Res. 2012;10:834–44. doi: 10.1158/1541-7786.MCR-12-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laman H, Funes JM, Ye H, Henderson S, Galinanes-Garcia L, Hara E, et al. Transforming activity of Fbxo7 is mediated specifically through regulation of cyclin D/cdk6. EMBO J. 2005;24:3104–16. doi: 10.1038/sj.emboj.7600775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman NL, Verschuren EW, Hsu JY, Cherry AM, Jackson PK. Overexpression of the anaphase promoting complex/cyclosome inhibitor Emi1 leads to tetraploidy and genomic instability of p53-deficient cells. Cell Cycle. 2006;5:1569–73. doi: 10.4161/cc.5.14.2925. [DOI] [PubMed] [Google Scholar]
- Lens SM, Voest EE, Medema RH. Shared and separate functions of polo-like kinases and aurora kinases in cancer. Nat Rev Cancer. 2010;10:825–41. doi: 10.1038/nrc2964. [DOI] [PubMed] [Google Scholar]
- Li D, Xie P, Zhao F, Shu J, Li L, Zhan Y, et al. F-box protein Fbxo3 targets Smurf1 ubiquitin ligase for ubiquitination and degradation. Biochem Biophys Res Commun. 2015;458:941–5. doi: 10.1016/j.bbrc.2015.02.089. [DOI] [PubMed] [Google Scholar]
- Li P, Li C, Zhao X, Zhang X, Nicosia SV, Bai W. p27(Kip1) stabilization and G(1) arrest by 1,25-dihydroxyvitamin D(3) in ovarian cancer cells mediated through down-regulation of cyclin E/cyclin-dependent kinase 2 and Skp1-Cullin-F-box protein/Skp2 ubiquitin ligase. J Biol Chem. 2004;279:25260–7. doi: 10.1074/jbc.M311052200. [DOI] [PubMed] [Google Scholar]
- Li W, Bengtson MH, Ulbrich A, Matsuda A, Reddy VA, Orth A, et al. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle's dynamics and signaling. PLoS One. 2008;3:e1487. doi: 10.1371/journal.pone.0001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichner Z, Pall E, Kerekes A, Pallinger E, Maraghechi P, Bosze Z, et al. The miR-290-295 cluster promotes pluripotency maintenance by regulating cell cycle phase distribution in mouse embryonic stem cells. Differentiation. 2011;81:11–24. doi: 10.1016/j.diff.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Limon-Mortes MC, Mora-Santos M, Espina A, Pintor-Toro JA, Lopez-Roman A, Tortolero M, et al. UV-induced degradation of securin is mediated by SKP1-CUL1-beta TrCP E3 ubiquitin ligase. J Cell Sci. 2008;121:1825–31. doi: 10.1242/jcs.020552. [DOI] [PubMed] [Google Scholar]
- Lin DI, Barbash O, Kumar KG, Weber JD, Harper JW, Klein-Szanto AJ, et al. Phosphorylation-dependent ubiquitination of cyclin D1 by the SCF(FBX4-alphaB crystallin) complex. Mol Cell. 2006;24:355–66. doi: 10.1016/j.molcel.2006.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin P, Fu J, Zhao B, Lin F, Zou H, Liu L, et al. Fbxw8 is involved in the proliferation of human choriocarcinoma JEG-3 cells. Mol Biol Rep. 2011;38:1741–7. doi: 10.1007/s11033-010-0288-7. [DOI] [PubMed] [Google Scholar]
- Lindqvist A, Rodriguez-Bravo V, Medema RH. The decision to enter mitosis: feedback and redundancy in the mitotic entry network. J Cell Biol. 2009;185:193–202. doi: 10.1083/jcb.200812045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Lear T, Zhao Y, Zhao J, Zou C, Chen BB, et al. F-box protein Fbxl18 mediates polyubiquitylation and proteasomal degradation of the pro-apoptotic SCF subunit Fbxl7. Cell Death Dis. 2015;6:e1630. doi: 10.1038/cddis.2014.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockwood WW, Chandel SK, Stewart GL, Erdjument-Bromage H, Beverly LJ. The novel ubiquitin ligase complex, SCF(Fbxw4), interacts with the COP9 signalosome in an F-box dependent manner, is mutated, lost and under-expressed in human cancers. PLoS One. 2013;8:e63610. doi: 10.1371/journal.pone.0063610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomonosov M, Meziane el K, Ye H, Nelson DE, Randle SJ, Laman H. Expression of Fbxo7 in haematopoietic progenitor cells cooperates with p53 loss to promote lymphomagenesis. PLoS One. 2011;6:e21165. doi: 10.1371/journal.pone.0021165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Jackson MW, Singhi AD, Kandel ES, Yang M, Zhang Y, et al. Validation-based insertional mutagenesis identifies lysine demethylase FBXL11 as a negative regulator of NFkappaB. Proc Natl Acad Sci U S A. 2009;106:16339–44. doi: 10.1073/pnas.0908560106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Jackson MW, Wang B, Yang M, Chance MR, Miyagi M, et al. Regulation of NF-kappaB by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc Natl Acad Sci U S A. 2010;107:46–51. doi: 10.1073/pnas.0912493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Li J, Cheng D, Parameswaran B, Zhang S, Jiang Z, et al. The F-box protein FBXO44 mediates BRCA1 ubiquitination and degradation. J Biol Chem. 2012;287:41014–22. doi: 10.1074/jbc.M112.407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado-Oliveira G, Guerreiro E, Matias AC, Facucho-Oliveira J, Pacheco-Leyva I, Braganca J. FBXL5 modulates HIF-1alpha transcriptional activity by degradation of CITED2. Arch Biochem Biophys. 2015;576:61–72. doi: 10.1016/j.abb.2015.04.012. [DOI] [PubMed] [Google Scholar]
- Mallampalli RK, Kaercher L, Snavely C, Pulijala R, Chen BB, Coon T, et al. Fbxl12 triggers G1 arrest by mediating degradation of calmodulin kinase I. Cell Signal. 2013;25:2047–59. doi: 10.1016/j.cellsig.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malonia SK, Dutta P, Santra MK, Green MR. F-box protein FBXO31 directs degradation of MDM2 to facilitate p53-mediated growth arrest following genotoxic stress. Proc Natl Acad Sci U S A. 2015;112:8632–7. doi: 10.1073/pnas.1510929112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9:153–66. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- Margottin-Goguet F, Hsu JY, Loktev A, Hsieh HM, Reimann JD, Jackson PK. Prophase destruction of Emi1 by the SCF(betaTrCP/Slimb) ubiquitin ligase activates the anaphase promoting complex to allow progression beyond prometaphase. Dev Cell. 2003;4:813–26. doi: 10.1016/s1534-5807(03)00153-9. [DOI] [PubMed] [Google Scholar]
- Metzger MB, Hristova VA, Weissman AM. HECT and RING finger families of E3 ubiquitin ligases at a glance. J Cell Sci. 2012;125:531–7. doi: 10.1242/jcs.091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meziane el K, Randle SJ, Nelson DE, Lomonosov M, Laman H. Knockdown of Fbxo7 reveals its regulatory role in proliferation and differentiation of haematopoietic precursor cells. J Cell Sci. 2011;124:2175–86. doi: 10.1242/jcs.080465. [DOI] [PubMed] [Google Scholar]
- Michel JJ, Xiong Y. Human CUL-1, but not other cullin family members, selectively interacts with SKP1 to form a complex with SKP2 and cyclin A. Cell Growth Differ. 1998;9:435–49. [PubMed] [Google Scholar]
- Minella AC, Grim JE, Welcker M, Clurman BE. p53 and SCFFbw7 cooperatively restrain cyclin E-associated genome instability. Oncogene. 2007;26:6948–53. doi: 10.1038/sj.onc.1210518. [DOI] [PubMed] [Google Scholar]
- Nakayama K, Hatakeyama S, Maruyama S, Kikuchi A, Onoe K, Good RA, et al. Impaired degradation of inhibitory subunit of NF-kappa B (I kappa B) and beta-catenin as a result of targeted disruption of the beta-TrCP1 gene. Proc Natl Acad Sci U S A. 2003;100:8752–7. doi: 10.1073/pnas.1133216100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K, Nagahama H, Minamishima YA, Miyake S, Ishida N, Hatakeyama S, et al. Skp2-mediated degradation of p27 regulates progression into mitosis. Dev Cell. 2004;6:661–72. doi: 10.1016/s1534-5807(04)00131-5. [DOI] [PubMed] [Google Scholar]
- Nakayama KI, Hatakeyama S, Nakayama K. Regulation of the cell cycle at the G1-S transition by proteolysis of cyclin E and p27Kip1. Biochem Biophys Res Commun. 2001;282:853–60. doi: 10.1006/bbrc.2001.4627. [DOI] [PubMed] [Google Scholar]
- Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer. 2006;6:369–81. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- Nalepa G, Rolfe M, Harper JW. Drug discovery in the ubiquitin-proteasome system. Nat Rev Drug Discov. 2006;5:596–613. doi: 10.1038/nrd2056. [DOI] [PubMed] [Google Scholar]
- Okabe H, Lee SH, Phuchareon J, Albertson DG, McCormick F, Tetsu O. A critical role for FBXW8 and MAPK in cyclin D1 degradation and cancer cell proliferation. PLoS One. 2006;1:e128. doi: 10.1371/journal.pone.0000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagan J, Pagano M. FBXW5 controls centrosome number. Nat Cell Biol. 2011;13:888–90. doi: 10.1038/ncb2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagan JK, Marzio A, Jones MJ, Saraf A, Jallepalli PV, Florens L, et al. Degradation of Cep68 and PCNT cleavage mediate Cep215 removal from the PCM to allow centriole separation, disengagement and licensing. Nat Cell Biol. 2015;17:31–43. doi: 10.1038/ncb3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pateras IS, Apostolopoulou K, Koutsami M, Evangelou K, Tsantoulis P, Liloglou T, et al. Downregulation of the KIP family members p27(KIP1) and p57(KIP2) by SKP2 and the role of methylation in p57(KIP2) inactivation in nonsmall cell lung cancer. Int J Cancer. 2006;119:2546–56. doi: 10.1002/ijc.22214. [DOI] [PubMed] [Google Scholar]
- Perciavalle RM, Opferman JT. Delving deeper: MCL-1's contributions to normal and cancer biology. Trends Cell Biol. 2013;23:22–9. doi: 10.1016/j.tcb.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschiaroli A, Dorrello NV, Guardavaccaro D, Venere M, Halazonetis T, Sherman NE, et al. SCFbetaTrCP-mediated degradation of Claspin regulates recovery from the DNA replication checkpoint response. Mol Cell. 2006;23:319–29. doi: 10.1016/j.molcel.2006.06.013. [DOI] [PubMed] [Google Scholar]
- Peschiaroli A, Scialpi F, Bernassola F, Pagano M, Melino G. The F-box protein FBXO45 promotes the proteasome-dependent degradation of p73. Oncogene. 2009;28:3157–66. doi: 10.1038/onc.2009.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–56. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–33. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Postow L, Funabiki H. An SCF complex containing Fbxl12 mediates DNA damage-induced Ku80 ubiquitylation. Cell Cycle. 2013;12:587–95. doi: 10.4161/cc.23408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puklowski A, Homsi Y, Keller D, May M, Chauhan S, Kossatz U, et al. The SCF FBXW5 E3-ubiquitin ligase is regulated by PLK4 and targets HsSAS-6 to control centrosome duplication. Nat Cell Biol. 2011;13:1004–9. doi: 10.1038/ncb2282. [DOI] [PubMed] [Google Scholar]
- Rajagopalan H, Jallepalli PV, Rago C, Velculescu VE, Kinzler KW, Vogelstein B, et al. Inactivation of hCDC4 can cause chromosomal instability. Nature. 2004;428:77–81. doi: 10.1038/nature02313. [DOI] [PubMed] [Google Scholar]
- Ren Y, Wu L, Li X, Li W, Yang Y, Zhang M. FBXL10 contributes to the progression of nasopharyngeal carcinoma via involving in PI3K/mTOR pathway. Neoplasma. 2015;62:925–31. doi: 10.4149/neo_2015_112. [DOI] [PubMed] [Google Scholar]
- Rossi M, Duan S, Jeong YT, Horn M, Saraf A, Florens L, et al. Regulation of the CRL4(Cdt2) ubiquitin ligase and cell-cycle exit by the SCF(Fbxo11) ubiquitin ligase. Mol Cell. 2013;49:1159–66. doi: 10.1016/j.molcel.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol. 2007;8:798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- Saiga T, Fukuda T, Matsumoto M, Tada H, Okano HJ, Okano H, et al. Fbxo45 forms a novel ubiquitin ligase complex and is required for neuronal development. Mol Cell Biol. 2009;29:3529–43. doi: 10.1128/MCB.00364-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath SC, Ohi R, Leismann O, Salic A, Pozniakovski A, Funabiki H. The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell. 2004;118:187–202. doi: 10.1016/j.cell.2004.06.026. [DOI] [PubMed] [Google Scholar]
- Santra MK, Wajapeyee N, Green MR. F-box protein FBXO31 mediates cyclin D1 degradation to induce G1 arrest after DNA damage. Nature. 2009;459:722–5. doi: 10.1038/nature08011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarikas A, Hartmann T, Pan ZQ. The cullin protein family. Genome Biol. 2011;12:220. doi: 10.1186/gb-2011-12-4-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasajima H, Nakagawa K, Kashiwayanagi M, Yokosawa H. Polyubiquitination of the B-cell translocation gene 1 and 2 proteins is promoted by the SCF ubiquitin ligase complex containing betaTrCP. Biol Pharm Bull. 2012;35:1539–45. doi: 10.1248/bpb.b12-00330. [DOI] [PubMed] [Google Scholar]
- Seki A, Coppinger JA, Du H, Jang CY, Yates JR, 3rd, Fang G. Plk1- and beta-TrCP-dependent degradation of Bora controls mitotic progression. J Cell Biol. 2008;181:65–78. doi: 10.1083/jcb.200712027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi D, Tan Z, Lu R, Yang W, Zhang Y. MicroRNA-218 inhibits the proliferation of human choriocarcinoma JEG-3 cell line by targeting Fbxw8. Biochem Biophys Res Commun. 2014;450:1241–6. doi: 10.1016/j.bbrc.2014.06.094. [DOI] [PubMed] [Google Scholar]
- Sidow A, Bulotsky MS, Kerrebrock AW, Birren BW, Altshuler D, Jaenisch R, et al. A novel member of the F-box/WD40 gene family, encoding dactylin, is disrupted in the mouse dactylaplasia mutant. Nat Genet. 1999;23:104–7. doi: 10.1038/12709. [DOI] [PubMed] [Google Scholar]
- Siepka SM, Yoo SH, Park J, Song W, Kumar V, Hu Y, et al. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell. 2007;129:1011–23. doi: 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu KT, Rosner MR, Minella AC. An integrated view of cyclin E function and regulation. Cell Cycle. 2012;11:57–64. doi: 10.4161/cc.11.1.18775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar JR, Pagan JK, Pagano M. Mechanisms and function of substrate recruitment by F-box proteins. Nat Rev Mol Cell Biol. 2013;14:369–81. doi: 10.1038/nrm3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar JR, Pagan JK, Pagano M. SCF ubiquitin ligase-targeted therapies. Nat Rev Drug Discov. 2014;13:889–903. doi: 10.1038/nrd4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery SD, Moore RV, Brinkley BR, Hall RM. Aurora-C and Aurora-B share phosphorylation and regulation of CENP-A and Borealin during mitosis. Cell Cycle. 2008;7:787–95. doi: 10.4161/cc.7.6.5563. [DOI] [PubMed] [Google Scholar]
- Spruck CH, Won KA, Reed SI. Deregulated cyclin E induces chromosome instability. Nature. 1999;401:297–300. doi: 10.1038/45836. [DOI] [PubMed] [Google Scholar]
- Suzuki S, Fukasawa H, Misaki T, Togawa A, Ohashi N, Kitagawa K, et al. The amelioration of renal damage in Skp2-deficient mice canceled by p27 Kip1 deficiency in Skp2−/− p27−/− mice. PLoS One. 2012;7:e36249. doi: 10.1371/journal.pone.0036249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan MK, Lim HJ, Harper JW. SCF(FBXO22) regulates histone H3 lysine 9 and 36 methylation levels by targeting histone demethylase KDM4A for ubiquitin-mediated proteasomal degradation. Mol Cell Biol. 2011;31:3687–99. doi: 10.1128/MCB.05746-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedesco D, Lukas J, Reed SI. The pRb-related protein p130 is regulated by phosphorylation-dependent proteolysis via the protein-ubiquitin ligase SCF(Skp2). Genes Dev. 2002;16:2946–57. doi: 10.1101/gad.1011202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng CL, Hsieh YC, Phan L, Shin J, Gully C, Velazquez-Torres G, et al. FBXW7 is involved in Aurora B degradation. Cell Cycle. 2012;11:4059–68. doi: 10.4161/cc.22381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetzlaff MT, Bai C, Finegold M, Wilson J, Harper JW, Mahon KA, et al. Cyclin F disruption compromises placental development and affects normal cell cycle execution. Mol Cell Biol. 2004;24:2487–98. doi: 10.1128/MCB.24.6.2487-2498.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsumi T, Kuwabara H, Arai T, Xiao Y, Decaprio JA. Disruption of the Fbxw8 gene results in pre- and postnatal growth retardation in mice. Mol Cell Biol. 2008;28:743–51. doi: 10.1128/MCB.01665-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzatsos A, Paskaleva P, Ferrari F, Deshpande V, Stoykova S, Contino G, et al. KDM2B promotes pancreatic cancer via Polycomb-dependent and -independent transcriptional programs. J Clin Invest. 2013;123:727–39. doi: 10.1172/JCI64535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzatsos A, Paskaleva P, Lymperi S, Contino G, Stoykova S, Chen Z, et al. Lysine-specific demethylase 2B (KDM2B)-let-7-enhancer of zester homolog 2 (EZH2) pathway regulates cell cycle progression and senescence in primary cells. J Biol Chem. 2011;286:33061–9. doi: 10.1074/jbc.M111.257667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida S, Watanabe N, Kudo Y, Yoshioka K, Matsunaga T, Ishizaka Y, et al. SCFbeta(TrCP) mediates stress-activated MAPK-induced Cdc25B degradation. J Cell Sci. 2011;124:2816–25. doi: 10.1242/jcs.083931. [DOI] [PubMed] [Google Scholar]
- Ungermannova D, Gao Y, Liu X. Ubiquitination of p27Kip1 requires physical interaction with cyclin E and probable phosphate recognition by SKP2. J Biol Chem. 2005;280:30301–9. doi: 10.1074/jbc.M411103200. [DOI] [PubMed] [Google Scholar]
- Van Rechem C, Black JC, Abbas T, Allen A, Rinehart CA, Yuan GC, et al. The SKP1-Cul1-F-box and leucine-rich repeat protein 4 (SCF-FbxL4) ubiquitin ligase regulates lysine demethylase 4A (KDM4A)/Jumonji domain-containing 2A (JMJD2A) protein. J Biol Chem. 2011;286:30462–70. doi: 10.1074/jbc.M111.273508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. The ubiquitin system, an immense realm. Annu Rev Biochem. 2012;81:167–76. doi: 10.1146/annurev-biochem-051910-094049. [DOI] [PubMed] [Google Scholar]
- Vinas-Castells R, Frias A, Robles-Lanuza E, Zhang K, Longmore GD, Garcia de Herreros A, et al. Nuclear ubiquitination by FBXL5 modulates Snail1 DNA binding and stability. Nucleic Acids Res. 2014;42:1079–94. doi: 10.1093/nar/gkt935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner KW, Alam H, Dhar SS, Giri U, Li N, Wei Y, et al. KDM2A promotes lung tumorigenesis by epigenetically enhancing ERK1/2 signaling. J Clin Invest. 2013;123:5231–46. doi: 10.1172/JCI68642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Chen Y, Lin P, Li L, Zhou G, Liu G, et al. The CUL7/F-box and WD repeat domain containing 8 (CUL7/Fbxw8) ubiquitin ligase promotes degradation of hematopoietic progenitor kinase 1. J Biol Chem. 2014a;289:4009–17. doi: 10.1074/jbc.M113.520106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Dai X, Zhong J, Inuzuka H, Wan L, Li X, et al. SCF(beta-TRCP) promotes cell growth by targeting PR-Set7/Set8 for degradation. Nat Commun. 2015;6:10185. doi: 10.1038/ncomms10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Liu P, Inuzuka H, Wei W. Roles of F-box proteins in cancer. Nat Rev Cancer. 2014b;14:233–47. doi: 10.1038/nrc3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wei W. Fbxo45 joins the 'Par-4'ty in controlling apoptosis of cancer cells. Cell Death Differ. 2014;21:1508–10. doi: 10.1038/cdd.2014.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N, Arai H, Nishihara Y, Taniguchi M, Watanabe N, Hunter T, et al. M-phase kinases induce phospho-dependent ubiquitination of somatic Wee1 by SCFbeta-TrCP. Proc Natl Acad Sci U S A. 2004;101:4419–24. doi: 10.1073/pnas.0307700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S, Yang HC, Chuang HC, Yang J, Kulp SK, Lu PJ, et al. A novel mechanism by which thiazolidinediones facilitate the proteasomal degradation of cyclin D1 in cancer cells. J Biol Chem. 2008;283:26759–70. doi: 10.1074/jbc.M802160200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Ayad NG, Wan Y, Zhang GJ, Kirschner MW, Kaelin WG., Jr. Degradation of the SCF component Skp2 in cell-cycle phase G1 by the anaphase-promoting complex. Nature. 2004;428:194–8. doi: 10.1038/nature02381. [DOI] [PubMed] [Google Scholar]
- Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- Werner A, Disanza A, Reifenberger N, Habeck G, Becker J, Calabrese M, et al. SCFFbxw5 mediates transient degradation of actin remodeller Eps8 to allow proper mitotic progression. Nat Cell Biol. 2013;15:179–88. doi: 10.1038/ncb2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz IE, Kusam S, Lam C, Okamoto T, Sandoval W, Anderson DJ, et al. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature. 2011;471:110–4. doi: 10.1038/nature09779. [DOI] [PubMed] [Google Scholar]
- Wu W, Ding H, Cao J, Zhang W. FBXL5 inhibits metastasis of gastric cancer through suppressing Snail1. Cell Physiol Biochem. 2015;35:1764–72. doi: 10.1159/000373988. [DOI] [PubMed] [Google Scholar]
- Wu X, Johansen JV, Helin K. Fbxl10/Kdm2b recruits polycomb repressive complex 1 to CpG islands and regulates H2A ubiquitylation. Mol Cell. 2013;49:1134–46. doi: 10.1016/j.molcel.2013.01.016. [DOI] [PubMed] [Google Scholar]
- Xing W, Busino L, Hinds TR, Marionni ST, Saifee NH, Bush MF, et al. SCF(FBXL3) ubiquitin ligase targets cryptochromes at their cofactor pocket. Nature. 2013;496:64–8. doi: 10.1038/nature11964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Bernaudo S, Fu G, Lee DY, Yang BB, Peng C. Cyclin G2 is degraded through the ubiquitin-proteasome pathway and mediates the antiproliferative effect of activin receptor-like kinase 7. Mol Biol Cell. 2008;19:4968–79. doi: 10.1091/mbc.E08-03-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yam CH, Ng RW, Siu WY, Lau AW, Poon RY. Regulation of cyclin A-Cdk2 by SCF component Skp1 and F-box protein Skp2. Mol Cell Biol. 1999;19:635–45. doi: 10.1128/mcb.19.1.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Nan H, Ma J, Jiang L, Guo Q, Han L, et al. High Skp2/Low p57(Kip2) Expression is Associated with Poor Prognosis in Human Breast Carcinoma. Breast Cancer (Auckl) 2015;9:13–21. doi: 10.4137/BCBCR.S30101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CS, Yu C, Chuang HC, Chang CW, Chang GD, Yao TP, et al. FBW2 targets GCMa to the ubiquitin-proteasome degradation system. J Biol Chem. 2005;280:10083–90. doi: 10.1074/jbc.M413986200. [DOI] [PubMed] [Google Scholar]
- Yao I, Takagi H, Ageta H, Kahyo T, Sato S, Hatanaka K, et al. SCRAPPER-dependent ubiquitination of active zone protein RIM1 regulates synaptic vesicle release. Cell. 2007;130:943–57. doi: 10.1016/j.cell.2007.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaron A, Hatzubai A, Davis M, Lavon I, Amit S, Manning AM, et al. Identification of the receptor component of the IkappaBalpha-ubiquitin ligase. Nature. 1998;396:590–4. doi: 10.1038/25159. [DOI] [PubMed] [Google Scholar]
- Yeh KH, Kondo T, Zheng J, Tsvetkov LM, Blair J, Zhang H. The F-box protein SKP2 binds to the phosphorylated threonine 380 in cyclin E and regulates ubiquitin-dependent degradation of cyclin E. Biochem Biophys Res Commun. 2001;281:884–90. doi: 10.1006/bbrc.2001.4442. [DOI] [PubMed] [Google Scholar]
- Yoo SH, Mohawk JA, Siepka SM, Shan Y, Huh SK, Hong HK, et al. Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell. 2013;152:1091–105. doi: 10.1016/j.cell.2013.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K. Characterization of estrogen-induced F-box protein FBXO45. Oncol Rep. 2005;14:531–5. [PubMed] [Google Scholar]
- Yu ZK, Gervais JL, Zhang H. Human CUL-1 associates with the SKP1/SKP2 complex and regulates p21(CIP1/WAF1) and cyclin D proteins. Proc Natl Acad Sci U S A. 1998;95:11324–9. doi: 10.1073/pnas.95.19.11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yumimoto K, Muneoka T, Tsuboi T, Nakayama KI. Substrate binding promotes formation of the Skp1-Cul1-Fbxl3 (SCF(Fbxl3)) protein complex. J Biol Chem. 2013;288:32766–76. doi: 10.1074/jbc.M113.511303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachara NE, Hart GW. Cell signaling, the essential role of O-GlcNAc! Biochim Biophys Acta. 2006;1761:599–617. doi: 10.1016/j.bbalip.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Zhang C, Li X, Adelmant G, Dobbins J, Geisen C, Oser MG, et al. Peptidic degron in EID1 is recognized by an SCF E3 ligase complex containing the orphan F-box protein FBXO21. Proc Natl Acad Sci U S A. 2015;112:15372–7. doi: 10.1073/pnas.1522006112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Kong Y, Xu X, Xing H, Zhang Y, Han F, et al. F-box protein FBXO31 is down-regulated in gastric cancer and negatively regulated by miR-17 and miR-20a. Oncotarget. 2014;5:6178–90. doi: 10.18632/oncotarget.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YW, Brognard J, Coughlin C, You Z, Dolled-Filhart M, Aslanian A, et al. The F box protein Fbx6 regulates Chk1 stability and cellular sensitivity to replication stress. Mol Cell. 2009;35:442–53. doi: 10.1016/j.molcel.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–9. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- Zhu J, Deng S, Duan J, Xie X, Xu S, Ran M, et al. FBXL20 acts as an invasion inducer and mediates E-cadherin in colorectal adenocarcinoma. Oncol Lett. 2014;7:2185–91. doi: 10.3892/ol.2014.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Li K, Dong L, Chen Y. Role of FBXL20 in human colorectal adenocarcinoma. Oncol Rep. 2012;28:2290–8. doi: 10.3892/or.2012.2065. [DOI] [PubMed] [Google Scholar]
- Zhuang XW, Li J, Brost BC, Xia XY, Chen HB, Wang CX, et al. Decreased expression and altered methylation of syncytin-1 gene in human placentas associated with preeclampsia. Curr Pharm Des. 2014;20:1796–802. doi: 10.2174/13816128113199990541. [DOI] [PubMed] [Google Scholar]


