Abstract
TEA domain (TEAD) transcription factors bind to the co-activator YAP/TAZ, and regulate the transcriptional output of Hippo pathway, playing critical roles in organ size control and tumorigenesis. Protein S-palmitoylation attaches fatty acid (palmitate) to cysteine residues, and regulates protein trafficking, membrane localization and signaling activities. Using activity-based chemical probes, we discovered that human TEADs possess intrinsic palmitoylating enzyme-like activities, and undergo autopalmitoylation at evolutionarily conserved cysteine residues under physiological conditions. We determined the crystal structures of lipid-bound TEADs, and found that the lipid chain of palmitate inserts into a conserved deep hydrophobic pocket. Strikingly, palmitoylation is required for TEAD’s binding to YAP/TAZ, but dispensable for the binding to Vgll4 tumor suppressor. In addition, palmitoylation does not alter TEAD’s localization. Moreover, TEAD palmitoylation-deficient mutants impaired TAZ-mediated muscle differentiation in vitro, and Yorkie-mediated tissue overgrowth in Drosophila in vivo. Our study directly linked autopalmitoylation to the transcriptional regulation of Hippo pathway.
INSTRODUCTION
Hippo signaling plays key roles in organ size control and tumor suppression1,2. The signal transduction involves a core kinase cascade, including MST1/2 and Lats1/2 kinases, leading to YAP/TAZ phosphorylation, cytoplasmic retention and inhibition3. Physiological or pathological inactivation of these kinases leads to YAP/TAZ dephosphorylation and nuclear accumulation. Subsequently, nuclear YAP/TAZ binds to the TEA domain transcription factors (TEAD1–4 in mammals, and Scalloped in Drosophila) to mediate the target genes expression3,4. The TEAD–YAP complex regulates normal development of skin, muscle, lung and liver, and are also oncogenic factor amplified in many human cancers5,6. TEADs can also bind to Vgll4, which has been implicated as a tumor suppressor by competing with YAP/TAZ for TEADs binding7,8. Therefore, TEADs are essential in regulating the transcriptional output of Hippo pathway. Although targeting TEAD–YAP could be a promising therapeutic approach for diseases with deregulated Hippo pathway9, it remains challenging to directly inhibit transcription factors with small molecules. Therefore, understanding the regulation of TEADs might reveal new therapeutic opportunities for drug discovery.
Post-translational S-palmitoylation attaches a 16-carbon palmitate to the cysteine residue through a reversible thioester bond. A large number of palmitoylated proteins have been identified through proteomic studies10–14. Dynamic S-palmitoylation plays critical roles regulating the trafficking, membrane localization and functions of many proteins, including Src-family kinases, GTPases, and synaptic adhesion molecules15,16. Asp-His-His-Cys (DHHC) family proteins are evolutionarily conserved protein palmitoyl acyltransferases (PATs)10, mediating enzymatic S-palmitoylation17. In addition, some proteins could bind to palmitoyl-Coenzyme A (CoA) directly, and undergo PAT-independent autopalmitoylation18. However, autopalmitoylation is poorly characterized. Most of the reported examples of autopalmitoylation are observed under non-physiological, high concentration of palmitoyl-CoA (>100 µM)19. To date, only a few proteins, including yeast transporter protein Bet3, are autopalmitoylated under physiological concentrations of palmitoyl-CoA (1–10 µM)18,20. Therefore, it is important to reveal additional autopalmitoylated proteins and to understand their regulations and functions.
Towards this end, we have developed activity-based chemical probes based on irreversible inhibitors of PATs, 2-bromopalmitate (2-BP) and cerulenin, which inhibit palmitoylating activities by alkylating the active site cysteines of the enzymes or autopalmitoylated proteins21. We have synthesized analogues of 2-BP and cerulenin with an alkyne tail, which serve as bioorthogonal chemical reporters for covalently labeling and profiling PATs and autopalmitoylated proteins22,23. Through proteomic and biochemical studies, we have identified that the TEAD transcription factors are palmitoylated at evolutionarily conserved cysteine residues. We found that TEADs undergo PAT-independent autopalmitoylation, under physiological concentrations of palmitoyl-CoA. We determined the crystal structures of the lipid-bound TEADs, and revealed a new ligand-binding site in TEADs. Furthermore, autopalmitoylation plays critical roles in regulating TEAD–YAP association and their physiological functions in vitro and in vivo. Therefore, palmitoylation of TEADs plays important roles in regulating Hippo pathway transcriptional complexes.
RESULTS
TEAD transcription factors are palmitoylated
To detect protein palmitoylation, analogues of palmitate, such as 15-hexadecynoic acid (Fig. 1a, 1), have been widely used as chemical reporters to metabolically label palmitoylated proteins (substrates)24,25. To explore PATs and autopalmitoylated proteins, we synthesized the activity-based chemical probes, 2-bromohexadec-15-ynoic acid (2) and cis-2,3-epoxy-4-oxooctadec-17-ynamide (3), respectively22,23. We have performed labeling, enrichment and proteomic analysis of the probe-labeled proteins and found that 2 and 3 can covalently label >300 proteins, including several known PATs and acyltransferases22,23.
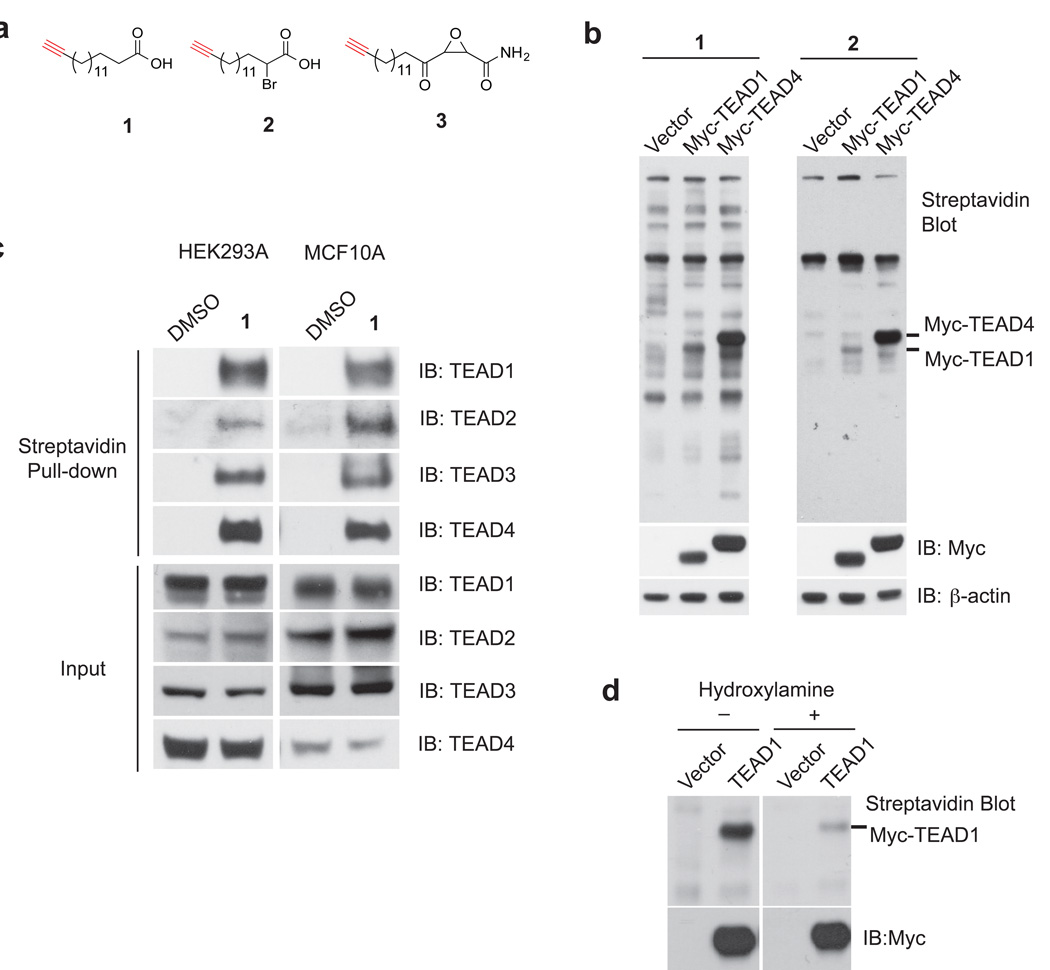
Figure 1. Chemical approaches reveal that TEA domain (TEAD) transcription factors are palmitoylated.
(a) Structures of the chemical reporter of palmitoylation (1), and the activity-based chemical probes for palmitoyl acyltransferases (PATs) and autopalmitoylated proteins (2 and 3).
(b) 1 and 2 labeled myc-TEAD1 and myc-TEAD4 in HEK293A cells. The streptavidin blot showed the palmitoylation of TEADs.
(c) Endogenous human TEAD1– 4 are all palmitoylated. The palmitoylated proteome of HEK293A and MCF10A cells was labeled by 1, and enriched by streptavidin beads. Western blots of TEAD1– 4 were carried out in the pull-down samples using anti-TEAD1, 2, 3, 4 antibodies. See Supplementary Fig. 10 for the full image of the blots.
(d) TEAD1 is S-palmitoylated and hydroxylamine treatment dramatically decreased its palmitoylation levels. See Supplementary Fig. 10 for the full image of the blots.
Among the hits from chemoproteomic studies, we identified the TEA domain (TEAD/TEF) transcription factors (TEAD1 and TEAD3), with multiple unique matching peptides in proteomic studies (Supplementary Results, Supplementary Fig. 1a). TEADs bind to the transcription co-activator YAP/TAZ, and regulate the transcriptional output of Hippo pathway4–6,26, which plays critical roles in organ size control, regeneration and tumorigenesis1. To validate that TEADs are palmitoylated, we transfected Myc-TEAD1 and TEAD4 constructs in HEK293A cells; and then labeled cells with 50 µM of 1 or 2, followed by Cu-catalyzed 1,3-dipolar cycloaddition (Click reaction) with biotin-azide, and detection with streptavidin blots. Myc-TEAD1 and TEAD4 are indeed labeled by both probes (Fig. 1b), suggesting that TEADs are palmitoylated. To characterize whether endogenous TEAD proteins are palmitoylated, HEK293A and MCF10A cells were metabolically labeled with 1, followed by Click reaction with biotin-azide. The palmitoylated proteins were then enriched by streptavidin beads pull-down. We successfully detected all four endogenous human TEADs (TEAD1–4) in the pull-down samples by western blots (Fig. 1c), indicating that they were indeed palmitoylated in cells. TEAD2 and 4 were not among the hits in chemical proteomics studies, possibly due to their low abundance, and our stringent criteria for mass spectra analysis. Nevertheless, our detailed biochemical experiments confirmed that all TEADs should be palmitoylated in cells. Similarly, Drosophila Scalloped protein is palmitoylated (Supplementary Fig. 1b), suggesting that TEAD palmitoylation is evolutionarily conserved. In addition, TEAD1 can also be labeled by 3 (Supplementary Fig. 1c). Furthermore, treatment of hydroxylamine dramatically reduced the palmitoylation levels in TEAD1, suggesting that TEADs are S-palmitoylated through a reversible thioester linkage (Fig. 1d). Taken together, our results have revealed that TEAD family transcription factors are S-palmitoylated.
TEADs are palmitoylated at conserved cysteine residues
To identify the sites of palmitoylation in TEAD, we aligned sequences of TEAD family of proteins across different species, including human, Xenopus, zebra fish, Drosophila, and C. elegans. We found that three cysteine residues are evolutionarily conserved (Supplementary Fig. 2a). We speculated that these conserved cysteine residues might play roles in TEAD palmitoylation. We have mutated these residues to serine in human TEAD1 (C53S, C327S and C359S), and tested whether the mutation affects TEAD1 palmitoylation. C359S mutant showed the greatest loss of palmitoylation, and C327S and C53S also showed decreased palmitoylation (Fig. 2a). These results suggest that C359 plays a critical role in TEAD1 palmitoylation, and might be a major site of modification. Furthermore, combination mutation of all three cysteine residues, C53/327/359S (3CS), completely ablated TEAD1 palmitoylation (Fig. 2b), indicating that these residues are indeed involved for TEAD1 palmitoylation.
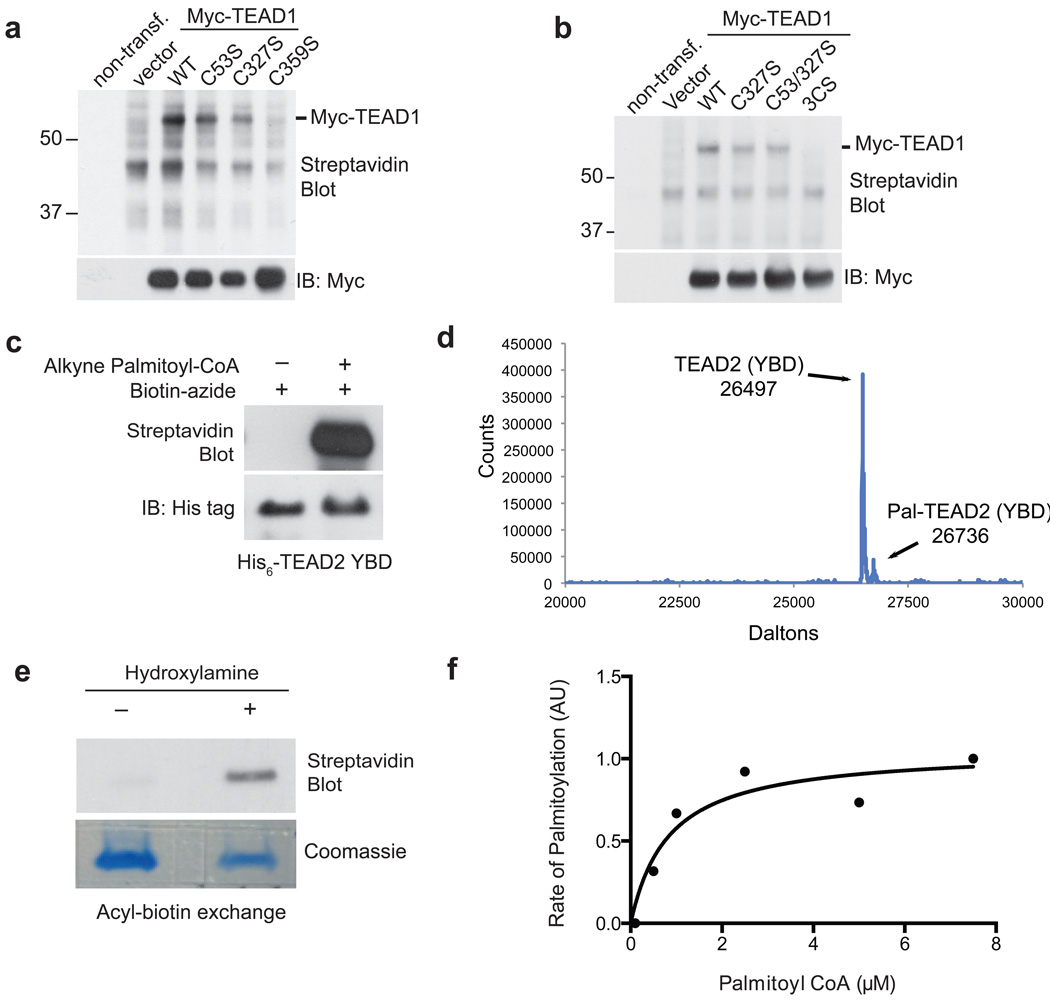
Figure 2. TEAD is autopalmitoylated at evolutionarily conserved cysteine residues under physiological palmitoyl-CoA concentrations.
(a) Mutation of the conserved cysteine residues (C53, C327 and C359) to serine residues individually or in combination (b) blocked palmitoylation of TEAD1. See Supplementary Figure 11 for the full image of the blots.
(c) Recombinant TEAD2 protein (YAP binding domain, YBD) is autopalmitoylated in vitro in the presence of alkyne palmitoyl-CoA. See Supplementary Fig. 11 for the full image of the blots.
(d) Mass spectrometry analysis of recombinant TEAD2 YBD reveals palmitoylation of TEAD2.
(e) Acyl-biotin exchange (ABE) assay confirmed autopalmitoylation of recombinant TEAD2 YBD. See Supplementary Fig. 11 for the full image of the blots.
(f) The Km value of palmitoyl-CoA in TEAD2 autopalmitoylation was estimated by plotting the reaction rate against the substrate concentration.
TEADs undergo PATs-independent autopalmitoylation
Since TEADs could be labeled by Probe 2 and 3 (Fig. 1b, Supplementary Fig. 1c), we hypothesized that TEADs might possess palmitoylating enzyme-like activities and undergo autopalmitoylation. We previously have purified recombinant TEAD2 protein27, allowing us to readily carry out in vitro experiments using TEAD2. We incubated recombinant hTEAD2 (full-length or YAP-binding domain (YBD): TEAD2217–447) with a clickable analogue of palmitoyl-CoA (15-hexadecynoic CoA) at neutral pH in vitro, followed by Click reaction with biotin-azide and streptavidin blot. Both TEAD2 full-length and the YBD are palmitoylated in vitro in the absence of PATs (Fig. 2c, Supplementary Fig. 2b). In addition, overexpression of each of the DHHC-family PATs did not significantly alter the palmitoylation levels of TEAD1 in cells (Supplementary Fig. 2c), confirming that TEAD palmitoylation is independent of PATs. We then carried out intact mass spectrometry analysis of the recombinant TEAD2-YBD. We have identified the peak corresponding to the unmodified TEAD2 (26497 Dalton). Interestingly, we have observed a small side peak (26736 Dalton) (Fig. 2d), consistent with a palmitate modification to the protein. These results suggest that a small fraction of the recombinant TEAD2-YBD is palmitoylated when expressed in bacteria. In addition, after incubating with palmitoyl-CoA in vitro, we observed that the abundance of the palmitoylated TEAD2 peak (26736 Dalton) increased significantly (Supplementary Fig. 3a), further confirming that TEAD2 can be autopalmitoylated. Moreover, autopalmitoylation of TEAD2 YBD was confirmed by acyl-biotin exchange (ABE) assay, which converts S-palmitoylation to stable biotinylation for detection (Fig. 2e). With 1 µM of palmitoyl-CoA at neutral pH, TEAD2 YBD was autopalmitoylated within 2 min, and the palmitoylation levels reached saturation after 10 min (Supplementary Fig. 3b). To determine dose-dependency of palmitoyl-CoA, recombinant TEAD2 YBD was incubated with various concentrations of alkyne palmitoyl-CoA for 3 min, and the reaction rate was determined by quantifying the intensities of streptavidin blots (Supplementary Fig. 3c). We estimated that the apparent Km of palmitoyl-CoA in TEAD2 autopalmitoylation is around 0.8 µM (Fig. 2f), which is comparable to the Km of DHHC-family PATs28. The physiological palmitoyl-CoA concentrations range from 100 nM to 10 µM in cells29. Therefore, our results suggested that TEAD palmitoylation indeed could happen under normal physiological conditions. To the best of our knowledge, TEADs are the first autopalmitoylated transcription factors, linking cellular palmitoyl-CoA levels directly to transcription factor regulation.
Structural analysis of palmitoylation of TEADs
To reveal the structural basis of lipid modification of TEADs, we carried out X-ray crystallography studies of TEAD2 YBD (residue 217–447). We expressed and purified native human TEAD2 YBD from bacteria, and determined its structure to a resolution of 2.0 Å (PDB code 5HGU) by molecular replacement with the selenomethionine-labeled TEAD2 YBD (PDB code 3L15)27 as the search model (Supplementary Table 1). We observed clear extra electron density in a deep hydrophobic pocket adjacent to C380 (corresponding to C359 of TEAD1), indicating that TEAD2 binds to an unknown small molecule ligand. Consistent with our results of TEAD2 palmitoylation by the chemical biology methods and mass spectrometry (Fig. 2d), we found that the extra electron density indeed corresponds to a 16-carbon fatty acid (palmitate, PLM) (Fig. 3a). The lipid chain of palmitate inserts deeply into the pocket, with the free carboxyl group pointing to, but not covalently attached to, C380 of TEAD2. We reasoned that the palmitate might initially be covalently attached to C380, but the labile thioester bond might be cleaved during purification and crystallization under slightly basic conditions. Consistently, surface drawing of TEAD2 reveals that the carboxyl group of palmitate is solvent accessible through an opening adjacent to C380 (Fig. 3b). This opening is also large enough to allow free palmitate to diffuse in and out of the pocket. Interestingly, a recent report of TEAD2 structure using a slightly different purification conditions resulted in higher yield of palmitoylated TEAD2, and the covalent bond can be observed in crystal structures30.
Figure 3. Structures of palmitate-bound human TEAD2 YBD and TEAD1–YAP complex.
The Fo – Fc omit electron density map for TEAD2 (a) and TEAD1–YAP (c) at the contour level of 2.5σ. Palmitate (PLM) is shown as yellow sticks, and surrounding residues are shown as cyan sticks. Palmitate is covalently linked to C359 of TEAD1 (c). Ribbon diagram (left) and electrostatic surface (right) of PLM-bound TEAD2 YBD (PDB code: 5HGU) (b) and TEAD1–YAP complex (d) are shown. TEADs are colored in cyan and YAP is colored in pink. Two conserved cysteine residues are shown. The surface opening in free TEAD2 and the corresponding position in TEAD1–YAP are indicated by red arrow. All structural figures were generated with PyMOL (https://www.pymol.org).
To explore whether covalent palmitoylation could be observed in other TEAD structures. We revisited the previously reported crystal structures of human TEAD1–YAP complex (PDB code 3KYS), mouse TEAD4-YAP (PDB code 3JUA), and human TEAD1-cyclic YAP (PDB code 4RE1)31–33. We have observed that similar lipid-like electron densities are present in all of the conserved deep pocket of these structures. In mTEAD4-YAP (3JUA), the electron density appeared to be covalently connected to C360 of TEAD4. However, the electron densities in 3JUA and 4RE1 are truncated, making it difficult to assign to PLM without prior knowledge of palmitoylation (Supplementary Fig.4a and 4b). The TEAD1-YAP complex (PDB code 3KYS), which was co-expressed in bacteria and purified as a complex, showed the highest quality of electron density in the hydrophobic pocket. We refined the structure, and found that the electron density indeed corresponds to a palmitate, covalently linked to C359 of TEAD1 (Fig. 3c). These results are consistent with our biochemical findings that TEAD1 C359 is palmitoylated. Interestingly, the surface opening observed in TEAD2 alone structure is blocked by the β1 segment of YAP peptide in the TEAD1–YAP complex (Fig. 3d). Therefore, the thioester bond is solvent inaccessible in the complex, together with the mild purification and crystallization conditions, it might help to preserve the covalent linkage. As there are no PATs present in bacteria, these findings also confirmed our results that TEAD1 is autopalmitoylated. Taken together, we have shown that TEADs have a conserved hydrophobic pocket occupied by a palmitate, revealing a new structural feature of these transcription factors. The lipid-binding pocket is highly conserved among other TEADs32. Therefore, palmitate-binding could be an important regulatory mechanism for all TEADs.
The structural studies suggested that TEAD1 C359 (corresponding to TEAD2 C380) palmitoylation is stable and can be crystalized. However, we cannot rule out that C327 (corresponding to TEAD2 C348) is partially or transiently palmitoylated in cells as shown in our in cell labeling experiments. We purified recombinant TEAD2 C380S and C348/380S (2CS) mutant. Consistent with the mutagenesis studies in Fig.2a and 2b, TEAD2 C380S mutant can still be autopalmitoylated in vitro, but TEAD2 2CS mutant cannot (Supplementary Fig. 5). These results suggest that both C348 and C380 are involved palmitoylation, and C380 palmitoylation is more stable.
It has been noted before that TEADs are structurally related to phosphodiesterase δ (PDEδ, PDB code 1KSHB and 3T5I)27,31,32,34, with two β-sheets packing against each other to form a β-sandwich motif. Interestingly, PDEδ has a similar hydrophobic pocket inside the β-sandwich motif, which binds to the farnesyl chain of GTPases35,36 (Supplementary Fig. 6). It is possible that such structural motif represents a common lipid-binding site, and other proteins with similar motif might also bind to lipid ligands. Interestingly, small molecule inhibitors of PDEδ can indeed bind to this pocket, and inhibit the association of PDEδ and farnesylated Ras proteins, leading to inhibition of Ras activities. Therefore, targeting such lipid-binding sites might lead to new small molecule inhibitors of important biological pathways.
Palmitoylation of TEAD regulates TEAD-YAP/TAZ association
Although all 4 TEAD proteins are palmitoylated, we focused on our functional studies using TEAD1, as it is one of the most abundant TEAD proteins ubiquitously expressed. As the palmitoylated cysteine (C359 of TEAD1) is located near the TEAD–YAP interface, we tested whether palmitoylation could allosterically regulate TEAD–YAP association. Indeed, we found that YAP could co-immunoprecipitate (co-IP) with WT TEAD1, but the association was significantly reduced with the palmitoylation-deficient mutants (C359S, C327/359S (2CS) or 3CS) (Fig. 4a). In addition, we tested TEAD–YAP/TAZ interaction using Gal4-TEAD1 or TEAD2 fusion protein, which can activate a Gal4-responsive luciferase reporter upon YAP or TAZ binding26,27,31. We found that Gal4-TEAD1/2 WT can activate the Gal4-repsonsive luciferase in the presence of YAP or TAZ, indicating of forming of active transcription complex. However, the palmitoylation-deficient mutants (C359S, 2CS and 3CS) have significantly reduced activities (Fig. 4b, Supplementary Fig. 7a–b), with TEAD 2CS and 3CS mutant lost most of the activities. Furthermore, a FRET-based binding assay (Alpha Screen) between TEAD1 and YAP also confirmed that TEAD mutant (C359S) had weaker association with YAP, and the palmitoylation-deficient mutants (2CS and 3CS) lost binding to YAP (Fig. 4c). Taken together, our results showed that palmitoylation of TEAD plays important roles in regulating its binding to transcription co-activators. We next examined the functional roles of TEAD palmitoylation. We observed that TEAD1 C359S mutant is partially defective in YAP-induced transcriptional activities. Consistently, TEAD1 2CS or 3CS mutant lost the activities in TEAD-binding element reporter (8×GTIIC-Luc) assays (Fig. 4d)37, suggesting that blocking TEAD palmitoylation impairs its transcriptional activity.
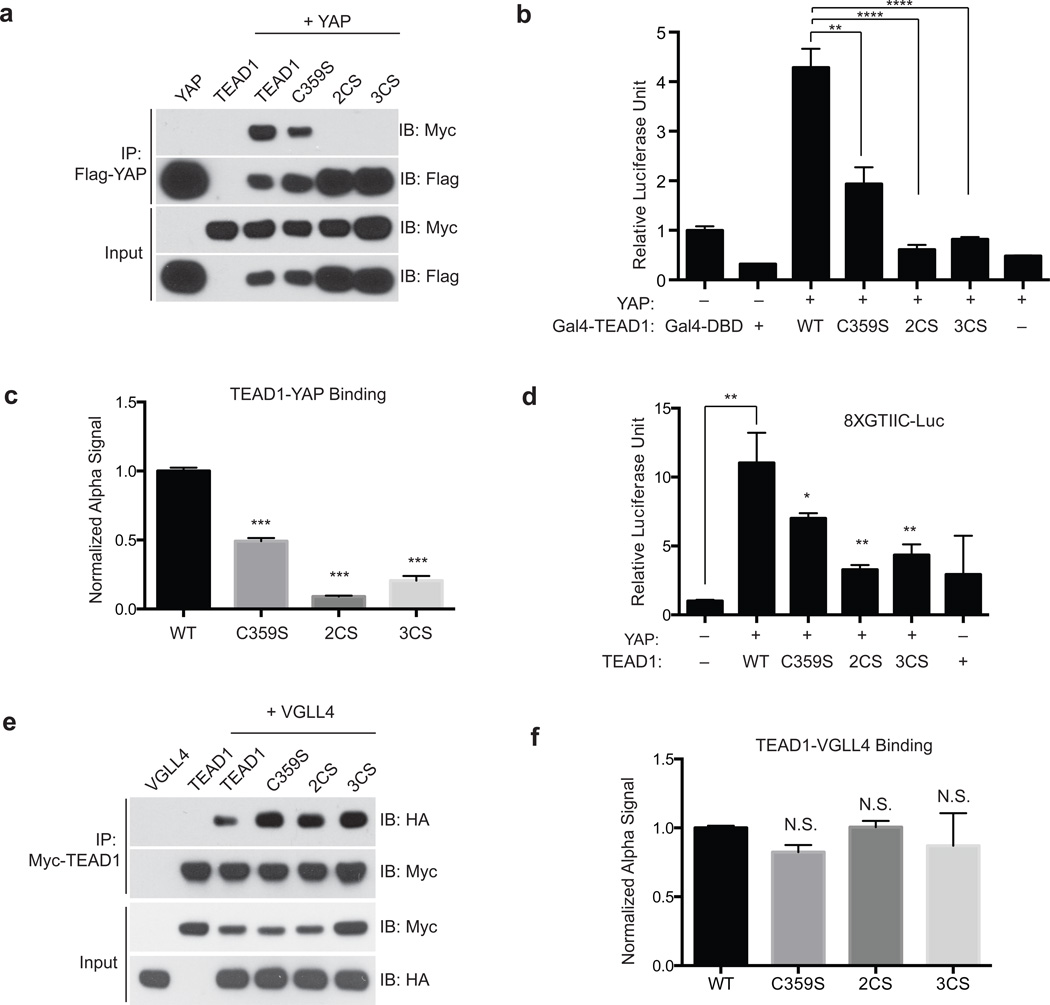
Figure 4. Palmitoylation of TEAD is required for its association with YAP/TAZ.
(a) Palmitoylation-deficient mutants of TEAD1 (C359S, C327/359S (2CS), and 3CS) have decreased association with YAP in co-immunoprecipitation (co-IP) experiments. See Supplementary Fig. 12 for the full image of the blots.
(b) YAP binds to and significantly activates Gal4-TEAD1 wild type (WT) in Gal4-responsive luciferase assay. The palmitoylation-deficient Gal4-TEAD1 mutants (C359S, 2CS and 3CS) significantly inhibits Gal4-responsive luciferase reporter. (Data are represented as mean ± SEM, n=3. P values were determined using two-tailed t-tests. ****, p<0.0001, **, p<0.005).
(c) FRET-based binding assay (Alpha Screen) showed that TEAD1 palmitoylation-deficient mutants (C359S, 2CS and 3CS) have decreased binding to YAP, comparing to TEAD1 WT. (Data are represented as mean ± SEM, n=3. P values were determined using two-tailed t-tests. ***, p<0.0005).
(d) Palmitoylation-deficient mutants of TEAD1 (C359S, 2CS, and 3CS) significantly decreased TEAD transcription activity shown in a TEAD-binding element driven luciferase reporter assay (8XGTIIC-luciferase). (Data are represented as mean ± SEM, n=3. P values were determined using two-tailed t-tests. *, p<0.05; **, p<0.005).
(e) Palmitoylation-deficient mutants of TEAD1 (C359S, 2CS, and 3CS) retain the binding to Vgll4 tumor suppressor in co-immunoprecipitation (co-IP) experiments. See Supplementary Fig. 12 for the full image of the blots.
(f) FRET-based binding assay (Alpha Screen) showed that TEAD1 palmitoylation-deficient mutants (C359S, 2CS and 3CS) and TEAD1 WT bind to Vgll4 similarly. (Data are represented as mean ± SEM, n=3. P values were determined using two-tailed t-tests. N.S., not significant)
In addition, both TEAD1 WT and 3CS mutant localized similarly in the nucleus (Supplementary Fig. 7c–e), suggesting that palmitoylation does not alter TEAD1 localization. These findings were consistent with our observations that palmitate binds to a deep pocket inside of TEAD. Unlike other palmitoylated proteins, palmitate might not serve as a membrane anchor for TEADs. Therefore, our results have uncovered new functions of protein palmitoylation in regulating transcription factor complexes.
Moreover, we found that TEAD2 2CS/3CS mutants were properly folded. It has been reported that TEADs can bind to Vgll4, a tumor suppressor that competes with YAP for TEAD binding, and consequently inhibits YAP oncogenic activity8. In the co-IP assay, we found that TEAD1 WT and the palmitoylation-deficient mutants (C359S, 2CS or 3CS mutants) are able to bind to Vgll4 (Fig. 4e). Consistently, in the FRET-based (Alpha Screen) binding assay, TEAD1 WT, C359S, 2CS and 3CS mutants all bind to Vgll4 similarly (Fig. 4f). Taken together, we showed that palmitoylation is required for TEAD1–YAP binding, but is dispensable for TEAD1–Vgll4 binding. In addition, as TEAD1 C359S, 2CS and 3CS mutants are still capable of binding to Vgll4, the loss of YAP binding are not due to misfolding.
In crystal structures, palmitate does not directly interact with YAP. Therefore, palmitate allosterically regulates YAP binding. It has been shown that YAP binds to TEAD through three interfaces31,32. Mutations of TEAD residues at interface III greatly inhibited YAP, not Vgll4 binding, suggesting interface III is more critical for YAP binding31,32. We hypothesize that palmitoylation allosterically changes the conformation of TEAD at or near interface III, thus regulating YAP binding, but not Vgll4 binding. Our results and a recent report have suggested that binding of palmitate rigidifies the structure of TEAD30. We speculate that it might affect the local side chain dynamics around the binding interface III, which was required for YAP binding. Further structural and protein side-chain dynamic studies using NMR spectrometry will provide more details about how palmitate allosterically regulates TEAD protein dynamics. Interestingly, fatty acylation has been shown before to allosterically regulate protein functions. For example, N-terminal myristoyl modification of c-Abl binds to the kinase domain and induces conformational changes of the protein, resulted in autoinhibition of c-Abl kinase activity38,39.
Palmitoylation regulates TEAD physiological functions
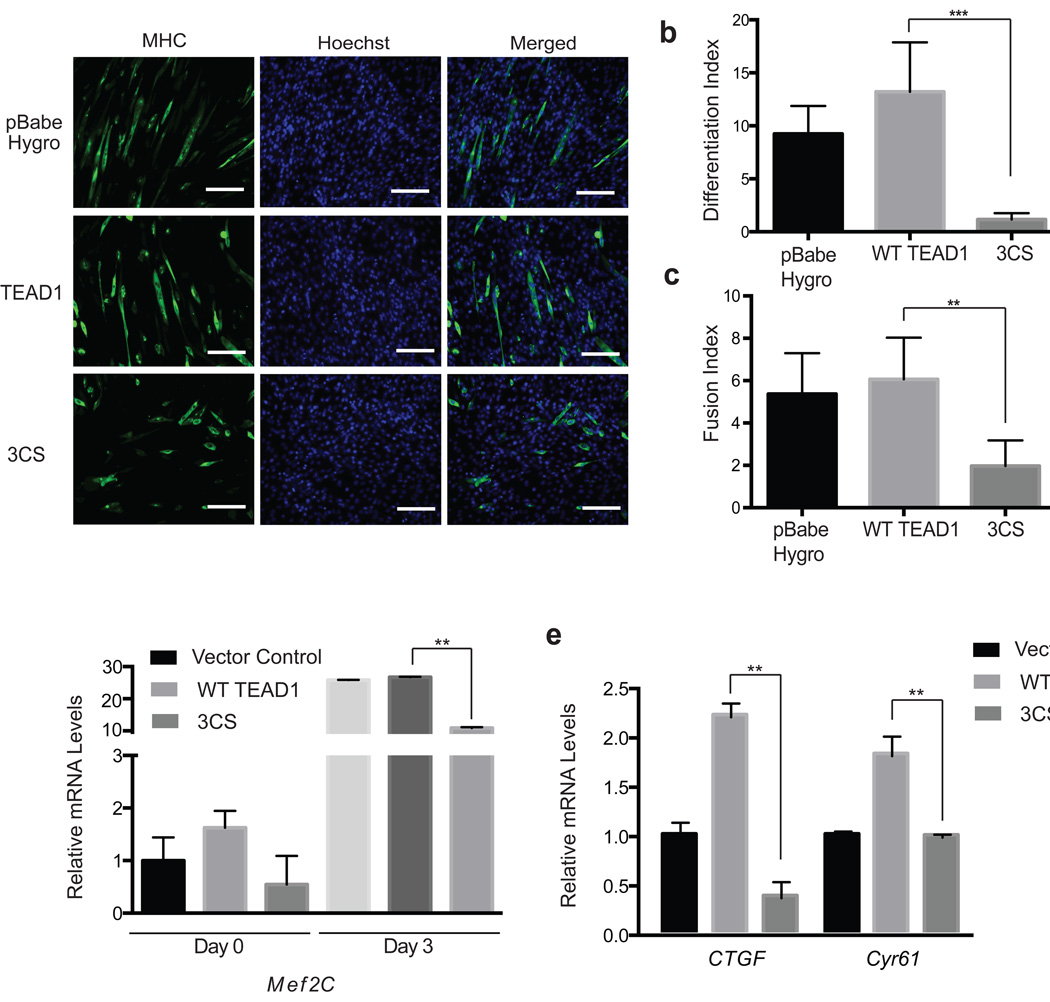
We next examined the physiological roles of TEAD palmitoylation. It has been shown that TAZ promotes terminal differentiation and myotube fusion of skeletal muscle cells through TEAD1 and TEAD440–42. A TEAD4 mutant (TEAD4-DBD), which lacks YAP/TAZ binding domain, functioned as a dominant negative mutant and inhibited C2C12 myoblast differentiation and myotube fusion40. Therefore, TEAD–TAZ association is critical for myogenesis. As TEAD palmitoylation is required for TAZ binding, we speculate that loss of TEAD palmitoylation might impair myogenesis. To test this hypothesis, we stably transfected C2C12 myoblast cells with TEAD1 WT or 3CS mutant, and then induced them to differentiate. We evaluated muscle differentiation by immunostaining of myosin heavy chain (MHC). TEAD1 3CS strongly inhibited muscle differentiation and myotube fusion, compared to vector control and TEAD1 WT (Fig. 5a, Supplementary Fig. 8a). C2C12 cells expressing TEAD1 3CS showed significantly lower differentiation index and fusion index (Fig. 5b–c). In addition, we observed that expression of TEAD1 3CS mutant blocked muscle differentiation gene (Mef2C, MyoG1, Myh4), as well as TEAD-specific target genes (CTGF and Cyr61) expression by qRT-PCR (Fig. 5d–e, Supplementary Fig. 8b–c). Taken together, our results suggested that palmitoylation is required for TEADs’ normal physiological functions in muscle differentiation in vitro.
Figure 5. Palmitoylation regulates TEAD functions in muscle cell differentiation in vitro.
(a) Representative images of myosin heavy chain (MHC, green) immunostaining of C2C12 cells. C2C12 cells stably expressing vector control (pBabe Hygro), TEAD1 WT or TEAD1 3CS mutant, were induced to differentiate for 3 days. Cell nuclei were stained with DAPI (blue). Scale bar: 100µm.
(b–c) TEAD1 3CS mutant significantly inhibited myogenic differentiation and myotube fusion. Differentiation and fusion indices were calculated by averaging the data obtained from five different fields. (Data are represented as mean ± SEM, n=5. P values were determined using two-tailed t-tests. **, p<0.005).
(d–f) TEAD 3CS mutant blocked the expression of myogenic markers Mef2C, and TEAD target genes (CTGF and Cyr61) in C2C12 cell. RNA samples of C2C12 stably expressed vector control, wild type and 3CS mutant of TEAD1 were collected and cDNA of each were synthesized. mRNA levels of each gene were determined by qRT-PCR using SYBR Green and normalized to GAPDH. (Data are represented as mean ± SEM, n=3. P values were determined using two-tailed t-tests. **, p<0.01)
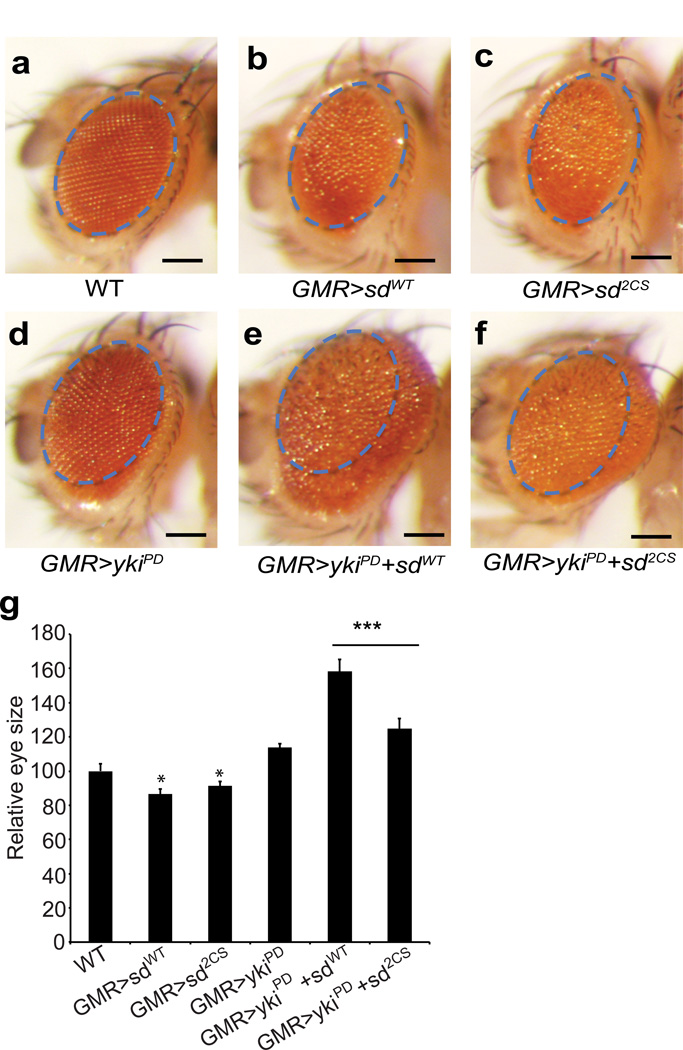
To further corroborate the functional significance of TEAD palmitoylation, we compared the ability of wild type Drosophila Scalloped (Sd) or palmitoylation-deficient (2CS) mutant (both constructs targeted to the same genetic locus to avoid positional effect of transgene insertion) to cooperate with Yorkie (Yki) in promoting tissue overgrowth using a sensitive in vivo assay. Differential splicing of Yki results in two isoforms containing two WW domains (Yki-PG and Yki-PF, Flybase) or a single WW domain (Yki-PD, Flybase). Unlike Yki-PG whose overexpression resulted in eye overgrowth5, overexpression of Yki-PD alone resulted in only slightly bigger eye sizes, but such changes are not statistically significant (Fig. 6a–d). Nevertheless, co-expression of Yki-PD and Sd (WT) caused a significant enlargement of eye size (Fig. 6e), providing a very sensitive assay for Sd–Yki complex in driving tissue overgrowth. Interestingly, this overgrowth phenotype was significantly compromised when Yki-PD was co-expressed with the palmitoylation-deficient Sd (2CS) mutant (Fig. 6f). We have quantified the eye sizes in all the flies, and have performed statistical analysis (Fig. 6g). In addition, the top views of the flies (Supplementary Fig. 9a–f) showed the size of eyes from a different angle. Interestingly, both Sd WT and 2CS mutant have statistically significant reduction of eye growth compared to WT flies (Fig. 6b, c, and g), which is consistent with the default repressor functions of Sd43. The difference between Sd WT and 2CS mutant is not statistically significant. Therefore, it is likely that loss of palmitoylation in Sd (2CS) does not affect its default repressor functions. This result is consistent with our findings in human cells, where TEAD1 (2CS) can still bind to Vgll4. To better evaluate the effects on target genes, we performed qRT-PCR analysis of Diap1 and Expanded in fly S2 cells with the expression of the 2CS mutant or WT Scalloped. Consistently, expression of Yki and WT Scalloped induced the expression of both genes, while expression of Yki and Scalloped 2CS mutant significantly compromised the target gene expression in fly cells (Supplementary Fig. 9g–h). Taken together, our results suggest that palmitoylation is required for the physiological function of the TEAD transcription factors.
Figure 6. Palmitoylation is required for the functions of Drosophila TEAD protein Scalloped (Sd) in vivo.
Images of compound eyes from the following genotypes: (a) GMR-gal4/+, (b) GMR-gal4/UAS-sdWT, (c) GMR-gal4/UAS-sd2CS, (d) GMR-gal4, UAS-ykiPD, (e) GMR-gal4, UAS-ykiPD/UAS-sdWT, (f) GMR-gal4, UAS-ykiPD/UAS-sd2CS. Scale bar: 150µm.
Note the overgrowth phenotype (enlarged eyes with rough surface) caused by co-expression of Yki-PD and Sd (WT) (e) is compromised when Yki-PD is co-expressed with the palmitoylation-deficient Sd (2CS) mutant (f). The images were taken with the same magnification. The size of the eye in wild type control flies is marked in blue dashed line, and the same area is shown in all images to facilitate comparison across all genotypes. (g) Relative sizes of the fly eyes are quantified in indicated genotypes. Sd WT and Sd 2CS flies are compared to the wild type, with statistically smaller eyes. (Data are represented as mean ± SEM, n=10 for each genotype. P values were determined using two-tailed t-tests. *, p<0.05; ***, p<0.001)
DISCUSSION
In summary, using chemical approaches, we have revealed that TEADs are specifically autopalmitoylated at evolutionarily conserved cysteine residues. Autopalmitoylation has been considered as a non-specific reaction of surface cysteine residues with high concentration of palmitoyl-CoA. However, our studies, together with the studies of yeast Bet3 protein, have shown that autopalmitoylation could happen under physiological conditions, with specific cysteine residues being modified. As there are only 23 DHHC-PATs, it is unlikely that they are responsible for all the palmitoylation activities in cells (more than 1000 protein substrates are S-palmitoylated). Therefore, it is possible that many S-palmitoylated proteins are modified through PAT-independent processes, and autopalmitoylation could be an important regulation for protein functions. Our studies have demonstrated for the first time to systematically identify autopalmitoylated proteins using chemical tools.
Palmitoylation has been commonly linked to membrane attachment and protein trafficking10,15. Our results have shown that palmitate binds to a hydrophobic pocket in the core of the protein, and does not regulate protein membrane binding. It has been noted that in the crystal structures of yeast Bet3 protein, the covalently attached palmitate also binds into a hydrophobic pocket in the protein44. Palmitoylation of Bet3 stabilizes the protein and is involved in regulating Bet3 degradation and co-factor binding18. Therefore, in addition to acting as a membrane-binding moiety, palmitoylation of proteins indeed has other important functions. Further studies of additional autopalmitoylated proteins will likely reveal new functions of protein palmitoylation.
We have observed that TEAD1 C359 is the major and stable site of modification, which is located at the opening of the lipid-binding pocket. We could not purify and crystalize palmitate-free TEAD2, and it is likely that binding of palmitate stabilized the conformation of TEAD, allowing the protein to be crystalized. Nevertheless, TEAD proteins might exist as palmitoylated and non-palmitoylated species in cells. We have also observed that TEAD2 C380S remains autopalmitoylated, but not the C348/380S mutant, consistent with the observation that TEAD 2CS/3CS mutant has more significant loss of activity than C380S. Although we did not observe the lipid modification of C348 in the crystal structures, which are only snapshots of the most stable conformations of the protein, both C348 and C380 should be involved in palmitoylation. It is possible that C348-palmitoylated TEAD2 has a different conformation, allowing palmitate to bind to the conserved deep pocket. In addition, another hydrophobic pocket near the surface is close to C348 in TEAD2 structure, which could accommodate the binding of hydrophobic ligands, such as bromofenamic acid (BFA)45. Further studies would be needed to reveal the detailed structures of C348-palmitoylated TEAD2.
As the levels of TEAD autopalmitoylation are highly relevant to the palmitoyl-CoA concentrations in cells. The cellular palmitoyl-CoA pool might be an upstream regulator of TEAD’s activities and Hippo pathway. Fatty acid synthase (FASN) is the key enzyme that synthesizes palmitoyl-CoA from acetyl-CoA and malonyl-CoA46. FASN has been proposed as a potential oncogene, which is upregulated in breast cancers and its expression is associated with poor prognosis46. High level of FASN might lead to high intracellular palmitoyl-CoA, thus promoting TEAD–YAP mediated oncogenic processes. Further studies would be needed to test whether TEAD–YAP activities are responsible for tumorigenesis in FASN-overexpressed cancer cells. Currently, we don’t have evidence that TEAD proteins can be palmitoylated and depalmitoylated in a dynamic fashion. Two potential depalmitoylating enzyme families, acylprotein thioesterases (APT1/2) and protein palmitoylthioesterases (PPT1/2) have been reported47,48. It would be interesting to investigate whether these enzymes contribute to TEAD depalmitoylation.
It remains challenging to develop potent and selective small molecule inhibitors to disrupt TEAD-YAP interaction, as the interaction interface is shallow and spans over a large area on the surface. Our results showed that the palmitate-binding pocket of TEADs is deep and hydrophobic, ideal for inhibitor binding. Indeed, a recent study has shown that this pocket is accessible by small molecules, although their potency and selectivity are not optimal45. Taken together, targeting autopalmitoylation of TEADs by small molecules could be a new strategy for drug discovery.
ONLINE METHODS
Labeling, Click reactions and streptavidin pull-down
HEK293A or MCF10A cells were labeled with DMSO or probe 1, 2 or 3 overnight. Cells were lysed in lysis buffer (50 mM TEA-HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.2% SDS, cOmplete EDTA-free protease inhibitors) followed by Click reaction with biotin-azide22. Proteins were precipitated with 9 volumes of 100% methanol for 2 h or overnight at −20°C. Proteins were recovered by centrifugation at 14,000 × g for 10 min and the precipitants were suspended in suspension buffer (50 mM Tris-HCl, pH 7.7, 150 mM NaCl, 10 mM EDTA, 1% SDS, 0.5% NP-40). Labeled cellular proteins were enriched using streptavidin agarose (Life technologies) at room temperature with rotation overnight. Protein-bound streptavidin agarose beads were washed three times with suspension buffer without NP-40 and bound proteins were eluted with elution buffer (30 mM D-Biotin, 2% SDS, 6M Urea). Samples were processed with SDS-PAGE sample buffer and proteins were resolved by SDS-PAGE. TEAD 1 – 4 in these samples were detected using TEAD-specific antibodies and streptavidin HRP. Blots were probes with anti-TEAD1 (#8526, 1:1000, Cell Signaling), anti-TEAD2 (#8870, 1:1000, Cell Signaling), anti-TEAD3 (#13224, 1:1000, Cell Signaling), anti-TEAD4 (ab58310, 1:1000, Abcam) and Streptavidin HRP (1:5000, Life technologies).
Cell culture
HEK293A, Phoenix, MCF10A and C2C12 cell lines (obtained from ATCC, Manassas, VA) were grown at 37°C with 5% CO2. HEK293A, Phoenix, and C2C12 cell lines were cultured in Dulbecco’s modified Eagles media (DMEM) (Life technologies) supplemented with 10% fetal bovine serum (FBS) (Thermo/Hyclone, Waltham, MA) and 50µg/mL penicillin/streptomycin. MCF10A cells were cultured in DMEM/F12 (Life technologies) supplemented with 5% horse serum, 20 ng/mL EGF, 0.5 µg/mL hydrocortisone, 100 ng/mL cholera toxin, 10 µg/mL insulin and 50 µg/mL penicillin/streptomycin. None of cell lines used in this paper listed in the database of commonly misidentified cell lines maintained by ICLAC. All cell lines are free of mycoplasma contamination.
Transfection and transduction
Plasmids were transfected with jetPRIME (Polyplus transfection) or XtremeGene HP (Roche) according to the manufacturer’s instructions.
For retrovirus production, Phoenix cells were transfected with VSV-G and empty pBabe hygro or pBabe hygro containing TEAD1 wild type or 3CS mutant cDNA. Supernatants were collected by centrifugation and filtered through a 0.45 µm syringe filter (Corning) 48 h post-transfection. Cells were infected with 2 mL viral supernatant in the presence of 10 µg/mL polybrene (Millipore). Cells were incubated for 24 – 48 h before splitting into selection medium.
Site-directed mutagenesis
Mutagenesis was performed using QuikChange II Site-Directed Mutagenesis kit (Agilent) following manufacturer’s instructions.
Co-immunoprecipitation
HEK-293A cells were transfected with the indicated constructs. After 48 h, cells were lysed with lysis buffer (50 mM Tris-HCl, pH 7.3, 150 mM NaCl, 0.5 mM EDTA, 1% Triton X-100, PhosSTOP phosphatase inhibitor cocktail, cOmplete EDTA-free protease inhibitors cocktail). Flag-YAP or Myc-TEAD1 was immunoprecipitated with anti-FLAG M2 magnetic beads (Sigma Aldrich) or anti-c-Myc antibody (M4439, Sigma Aldrich), respectively, overnight with rotation at 4°C. TEAD1 was captured using Protein A/G magnetic resins (Life technologies). Protein-bound resins were washed three times with lysis buffer and processed with SDS-PAGE sample buffer. Blots were probed with anti-c-Myc (Sigma Aldrich), anti-HA (Sigma Aldrich), anti-FLAG M2 (F1804, Sigma Aldrich).
FRET-based Alpha screen binding assay
Myc-TEAD1 and Flag-YAP or Flag-VGLL4 were transfected into HEK293A cells and 24 – 48 h post-transfection, cells were lysed with lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, PhosSTOP phosphatase inhibitor cocktail, cOmplete EDTA-free protease inhibitor). Anti-c-myc acceptor beads (Perkin Elmer) were added to each well and incubated for 2 h prior to addition of anti-FLAG donor beads (Perkin Elmer). Samples were incubated overnight in darkness and Alpha signals were recorded using Perkin Elmer EnVision plate reader.
Luciferase assay
Gal-UAS-Luc, YAP, Gal4-TEAD1, Gal4-DBD or Myc-TEAD1 and Renilla luciferase control constructs were transfected into 293T cells and 48 h post-transfection, cells were processed using the Dual-Glo luciferase assay system (Promega) following manufacturer’s instructions. Luminescence of Firefly and Renilla luciferase activities were quantified using Perkin Elmer EnVision plate reader.
Ni-NTA pull-down and acyl-biotin exchange
Recombinant His6TEAD2 was incubated with Ni-NTA resin (Life technologies) in PBS for 1 h at 4°C. Protein-bound resins were washed and then incubated with 50 µM alkyne palmitoyl-CoA for 2 h at 25°C. Resins were split into two equivalent reactions, washed with PBS and treated with 50 mM NEM (Thermo Scientific) overnight at 4°C. Samples were incubated with or without 0.5 M hydroxylamine (Sigma Aldrich) for 1 h at room temperature and then incubated with 1 µM Biotin-BMCC (Pierce Biotechnology, Inc.) for 1 h at room temperature. Samples were washed and processed with SDS-sample buffer. Proteins were resolved by SDS-PAGE and visualized by immunoblotting with streptavidin HRP, anti-His antibody (SAB1306085, Sigma Aldrich) or Coomassie blue staining.
C2C12 cells differentiation
C2C12 cells were transduced using retrovirus containing vector control (pBabe hygro), wild type or 3CS TEAD1. Stable expression was selected initially using 600 µg/mL Hygromycin B (Life technologies) and then decreased to 300 µg/mL for 2 weeks. To induce differentiation, the culture condition was replaced by differentiation medium (DMEM + 2% horse serum + 50 µg/mL penicillin/streptomycin) with medium change everyday.
Immunofluorescence
Cells were fixed with 4% paraformaldehyde and then permeabilized and blocked with 3% (w/v) BSA/PBS + 0.1% Triton X-100 at room temperature for 30 min. Cells were immunostained with anti-myosin (skeletal, fast) chain (M4276, 1:400, Sigma Aldrich), anti-Yap (1:1000, Abgent) or anti-c-myc (1:500, Sigma Aldrich) antibody overnight at 4°C. Cells were washed three times with PBS + 0.1% Triton X-100 followed by incubation with Alexafluor 488 conjugated anti-mouse secondary antibody (1:500, Life technologies) and Hoechst 33258 (1:2500, Life technologies) at room temperature for 2 h. Cells were washed again and images were captured using Nikon Digital Sight microscope.
Drosophila Genetics
UAS-ykiPD construct was generated by cloning the yki single WW domain isoform (Yki-PD) cDNA into the pUAST vector. UAS-sdWT and UAS-sd2CS constructs were generated by cloning wild-type scalloped (sd) or sd palmitoylation-deficient (2CS) mutant cDNA into the pUAST-attB vectors. UAS-ykiPD transgenic fly was created by conventional transposon-mediated transformation. UAS-sdWT and UAS-sd2CS transgenic flies were created by phiC31-mediated site-specific transformation, using the attP2 site at 51C. GMR-Gal4 was used for overexpression analysis. All crosses were done at 25°C. The quantification of fly eyes were carried out by analyzing the eye area in the images49, and normalized to the control wild type flies. n=10 for each genotypes. The qRT-PCR analysis of Diap-1 and Expanded was carried out using primer sequences previously reported49.
Protein Purification and Crystallization
The cDNA encoding human TEAD2 (residues 217–447, TEAD2217–447) was cloned into a pET29 vector (EMD Biosciences) that included a C-terminal His6-tag. The construct was verified by DNA sequencing. The pET29-TEAD2217–447 plasmid was transformed into the E. coli strain BL21(DE3)-T1R cells (Sigma) for protein expression. His6-tagged TEAD2217–447 was purified with Ni2+-NTA agarose resin (Qiagen) and then purified by anion exchange chromatography with a resource-Q column followed by size exclusion chromatography with a Superdex 75 column (GE Healthcare). Purified TEAD2217–447 was concentrated to 4 mg/ml in a buffer containing 20 mM Tris (pH 8.0), 100 mM NaCl, 2 mM MgCl2, 1 mM TCEP and 5% glycerol.
Crystals of TEAD2217–447 were grown at 20°C using the hanging-drop vapor-diffusion method with a reservoir solution containing 0.1 M Hepes (pH 7.2) and 2.4 M sodium formate. The crystals were cryo-protected with reservoir solution supplemented with 25% glycerol and then flash-cooled in liquid nitrogen.
In vitro Palmitoylation
Recombinant GST-TEAD2 or His6TEAD2 (500 ng) protein was incubated with the indicated concentrations of alkyne palmitoyl-CoA (Cayman Chemical) for 2 h or the indicated time in 50 mM MES, pH 6.4. The reaction was quenched with 1% SDS followed by Click reaction as described previously. Samples were analyzed by SDS-PAGE and streptavidin HRP. Bands intensity obtained from streptavidin blot were quantified using Image J (NIH) and the rate of palmitoylation in arbitrary unit was plotted against the concentration of palmitoyl-CoA. The data was fitted to the Michaelis-Menten equation using Prism v.6.0 (GraphPad). For mass spectrometry analysis, recombinant TEAD2 YBD (1 mg/ml) was incubated with 1 eq. of palmitoyl-CoA for 30 min at room temperature in a buffer containing 50 mM MES, pH 6.4.
Data Collection and Structure Determination
Diffraction data were collected at beamline 19-ID (SBC-CAT) at the Advanced Photon Source (Argonne National Laboratory) at the wavelength of 0.9791 Å at 100 K and processed with HKL3000. Phases were obtained by molecular replacement with Phaser using the crystal structure of human TEAD2 (PDB code: 3L15) as the search model. Iterative model building and refinements were carried out with COOT and Phenix, respectively. MolProbity was used for structure validation to show that all models have good geometry. Data collection and structure refinement statistics are summarized in Supplementary Table 1. Ramachandran statistics (Favored/allowed/outlier (%)) are 97.4/2.6/0.0. The crystal structure of palmitate-bound TEAD1–YAP was obtained by building two thioester-linked palmitate molecules into TEAD1–YAP (PDB code: 3KYS) with COOT using the electron density map calculated from 3KYS structure factor. The final model was refined with Phenix.
Statistical Analysis
No statistical method was used to predetermine sample size. The experiments were not randomized. For biochemical experiments we performed the experiments at least three independent times. Experiments for which we showed representative images were performed successfully at least 3 independent times. No samples or animal were excluded from the analysis. The investigators were not blinded to allocation during experiments and outcome assessment. All P values were determined using two-tailed t-tests and statistical significance was set at P = 0.05. The variance was similar between groups that we compared.
Database
PDB: 5HGU
Supplementary Material
Acknowledgments
This work was supported by Stewart Rahr-MRA (Melanoma Research Alliance) Young Investigator Award, Department of Defense (DoD) Career Development Award (W81XWH-13-1-0203), American Cancer Society (124929-RSG-13-291-01-TBE), and NIH/NCI R01CA181537 (to X.W.), NIH/NIDDK R01DK107651-01 (to X.W. and X. L.) and NIH/NIGMS R01GM107415 (to X. L.); D.P. is supported by Howard Hughes Medical Institute. We thank Drs. Nabeel Bardeesy, Masaki Fukata, Kun-Liang Guan, and Kristin White for constructs and cell lines; Drs. Jing-Ruey J. Yeh, Hongtao Yu and Nathanael Gray for discussion and critical comments of the manuscript; the Taplin Mass Spec Core facility at Harvard Medical School and the Proteomics Core at UTSW for proteomic studies. Use of the Argonne National Laboratory Structural Biology Center beamlines at the Advanced Photon Source was supported by the US DOE under contract DE-AC02-06CH11357.
Footnotes
Accession codes
Protein Data Bank (PDB): coordinates for the TEAD2-PLM complex have been deposited with accession code 5HGU.
Author contributions
X.W conceived the concepts, designed the experiments and supervised the studies. P.C. designed and performed the cell biology and biochemistry experiments with the help of M.D. X.H. performed protein purification, crystallization, and structure determination, and carried out mass spectrometry analysis of TEAD2 autopalmitoylation. B.Z. and G.J. synthesized the probes. B.Z. identified TEAD from mass spec studies and tested DHHC-family of PATs. J.Y, H.D, and D.P carried out Drosophila genetics experiments. X.L. contributed to experimental design and structure refinements of palmitate-bound TEAD2 and TEAD1-YAP complex. P.C., D.P, X.L and X.W. analyzed the data; P.C, D.P. X.L and X.W wrote the manuscript with input from all co-authors.
Competing financial interests
The authors declare no competing financial interests.
Any supplementary information, chemical compound information and source data are available in the online version of the paper.
REFERENCES
- 1.Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nature reviews. Cancer. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 2.Pan D. Hippo signaling in organ size control. Genes Dev. 2007;21:886–897. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]
- 3.Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ota M, Sasaki H. Mammalian Tead proteins regulate cell proliferation and contact inhibition as transcriptional mediators of Hippo signaling. Development. 2008;135:4059–4069. doi: 10.1242/dev.027151. [DOI] [PubMed] [Google Scholar]
- 5.Wu S, Liu Y, Zheng Y, Dong J, Pan D. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev Cell. 2008;14:388–398. doi: 10.1016/j.devcel.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 6.Zhao B, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes & development. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang W, et al. VGLL4 functions as a new tumor suppressor in lung cancer by negatively regulating the YAP-TEAD transcriptional complex. Cell Res. 2014;24:331–343. doi: 10.1038/cr.2014.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiao S, et al. A peptide mimicking VGLL4 function acts as a YAP antagonist therapy against gastric cancer. Cancer Cell. 2014;25:166–180. doi: 10.1016/j.ccr.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 9.Johnson R, Halder G. The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nature reviews. Drug discovery. 2014;13:63–79. doi: 10.1038/nrd4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smotrys JE, Linder ME. Palmitoylation of intracellular signaling proteins: regulation and function. Annu Rev Biochem. 2004;73:559–587. doi: 10.1146/annurev.biochem.73.011303.073954. [DOI] [PubMed] [Google Scholar]
- 11.Roth AF, et al. Global analysis of protein palmitoylation in yeast. Cell. 2006;125:1003–1013. doi: 10.1016/j.cell.2006.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wan J, Roth AF, Bailey AO, Davis NG. Palmitoylated proteins: purification and identification. Nat Protoc. 2007;2:1573–1584. doi: 10.1038/nprot.2007.225. [DOI] [PubMed] [Google Scholar]
- 13.Martin BR, Cravatt BF. Large-scale profiling of protein palmitoylation in mammalian cells. Nat Methods. 2009;6:135–138. doi: 10.1038/nmeth.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yount JS, et al. Palmitoylome profiling reveals S-palmitoylation-dependent antiviral activity of IFITM3. Nat Chem Biol. 2010;6:610–614. doi: 10.1038/nchembio.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Resh MD. Trafficking and signaling by fatty-acylated and prenylated proteins. Nat Chem Biol. 2006;2:584–590. doi: 10.1038/nchembio834. [DOI] [PubMed] [Google Scholar]
- 16.Fukata Y, Fukata M. Protein palmitoylation in neuronal development and synaptic plasticity. Nat Rev Neurosci. 2010;11:161–175. doi: 10.1038/nrn2788. [DOI] [PubMed] [Google Scholar]
- 17.Greaves J, Chamberlain LH. DHHC palmitoyl transferases: substrate interactions and (patho)physiology. Trends Biochem Sci. 2011;36:245–253. doi: 10.1016/j.tibs.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 18.Kummel D, Heinemann U, Veit M. Unique self-palmitoylation activity of the transport protein particle component Bet3: a mechanism required for protein stability. Proc Natl Acad Sci U S A. 2006;103:12701–12706. doi: 10.1073/pnas.0603513103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duncan JA, Gilman AG. Autoacylation of G protein alpha subunits. J Biol Chem. 1996;271:23594–23600. doi: 10.1074/jbc.271.38.23594. [DOI] [PubMed] [Google Scholar]
- 20.Yang J, et al. Submicromolar concentrations of palmitoyl-CoA specifically thioesterify cysteine 244 in glyceraldehyde-3-phosphate dehydrogenase inhibiting enzyme activity: a novel mechanism potentially underlying fatty acid induced insulin resistance. Biochemistry. 2005;44:11903–11912. doi: 10.1021/bi0508082. [DOI] [PubMed] [Google Scholar]
- 21.Resh MD. Use of analogs and inhibitors to study the functional significance of protein palmitoylation. Methods. 2006;40:191–197. doi: 10.1016/j.ymeth.2006.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng B, et al. 2-Bromopalmitate analogues as activity-based probes to explore palmitoyl acyltransferases. J Am Chem Soc. 2013;135:7082–7085. doi: 10.1021/ja311416v. [DOI] [PubMed] [Google Scholar]
- 23.Zheng B, Zhu S, Wu X. Clickable analogue of cerulenin as chemical probe to explore protein palmitoylation. ACS Chem Biol. 2015;10:115–121. doi: 10.1021/cb500758s. [DOI] [PubMed] [Google Scholar]
- 24.Hannoush RN. Profiling cellular myristoylation and palmitoylation using omega-alkynyl fatty acids. Methods Mol Biol. 2012;800:85–94. doi: 10.1007/978-1-61779-349-3_7. [DOI] [PubMed] [Google Scholar]
- 25.Hang HC, Linder ME. Exploring protein lipidation with chemical biology. Chem Rev. 2011;111:6341–6358. doi: 10.1021/cr2001977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu-Chittenden Y, et al. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes & development. 2012;26:1300–1305. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tian W, Yu J, Tomchick DR, Pan D, Luo X. Structural and functional analysis of the YAP-binding domain of human TEAD2. Proc Natl Acad Sci U S A. 2010;107:7293–7298. doi: 10.1073/pnas.1000293107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jennings BC, Linder ME. DHHC protein S-acyltransferases use similar ping-pong kinetic mechanisms but display different acyl-CoA specificities. J Biol Chem. 2012;287:7236–7245. doi: 10.1074/jbc.M111.337246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Faergeman NJ, Knudsen J. Role of long-chain fatty acyl-CoA esters in the regulation of metabolism and in cell signalling. Biochem J. 1997;323(Pt1):1–12. doi: 10.1042/bj3230001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noland CL, et al. Palmitoylation of TEAD Transcription Factors Is Required for Their Stability and Function in Hippo Pathway Signaling. Structure. 2016;24:1–8. doi: 10.1016/j.str.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 31.Li Z, et al. Structural insights into the YAP and TEAD complex. Genes & development. 2010;24:235–240. doi: 10.1101/gad.1865810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen L, et al. Structural basis of YAP recognition by TEAD4 in the hippo pathway. Genes Dev. 2010;24:290–300. doi: 10.1101/gad.1865310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou Z, et al. Targeting Hippo pathway by specific interruption of YAP-TEAD interaction using cyclic YAP-like peptides. Faseb J. 2015;29:724–732. doi: 10.1096/fj.14-262980. [DOI] [PubMed] [Google Scholar]
- 34.Ismail SA, et al. Arl2-GTP and Arl3-GTP regulate a GDI-like transport system for farnesylated cargo. Nat Chem Biol. 2011;7:942–949. doi: 10.1038/nchembio.686. [DOI] [PubMed] [Google Scholar]
- 35.Zhang H, et al. Photoreceptor cGMP phosphodiesterase delta subunit (PDEdelta) functions as a prenyl-binding protein. J Biol Chem. 2004;279:407–413. doi: 10.1074/jbc.M306559200. [DOI] [PubMed] [Google Scholar]
- 36.Chandra A, et al. The GDI-like solubilizing factor PDEdelta sustains the spatial organization and signalling of Ras family proteins. Nat Cell Biol. 2012;14:148–158. doi: 10.1038/ncb2394. [DOI] [PubMed] [Google Scholar]
- 37.Dupont S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 38.Nagar B, et al. Structural basis for the autoinhibition of c-Abl tyrosine kinase. Cell. 2003;112:859–871. doi: 10.1016/s0092-8674(03)00194-6. [DOI] [PubMed] [Google Scholar]
- 39.Hantschel O, et al. A myristoyl/phosphotyrosine switch regulates c-Abl. Cell. 2003;112:845–857. doi: 10.1016/s0092-8674(03)00191-0. [DOI] [PubMed] [Google Scholar]
- 40.Benhaddou A, et al. Transcription factor TEAD4 regulates expression of myogenin and the unfolded protein response genes during C2C12 cell differentiation. Cell Death Differ. 2012;19:220–231. doi: 10.1038/cdd.2011.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang Z, et al. Screening with a novel cell-based assay for TAZ activators identifies a compound that enhances myogenesis in C2C12 cells and facilitates muscle repair in a muscle injury model. Mol Cell Biol. 2014;34:1607–1621. doi: 10.1128/MCB.01346-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park GH, et al. Novel TAZ modulators enhance myogenic differentiation and muscle regeneration. Br J Pharmacol. 2014;171:4051–4061. doi: 10.1111/bph.12755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koontz LM, et al. The Hippo effector Yorkie controls normal tissue growth by antagonizing scalloped-mediated default repression. Dev Cell. 2013;25:388–401. doi: 10.1016/j.devcel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turnbull AP, et al. Structure of palmitoylated BET3: insights into TRAPP complex assembly and membrane localization. Embo J. 2005;24:875–884. doi: 10.1038/sj.emboj.7600565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pobbati AV, et al. Targeting the Central Pocket in Human Transcription Factor TEAD as a Potential Cancer Therapeutic Strategy. Structure. 2015;23:2076–2086. doi: 10.1016/j.str.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 2007;7:763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- 47.Wan J, Roth AF, Bailey AO, Davis NG. Palmitoylated proteins: purification and identification. Nature protocols. 2007;2:1573–1584. doi: 10.1038/nprot.2007.225. [DOI] [PubMed] [Google Scholar]
- 48.Fukata Y, Fukata M. Protein palmitoylation in neuronal development and synaptic plasticity. Nature reviews. Neuroscience. 2010;11:161–175. doi: 10.1038/nrn2788. [DOI] [PubMed] [Google Scholar]
- 49.Sorrentino G, et al. Metabolic control of YAP and TAZ by the mevalonate pathway. Nat Cell Biol. 2014;16:357–366. doi: 10.1038/ncb2936. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.