Abstract
Allostery provides a critical control over enzyme activity, biasing the catalytic site between inactive and active states. We find the Ciona intestinalis voltage-sensing phosphatase (Ci-VSP), which modifies phosphoinositide signaling lipids (PIPs), to have not one but two sequential active states with distinct substrate specificities, whose occupancy is allosterically controlled by sequential conformations of the voltage sensing domain (VSD). Using fast FRET reporters of PIPs to monitor enzyme activity and voltage clamp fluorometry to monitor conformational changes in the VSD, we find that Ci-VSP switches from inactive to a PIP3-preferring active state when the VSD undergoes an initial voltage sensing motion and then into a second PIP2-preferring active state when the VSD activates fully. This novel 2-step allosteric control over a dual specificity enzyme enables voltage to shape PIP concentrations in time, and provides a mechanism for the complex modulation of PIP-regulated ion channels, transporters, cell motility and endo/exocytosis.
Introduction
The allosteric regulation of an enzyme active site by the binding of a ligand, or another signal transduction event, at a different, non-overlapping site is a fundamental mechanism for feedback and feedforward regulation of cells1–5. Classically, allostery regulates the activity of the enzyme on a fixed set of substrates4,5. In the present study of the Ciona intestinalis voltage sensing phosphatase (Ci-VSP), we find that allosteric control can operate in more than one step – an initial step turns the enzyme on to one substrate-specific state and then a separate step switches the substrate specificity. This two-step sequence of enzyme activation and substrate switching is associated with a two-step sequence of conformational rearrangements in the allosteric regulatory domain.
VSPs are membrane-associated lipid phosphatases that dephosphorylate PIP signaling lipids in response to changes in membrane potential, thereby providing a mechanism for coupling the electrical activity of a cell to intracellular signaling6–8. Ci-VSP is a monomeric protein that contains a transmembrane VSD coupled to a cytosolic lipid phosphatase domain through a 16 amino-acid linker9–15. Although the phosphatase domain of Ci-VSP shares 44% identity with the catalytic domain of tumor suppressor protein ‘phosphatase and tensin homologue deleted on chromosome 10′ (PTEN), the enzymes exhibit different substrate preferences16–19. PTEN hydrolyzes phosphates from the 3-position of PI(3,4,5)P3 and PI(3,4)P2, but Ci-VSP removes both 3-phosphate from PI(3,4,5)P3 and PI(3,4)P2 as well as 5-phosphate from PI(3,4,5)P3 and PI(4,5)P220–22. Attempts to introduce PTEN specificity for 3-phosphate into VSP based on homology models had only partial success23,24, suggesting another cause of the broader substrate set of VSP. A possible clue came from the apparent separation in the voltage dependence of PIP2 production and destruction, but it was not clear whether this represented voltage regulation of enzyme activity or a differential effect of the electric field on substrates of different charge22. The notion that distinct conformations of the voltage sensor could place the enzyme domain into distinct and corresponding active states seemed both intriguing and plausible, given that VSDs of voltage-gated potassium channels and Ci-VSP transition between multiple conformations25–27, and a mutated zebrafish VSP appears to operate at different rates over voltage ranges corresponding to two phases of gating charge motion30.
Earlier studies on VSP activity relied primarily on either ion channel reporters of PI(4,5)P2, which may be influenced by voltage themselves, or soluble fluorescent PIP reporters that slowly partition between the cytoplasm and plasma membrane as membrane PIP concentrations change – the latter introducing kinetic delays that constrain the number of measurements that could be made, thereby hampering the determination of the voltage dependence of activity31–33. To overcome these problems we generated two fast genetically-encoded fluorescent reporters, one for PI(3,4)P2 and another for PI(4,5)P2, based on an earlier design34 that permanently targets the reporter to the membrane and reads out PIP binding as a change in Förster Resonance Energy Transfer (FRET). These reporters of enzyme activity were complemented by voltage clamp fluorometry, which monitors kinetics and voltage dependence of conformational changes in the VSD.
We find that small depolarizations of wildtype Ci-VSP produce accumulation of both PI(3,4)P2 and PI(4,5)P2 and large depolarizations evoke transient accumulation of PIP2 species followed by a more sustained depletion. Strikingly, a VSD mutation that stabilizes an intermediate VSD conformation enhances PIP3→PIP2 catalysis and blocks PIP2→PIP catalysis, whereas a mutation that makes it easier to enter the fully activated conformation of the VSD eases entry into the PIP2→PIP catalytic state. These data, taken with the recently published crystal structure of the Ci-VSD35, present strong evidence in support of a model wherein sequential transitions between three conformations of the VSD are coupled to transitions between three states of the enzyme domain: an inactive state at negative voltage to the PIP3-preferring state at intermediate voltage and then to the PIP2-preferring at strong depolarization. This intriguing mechanism of substrate switching enables the enzyme to operate in two distinct voltage-mediated modes, allowing for integration of voltage signals at the cell membrane into complex temporal changes in PIP signaling. Furthermore, the finding that sequential transitions in a multistate input domain can allosterically control corresponding transitions between multiple off and activated states of an enzyme output domain, provides a logic by which other allosterically regulated proteins may perform sophisticated operations.
Results
Fast PIP2 reporters
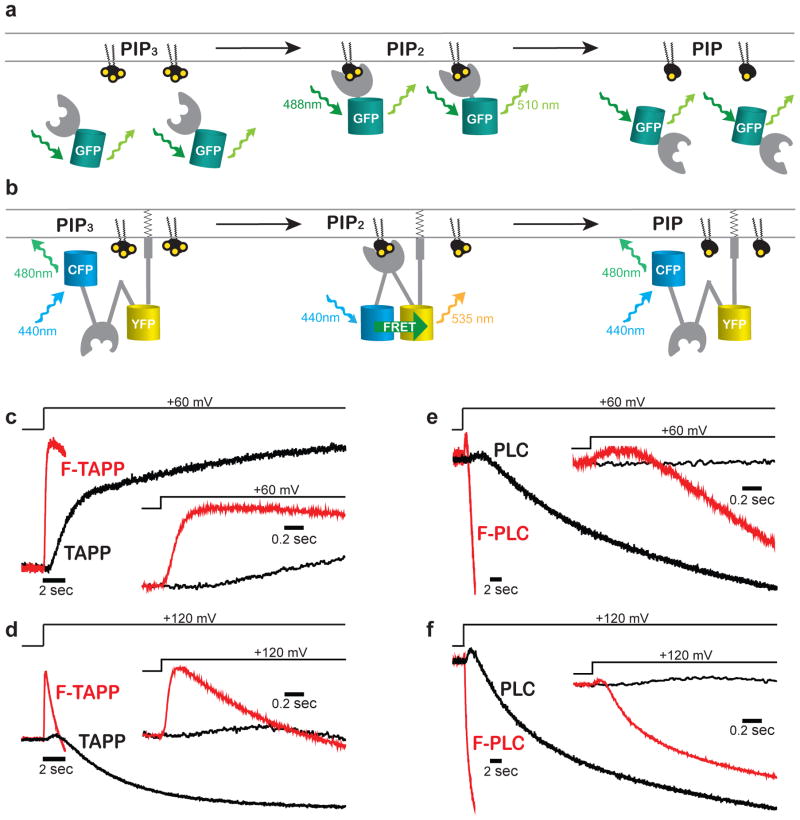
To measure the voltage dependence of Ci-VSP phosphatase activity, we needed a fast reporter of changes in PIP2 concentration that would enable kinetic analysis and measurement of responses to multiple voltage steps across a broad voltage range. The original class of soluble fluorescent protein-tagged PH (pleckstrin homology) domain protein reporters (Figure 1a) proved difficult to use in Xenopus oocytes because of their slow partitioning between the cytosol and the plasma membrane31–33. The long settling time poses several problems: a) it leads to a need of long voltage steps, resulting in PIP depletion and a consequent requirement of long recovery periods between voltage steps, with the upshot that one is limited in either the resolution or voltage range that can be examined, b) the long steps produce substantial photobleaching of the fluorescent protein tag, and c) the slow kinetics make it difficult to distinguish distinct components of activity.
Figure 1. Membrane-associated FRET reporters respond rapidly to changes in PIP2.
a,b) Cartoon depicting two classes of PIP2 reporter based on plextrin homology domains from TAPP [selectively binds PI(3,4)P2] or PLC [selectively binds for PI(4,5)P2]. a) Cytosolic reporters partition to and away from the plasma membrane as PIP2 accumulates and decreases, rate-limited by reporter diffusion time. b) Our FRET reporters “F-TAPP” and “F-PLC” pre-associate to the plasma membrane through CAAX prenylation and increase FRET when PIP2 binds, skipping the slow diffusion step of the reporter. c–f) Membrane-tethered F-TAPP and F-PLC YFP/CFP fluorescence response (red) is ~10x faster than changes in membrane fluorescence of cytosolic reporters (black). Insets show early phase of response on faster time base.
A membrane-tethered reporter is expected to respond to changes in PIP concentration more quickly by avoiding the slow equilibration between cytosol and plasma membrane. Such a reporter would need to detect PIP binding by a means other than re-localization. We turned to the CFP/YFP-based Förster Resonance Energy Transfer (FRET) reporter “Fllip-pm,” which is permanently targeted to the plasma membrane through CAAX prenylation and contains a PH domain from GRP134, which binds PI(3,4,5)P3 (Figure 1b). It had been shown previously that the specificity determining regions of the GRP1 PH domain of Fllip-pm can be replaced, for example with the PH domain of TAPP1 or PLC1δ, which selectively bind to either PI(3,4)P2 or PI(4,5)P2, respectively, to create reporters for these PIPs32,36,37. However, the speed of response of such reporters has not been characterized. Following this logic, we made two membrane-tethered reporters: FRET-TAPP and FRET-PLC, which we refer to hereafter as “F-TAPP” and “F-PLC”.
We excited CFP and simultaneously measured CFP and YFP fluorescence to generate YFP/CFP fluorescence signal (Supplementary Results, Supplementary Figure 1). Under the conditions of the experiment voltage steps over a wide range (from a holding potential of −100 mV to depolarizing steps of +210 mV) elicited no fluorescence responses from F-PLC in absence of Ci-VSP expression, but large responses when wildtype Ci-VSP was co-expressed (Supplementary Figure 2a,b). With F-TAPP in the absence of Ci-VSP a small and slow response was seen only at the strongest depolarizations, and these responses were dwarfed by much larger responses when wildtype Ci-VSP was co-expressed with the F-TAPP reporter (Supplementary Figure 2c,d). Similar to their “parental” cytosolic reporters, F-TAPP and F-PLC had an initial phase of fluorescence increase that, particularly at strong depolarization, was followed by a second phase of fluorescent decrease; however, in contrast to the cytosolic reporters, F-TAPP and F-PLC had much shorter delays and ~10-fold faster fluorescence changes (ΔFs) (Figure 1c–f, red traces) than the cytosolic reporters (Figure 1c–f, black traces).
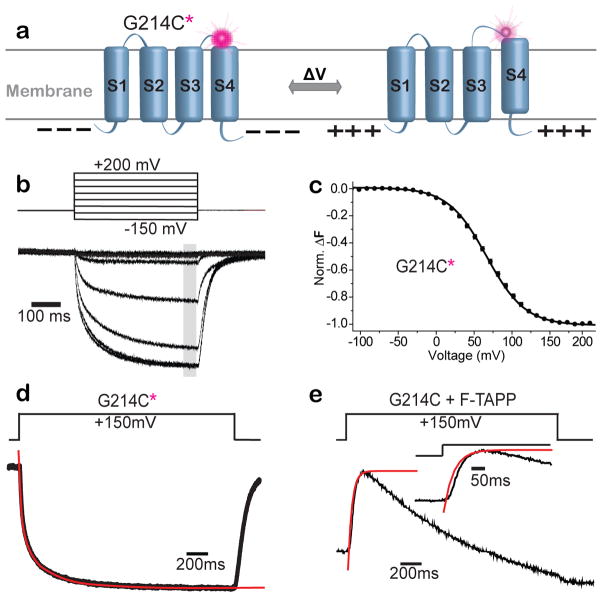
To assess the relationship between the timing of Ci-VSP activation and the change in PIP levels that result from its phosphatase activity, we compared the rate of change of PIP2 levels measured by the reporter to the timing of the voltage sensing rearrangement of Ci-VSP. To measure the protein motion of activation in the VSD, we attached the environmentally-sensitive fluorophore tetramethy-6-rhodamine-maleamide (TMRM) to a cysteine residue introduced into the VSD at residue 214 (G214C) – at the external top of the charge-containing S4 helix (Figure 2a) and performed Voltage Clamp Fluorometry. TMRM has been used extensively to detect voltage-driven conformational changes in VSDs38–43, including at the this position in Ci-VSP12,13,23,31,36. A depolarizing voltage step evoked a ΔF from 214C-TMRM from fluorescence quenching over a given voltage range (Figure 2b). Even though, the steady-state fluorescence-voltage relation (F-V) is well fit by a single Boltzmann relation (Fig. 2c, black symbols and curve), kinetic analysis reveals that it consists of two components with very different rates (Figure 2d and Supplementary Figure 3). The fast ΔF component has been shown to reflect the conformational change of gating charge displacement, the earliest step of Ci-VSP activation, and the slow ΔF component has been shown to be associated with a “relaxation” step29 that is analogous to C-type inactivation of potassium channels45–49.
Figure 2. PIP3→PIP2 activity occurs soon after VSD rearrangement associated with gating charge displacement.
a) Schematic of voltage clamp fluorometry with the environmentally-sensitive fluorophore, TMRM, attached to the G214C (G214C*) at the outer end of S4. b–d) Depolarizing voltage steps dim the fluorescence of G214C*. b) Fluorescence traces evoked by every fifth voltage steps (50mV increments from −150 to +200 mV, from VH = −100 mV). c) F-V relation from measurement at grey bar in (b) fit with a single Boltzmann relation (V1/2 = 67.7 mV ± 0.4; n=11). d) The fluorescence trace evoked by a step to +150 mV for G214C* labeled, catalytically inactive VSP (C363S) is well fit by a double exponential (τfast = 36.5 ± 7.5 s, τslow = 278.2 ± 12.8 s; n=8). e) The rapid increase in PI(3,4)P2 reported by F-TAPP (black) rapidly follows the τfast gating charge motion in the VSD (from (d)).
We examined the early phase of our membrane tethered PIP2 reporters and compared to the kinetics of S4 motion as reported by 214C-TMRM. We focused on the earlier phase of fluorescence increase of the F-TAPP response, which is similar to that of F-PLC, but larger (Figure 1c–f). This early fluorescence increase of F-TAPP followed the fast, gating charge-displacing component of 214C-TMRM with a remarkably short delay (Figure 2e). It was much faster than the slower, relaxation step reported by the 214C-TMRM ΔF (Supplementary Figure 3), indicating that the relaxation rearrangement of the VSD is too slow to account for the step that first turns on enzyme activity. Although we do not directly detect the phosphatase domain’s conformational change that turns on catalysis, this assay allows us to monitor catalytic activity with little delay.
Wildtype Ci-VSP transitions between two enzymatic states
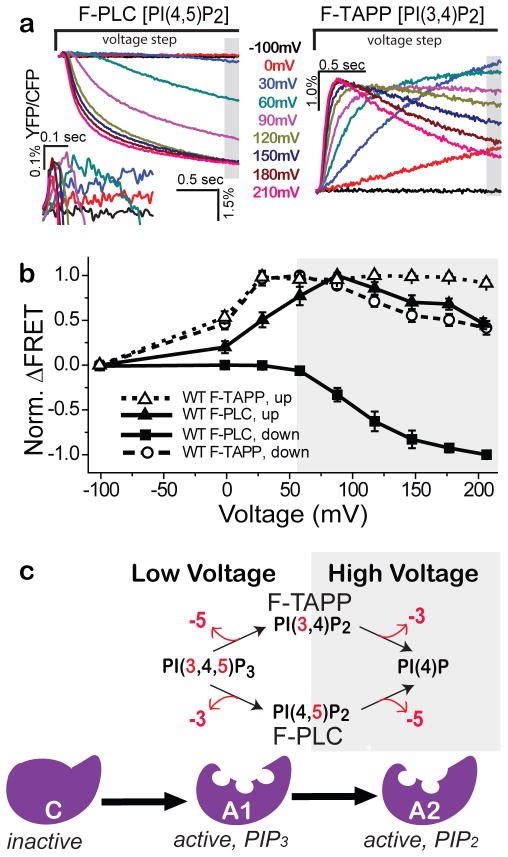
Wildtype Ci-VSP was co-expressed with either F-PLC or F-TAPP. Each cell was depolarized from a holding potential of −100 mV, where the enzyme is inactive13, to one of a series of command voltages for 2 seconds before returning to −100 mV for a 45 sec rest before the next voltage step. For F-PLC, depolarizing steps to +30mV and higher evoked a small and transient increase in FRET, which reached a maximum at about +90 mV (Figure 3a, left inset and b). Beginning with steps to about +60 mV, the initial increase in FRET was followed by a large decrease (Figure 3a, left and b). The two-phase behavior suggests that VSP first dephosphorylates PI(3,4,5)P3 at the 3-position to produce PI(4,5)P2 and then dephosphorylates PI(4,5)P2 at the 5-position to produce PI(4)P.
Figure 3. Wildtype Ci-VSP appears to have two active enzymatic states.
a) ΔFRET (ΔYFP/ΔCFP) for F-PLC (left) and F-TAPP (right) in response to 2 sec depolarizing steps from VH = −100 mV. Left inset) Blowup of early phase of ΔFRET for F-PLC. b) Normalized ΔFRET (mean ± s.e.m.; F-PLC: n=12; F-TAPP: n=17) at the end of a 2s voltage step, plotted against step voltage for F-PLC (closed squares) and F-TAPP (open circles). Also plotted is peak ΔFRET for F-PLC (closed triangles; see inset in (a)) and F-TAPP (open triangles). c) Schematic of working model of VSP enzyme with three sequential states; OFF/inactive (negative potentials), A1 selective for PIP3 (low depolarizing voltages), and A2 selective for PIP2 (high depolarizing voltages).
For F-TAPP, depolarizing steps to 0mV and higher evoked an increase in FRET, which reached a maximum at about +30 mV (Figure 3a, right and b). Beginning with steps to about +60 mV, the initial increase in FRET was followed by a decrease. This behavior suggests that VSP first dephosphorylates PI(3,4,5)P3 at the 5-position to produce PI(3,4)P2, and then dephosphorylates PI(3,4)P2 at the 3-position to produce PI(4)P.
Together, the F-PLC and F-TAPP data suggest a model of Ci-VSP where membrane depolarization drives Ci-VSP sequentially; first from an inactive state to an early catalytically active state that prefers PIP3 as a substrate (A1), and then into a later active state that prefers PIP2 as a substrate (A2) – with both A1 and A2 enzyme states dephosphorylating at either the 5-position or 3-position (Figure 3c). The early phase FRET increase seen with both reporters saturates at intermediate voltages, whereas the later phase FRET decrease grows stronger at larger depolarizations (Figure 3a, b), suggesting that A1 is favored at low depolarization and A2 at high depolarization. However, it is clear that the voltage ranges of the two components of activity overlap considerably in wildtype Ci-VSP.
Sequential VSD conformations and state-stabilization
To test the notion that depolarization turns Ci-VSP on in two phases of distinct phosphatase activity, we set out to better resolve these states by monitoring conformational changes in the VSD and making mutations to the VSD that shift the voltage dependence of these rearrangements and stabilize particular conformations. Our rationale was to determine if voltage-dependent changes in substrate preference reflected distinct voltage-dependent transitions in the VSD and, if so, to determine the relationship between the conformation of the VSD and the functional state of the enzyme domain.
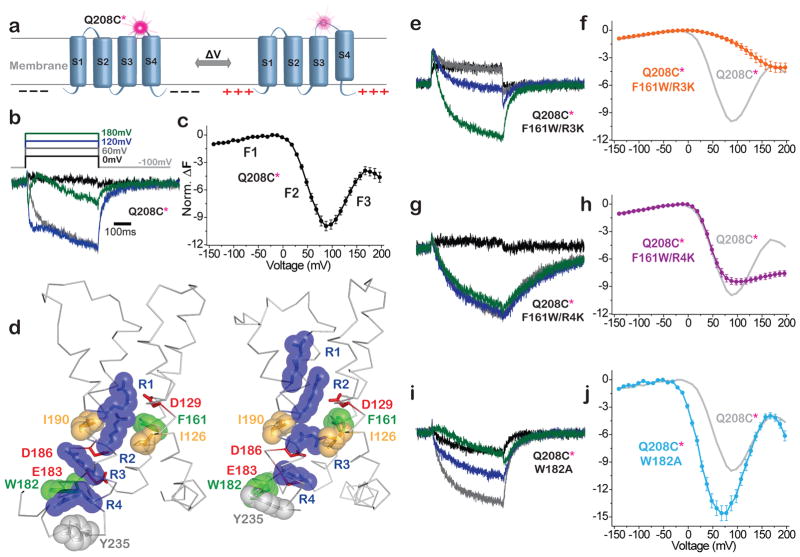
To monitor conformational rearrangements in the VSD we turned to voltage clamp fluorometry, using a cysteine substitution at residue 208 (Q208C) in the S3–S4 loop as an attachment site for TMRM (Figure 4a). As shown earlier12, Q208C-TMRM has a triphasic ΔF indicating three conformational changes: a small and fast increase in fluorescence between −100 and −20 mV (F1) where the enzyme is inactive, and two fluorescent components over the voltage range where the enzyme is active: i) an intermediate speed fluorescence decrease between −20 and +90 mV (F2), and a late slow fluorescence increase at more positive voltages (F3) (Figure 4b,c).
Figure 4. VSD mutants stabilize discrete VSD conformations.
a) Schematic of voltage clamp fluorometry with TMRM at Q208C (208*) in S3–S4 loop. b,c,e–j) Deplarization-induced ΔF traces (b,e,g,i) and corresponding average F-V relations (c,f,h,j) calculated from fluorescence measured at end of each 500ms step (mean + s.e.m.). b,c) WT (Q208C*) (n=7) has three fluorescence components: F1, F2 and F3. d) Crystal structures of VSD from WT Ci-VSP (resting at zero voltage; S4 “down”; PDB: 4G80) and the R217E mutant (activated at zero voltage; S4 “up”; PDB: 4G7V)43. Deduced activation transition moves R1–R4 (blue) outward (up), with R2 crossing hydrophobic plug (I126, I190 and F161) and R4 crossing hydrophobic residue W182. e,f) F161W/R3K (n=6) shifts F2 to more positive voltage (i.e. stabilizes conformation between F1 and F2). g,h) F161W/R4K (n=5) has relatively unperturbed F1 and F2 components, but F3 is suppressed (i.e. stabilizes the conformation between F2 and F3. i,j) W182A (n=7) shifts F2 and F3 to more negative voltages. Amplitudes of mutant F-Vs (f,h,j) normalized to WT using the F1 component.
In an effort to stabilize discrete conformations of the VSD, we considered that, in ion channels as S4 moves outward in response to membrane depolarization, its arginines ratchet between electrostatic interactions with acidic partners and cross barriers made of bulky hydrophobic side chains50–55. Increasing the bulk of these hydrophobic side chains can increase the barrier to S4 motion56, as can substitution of an arginine side chain by a longer lysine side chain57,58. The crystal structures of Ci-VSP show two potential hydrophobic barriers that face the arginines of S4, one near the middle of the membrane formed by I126, F161 and I190, and the other at the internal end of S4, formed by W18235 (Figure 4d). In an attempt to stabilize an intermediate conformation of the VSD, we sought to obstruct the outward motion of S4 by substituting the phenylalanine at position 161 with a bulkier tryptophan (F161W) and by individually replacing two arginines with lysine: one that interacts with this middle barrier – R229K (R3K), and one that crosses the internal barrier – R232K (R4K). These single arginine mutations were combined with the F161W mutation to maximize the potential effect.
The F161W/R3K double mutant did not affect F1, but shifted F2 in the positive direction to overlap with F3 (Figure 4e,f), suggesting stabilization of the inactive conformation between F1 and F2. In contrast, the F161W/R4K double mutant strongly suppressed F3 and had minimal effect on F1 and F2 (Figure 4g,h), suggesting stabilization of an intermediate activated conformation of the VSD between F2 and F3. In an effort to exert the opposite effect on activation, we attempted to eliminate a barrier to outward S4 motion. We focused on an inner hydrophobic residue, W182, which we substituted with a much smaller alanine (W182A). The W182A mutation shifted both F2 and F3 in the negative direction (Figure 4i,j), consistent with easier entry into both the partially and fully activated conformations of the VSD.
Having made one mutant, F161W/R4K, that stabilizes an intermediate activated state of the VSD and another, W182A, that eases activation, we next returned to our FRET reporter assay to analyze the effects of these mutations on enzyme activity.
VSD mutants alter transitions between enzyme states
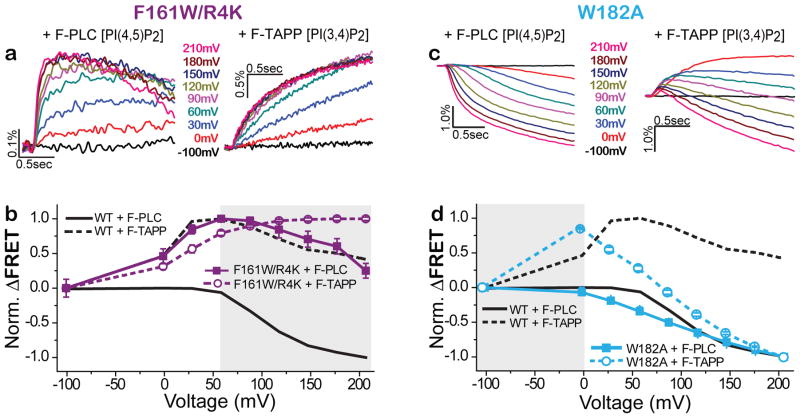
To test the effect on enzyme activity of mutants that shift the voltage dependence by altering conformational transitions in the VSD, we used F-TAPP and F-PLC to monitor the voltage-triggered production and consumption of PI(3,4)P2 and PI(4,5)P2, respectively. We first tested the F161W/R4K double mutant, which stabilized the intermediate activated conformation of the VSD. Using the F-TAPP reporter to track PI(3,4)P2, we found that the early phase of fluorescence increase seen with WT Ci-VSP (Figure 3a, right, and b, open symbols) was preserved in the F161W/R4K double mutant, but the late phase decrease was eliminated (Figure 5a, right, and b, open symbols). Using the F-PLC reporter to track PI(4,5)P2, we found that the small early phase of fluorescence increase seen with WT Ci-VSP (Figure 3a, left, and b, closed symbols) was, in the F161W/R4K double mutant, sustained with a substantial increase in amplitude relative to the late phase of fluorescence decrease (Figure 5a, left, and b, closed symbols). These observations suggest that F161W/R4K retains the PIP3 dephosphorylating A1 state, but is largely blocked from entry into the PIP2 dephosphorylating A2 state, even at strong depolarization. This finding is consistent with the impact of the F161W/R4K double mutant on VSD motion as reported by 208C-TMRM, which showed entry into the intermediate activated state, but suppression of entry into the fully activated state (Figure 4g,h). The enzymatic behavior of the WT and F161W/R4K mutant were similar whether the measured in Ci-VSP containing the cysteine labeling site for TMRM at G214C or Q208C (Supplementary Figure 4).
Figure 5. VSD mutants stabilize discrete enzyme activity states.
a,b) F161W/R4K mutant favors the A1 (PIP3→PIP2) enzyme activity state. F-PLC detects that F161W/R4K (a, traces; b, solid squares, n=14) augments the accumulation of PI(4,5)P2 (increased FRET due to dephosphorylation of the 3-position phosphate of PI(3,4,5)P3) and suppresses the depletion of PI(4,5)P2 (decreased FRET due to 5-position dephosphorylation of PI(4,5)P2) that are characteristic of WT Ci-VSP (solid grey line). F-TAPP detects that F161W/R4K (a, traces; b, open circles, n=12) augments the accumulation of PI(3,4)P2 (increased FRET due to dephosphorylation of the 5-position phosphate of PI(3,4,5)P3) and suppresses the depletion of PI(3,4)P2 (decreased FRET due to 5-position dephosphorylation of PI(3,4,5)P3) that are characteristic of WT Ci-VSP (dashed grey line). c,d) W182A shifts to more negative voltage the transition from the inactive state to the A1 active state that produces accumulation of PIP2 as well as between A1 and the A2 state that depletes of PIP2, as seen in both F-PLC (c, traces; d, solid squares, n=12) and F-TAPP (c, traces; d, open circles, n=14).
We next turned to W182 at the internal hydrophobic plug, which R4 crosses when S4 moves outward during activation35 (Figure 4d), we found that mutation of this residue to alanine (W182A) shifted entry into both the intermediate and fully activated conformations of the VSD to more negative voltages (Figure 4i,j). Using the F-TAPP reporter to track PI(3,4)P2, we found that the early phase of fluorescence increase was shifted to more negative voltages in the W182A mutant, the late phase fluorescence decrease for this mutant was also shifted to more negative voltages – with the decrease fast enough at strong depolarization to blunt the amplitude of the early phase of fluorescence increase (Figure 5c, right, and d, open symbols). Similarly, using the F-PLC reporter to track PI(4,5)P2, we found that the small early phase of fluorescence increase was reduced in amplitude in the W182A mutant relative to the late phase of fluorescence decrease and that both were shifted to more negative voltages (Figure 5c, left, and d, closed symbols).
These observations indicate that a double mutation in the VSD that strongly inhibits entry into the fully activated conformation of the VSD also strongly inhibits entry of the enzyme domain into the PIP2 dephosphorylating A2 state, limiting Ci-VSP to operate almost entirely in the A1 state as a PIP3 dephosphorylating enzyme. In contrast, a mutation in the VSD that shifts to negative voltages entry into the both intermediate and fully activated conformations of the VSD also shifts to more negative voltages both the transitions from inactive to A1 and from A1 to A2 state, enabling the enzyme to operate as a PIP2 dephosphorylating enzyme at smaller depolarization.
VSD conformations in inactive and active enzyme states
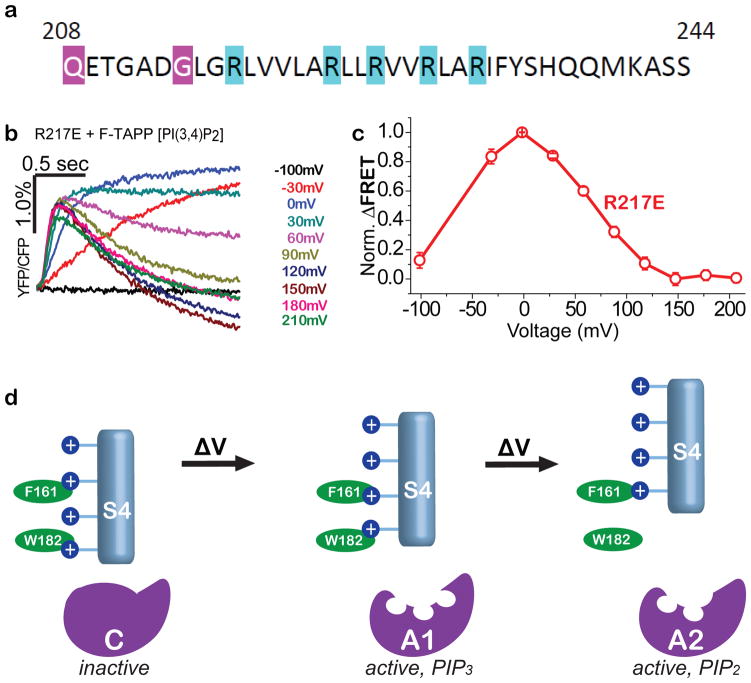
The recent crystal structures of the isolated VSD from Ci-VSP revealed two conformations: one of the wildtype protein, whose VSD is favored to be at rest under the zero voltage conditions of crystallization, and the other from a point mutant of an arginine to glutamate at the outer end of S4 (R217E) (Figure 6a), which shifts the voltage dependence of gating charge motion in the negative direction so that the VSD is favored to be activated at zero voltage35,59.
Figure 6. Two-step VSD conformational control over VSP phosphatase with two active states.
a) S4 sequence of with arginines (blue), including R217, whose mutation to glutamate stabilizes an activated conformation of the VSD for crystallography43,67. b,c) F-TAPP traces (b) and F-V (c) shows that the R217E mutant is at peak A1 PI(3,4,5)P3→PI(3,4)P2 activity at zero voltage, suggesting that “up” VSD structure in Fig. 4d corresponds to A1 enzyme state. d) Model of sequential depolarization-driven transitions in VSD that sequentially transition the phosphatase domain from inactive to the PIP3-preferring A1 active state and then to the PIP2-prefering A2 active state.
We asked which activity state is reached in R217E at zero voltage. To address this we turned to the F-TAPP reporter for its ability to robustly track both the PIP3-preferring A1 catalytic state, which produces PI(3,4)P2, and the PIP2-preferring A2 catalytic state, which consumes PI(3,4)P2. We found that the R217E mutation shifts the voltage dependence of activity in such a way that zero voltage corresponds to the voltage with the maximal increase in PI(3,4)P2 (Figure 6b,c) and PI(4,5)P2 (Supplementary Figure 5). These results suggest that the “up” conformation of the VSD seen in the Ci-VSP R217E crystal structure is the state that favors the A1 activity state of the enzyme domain.
DISCUSSION
Using a pair of fast reporters for two species of PIP2, which we generated to monitor lipid phosphatase enzyme activity, we find that Ci-VSP has two distinct activity states: a low-voltage A1 state with substrate preference for PIP3, and a high-voltage A2 state with preference for PIP2. In wildtype Ci-VSP voltage steps to around the amplitude of the peak of the action potential elicit a sequential transition from the inactive state at negative voltage first into the A1 state and then into the A2 state, yielding a transient rise in PIP2 followed by a reduction.
Since the VSD undergoes a series of conformational changes in response to membrane depolarization25–30, we focused on mutations that stabilize the VSD in different conformations by increasing or decreasing barriers for the outward motion of S4. Voltage clamp fluorometry measurement of the conformational changes in the VSD identified two mutants that stabilize different VSD conformations. One of these (F161W/R4K), which we designed to increase the barrier to outward S4 motion both by increasing the bulk of hydrophobic plug residue 161 from a phenylalanine to a tryptophan and by substituting the fourth arginine of S4 with the longer lysine, stabilized an intermediate conformation of the VSD. The other (W182A), which we designed to reduce the barrier to S4 motion by substituting S4-facing tryptophan 182 with a much smaller alanine, stabilized the fully activated conformation. We find that the first mutant allows the enzyme to enter the A1 active state, but strongly suppresses its ability to enter the A2 state, consistent with the observed strong stabilization of the intermediate activated conformation of the VSD. Moreover, we find that the second mutant shifts in the negative direction the voltage dependence of entry into both the A1 and A2 states, consistent with a shift in the negative direction of the voltage dependence of the entire set of activation motion of the VSD.
A recent observation of separation in the voltage dependencies of PI(3,4)P2 production and destruction in WT Ci-VSP was proposed to be due to either distinct states of the enzyme or differential effects of the electric field on the substrate because of the differences in charge between PIP3 and PIP222. Our finding that mutations in the VSD produce parallel shifts in the voltage dependence of VSD voltage sensing rearrangements and the two forms of substrate preference indicate that the enzyme domain assumes distinct activity states at intermediate and strong depolarization, independent of substrate charge.
While the voltage clamp fluorometry measurements reveal multiple conformational transitions in the VSD of the protein, crystal structures have been obtained for only two conformations, which differ by only “one click,” i.e. where S4 is displaced by one arginine register35. One of the structures was of the wildtype Ci-VSP whose VSD is favored to be at rest in the absence of a membrane electric field, and the other was of the R217E mutant which shifts the voltage dependence of gating charge displacement in the negative direction so that an activated state in the VSD is favored at zero voltage. Whereas the wildtype Ci-VSP is inactive at zero voltage, we find that the R217E mutant is in the A1 state at zero voltage. This suggests that for Ci-VSP to reach the A2 activity state its VSD must undergo an additional voltage-sensing (gating charge displacing) rearrangement, possibly a “second click” (Figure 6d). A second gating charge displacing step of this kind, in the voltage range of +100 to +170 mV in WT Ci-VSP (see Figure 4c) has not been reported; however, such a gating charge step would be expected to be much harder to detect than the fast gating current that is detected in the 0 to +100 mV range35,59 given the slower kinetics of the VSD conformational change of the F3 transition as compared to the F2 transition reported by 208C-TMRM (see Figure 4b).
In summary, we find that Ci-VSP has two different active enzyme states and with sequential conformational changes in the VSD induced by membrane depolarization allosterically controlling a parallel set of transitions in the enzyme domain from a state where the enzyme is inactive to a first active state with PIP3 specificity and then a second active state with PIP2 specificity. Our analysis enables us to assign a known activated structure of the VSD to the initial low voltage active state of the enzyme, and to propose a model for the VSD conformation associated with the subsequent high voltage active state of the enzyme. This novel 2-step allosteric control of a multiple specificity lipid phosphatase enables voltage to shape PIP concentrations in time. We propose that multi-state regulation may not be unique to Ci-VSP, and suggest that other enzymes may also employ allosteric regulatory domains that assume multiple conformations to drive enzyme sites into multiple distinct catalytic states.
ONLINE METHODS
Molecular Biology
The plasmid containing Ci-VSP in the pSD64TF vector was kindly provided by Y. Okamura (Osaka University). Ci-VSP was modified using the quick change protocol for site-specific mutagenesis which introduced the reported mutations in the voltage sensor and/or active site of the construct; mRNA produced by linearization with Xba1 and SP6 transcription.
The Fllip-pm FRET reporter construct kindly provided by Michiyuki Matsuda (Kyoto University), was subcloned into the pGEMHE vector. The GRP1-PH core domain region of the Fllip-pm construct was identified using the Conserved Domain Database (CDD)60 which confirmed that the construct contained two restriction enzyme sites, BspE1 and Kpn1, flanking the GRP1-PH core domain34. Similarly, we used the CDD and multiple sequence alignments of the PH-like superfamily to identify analogous core domains from TAPP1-PH kindly provided by Tamas Balla (NIH) and PLCδ1-PH kindly provided by Tobias Meyer (Stanford University). The identified TAPP1-PH and PLCδ1-PH core domains were cloned out using PCR with a pair of primers that introduced the BspE1 and Kpn1 restriction sites. Finally, the GRP1-PH core domain was digested out of the Fllip-pm backbone, and a standard ligation protocol was used to replace it with either the TAPP1-PH core domain to produce F-TAPP, or the PLCδ1-PH core domain to produce F-PLC. The resulting pGEMHE vector containing F-TAPP and F-PLC was linearization by NheI and T7 transcribed to produce mRNA for co-expression of the reporters with Ci-VSP. In response to either PI(3,4)P2 or PI(4,5)P2 depletion the two resulting FRET probes, F-TAPP or F-PLC respectively, produced detectable decreases in CFP fluorescence that corresponded to synchronized increases in YFP fluorescence.
Fluorescence Measurement of Activity
Ci-VSP (G214C, G214C F161W/R4K, G214C W182A) mRNA and FRET reporter (F-TAPP or F-PLC) mRNA were combined at a ratio 2:1 (0.8μg μl−1 VSP: 0.4μg μl−1 F-TAPP FRET reporter) or 1:1 (0.8μg μl−1 VSP: 0.8μg μl−1 F-PLC FRET reporter). Ci-VSP with R217E mRNA was expressed with F-TAPP FRET reporter mRNA combined at a ratio 1:1 (0.8μg μl−1 VSP: 0.8μg μl−1 F-TAPP FRET reporter); 50nL of this mRNA mixture (~1.2 μg μl−1) was injected in Xenopus laevis oocytes which were incubated in ND-96 (96mM NaCl, 2mM KCl, 1.8mM CaCl2, 1mM MgCl2, 50mg ml−1 gentamicin, 2.5mM Na pyruvate and 5mM HEPES) media for 36–48 hours at 18° C. To limit leak during the experiment the oocytes were perfused with an NMG buffered solution (110mM N-methyl-D-glucamine (NMG) methanesulfonic acid (MS), 2mM KMS, 2mM Ca(MS)2, 10mM HEPES, pH 7.5). Oocytes were pre-treated and recordings were performed in 8–10μM human recombinant Insulin (Gibco, Invitrogen) to activate PI3 kinase and increase PIP3 concentrations in the cell membrane, thereby making PIP3 dephosphorylation easier to detect (Supplementary Figure 6).
Fluorescence detection was made through an Olympus IX-71 inverted fluorescence microscope equipped with a 20X 0.75 NA fluorescence objective (Olympus UApo/340), Uniblitz shutter (Vincent Associates), Dagan CA-1B amplifier, and a Chroma S-011335 filter cube (Olympus U-MF2) containing excitation filter (ET420nm/40nm) and dichroic (440dcrxu).The samples were illuminated by a 100W mercury arc lamp (Hamamatsu) filtered down to 25% with a neutral density filter (ND 0.6), and the emitted light was detected by a pair of PMT-100 Photomultipliers (Applied Scientific Instrumentation) attached to the left side part of the Olympus IX-71 scope through a Photoport Bean Splitter (Applied Scientific Instrumentation) equipped with a dichroic (495dcsp, Chroma Technology) and two emission filters – HQ470nm/20nm (Chroma Technology) for the CFP channel and ET535nm/30nm (Chroma Technology) for the YFP channel. A low noise acquisition system (Molecular Devices) consisting of a digitizer (Digidata-1440A) and the pClamp 10 software suite was used to control the shutter, photomultiplier tubes and the amplifier. The YFP and CFP signals from the photomultiplier tubes were low-pass filtered at 500Hz using an eight-pole Bessel filter before being recorded by the Clampex 10 application (pClamp 10, Molecular Devices).
Voltage Clamp Fluorometry
Voltage clamp fluorometry was conducted as described in previous works12,13,23,41. Xenopus laevis oocytes were injected with 50nL of Ci-VSP mRNA at 0.4–0.8μg μl−1 and incubated in ND-96 (96mM NaCl, 2mM KCl, 1.8mM CaCl2, 1mM MgCl2, 50mg ml−1 gentamicin, 2.5mM Na pyruvate and 5mM HEPES) media for 24–48 hours at 18° C. On the day of the experiments the oocytes were incubated for 30 minutes in a high-potassium solution (92mM KCl, 0.75mM CaCl2, 1mM MgCl2, 10mM HEPES, pH 7.5) with 12uM tetramethylrhodamine-6-maleimide (Invitrogen). Following incubation the oocytes were subjected to multiple washing steps in ND-96 and stored in darkness at 12° C until tested.
Fluorescence detection was made through an Olympus IX-71 inverted fluorescence microscope equipped with a 20X 0.75 NA fluorescence objective (Olympus UApo/340), Uniblitz shutter (Vincent Associates), Dagan CA-1B amplifier, and Chroma U-N41002a filter cube (Chroma technology) containing an excitation filter (HQ535nm/50nm), emission filter (HQ620nm/60nm) and dichroic (Q565nm-LP). The samples were illuminated by a 100W mercury arc lamp (Hamamatsu) filtered down to 5% with a neutral density filter (ND 1.3), and the emitted light was detected by Hamamatsu HC120-05 photomultiplier tube attached to the right side part of the Olympus IX-71 scope. The Digidata-1440 low-noise acquisition system with the pClamp 10 analysis software suite (Axon Instruments) was used to control the shutter, photomultiplier tube and amplifier. The signal from the photomultiplier tube was sent through an eight-pole Bessel filter for low-pass filtering at 1kHz and collected in the Clampex 10 application within PClamp 10.
Data Analysis
F-TAPP and F-PLC data was collected in Clampex10. In Clampfit, the YFP signal was divided by the CFP signal. The baseline for each of the voltage traces was manually corrected for bleach by assigning the baseline to the pre-voltage step decrease in fluorescence, and a final baseline subtraction was done to set the baselines of all voltage traces to a shared baseline for comparison. The 2 second voltage range was selected and the traces were exported into OriginPro8 for plotting, and then exported to Adobe Illustrator CS5.1 to generate figures. TMRM data was collected in Clampex.
VCF data was collected in Clampex10, and mean fluorescence was calculated in Clampfit then exported to OriginPro 8 for plotting and fitting. Raw fluorescent traces for 214* with the catalytically inactive C363S background were exported from Clampfit, and fit in OriginPro 8 using a simple sum of two exponentials; y (t)= y0 + A1*exp(−k1(t−t0)) + A2*exp(−k2(t−t0)). The two amplitudes (A1 and A2) and the two tau values (τ1 and τ 2) were fit freely, thus no assumptions were made about the concerted nature of the two events. Fits were from raw traces of high expression catalytically inactive mutants (C363S). Holding potential was −100mV with 12 voltage steps of 500ms duration to voltages from −30mV to 180mV in 30mV increments. VCF data was exported from OriginPro 8 and all figures were constructed in Adobe IllustratorCS5.1.
Supplementary Material
Acknowledgments
We thank Y. Okamura (Osaka University) for Ci-VSP, M. Matsuda (Kyoto University) for Fllip-pm, T. Meyer (Stanford University) for GFP-PLC-PH and T. Balla (NIH) for GFP-TAPP-PH. We thank Hitomi Okada, Cherise Stanley and Zhu Fu for technical support, as well as Andreas Reiner, Shashank Bharill, Susy Kohout, Elizabeth Carroll and other current and former members of the Isacoff lab for guidance on analysis and helpful discussions. This work was supported by the National Institutes of Health (R01GM117051, T32GM008295) (E.Y.I.) as well as fellowship support for provided by the UC Berkeley Chancellors Fellowship for Graduate Study (S.S.G).
Footnotes
AUTHOR CONTRIBUTIONS
E.Y.I and S.S.G. conceived the study, analyzed the data, directed the evolution of the project and wrote the paper. S.S.G. conducted the experiments.
COMPETING FINANCIAL INTERESTS
The authors have no competing financial interests.
References
- 1.Changeux JP. The feedback control mechanisms of biosynthetic L-threonine deaminase by L-isoleucine. Cold Spring Harb Symp Quant Biol. 1961;26:313–8. doi: 10.1101/sqb.1961.026.01.037. [DOI] [PubMed] [Google Scholar]
- 2.Monod J, Jacob F. General Conclusions - Teleonomic Mechanisms in Cellular Metabolism, Growth, and Differentiation. Cold Spring Harb Symp Quant Biol. 1961;26:389. doi: 10.1101/sqb.1961.026.01.048. [DOI] [PubMed] [Google Scholar]
- 3.Monod J, Wyman J, Changeux JP. On the Nature of Allosteric Transitions: A Plausible Model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 4.Hilser VJ. An Ensemble View of Allostery. Science. 2010;327:653–654. doi: 10.1126/science.1186121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Motlagh HN, Wrabl JO, Li J, Hilser VJ. The ensemble nature of allostery. Nature. 2014;508:331–339. doi: 10.1038/nature13001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murata Y, Iwasaki H, Sasaki M, Inaba K, Okamura Y. Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature. 2005;435:1239–1243. doi: 10.1038/nature03650. [DOI] [PubMed] [Google Scholar]
- 7.Suh BC, Hille B. PIP2 is a necessary cofactor for ion channel function: how and why? Annu Rev Biophys. 2008;37:175–95. doi: 10.1146/annurev.biophys.37.032807.125859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Worby CA, Dixon JE. Phosphoinositide phosphatases: emerging roles as voltage sensors? Mol Interv. 2005;5:274–7. doi: 10.1124/mi.5.5.5. [DOI] [PubMed] [Google Scholar]
- 9.Hobiger K, Utesch T, Mroginski MA, Friedrich T. Coupling of Ci-VSP Modules Requires a Combination of Structure and Electrostatics within the Linker. Biophys J. 2012;102:1313–1322. doi: 10.1016/j.bpj.2012.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hobiger K, Utesch T, Mroginski MA, Seebohm G, Friedrich T. The Linker Pivot in Ci-VSP: The Key to Unlock Catalysis. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0070272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hossain MI, et al. Enzyme domain affects the movement of the voltage sensor in ascidian and zebrafish voltage-sensing phosphatases. J Biol Chem. 2008;283:18248–59. doi: 10.1074/jbc.M706184200. [DOI] [PubMed] [Google Scholar]
- 12.Kohout SC, et al. Electrochemical coupling in the voltage-dependent phosphatase Ci-VSP. Nat Chem Biol. 2010;6:369–75. doi: 10.1038/nchembio.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kohout SC, Ulbrich MH, Bell SC, Isacoff EY. Subunit organization and functional transitions in Ci-VSP. Nat Struct Mol Biol. 2008;15:106–8. doi: 10.1038/nsmb1320. [DOI] [PubMed] [Google Scholar]
- 14.Murata Y, Okamura Y. Depolarization activates the phosphoinositide phosphatase Ci-VSP, as detected in Xenopus oocytes coexpressing sensors of PIP2. J Physiol (Lond) 2007;583:875–89. doi: 10.1113/jphysiol.2007.134775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Villalba-Galea CA, Miceli F, Taglialatela M, Bezanilla F. Coupling between the voltage-sensing and phosphatase domains of Ci-VSP. J Gen Physiol. 2009;134:5–14. doi: 10.1085/jgp.200910215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hobiger K, Friedrich T. Voltage sensitive phosphatases: emerging kinship to protein tyrosine phosphatases from structure-function research. Front Pharmacol. 2015;6:20. doi: 10.3389/fphar.2015.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalli AC, Devaney I, Sansom MS. Interactions of phosphatase and tensin homologue (PTEN) proteins with phosphatidylinositol phosphates: insights from molecular dynamics simulations of PTEN and voltage sensitive phosphatase. Biochemistry. 2014;53:1724–32. doi: 10.1021/bi5000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lacroix J, et al. Controlling the activity of a phosphatase and tensin homolog (PTEN) by membrane potential. J Biol Chem. 2011;286:17945–53. doi: 10.1074/jbc.M110.201749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okamura Y, Dixon JE. Voltage-Sensing Phosphatase: Its Molecular Relationship With PTEN. Physiology. 2011;26:6–13. doi: 10.1152/physiol.00035.2010. [DOI] [PubMed] [Google Scholar]
- 20.Halaszovich CR, Schreiber DN, Oliver D. Ci-VSP is a depolarization-activated phosphatidylinositol-4,5-bisphosphate and phosphatidylinositol-3,4,5-trisphosphate 5′-phosphatase. J Biol Chem. 2009;284:2106–13. doi: 10.1074/jbc.M803543200. [DOI] [PubMed] [Google Scholar]
- 21.Iwasaki H, et al. A voltage-sensing phosphatase, Ci-VSP, which shares sequence identity with PTEN, dephosphorylates phosphatidylinositol 4,5-bisphosphate. PNAS. 2008;105:7970–5. doi: 10.1073/pnas.0803936105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurokawa T, et al. 3′ Phosphatase activity toward phosphatidylinositol 3,4-bisphosphate [PI(3,4)P2] by voltage-sensing phosphatase (VSP) Proc Natl Acad Sci USA. 2012;109:10089–94. doi: 10.1073/pnas.1203799109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu L, et al. A glutamate switch controls voltage-sensitive phosphatase function. Nat Struct Mol Biol. 2012;19:633–41. doi: 10.1038/nsmb.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuda M, et al. Crystal structure of the cytoplasmic phosphatase and tensin homolog (PTEN)-like region of Ciona intestinalis voltage-sensing phosphatase provides insight into substrate specificity and redox regulation of the phosphoinositide phosphatase activity. J Biol Chem. 2011;286:23368–77. doi: 10.1074/jbc.M110.214361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baker OS, Larsson HP, Mannuzzu LM, Isacoff EY. Three transmembrane conformations and sequence-dependent displacement of the S4 domain in shaker K+ channel gating. Neuron. 1998;20:1283–1294. doi: 10.1016/s0896-6273(00)80507-3. [DOI] [PubMed] [Google Scholar]
- 26.Pathak M, Kurtz L, Tombola F, Isacoff E. The cooperative voltage sensor motion that gates a potassium channel. J Gen Physiol. 2005;125:57–69. doi: 10.1085/jgp.200409197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schoppa NE, Sigworth FJ. Activation of Shaker potassium channels. III. An activation gating model for wild-type and V2 mutant channels. J Gen Physiol. 1998;111:313–42. doi: 10.1085/jgp.111.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seoh SA, Sigg D, Papazian DM, Bezanilla F. Voltage-sensing residues in the S2 and S4 segments of the Shaker K+ channel. Neuron. 1996;16:1159–1167. doi: 10.1016/s0896-6273(00)80142-7. [DOI] [PubMed] [Google Scholar]
- 29.Villalba-Galea CA, Sandtner W, Starace DM, Bezanilla F. S4-based voltage sensors have three major conformations. Proc Natl Acad Sci USA. 2008;105:17600–17607. doi: 10.1073/pnas.0807387105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakata S, Okamura Y. Phosphatase activity of the voltage-sensing phosphatase, VSP, shows graded dependence on the extent of activation of the voltage sensor. J Physiol (Lond) 2014;592:899–914. doi: 10.1113/jphysiol.2013.263640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Idevall-Hagren O, De Camilli P. Detection and manipulation of phosphoinositides. Biochim Biophys Acta. 2015;1851:736–745. doi: 10.1016/j.bbalip.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mavrantoni A, Thallmair V, Leitner MG, Schreiber DN, Oliver D, Halaszovich CR. A method to control phosphoinositides and to analyze PTEN function in living cells using voltage sensitive phosphatases. Front Pharmacol. 2015;6:68. doi: 10.3389/fphar.2015.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakata S, Hossain MI, Okamura Y. Coupling of the phosphatase activity of Ci-VSP to its voltage sensor activity over the entire range of voltage sensitivity. J Physiol (Lond) 2011;589:2687–2705. doi: 10.1113/jphysiol.2011.208165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sato M, Ueda Y, Takagi T, Umezawa Y. Production of PtdInsP(3) at endomembranes is triggered by receptor endocytosis. Nat Cell Biol. 2003;5:1016–1022. doi: 10.1038/ncb1054. [DOI] [PubMed] [Google Scholar]
- 35.Li QF, et al. Structural mechanism of voltage-dependent gating in an isolated voltage-sensing domain. Nat Struct Mol Biol. 2014;21:244–252. doi: 10.1038/nsmb.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishioka T, et al. Rapid Turnover Rate of Phosphoinositides at the Front of Migrating MDCK Cells. Molecular Biology of the Cell. 2008;19:4213–4223. doi: 10.1091/mbc.E08-03-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshizaki H, Mochizuki N, Gotoh Y, Matsuda M. Akt-PDK1 Complex Mediates Epidermal Growth Factor-induced Membrane Protrusion through Ral Activation. Molecular Biology of the Cell. 2007;18:119–128. doi: 10.1091/mbc.E06-05-0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cha A, Bezanilla F. Characterizing voltage-dependent conformational changes in the Shaker K+ channel with fluorescence. Neuron. 1997;19:1127–1140. doi: 10.1016/s0896-6273(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 39.Gandhi CS, Loots E, Isacoff EY. Reconstructing voltage sensor-pore interaction from a fluorescence scan of a voltage-gated K+ channel. Neuron. 2000;27:585–595. doi: 10.1016/s0896-6273(00)00068-4. [DOI] [PubMed] [Google Scholar]
- 40.Koch HP, et al. Multimeric nature of voltage-gated proton channels. Proc Natl Acad Sci USA. 2008;105:9111–6. doi: 10.1073/pnas.0801553105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mannuzzu LM, Moronne MM, Isacoff EY. Direct physical measure of conformational rearrangement underlying potassium channel gating. Science. 1996;271:213–6. doi: 10.1126/science.271.5246.213. [DOI] [PubMed] [Google Scholar]
- 42.Pathak MM, et al. Closing in on the resting state of the Shaker K(+) channel. Neuron. 2007;56:124–40. doi: 10.1016/j.neuron.2007.09.023. [DOI] [PubMed] [Google Scholar]
- 43.Tombola F, Pathak MM, Isacoff EY. How far will you go to sense voltage? Neuron. 2005;48:719–25. doi: 10.1016/j.neuron.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 44.Castle PM, Zolman KD, Kohout SC. Voltage-sensing phosphatase modulation by a C2 domain. Front Pharmacol. 2015;6:63. doi: 10.3389/fphar.2015.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Catterall WA. Molecular-Properties of Voltage-Sensitive Sodium-Channels. Annu Rev Biochem. 1986;55:953–985. doi: 10.1146/annurev.bi.55.070186.004513. [DOI] [PubMed] [Google Scholar]
- 46.Guy HR, Seetharamulu P. Molecular-Model of the Action-Potential Sodium-Channel. Proc Natl Acad Sci USA. 1986;83:508–512. doi: 10.1073/pnas.83.2.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Papazian DM, et al. Electrostatic interactions of S4 voltage sensor in Shaker K+ channel. Neuron. 1995;14:1293–301. doi: 10.1016/0896-6273(95)90276-7. [DOI] [PubMed] [Google Scholar]
- 48.Papazian DM, Timpe LC, Jan YN, Jan LY. Alteration of voltage-dependence of Shaker potassium channel by mutations in the S4 sequence. Nature. 1991;349:305–10. doi: 10.1038/349305a0. [DOI] [PubMed] [Google Scholar]
- 49.Tombola F, Pathak MM, Isacoff EY. How does voltage open an ion channel? Annu Rev Cell Dev Biol. 2006;22:23–52. doi: 10.1146/annurev.cellbio.21.020404.145837. [DOI] [PubMed] [Google Scholar]
- 50.Campos FV, Chanda B, Roux B, Bezanilla F. Two atomic constraints unambiguously position the S4 segment relative to S1 and S2 segments in the closed state of Shaker K channel. Proc Natl Acad Sci USA. 2007;104:7904–9. doi: 10.1073/pnas.0702638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chamberlin A, et al. Hydrophobic plug functions as a gate in voltage-gated proton channels. Proc Natl Acad Sci USA. 2014;111:E273–82. doi: 10.1073/pnas.1318018111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lacroix JJ, Bezanilla F. Control of a final gating charge transition by a hydrophobic residue in the S2 segment of a K+ channel voltage sensor. Proc Natl Acad Sci USA. 2011;108:6444–9. doi: 10.1073/pnas.1103397108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lacroix JJ, Hyde HC, Campos FV, Bezanilla F. Moving gating charges through the gating pore in a Kv channel voltage sensor. Proc Natl Acad Sci USA. 2014;111:E1950–E1959. doi: 10.1073/pnas.1406161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pless SA, Galpin JD, Niciforovic AP, Ahern CA. Contributions of countercharge in a potassium channel voltage-sensor domain. Nat Chem Biol. 2011;7:617–23. doi: 10.1038/nchembio.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Starace DM, Bezanilla F. A proton pore in a potassium channel voltage sensor reveals a focused electric field. Nature. 2004;427:548–553. doi: 10.1038/nature02270. [DOI] [PubMed] [Google Scholar]
- 56.Lacroix JJ, et al. Intermediate state trapping of a voltage sensor. J Gen Physiol. 2012;140:635–52. doi: 10.1085/jgp.201210827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perozo E, Santacruz-Toloza L, Stefani E, Bezanilla F, Papazian DM. S4 mutations alter gating currents of Shaker K channels. Biophys J. 1994;66:345–54. doi: 10.1016/s0006-3495(94)80783-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tao X, Lee A, Limapichat W, Dougherty DA, MacKinnon R. A Gating Charge Transfer Center in Voltage Sensors. Science. 2010;328:67–73. doi: 10.1126/science.1185954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Villalba-Galea CA, Frezza L, Sandtner W, Bezanilla F. Sensing charges of the Ciona intestinalis voltage-sensing phosphatase. J Gen Physiol. 2013;142:543–555. doi: 10.1085/jgp.201310993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marchler-Bauer A, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2010;39:D225–9. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.