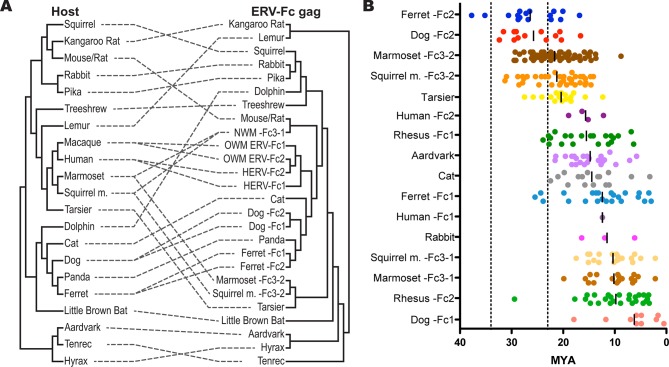
Figure 5. ERV-Fc has a multimillion-year history of replication with multiple cross-species transmissions.
(A) Tanglegram comparison of host (left) and ERV-Fc phylogenies (right); dashed lines match species and the ERV-Fc found within their genome. The host phylogeny was adapted from (Bininda-Emonds et al., 2007), while the ERV-Fc phylogeny is a supertree generated using Matrix Representation Parsimony (MRP) based on CA and Gag amino acid phylogenies. (B) LTR-derived age estimates of ERV-Fc loci derived by applying a neutral evolution rate of 4.5×10-9 substitutions per site per year to the nucleotide divergence between the 5’ and 3’ LTRs. Each plotted point represents the age estimate of a single genomic locus. Loci that show clear signatures of gene conversion or recombination have been omitted from this analysis. The average age is indicated by black vertical lines. Dotted lines indicate the approximate boundaries of the Oligocene epoch (~33.9 to ~23 MYA). ERV, endogenous retrovirus.
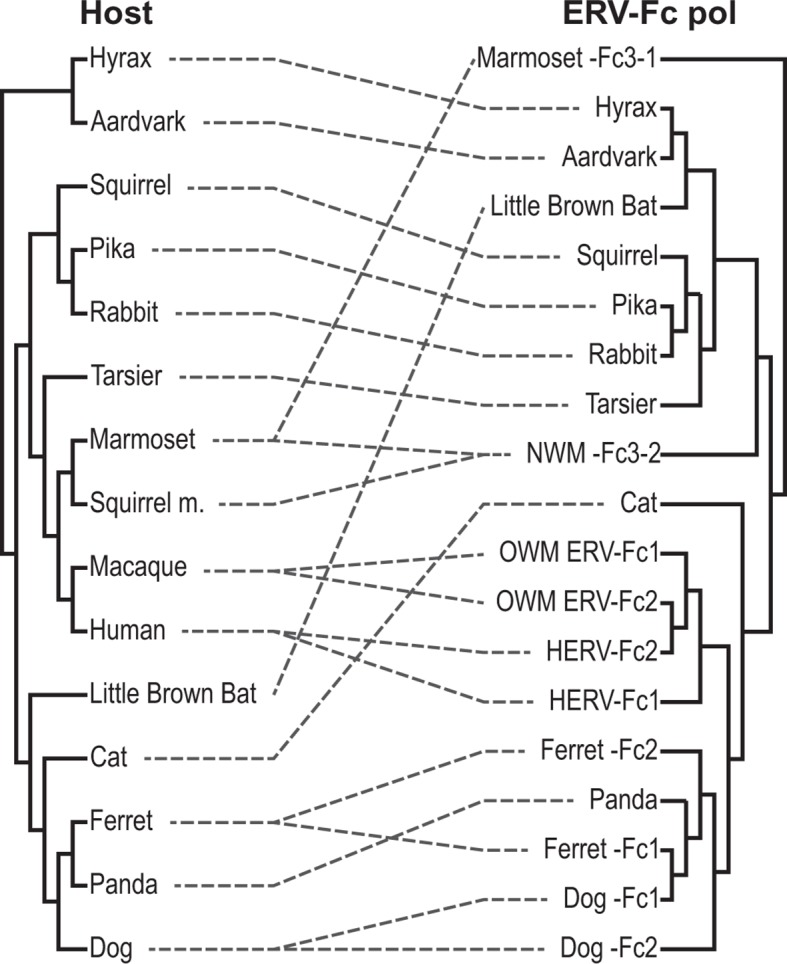
Figure 5—figure supplement 1. Tanglegram comparison of host (left) and ERV-Fc phylogenies (right).