Abstract
Key message
TransgenicNicotiana benthamianalines with constitutive expression of an Arabidopsis lectin receptor kinase gene (LecRK-I.9orLecRK-IX.1) show enhanced resistance toPhytophthorapathogens, demonstrating conserved gene functionality after interfamily transfer.
Abstract
In plants, cell surface receptors mediate the first layer of innate immunity against pathogenic microbes. In Arabidopsis several L-type lectin receptor kinases (LecRKs) were previously found to function as Phytophthora resistance components. In this study, we determined the functionality of Arabidopsis LecRK-I.9 or LecRK-IX.1 in Phytophthora resistance when transferred into the Solanaceous plant Nicotiana benthamiana. Multiple transgenic lines were generated for each LecRK gene and molecular analyses revealed variation in transgene copy number, transgene expression levels and LecRK protein accumulation. Infection assays showed that transgenic N. benthamiana plants expressing either Arabidopsis LecRK-I.9 or LecRK-IX.1 are more resistant to Phytophthora capsici and to Phytophthora infestans. These results demonstrate that Arabidopsis LecRK-I.9 and LecRK-IX.1 retained their Phytophthora resistance function when transferred into N. benthamiana. Therefore, these LecRKs have the potential to function as a complementary Phytophthora resistance resource in distantly related plant species next to the canonical Phytophthora resistance genes encoding nucleotide-binding leucine-rich repeat proteins.
Keywords: L-type lectin receptor kinases, LecRK, Phytophthora, Disease resistance, Nicotiana benthamiana, Interfamily gene transfer
Introduction
Plant diseases caused by Phytophthora species are widespread and cause enormous economic losses on a large variety of crops (Tyler 2007; Fry 2008; Lamour et al. 2012). Under favorable conditions, Phytophthora pathogens reproduce rapidly and become epidemic within a short period of time. Phytophthora disease control is costly and often depends on excessive application of preventive fungicides. Hence, development of plant cultivars with durable resistance against different Phytophthora species is under high demand. Currently, breeding programs are mainly focused on the exploitation of resistance (R) genes that encode intracellular nucleotide-binding leucine-rich repeat (NLR) proteins to restrict Phytophthora pathogens (Vleeshouwers et al. 2011). However, these attempts are often hampered by the quick adaptation of Phytophthora pathogens that leads to evasion of the R-gene mediated recognition (Fry 2008; Vleeshouwers et al. 2011; Rodewald and Trognitz 2013).
Plants also respond to pathogens by activation of plasma membrane-localized receptor-like kinases (RLKs) that function as pattern recognition receptors (PRRs) to initiate defense (Zipfel 2014). Plant resistance mediated by PRRs has been hypothesized to confer broad-spectrum resistance against plant pathogens, but thus far received little attention in resistance breeding of crop plants. One of the largest classes of RLKs comprising potential PRRs are the L-type lectin receptor kinases (LecRKs). LecRKs are ubiquitously present throughout the plant kingdom. Arabidopsis has 45 LecRK genes, several of which belong to evolutionary conserved clades whereas others are species- or genus-specific (Bouwmeester and Govers 2009; Hofberger et al. 2015; Wang et al. 2015a).
Thus far, several LecRKs have been found to play a role in plant resistance to different Phytophthora pathogens. Arabidopsis LecRK-I.9 was the first one described to function as a Phytophthora resistance component (Bouwmeester et al. 2011). To unravel the function of other Arabidopsis LecRKs, a large set of T-DNA insertion mutants covering 36 of the 45 LecRKs was analysed (Wang et al. 2014) and infection assays revealed that mutants of 13 LecRKs showed compromised Phytophthora resistance. These included mutants of the previously identified LecRK-I.9 and of the two members of clade IX, namely LecRK-IX.1 and LecRK-IX-2. More recently, the latter two were analysed in more detail and this confirmed that they both function as Phytophthora resistance component in Arabidopsis (Wang et al. 2015b).
Engineering plants via interfamily transfer of resistance components has the potential to improve disease resistance in crop plants. A successful example is the transfer of Arabidopsis EFR into Solanaceous plants (Lacombe et al. 2010). EFR is a receptor of bacterial elongation factor EF-TU and is restricted to the Brassicaceae family (Kunze et al. 2004; Zipfel et al. 2006). Nicotiana benthamiana and tomato transgenic plants expressing EFR gained the capacity to respond to EF-Tu and showed enhanced resistance to various bacterial pathogens (Lacombe et al. 2010). In a similar way, Arabidopsis LecRK-I.9 as transgene in potato was shown to confer enhanced resistance to Phytophthora infestans (Bouwmeester et al. 2014). Consistently, transient expression of LecRK-I.9 in N. benthamiana also resulted in increased resistance to Phytophthora pathogens (Bouwmeester et al. 2014) and the same holds for LecRK-IX.1 and LecRK-IX.2 (Wang et al. 2015b). Likewise, Arabidopsis LecRK-VI.2 maintained its function in bacterial resistance when expressed in N. benthamiana (Huang et al. 2014).
The objective of this work was to check whether the Arabidopsis lectin receptor kinase genes LecRK-I.9 and LecRK-IX.1 maintain their functionality in Phytophthora resistance when stably integrated as transgene in the distantly related species N. benthamiana. To this end, transgenic N. benthamiana plants ectopically expressing either Arabidopsis LecRK-I.9 or LecRK-IX.1 were generated using Agrobacterium-mediated transformation. The obtained transgenic lines were subjected to molecular analyses to determine transgene copy number, transgene expression level and LecRK protein accumulation. Thereafter, we monitored the phenotypic changes of these transgenic N. benthamiana lines in growth and response to different Phytophthora pathogens. Since N. benthamiana is a model plant amenable for virus-induced gene silencing and is widely used for studying plant-pathogen interactions and protein–protein interactions, these transgenic plants are valuable as experimental tool for further functional analysis of LecRKs.
Materials and methods
Plant growth conditions and infection assays
Nicotiana benthamiana seeds were surface-sterilized by 70 % ethanol and 1 % NaClO, and grown on MS medium (4.4 g MS salt, 20 g sucrose and 8 g agar) or MS medium supplemented with antibiotics in a conditioned growth chamber at 19–21 °C, with a 16 h photoperiod and a relative humidity of 75–80 %. Plants grown in soil were kept in a greenhouse with similar settings. Supplementary light (100 W m−2) was applied when the light intensity dropped below 150 W m−2.
Phytophthora capsici isolates LT263 and LT3239 were maintained in the dark on V8 plates at 25 °C (Wang et al. 2013), and P. infestans isolate 14-3-GFP on rye sucrose agar at 18 °C (Bouwmeester et al. 2014). P. infestans zoospores were obtained by treating sporulating mycelia with cold water for 3–4 h. For detached-leaf assays, leaves from 5-week-old N. benthamiana plants were collected and the abaxial sides were inoculated with P. capsici mycelial plugs (Ø 0.5 cm) or 10 µL droplets of a P. infestans zoospore suspension with a concentration of 5 × 105 zoospores mL−1. Inoculated leaves were kept in transparent plastic boxes with high humidity in the dark overnight and thereafter exposed to a condition with a 12 h photoperiod and appropriate temperature settings. Disease severity was monitored by measuring lesion sizes (Vleeshouwers et al. 1999) 3 and 6 days after inoculation with P. capsici and P. infestans, respectively.
Agrobacterium-mediated transformation of N. benthamiana
A. tumefaciens GV3101 carrying the binary vectors pBIN-KS-35S::AtLecRK-I.9-eGFP, pBIN-KS-35S::AtLecRK-IX.1-eGFP (Wang et al. 2015b) and pBIN61-35S::GFP (Fig. 2a) were grown overnight at 28 °C in Yeast Extract Broth with appropriate antibiotics. A. tumefaciens cells were pelleted, resuspended and incubated in MMA induction medium (10 mM MES, 10 mM MgCl2, 50 μM acetosyringone, pH 5.6) for 3 h. A. tumefaciens cells were collected by centrifugation and resuspended in MS broth supplemented with 150 μM acetosyringone. Leaf pieces (2–3 cm2) were cut from 5-week-old N. benthamiana leaves and incubated with A. tumefaciens cells for 30 min. Thereafter, leaf discs were dried on filter paper to remove excess A. tumefaciens and incubated on regeneration medium consisting of MS salt, 1 mg/L cytokinins 6-BAP (6-benzyl amino purine), 0.1 mg/L auxin NAA (1-naphthaleneacetic acid) and 0.8 % agar for 2 days at 19–21 °C. Leaf pieces were transferred every week to fresh regeneration medium supplemented with 400 mg/L cefotaxime, 200 mg/L vancomycin and 200 mg/L kanamycin until the appearance of plantlets. The generated plantlets were transferred to MS medium containing 200 mg/L kanamycin to allow root development. Upon root generation, plantlets were transferred into soil and kept in the greenhouse for seed harvesting.
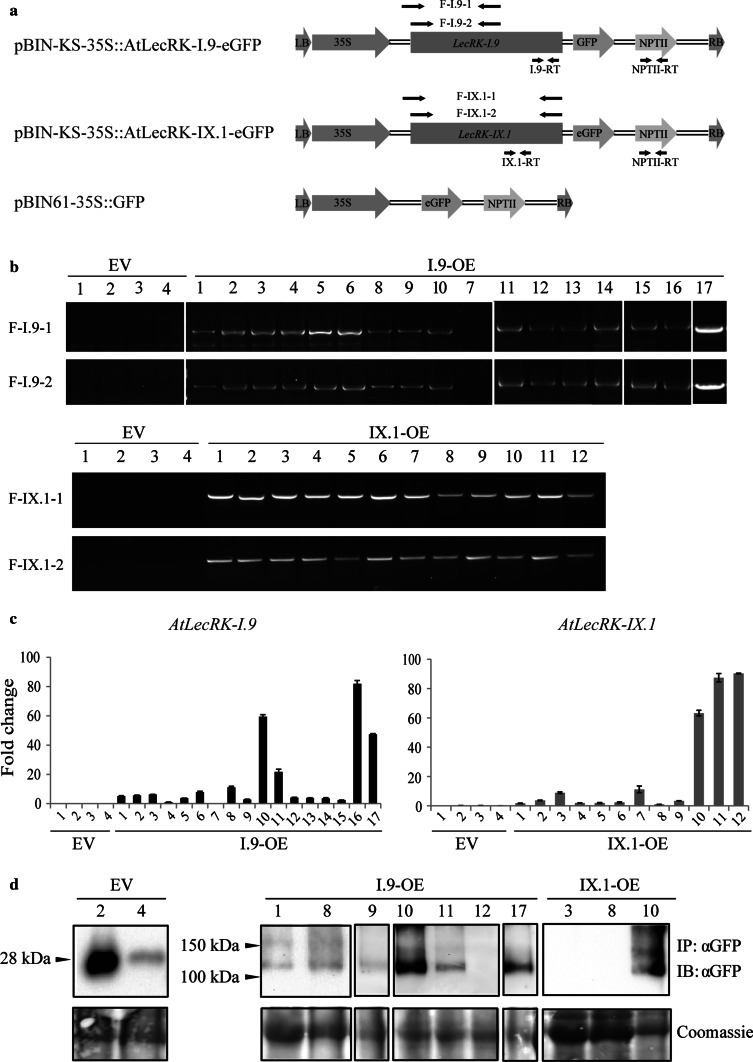
Fig. 2.
Molecular characterization of transgenic N. benthamiana lines. a Schematic representation of T-DNA regions of the vectors used for N. benthamiana transformation. PCR amplified fragments and position of the primers are indicated by the head-to-head arrow pairs. The fragments F-I.9-1, F-I.9-2, F-IX.1-1, F-IX.1-2 and NPTII-RT were amplified to determine transgene presence in transgenic lines, while fragments I.9-RT and IX.1-RT were amplified to monitor transgene mRNA levels. b Presence of AtLecRK-I.9 or AtLecRK-IX.1 in transgenic N. benthamiana lines. Genomic DNA from each line was used as template for amplification with primers indicated in (a). c Relative quantification of transgene expression levels in transgenic N. benthamiana lines. Transcript levels were normalized to NbActin and values are expressed as mean fold changes (±SD) relative to the transcript level of AtLecRK-I.9 in I.9-OE-4 or the transcript level of AtLecRK-IX.1 in IX.1-OE-8 that was arbitrarily set as 1. d GFP, LecRK-I.9-eGFP and LecRK-IX.1-eGFP accumulation in transgenic N. benthamiana lines. Proteins were immunodetected using anti-GFP-HRP. Coomassie staining was used to indicate the amount of loading in each lane
Transgene detection in transgenic N.benthamiana
Genomic DNA was isolated using CTAB buffer (0.02 % CTAB, 100 mM Tris–HCl pH 8.0, 20 mM EDTA pH 8.0, 1.4 M NaCl and 1 % PVP) followed by precipitation with isopropanol. RNA was removed by RNaseA. Transgene presence was checked by PCR using specific primers for AtLecRK-I.9 and AtLecRK-IX.1 and a primer matching a flanking sequence in the binary vector (Table 1; Fig. 2a).
Table 1.
Primers used in this study
| Primer | Sequence 5′–3′ | Used to/for | |
|---|---|---|---|
| NPTII-RT-F | GGAGAGGCTATTCGGCTATG | Check presence of NPTII | |
| NPTII-RT-R | TCGTCCTGCAGTTCATTCAG | Check presence of NPTII | |
| Nbactin-F | TATGGAAACATTGTGCTCAGTGG | Endogenous control | |
| Nbactin-R | CCAGATTCGTCATACTCTGCC | Endogenous control | |
| Oligo-dT | GACTCGAGTCGACATCGATTTTTTTTTTTTTTT | cDNA synthesis | |
| pGRAB-F1 | CCCACTATCCTTCGCAAGACCCTTCC | Check presence of T-DNA | |
| IX.1-RT-F | CAAGGCGAGTAATGTGATGCT | Check presence of AtLecRK-IX.1; qRT-PCR | |
| IX.1-RT-R | TAACCAAATGTTCCTGCTAACC | qRT-PCR | |
| IX.1-F | TCAAGCCTGGAGCTAATAG | Check presence of AtLecRK-IX.1 | |
| IX.1-R | ACGACCATGTTGAGCACTTG | Check presence of AtLecRK-IX.1 | |
| I.9-RT-F | TTTGCCAGATTTCTCACCATACAC | qRT-PCR | |
| I.9-RT-R | TCTGTTGACTGCCAAGCGTAAG | qRT-PCR | |
| I.9-F | ATGGCTCGTTGGTTGCTTCAG | Check presence of AtLecRK-I.9 | |
| I.9-R | GCTTTGACATCTCGGTGCAGAAC | Check presence of AtLecRK-I.9 | |
Transgene copy number in T0 transgenic lines was determined according to Honda et al. (2002). Briefly, qPCR was performed using genomic DNA as template with specific primers for AtLecRK-I.9, AtLecRK-IX.1, the neomycin phosphotransferase II gene (NPTII) or NbActin (Table 1). The copy number was calculated by normalizing the amplification of AtLecRK-I.9, AtLecRK-IX.1 or NPTII to NbActin.
RNA isolation and qRT-PCR
Total RNA was isolated from leaves collected from 6-week-old T0 transgenic N. benthamiana plants with a NucleoSpin RNA plant Kit (Macherey–Nagel) and thereafter used as template for cDNA synthesis with an oligo-dT primer and a M-MLV reverse transcriptase kit (Promega). qRT-PCR was performed as previously described (Wang et al. 2014). Transcript levels of AtLecRK-I.9 and AtLecRK-IX.1 were normalized to NbActin.
Protein extraction, immunoprecipitation and western blotting
Leaves collected from 6-week-old T0 transgenic N. benthamiana plants were ground in liquid nitrogen and subsequently incubated for 30 min in an extraction buffer containing 150 mM NaCl, 50 mM Tris–HCl pH 8.0, 1.0 % IGEPAL® CA-630 (Sigma) and one protease inhibitor cocktail tablet per 50 mL (Roche). The homogenate was centrifuged at 18,000 rpm for 20 min and the supernatant was incubated with GFP-trap_A® beads (Chromotek) at 4 °C for 1–2 h. After incubation, GFP-beads were spinned down and washed six times with extraction buffer. Proteins were eluted from GFP-trap_A® beads by boiling for 5 min, separated on an 8 % SDS-PAGE gel and electroblotted onto PVDF membrane (Bio-Rad). Accumulation of eGFP-tagged LecRK-I.9 and LecRK-IX.1 was detected by immunoblotting using anti-GFP-HRP (Miltenyi Biotec). Supersignal West Femto Chemiluminescent Substrate (Thermo Scientific) was used for signal development. Coomassie brilliant blue staining was used to indicate the amount of loading.
Results and discussion
Generation of transgenic N. benthamiana ectopically expressing Arabidopsis LecRK-I.9 or LecRK-IX.1
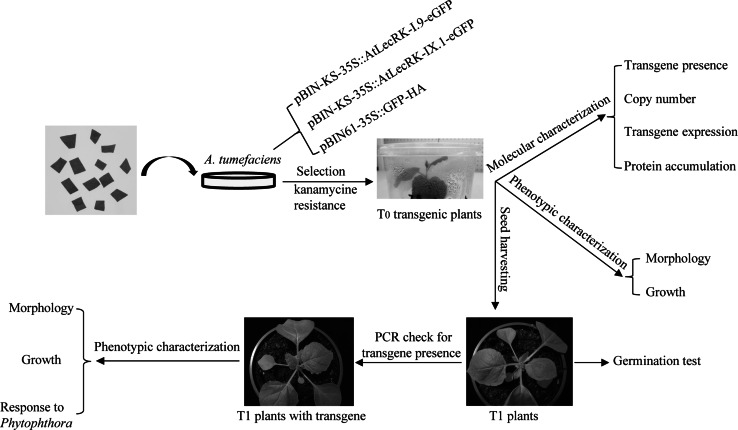
Agrobacterium-mediated transformation of N. benthamiana with the binary vectors pBIN-KS-35S::AtLecRK-I.9-eGFP, pBIN-KS-35S::AtLecRK-IX.1-eGFP and pBIN61-35S::GFP (Fig. 2a) resulted in 17, 12 and 4 T0 kanamycin-resistant lines, respectively. These were named as I.9-OE-1–17, IX.1-OE-1–12 and EV-1–4 (Table 2). The flowchart in Fig. 1 gives an overview of the various steps in the process of selection and analysis of the T0 and T1 transgenic lines. For the molecular characterization, we first determined whether AtLecRK-I.9 and AtLecRK-IX.1 were successfully transferred into N. benthamiana by PCR using gene-specific primers (Fig. 2a). Of the selected kanamycin-resistant plants, the four empty vector transgenic lines (EV-1 to EV-4) and one LecRK-I.9 line, i.e. I.9-OE-7, gave no PCR product (Fig. 2b).
Table 2.
Transgenic N. benthamiana lines vary in transgene copy number, gene expression and growth
| Line no. | Transgene copy number | Relative transgene expressiona | Morphology (T0 transformants vs. wild type)b | Germination ratec | ||
|---|---|---|---|---|---|---|
| AtLecRK-I.9/IX.1 | NPTII | AtLecRK-I.9 | AtLecRK-IX.1 | |||
| EV− | ||||||
| 1 | 0 | 1 | n.d. | n.d. | – | 100 |
| 2 | 0 | 2 | n.d. | n.d. | –e | 100 |
| 3 | 0 | 5 | n.d. | n.d. | – | 100 |
| 4 | 0 | 2 | n.d. | n.d. | –e | 100 |
| I.9-OE− | ||||||
| 1 | 1 | 1 | 5.0 | Slightly smallere | 32 | |
| 2 | 2 | 2 | 5.6 | Smaller plants with curly leaves | 60 | |
| 3 | 3 | 3 | 6.2 | Smaller plants with compacted rosettee | 40 | |
| 4 | 3 | 3 | 1.0 | – | 60 | |
| 5 | 1 | 1 | 3.7 | –e | 100 | |
| 6 | 4 | 4 | 7.8 | – | 55 | |
| 7 | 0 | 1 | n.d. | –e | /d | |
| 8 | 1 | 1 | 11.1 | – | 60 | |
| 9 | 1 | 1 | 2.8 | Smallere | 85 | |
| 10 | 5 | 6 | 59.4 | Smallere | 95 | |
| 11 | 4 | 6 | 21.7 | Smallere | 65 | |
| 12 | 1 | 1 | 4.1 | Smaller plants with compacted rosette, thick leavese | 55 | |
| 13 | 2 | 2 | 3.9 | – | 50 | |
| 14 | 2 | 2 | 3.6 | – | 70 | |
| 15 | 1 | 1 | 2.3 | Smallere | 60 | |
| 16 | 1 | 1 | 81.9 | Smaller plants with compacted rosette | 35 | |
| 17 | 3 | 3 | 47.3 | Slightly smaller and curly leaves | 85 | |
| IX.1-OE− | ||||||
| 1 | 2 | 2 | 1.7 | –e | 100 | |
| 2 | 1 | 2 | 3.7 | Smaller plants with curly round leavese | 100 | |
| 3 | 1 | 2 | 9.0 | Smallere | 100 | |
| 4 | 2 | 2 | 2.0 | –e | 100 | |
| 5 | 2 | 3 | 2.0 | –e | 100 | |
| 6 | 1 | 2 | 2.4 | – | 100 | |
| 7 | 1 | 1 | 11.2 | Smallere | 100 | |
| 8 | 2 | 2 | 1.0 | – | 100 | |
| 9 | 1 | 1 | 3.4 | – | 100 | |
| 10 | 1 | 1 | 63.2 | Smaller plants with old leaves showing necrosise | 100 | |
| 11 | 1 | 1 | 87.3 | Smaller plants with old leaves showing necrosis | 100 | |
| 12 | 1 | 1 | 90.3 | Smaller plants with old leaves showing necrosise | 100 | |
a n.d. not detectable
b –, no difference compared with wild-type N. benthamiana
cPercentage of germinated seeds of T1 progeny lines (n = 18–24) after 3 days on MS
d / not tested
eSimilar morphology in T1 progeny lines harboring the transgene
Fig. 1.
Flowchart of the generation, selection and characterization of transgenic N. benthamiana lines harboring Arabidopsis LecRK-I.9 or LecRK-IX.1
To determine transgene copy number, we performed qPCR analysis. For both AtLecRK-I.9 and AtLecRK-IX.1, the copy number in transgenic lines ranged from 0 to 5 and this number was not always consistent with that of the determined NPTII copy number (Table 2). In the transgenic lines I.9-OE-7, I.9-OE-10, I.9-OE-11, IX.1-OE-2, IX.1-OE-3, IX.1-OE-5 and IX.1-OE-6, more copies were detected for NPTII than for LecRKs, indicating the presence of truncated T-DNA fragments. This likely explains why the T0 line I.9-OE-7 lacks AtLecRK-I.9 but is still kanamycin-resistant.
Expression of AtLecRK-I.9 and AtLecRK-IX.1 in the T0 plants was determined by quantifying mRNA levels using qRT-PCR. For both AtLecRK-I.9 and AtLecRK-IX.1, the expression level varied among individual transgenic lines (Fig. 2c). Variations in transgene expression level in stable transgenic lines have often been attributed to the site(s) of transgene insertion and transgene copy number (Kole et al. 2010). In T0 plants, however, no correlation was found between copy number and transgene expression level. Transgene expression level varied among individual transgenic lines with the same transgene copy number. Some of the transgenic lines with a single copy of the transgene showed even higher expression levels than those with two or more copies. For example, line I.9-OE-16 contains a single AtLecRK-I.9 copy but showed the highest transgene expression level of all I.9-OE lines. Even transgenic lines with the same transgene number, e.g. line IX.1-OE-3 and IX.1-OE-11, showed quite different expression levels (Fig. 2c). To determine whether the transgenic lines produce the LecRK proteins, we isolated proteins and performed western blot analysis using GFP antibody. For this analysis we selected a subset of seven I.9-OE lines and three IX.1-OE lines that varied in transgene copy number and expression. In two EV lines (i.e. EV-2 and EV-4), different amounts of GFP with the expected size around 28 kDa were detected. In the I.9-OE lines, variable amounts of LecRK-I.9-eGFP were detected (Fig. 2d) and comparison with the expression levels (Fig. 2c) suggests that the accumulation of the LecRK-I.9 is correlated with the transgene expression level. LecRK-IX.1-eGFP was only detected in one of the three IX.1-OE lines, and this is the one, IX.1-OE-10, with the highest transgene expression level of the three selected IX.1-OE lines (Fig. 2d).
Morphology and growth alterations of I.9-OE and IX.1-OE lines
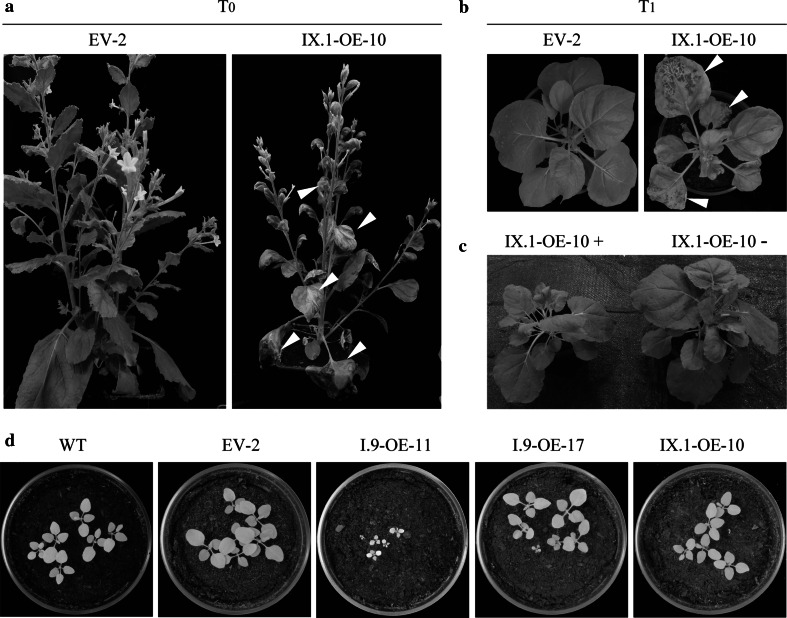
In a previous study, it was reported that overexpression of AtLecRK-I.9 in Arabidopsis led to more compact rosettes with smaller and slightly wrinkled leaves (Bouwmeester et al. 2011). In line with this, ectopic expression of AtLecRK-I.9 in potato also led to developmental defects, such as aberrant leaf morphology (Bouwmeester et al. 2014). In this study, the T0 transgenic N. benthamiana plants were monitored for growth and developmental alterations starting from 1-week after transfer into soil until seed set (Table 2; Fig. 1). All the four EV lines showed normal development; i.e. leaf morphology, branching and plant height were similar to untransformed N. benthamiana. In contrast, several I.9-OE lines were smaller in size and displayed more compact rosettes or curly leaves. These phenotypic changes, however, were not correlated with AtLecRK-I.9 expression levels, but this could be due to a combined effect of varying transgene copy numbers, transgene insertion sites and transgene expression levels in the different transgenic lines. Three out of the twelve IX.1-OE lines, namely IX.1-OE-10, IX.1-OE-11 and IX.1-OE-12, showed spontaneous cell death in leaves of over 5-week-old plants (Fig. 3a), a phenomenon that was not observed in any of the N. benthamiana lines expressing LecRK-I.9. Considering that these three IX.1-OE lines showed much higher AtLecRK-IX.1 expression levels than the rest, the cell death phenotype might be attributed to the elevated AtLecRK-IX.1 expression. This is supported by our previous observations reported in Wang et al. (2015b) that transient expression of AtLecRK-IX.1 in N. benthamiana also enhances cell death. Moreover, a similar cell death phenotype was found when AtLecRK-IX.1 was overexpressed in Arabidopsis, and this was also shown to be correlated with AtLecRK-IX.1 expression levels (Wang et al. 2015b).
Fig. 3.
Morphology of transgenic N. benthamiana lines. a The T0 N. benthamiana IX.1-OE-10 plant, but not the T0 EV-2 plant displayed cell death. Ten-week-old plants were photographed. b The T1 N. benthamiana IX.1-OE-10 plant, but not the EV-2 plant developed cell death. Six-week-old plants were photographed. c T1 progeny of the IX.1-OE-10 line harboring AtLecRK-IX.1 (+) is smaller in size than the T1 progeny without AtLecRK-IX.1 (−). Six-week-old plants were photographed. d Germination of seeds harvested from untransformed and transgenic N. benthamiana plants. Six seeds were sown in each pot. Two-week-old seedlings were photographed
To determine whether the observed LecRK-mediated phenotypes are maintained in the offspring, two EV lines, nine I.9-OE lines and eight IX.1-OE lines were propagated and their segregating progeny (T1 plants) was assayed for transgene presence and alteration in morphology and development of spontaneous cell death. As indicated in Table 2, the phenotypes in T1 plants containing the transgene were similar to those observed in T0 lines. Spontaneous cell death was only found in progeny of IX.1-OE lines that has high transgene expression (Fig. 3b; Table 2). These plants also showed retarded growth when compared to those without AtLecRK-IX.1 (Fig. 3c) or with low transgene expression (Table 2). Based on these observations, we conclude that the observed morphological and cell death phenotypes in these lines are due to the presence of the AtLecRK-IX.1 transgene and not to a random gene insertion effect. In a previous study, we showed that mutation of the catalytic RD-motif within LecRK-IX.1 abolishes induction of cell death and pathogen resistance (Wang et al. 2015b). Hence, we anticipate that the induced cell death is either directly mediated by LecRK-IX or indirectly through constitutive activation of defense.
For all the EV and IX.1-OE lines, seeds harvested from T0 plants have a similar germination efficiency as untransformed N. benthamiana when grown in soil or on MS medium (Table 2; Fig. 3d). However, 10 out of the 16 I.9-OE lines showed severe defects in seed germination, with a germination rate of 60 % or lower (Table 2). Also here, the severity of the phenotype does not correlate with AtLecRK-I.9 expression levels in the various transgenic lines.
I.9-OE and IX.1-OE lines show enhanced resistance to Phytophthora pathogens
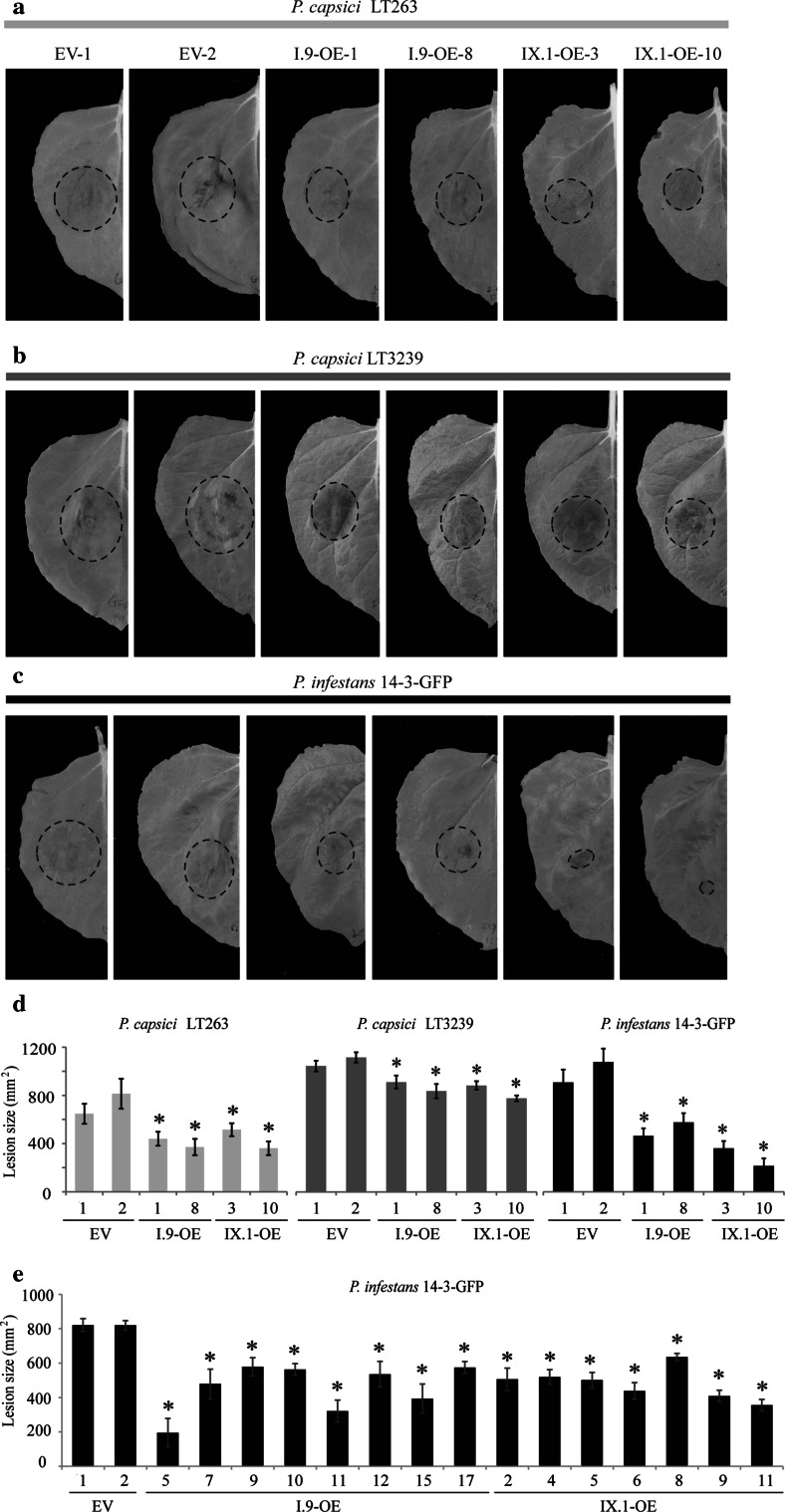
Both LecRK-I.9 and LecRK-IX.1 were previously shown to function in Phytophthora resistance in Arabidopsis (Bouwmeester et al. 2011; Wang et al. 2015b). In order to determine whether both LecRKs retain their function in Phytophthora resistance, we checked the response of N. benthamiana I.9-OE and IX.1-OE lines upon inoculation with P. capsici and P. infestans. Leaves from T1 progenies harboring the transgenes were used for infection assays. It has to be noted that for lines with a cell death phenotype only leaves without any visible cell death symptoms were used for infection assays. Upon plug-inoculation with P. capsici LT263 or LT3239, smaller lesions were found on the I.9-OE and IX.1-OE lines when compared to EV-1 and EV-2 (Fig. 4a, b), indicating that constitutive expression of AtLecRK-I.9 or AtLecRK-IX.1 in N. benthamiana enhances resistance to different isolates of P. capsici. This increased resistance was also found in I.9-OE and IX.1-OE lines when inoculated with P. infestans (Fig. 4c, d). As shown in Fig. 4e, all tested transgenic lines showed reduced lesion sizes when compared with those on EV control plants. There was however, no indication for a correlation between lesion size and the level of transgene expression (Fig. 2; Table 2). For example, lesion sizes on lines with high transgene expression levels (e.g. I.9-OE-10 and I.9-OE-17) were found to be comparable with those displayed on lines that have low transgene expression levels (e.g. I.9-OE-9). On the other hand lines that have one transgene copy with similar transgene expression levels were found to vary significantly in lesions sizes (e.g. I.9-OE-5 and I.9-OE-7). These findings show that the resistance phenotype in these transgenic lines is regulated at different levels and cannot be entirely attributed to the level of transgene expression or the transgene copy number.
Fig. 4.
Infection assays on transgenic N. benthamiana lines with different Phytophthora pathogens. a–c Disease symptoms on transgenic N. benthamiana EV, I.9-OE and IX.1-OE lines 3 days after plug-inoculation with P. capsici LT263 (a) or LT3239 (b), or 6 days after zoospore-inoculation with P. infestans 14-3-GFP (c). Lesions are indicated by black circles. d Average lesion sizes on N. benthamiana plants upon inoculation with Phytophthora pathogens. Each experiment included 12–20 inoculation sites. Bars represent the mean lesion sizes (±SE). Asterisks indicate significant difference in lesion sizes (p < 0.01) compared to the EV lines based on One-way ANOVA with Tukey’s HSD test. Infection assays were repeated three times with both P. capsici isolates and twice with P. infestans with similar results. e Average lesion sizes on N. benthamiana plants inoculated with P. infestans. Each experiment included 12–20 inoculation sites. Bars represent the mean lesion sizes (±SE). Asterisks indicate significant difference in lesion sizes (p < 0.01) compared to the EV lines based on One-way ANOVA with Tukey’s HSD test. Infection assays were repeated twice with similar results
Conclusions
In this study, multiple transgenic N. benthamiana lines with constitutive expression of Arabidopsis LecRK-I.9 or LecRK-IX.1 were obtained. Transgenic lines varied in transgene copy number, transgene expression level and protein accumulation. Ectopic expression of either AtLecRK-I.9 or AtLecRK-IX.1 in N. benthamiana increased the resistance to different Phytophthora species. Our results suggest that Arabidopsis LecRK-I.9 and LecRK-IX.1 maintained their function in Phytophthora resistance when transferred into N. benthamiana, which is in line with results that we obtained previously with transgenic potato plants expressing LecRK-I.9 (Bouwmeester et al. 2014). These findings suggest that LecRKs could be used as complementary resistance components, in combination with canonical NLR-encoding R genes, for engineering broad-spectrum disease resistance to Phytophthora pathogens. However, ectopic expression of both LecRKs also caused several adverse effects on plant fitness, such as curly leaves, leaf necrosis or reduced plant size. In the case of LecRK-IX.1, its function in Phytophthora disease resistance is independent of that in plant cell death induction (Wang et al. 2015b). Therefore, we anticipate that it should be possible to optimize the receptors in such a way that downstream signaling does no longer cause plant growth alterations while the LecRK-mediated disease resistance is maintained. The transgenic N. benthamiana lines that we describe in this study can be used as a valuable experimental tool for further analysis of the components required for LecRK-mediated resistance and plant growth alterations, for example via the virus-induced gene silencing or protein complex pull-down assays.
Author contribution statement
Y.W., F.G. and K.B. conceived and designed research. Y.W., D.L.N. and H.M.J. carried out the experiments and analysed data. Y.W., F.G. and K.B. wrote the manuscript. All authors read and approved the manuscript.
Acknowledgments
We thank Bert Essenstam from Unifarm for plant care. This project was financially supported by a Wageningen University sandwich-PhD fellowship (Y.W.), a Huygens scholarship (Y.W.), the Netherlands Fellowship Program (D.L.N. and H.M.J.), a VENI grant (K.B.) from the Netherlands Organization for Scientific Research (Technology Foundation NWO-STW), and the Food-for-Thought campaign of the Wageningen University fund.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
F. Govers and K. Bouwmeester have contributed equally to this work.
References
- Bouwmeester K, Govers F. Arabidopsis L-type lectin receptor kinases: phylogeny, classification, and expression profiles. J Exp Bot. 2009;60:4383–4396. doi: 10.1093/jxb/erp277. [DOI] [PubMed] [Google Scholar]
- Bouwmeester K, de Sain M, Weide R, Gouget A, Klamer S, Canut H, Govers F. The lectin receptor kinase LecRK-I.9 is a novel Phytophthora resistance component and a potential host target for a RXLR effector. PLoS Pathog. 2011;7:e1001327. doi: 10.1371/journal.ppat.1001327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwmeester K, Han M, Blanco-Portales R, Song W, Weide R, Guo L, van der Vossen EAG, Govers F. The Arabidopsis lectin receptor kinase LecRK-I.9 enhances resistance to Phytophthora infestans in Solanaceous plants. Plant Biotechnol J. 2014;12:10–16. doi: 10.1111/pbi.12111. [DOI] [PubMed] [Google Scholar]
- Fry W. Phytophthora infestans: the plant (and R gene) destroyer. Mol Plant Pathol. 2008;9:385–402. doi: 10.1111/j.1364-3703.2007.00465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofberger JA, Nsibo DL, Govers F, Bouwmeester K, Schranz ME. A complex interplay of tandem- and whole genome duplication drives expansion of the L-type lectin receptor kinase gene family in the Brassicaceae. Genome Biol Evol. 2015;7:720–734. doi: 10.1093/gbe/evv020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda M, Muramoto Y, Kuzuguchi T, Sawano S, Machida M, Koyama H. Determination of gene copy number and genotype of transgenic Arabidopsis thaliana by competitive PCR. J Exp Bot. 2002;53:1515–1520. doi: 10.1093/jexbot/53.373.1515. [DOI] [PubMed] [Google Scholar]
- Huang PY, Yeh YH, Liu AC, Chien CP, Zimmerli L. The Arabidopsis LecRK-VI.2 associates with the pattern-recognition receptor FLS2 and primes Nicotiana benthamiana pattern-triggered immunity. Plant J. 2014;79:243–255. doi: 10.1111/tpj.12557. [DOI] [PubMed] [Google Scholar]
- Kole C, Michler C, Abbott A, Hall T, Kohli A, Miro B, Twyman R. Transgene integration, expression and stability in plants: strategies for improvements. In Transgenic crop plants. Berlin: Springer; 2010. pp. 201–237. [Google Scholar]
- Kunze G, Zipfel C, Robatzek S, Niehaus K, Boller T, Felix G. The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell. 2004;16:3496–3507. doi: 10.1105/tpc.104.026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacombe S, Rougon-Cardoso A, Sherwood E, Peeters N, Dahlbeck D, van Esse HP, Smoker M, Rallapalli G, Thomma BPHJ, Staskawicz BJ, Jones JDG, Zipfel C. Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nat Biotechnol. 2010;28:365–369. doi: 10.1038/nbt.1613. [DOI] [PubMed] [Google Scholar]
- Lamour KH, Stam R, Jupe J, Huitema E. The oomycete broad-host-range pathogen Phytophthora capsici. Mol Plant Pathol. 2012;13:329–430. doi: 10.1111/j.1364-3703.2011.00754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodewald J, Trognitz B. Solanum resistance genes against Phytophthora infestans and their corresponding avirulence genes. Mol Plant Pathol. 2013;14:740–757. doi: 10.1111/mpp.12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler BM. Phytophthora sojae: root rot pathogen of soybean and model oomycete. Mol Plant Pathol. 2007;8:1–8. doi: 10.1111/j.1364-3703.2006.00373.x. [DOI] [PubMed] [Google Scholar]
- Vleeshouwers VGAA, van Dooijeweert W, Keizer LCP, Sijpkes L, Govers F, Colon LT. A laboratory assay for Phytophthora infestans resistance in various Solanum species reflects the field situation. Eur J Plant Pathol. 1999;10:241–250. doi: 10.1023/A:1008710700363. [DOI] [Google Scholar]
- Vleeshouwers VGAA, Raffaele S, Vossen JH, Champouret N, Oliva R, Segretin ME, Rietman H, Cano LM, Lokossou A, Kessel G, Pel MA, Kamoun S. Understanding and exploiting late blight resistance in the age of effectors. Annu Rev Phytopathol. 2011;49:507–531. doi: 10.1146/annurev-phyto-072910-095326. [DOI] [PubMed] [Google Scholar]
- Wang Y, Bouwmeester K, van de Mortel JE, Shan W, Govers F. A novel Arabidopsis-oomycete pathosystem: differential interactions with Phytophthora capsici reveal a role for camalexin, indole glucosinolates and salicylic acid in defence. Plant Cell Environ. 2013;36:1192–1203. doi: 10.1111/pce.12052. [DOI] [PubMed] [Google Scholar]
- Wang Y, Bouwmeester K, Beseh P, Shan W, Govers F. Phenotypic analyses of Arabidopsis T-DNA insertion lines and expression profiling reveal that multiple L-type lectin receptor kinases are involved in plant immunity. Mol Plant-Microbe Interact. 2014;27:1390–1402. doi: 10.1094/MPMI-06-14-0191-R. [DOI] [PubMed] [Google Scholar]
- Wang Y, Weide R, Govers F, Bouwmeester K. L-type lectin receptor kinases in Nicotiana benthamiana and tomato and their role in Phytophthora resistance. J Exp Bot. 2015;66:6731–6743. doi: 10.1093/jxb/erv379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cordewener JHG, America AHP, Shan W, Bouwmeester K, Govers F. Arabidopsis lectin receptor kinases LecRK-IX.1 and LecRK-IX.2 are functional analogs in regulating Phytophthora resistance and plant cell death. Mol Plant-Microbe Interact. 2015;28:1032–1048. doi: 10.1094/MPMI-02-15-0025-R. [DOI] [PubMed] [Google Scholar]
- Zipfel C. Plant pattern-recognition receptors. Trends Immunol. 2014;35:345–351. doi: 10.1016/j.it.2014.05.004. [DOI] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JDG, Boller T, Felix G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125:749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]