SUMMARY
Repair of DNA double-strand breaks (DSBs) is essential for genomic stability. The most common DSB repair mechanism in human cells, non-homologous end joining (NHEJ), rejoins broken DNA ends by direct ligation. It remains unclear how components of the NHEJ machinery assemble a synaptic complex that bridges DNA ends. Here, we use single-molecule imaging in a vertebrate cell-free extract to show that synapsis of DNA ends occurs in at least two stages that are controlled by different NHEJ factors. DNA ends are initially tethered in a long-range complex whose formation requires the Ku70/80 heterodimer and the DNA-dependent protein kinase catalytic subunit (DNA-PKcs). The ends are then closely aligned, which requires XLF, a non-catalytic function of XRCC4-LIG4, and DNA-PK activity. These results reveal a structural transition in the synaptic complex that governs alignment of DNA ends. Our approach provides a means of studying physiological DNA double-strand break repair at single-molecule resolution.
Graphical Abstract
INTRODUCTION
Most DNA double-strand breaks (DSBs) in human cells are repaired by non-homologous end joining (NHEJ), a mechanism that directly ligates broken DNA ends (Chiruvella et al., 2013; Radhakrishnan et al., 2014). By employing a range of DNA processing enzymes, NHEJ can join a variety of damaged or mismatched substrates (Ma et al., 2005; Waters et al., 2014a). A drawback of this versatility is the potential to generate mutations, either by inserting or deleting nucleotides during processing or by joining the wrong pairs of ends. Understanding how cells minimize such errors, while ensuring timely repair of double-strand breaks, requires a detailed picture of the protein complex that holds together DNA ends to be processed and ligated.
Broken DNA ends are first bound by the basket-shaped Ku70/80 heterodimer, which recruits the 469 kDa DNA-dependent protein kinase catalytic subunit (DNA-PKcs) to form the DNA-PK holoenzyme (Carter et al., 1990; Dvir et al., 1992, 1993; Gottlieb and Jackson, 1993; Lees-Miller et al., 1990). DNA-PKcs phosphorylates several NHEJ factors, including itself (Dobbs et al., 2010), and its kinase activity is essential for NHEJ (Dobbs et al., 2010; Jette and Lees-Miller, 2015; Jiang et al., 2015). During classical NHEJ (c-NHEJ), DNA ends are ligated by a complex of DNA ligase IV (LIG4) and XRCC4 (Critchlow et al., 1997; Grawunder et al., 1997). The XRCC4 paralog XLF (XRCC4-like factor) stimulates the activity of the XRCC4-LIG4 complex in vitro and is important for NHEJ in vivo (Ahnesorg et al., 2006; Buck et al., 2006; Gu et al., 2007; Lu et al., 2007; Tsai et al., 2007; Zha et al., 2007). Another recently discovered paralog of XRCC4 and XLF, PAXX, has been implicated in NHEJ, although its function is unclear (Craxton et al., 2015; Ochi et al., 2015; Xing et al., 2015).
Almost all of the factors described above have been proposed to play a role in bridging DNA ends. Early work reported DNA bridging by purified Ku70/80 protein (Ramsden and Gellert, 1998, but see Cottarel et al., 2013). In addition, DNA-PK holoenzyme complexes assembled with purified Ku70/80 and DNA-PKcs can dimerize to bridge DNA ends (Cary et al., 1997; DeFazio et al., 2002; Hammel et al., 2010; Spagnolo et al., 2006). Similar DNA pull-down experiments in a human cell-free extract support a role for Ku and DNA-PKcs in synapsis of DNA ends and additionally implicate LIG4, independent of its catalytic activity (Cottarel et al., 2013). Purified XLF and XRCC4 interact to form long, alternating oligomers capable of bridging DNA molecules in vitro (reviewed in (Mahaney et al., 2013)). However, a recent report that XRCC4-XLF interactions are dispensable for NHEJ in some cell types (Roy et al., 2015) suggests that XLF-XRCC4 filaments are not universally required for synapsis. Collectively, these observations have not coalesced into a coherent model of physiological synaptic complex assembly. Specifically, the steps in this process and the roles of individual NHEJ factors are unknown.
Here, we address these questions by visualizing joining of fluorescently labeled DNA ends in X. laevis egg extracts, which support highly efficient NHEJ. We first demonstrate that ligation in this system requires Ku70/80, DNA-PKcs, DNA-PKcs kinase activity, XLF, and XRCC4-LIG4, indicating that it occurs by a physiological mechanism. Next, we present a single-molecule FRET assay that reveals two conformational stages in end synapsis: 1) a long-range complex in which DNA ends are tethered but too far apart to detect FRET between end-proximal dyes, and 2) a short-range complex in which DNA ends are closely apposed. Using small-molecule inhibitors, immunodepletion, and rescue with purified proteins, we define the roles of NHEJ factors at these two stages of synapsis. We find that long-range complex formation requires Ku70/80 and DNA-PKcs, but not DNA-PK catalytic activity. Subsequent transition to the short-range complex requires DNA-PK catalytic activity, XLF, and XRCC4-LIG4, but not LIG4 catalytic activity. These results define the molecular requirements for physiological NHEJ synaptic complex assembly and reveal that a programmed rearrangement of this complex is required for close alignment of DNA ends.
RESULTS
Validation of an In Vitro Non-Homologous End Joining System
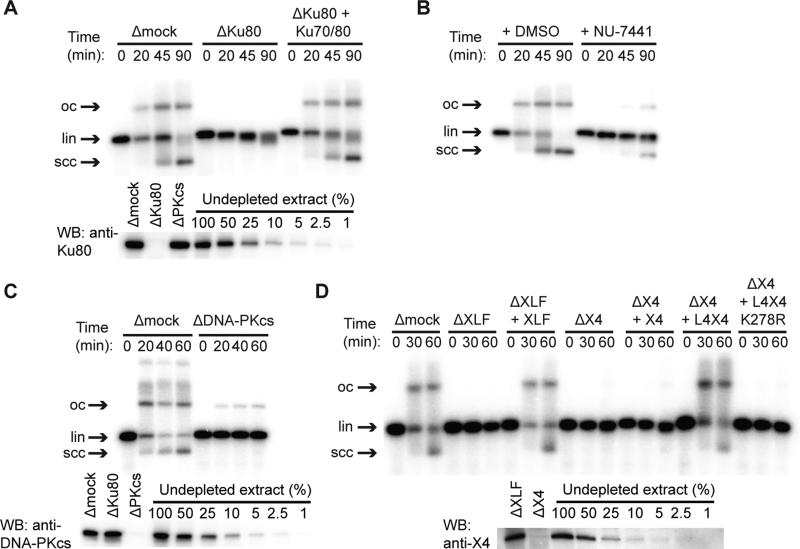
To study NHEJ in vitro under physiological conditions, we used a cell-free extract of Xenopus laevis eggs, which contains the entire soluble proteome and packages added DNA into nucleosomes (Laskey et al., 1977). Previous work showed that egg extract efficiently joins both compatible and incompatible DNA ends in a manner that depends on Ku70/80 and DNA-PK activity (Labhart, 1999; Postow et al., 2008; Thode et al., 1990; Di Virgilio and Gautier, 2005). Similarly, we observed that joining of blunt-ended linear DNAs was inhibited by immunodepletion of Ku (Fig. 1A) and restored with recombinant X. laevis Ku70/80 (Fig. 1A). End joining was also inhibited by small-molecule inhibitors or immunodepletion of DNA-PKcs, indicating that not only the presence but also the kinase activity of DNA-PKcs is required for end joining (Figs. 1B-C and S1A). Immunodepletion of XLF likewise abolished end joining, which was restored by recombinant X. laevis XLF protein (Fig. 1D). Consistent with previous results suggesting a stable complex between XRCC4 and LIG4 (Bryans et al., 1999), anti-XRCC4 immunoprecipitates contained an adenylated protein of the molecular weight expected for X. laevis LIG4 (Fig. S1B). Immunodepletion of extract with anti-XRCC4 antibody eliminated end joining, and end joining was restored by recombinant X. laevis LIG4:XRCC4 complex but not catalytically inactive LIG4K278R:XRCC4 (Cottarel et al., 2013), XRCC4 alone, or XLF (Fig. 1D and Fig. S1D). Taken together, these results demonstrate that X. laevis egg extract joins DNA ends efficiently in a manner that requires the c-NHEJ factors Ku70/80, DNA-PKcs, XLF, and LIG4:XRCC4, as well as DNA-PK catalytic activity.
Figure 1. End Joining in Xenopus Egg Extract Depends on Classical NHEJ Factors.
(A) Inhibition of end joining by immunodepletion of Ku70/80 with αKu80 antibody, and rescue with recombinant X. laevis Ku70/80. lin, linear DNA substrate; scc, supercoiled closed-circular products; oc, open-circular products. In other conditions, Ku immunodepletion selectively inhibited circularization, as previously reported (Fig. S1C) (Labhart, 1999; Di Virgilio and Gautier, 2005).
(B) Inhibition of end joining by the DNA-PK inhibitor NU-7441.
(C) DNA-PKcs immunodepletion.
(D) Immunodepletion of XLF (ΔXLF) or XRCC4 (ΔX4) and rescue with recombinant X. laevis XLF, XRCC4 (X4), wild-type LIG4:XRCC4 (L4X4), or catalytically inactive LIG4K278R:XRCC4 (L4X4 K278R). Lower panels in (A), (B), and (D) are western blots of immunodepleted extract with indicated antibodies. Uncropped blots are shown in Fig. S1F-H. XLF was not clearly visible in western blots of extract, but immunoprecipitated XLF was detected by western blotting and mass spectrometry (Fig. S1J-K).
See also Figure S1.
Two-Stage Synapsis of DNA Ends during NHEJ
To address which NHEJ factors are required for synapsis of DNA ends in this system, we employed a combination of single-molecule co-localization and Förster resonance energy transfer (FRET). A blunt-ended 100 bp DNA duplex labeled with Cy3 near one end and biotinylated at the other end (Cy3-DNA) was attached to a streptavidin-coated glass coverslip within a microfluidic channel (Fig. 2A). Next, a blunt-ended 100 bp DNA duplex labeled near both ends with Cy5 (Cy5-DNA) was added to egg extract and drawn into the channel. In each case, Cy3 and Cy5 labels were placed 7 nucleotides from the DNA end, which did not disrupt end joining (Fig. S2A-B). Surface-tethered DNAs were imaged using total internal reflection (TIR) illumination, alternating between excitation of Cy3 with a 532 nm laser and Cy5 with a 641 nm laser. Using a frame integration time of 1 s, only Cy5-DNAs tethered to Cy3-DNAs appeared as discrete spots (Fig. S2C). FRET between Cy3 and Cy5 indicated close association between the two dyes.
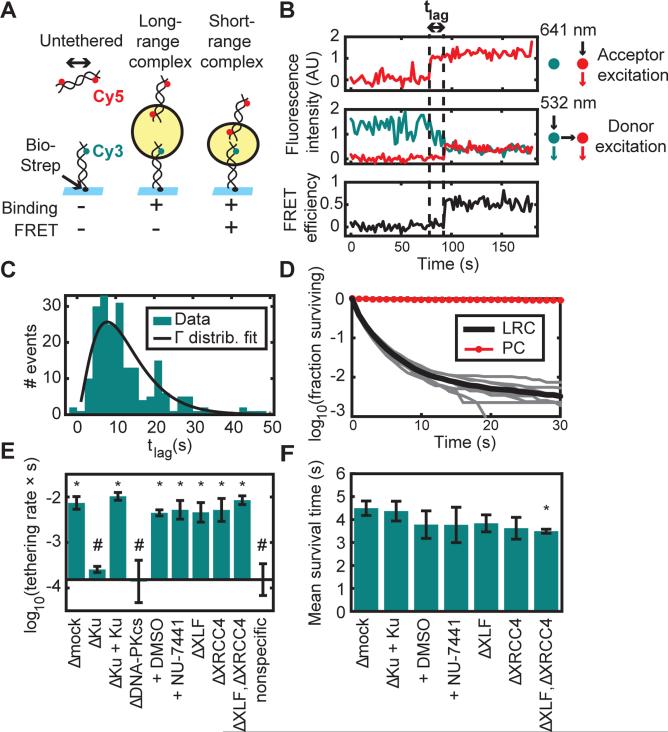
Figure 2. Two-Stage Synapsis of DNA Ends.
(A) Schematic of the intermolecular tethering assay. A 100-base pair (bp) DNA duplex biotinylated at one end (Bio) and labeled 7 bp from the other end with Cy3 was attached to a glass surface coated with streptavidin (Strep). A second 100-bp duplex labeled 7 bp from each end with Cy5 was added to egg extract and introduced into the flowcell. Cy5-DNA binding and FRET were monitored by alternating excitation of Cy3 and Cy5. The yellow circle represents the protein complex that bridges DNA ends.
(B) Example trajectory showing a lag time (tlag) between Cy5-DNA binding and transition to a high-FRET state. Upper panel, Cy5 signal with Cy5 (641 nm) excitation, which appears upon Cy5-DNA binding to the stationary Cy3-DNA. Middle panel, Cy3 (cyan) and Cy5 (red) signal with Cy3 (532 nm) excitation. Lower panel, calculated FRET efficiency.
(C) Histogram of lag times between binding and transition to high FRET (N = 207 trajectories). A fit to a gamma distribution (k = 2.7, θ = 4.7 s) suggests that multiple steps are required for the long- to short-range transition, each with a time constant on the order of seconds.
(D) Survival curve of long-range complexes (LRC) in mock-immunodepleted extract (N = 57921 events) and biotinylated Cy5-DNAs as a photostability control (PC; N = 3907 molecules). The thick black line shows the mean of nine experimental replicates (thin gray lines).
(E) Long-range complex formation rate in extract immunodepleted of different factors or treated with DNA-PK inhibitor (NU-7441) or vehicle (DMSO). Elements of the two sets of conditions labeled * and # are significantly different from elements of the other set but not from elements of the same set (p < 10−5, ANOVA with Tukey's post-hoc test). Error bars: ±2*S.E.M. See Table S1 for sample sizes.
(F) Mean survival time of tethered complexes was similar among different conditions, although slightly reduced by XLF/XRCC4 double-depletion (p = 0.03 compared to Δmock, ANOVA with Tukey's post-hoc test). Error bars: ±2*S.E.M. XLF and XRCC4 depletion also appeared to reduce the fraction of long-range complexes that were extremely long-lived (see Fig. S2D).
See also Figure S2 and Table S1.
Single-molecule intensity traces revealed a time delay between Cy5-DNA binding to Cy3-DNAs and the appearance of FRET (“tlag” in Figs. 2B and S2C). We refer to the Cy5-DNA-bound, low-FRET state as a “long-range” synaptic complex, because the dyes are outside of the ~100 Å maximum FRET radius, and the subsequent high-FRET state as a “short-range” synaptic complex. The lag time between long-range and short-range complex formation was not exponentially distributed, implying that this transition involves more than a single rate-limiting step (Fig. 2C). Most long-range complexes were short-lived, with 59% lasting only a single Cy5 excitation frame (~2 s; see Fig. 2D). Among the 14% of long-range complexes that survived at least four Cy5 excitation frames (~8 s), about 1% progressed to a high-FRET state, while the rest dissociated (see Supplemental Experimental Procedures and Fig. S4B). The low transition probability from the long-range to the short-range complex suggests that broken DNA ends typically interact many times before stably associating. Such repeated interactions would likely be facilitated by constrained diffusion of broken DNA ends within chromosomes (see also below) (Jakob et al., 2009; Kruhlak et al., 2006; Lucas et al., 2014; Soutoglou et al., 2007).
Molecular Requirements for Initial Tethering of DNA Ends
We next investigated the requirements for long-range synaptic complex formation. Immunodepletion of Ku70/80 or DNA-PKcs reduced the rate of long-range complex formation >30-fold, to a level that was indistinguishable from nonspecific Cy5-DNA binding to the surface in the absence of Cy3-DNAs (Fig. 2E). The defect in Ku-depleted extract was reversed by addition of recombinant Ku70/80. In contrast, the rate of long-range complex formation was unaffected by kinase inhibitors of DNA-PK or immunodepletion of XRCC4-LIG4, XLF, or both (Fig. 2E). The average survival time of long-range complexes was also similar among these conditions (Fig. 2F). These results argue that the DNA-PK holoenzyme, independent of its catalytic activity, forms the initial long-range bridge between broken DNA ends.
Requirements for Short-Range Synapsis of DNA Ends
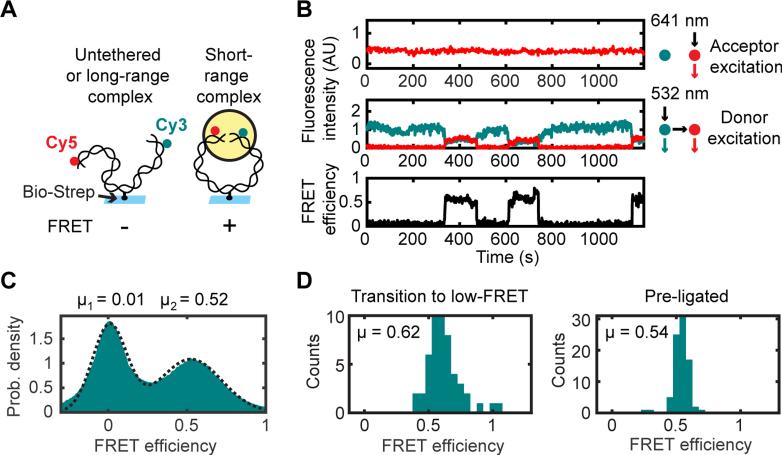
We next addressed which factors are required for the formation of the short-range synaptic complex. Because of the relatively low yield of high-FRET complexes in our intermolecular tethering assay, we designed an intramolecular NHEJ substrate to increase the frequency of collisions between DNA ends. A 2000 bp PCR product with Cy3 and Cy5 labels incorporated 7 bp from either blunt end was bound to a coverslip by an internal biotin-streptavidin attachment (Fig. 3A). Addition of egg extract led to the appearance of high-FRET complexes (Fig. 3B and S3), some of which transitioned back to a low-FRET state (Fig. 3B and S3H-O). Data compiled for many substrates showed a bimodal distribution of FRET efficiencies (Fig. 3C). The center of the high-FRET peak in this distribution (EFRET = 0.52) was similar to the FRET efficiency of DNA molecules that were pre-ligated with T4 DNA ligase before imaging in egg extract (EFRET = 0.54), indicating close alignment of DNA ends within high-FRET complexes (Fig. 3C-D). A similar mean FRET efficiency was seen for the subset of high-FRET molecules that subsequently reverted to a low-FRET state (EFRET = 0.62). These results indicate that within the short-range complex, DNA ends are closely juxtaposed, even before ligation (Fig. 3D).
Figure 3. Single-Molecule Circularization Assay.
(A) A 2-kilobase pair (kb), blunt-ended DNA labeled 7 bp from one end with Cy3 and 7 bp from the other end with Cy5 was tethered by an internal biotin-streptavidin attachment to a glass coverslip, and FRET between Cy3 and Cy5 was measured after egg extract was added to the flowcell.
(B) Multiple transitions on a single substrate between low and high FRET. Top panel, Cy5 fluorescence intensity with direct (641 nm) excitation. Middle panel, Cy3 (cyan) and Cy5 (red) fluorescence with Cy3 (532 nm) excitation. Lower panel, FRET efficiency.
(C) Histogram of FRET efficiencies in mock-immunodepleted extract, compiled over all substrates and all frames in three 30-min timecourse experiments (n = 434397 substrates * frames).
(D) Histograms of average FRET efficiencies from high-FRET segments of long timecourse trajectories. Left panel, high-FRET complexes in extract that subsequently transitioned to a low-FRET state (indicating that they were unligated; N = 50 trajectory segments). Right panel, gel-purified circular standard generated by T4 DNA ligase (N = 83 trajectory segments).
See also Figure S3.
To measure the kinetics of short-range synapsis while avoiding photobleaching, we sampled fresh fields of view every ~18 s after extract addition and obtained FRET efficiency histograms at each time point, which are displayed as kymographs in Fig. 4A-B. In unperturbed extract, substrates transitioned from an initial zero-FRET state to a final state with FRET efficiency ~0.5 (Fig. 4A). The kinetics of this short-range synapsis were quantified by plotting the fraction of substrates with FRET efficiency > 0.25 as a function of time (e.g., black curve in Fig. 4E summarizes Fig. 4A). As expected from a sequential mechanism of synaptic complex assembly, immunodepletion of Ku70/80 or DNA-PKcs, which disrupts formation of the long-range synaptic complex (Fig. 2E), also prevented formation of the short-range synaptic complex (Fig. 4C-D). In contrast to the long-range synaptic complex, the short-range synaptic complex was abolished by small-molecule inhibitors of DNA-PK or immunodepletion of XLF or XRCC4-LIG4 (Fig. 4E-F and Fig. 5A). Purified XLF protein rescued short-range synapsis in XLF-depleted extract (Fig. 4F). Analogous results were seen for short-range complex formation in the intermolecular synapsis assay, although not all comparisons were statistically significant (Fig. S4). Collectively, these results show that in addition to the factors required to form the long-range complex (Ku70/80 and DNA-PKcs), formation of the short-range complex requires DNA-PK catalytic activity, XLF, and XRCC4-LIG4.
Figure 4. Requirements for Short-Range Synapsis of DNA Ends.
(A, B) Time-resolved FRET efficiency histograms for a population of substrates in extract treated with NU-7441 (B) or DMSO solvent control (A).
(C-F) Fraction of circularization substrates with a FRET efficiency greater than 0.25 as a function of time after extract addition. Mean (thick line) and range (error bars) of all replicates. See Table S2 for sample sizes. (C) Ku-immunodepleted extract or Ku-immunodepleted extract supplemented with purified Ku70/80 (p = 0.003, t-test of overall fraction FRET positive by replicate). (D) DNA-PKcs-immunodepleted extract and mock depletion with pre-immune IgG. (p = 0.008, t-test of overall fraction FRET positive by replicate) (E) Treatment with NU-7441, another DNA-PK inhibitor (PIK-75) or DMSO solvent (p = 0.002 for DMSO vs. NU-7441, p = 0.014 for DMSO vs. PIK-75, NU-7441 vs. PIK-75 comparison not significant; t-test with Bonferroni correction of overall fraction FRET positive by replicate). Black and red curves are derived from the histograms in (A) and (B), respectively. (F) Immunodepletion of XLF and rescue with recombinant XLF protein (p = 0.0007 for ΔMock vs. ΔXLF, p = 0.0014 for ΔXLF vs. ΔXLF + XLF, ΔMock vs. ΔXLF + XLF comparison not significant, t-test with Bonferroni correction of overall fraction FRET positive by replicate).
See also Figure S4 and Table S2.
Figure 5. A Non-Catalytic Role of LIG4 in Short-Range Synapsis.
(A) Kinetics of circularization, as in Fig. 4, for extract immunodepleted of XRCC4 (ΔX4). Mean (thick line) and range (error bars) of all replicates. Successful rescue with recombinant wild-type or catalytically inactive (K278R) LIG4:XRCC4 complex (L4:X4) but not XRCC4 alone (X4). p = 0.0004 for ΔX4 vs. ΔX4 + L4:X4, p = 0.0009 for ΔX4 vs. ΔX4 + L4K278R:X4, ΔX4 vs. ΔX4 + X4 comparison not significant, t-test with Bonferroni correction of overall fraction FRET positive by replicate.
(B) Kaplan-Meier survival curves of high-FRET complexes formed in undepleted extract (N = 124) or XRCC4-depleted extract supplemented with LIG4K278R:XRCC4 (N = 180). The two curves differ significantly (p = 0.01, two-tailed log rank test).
(C) FRET efficiency histograms before and after a 1% sodium dodecyl sulfate (SDS) wash of substrates incubated for 30 min in XRCC4-depleted extract supplemented with either wild-type or catalytically inactive (K278R) LIG4:XRCC4. In the left panel, the apparent increase in the high-FRET population after SDS wash is likely due to the fact that the pre-wash histogram is compiled from a timelapse movie in extract, during which the high-FRET population was increasing.
See also Figures S4 and S5 and Table S2.
A non-catalytic role for LIG4 in short-range synapsis
Interestingly, short-range synapsis in XRCC4-depleted extract was rescued by LIG4:XRCC4 but not by XRCC4 alone (Fig. 5A and S1I), indicating that LIG4 is required for short-range synapsis of DNA ends. Catalytically inactive LIG4K278R:XRCC4 also rescued short-range complex formation, although high-FRET complexes accumulated to a lower level (Fig. 5A), were shorter-lived than high-FRET complexes formed in undepleted extract (Fig. 5B), and were dissociated by 1% SDS (Fig. 5C), consistent with synapsis without ligation. Short-range complexes formed in the presence of catalytically inactive LIG4:XRCC4 were nonetheless much longer-lived than the long-range synaptic complex (compare Fig. 2D and 5B). These results reveal that the presence of LIG4, independent of its catalytic activity, is required for a conformational transition in the synaptic complex that aligns DNA ends and stabilizes their association.
DISCUSSION
Here, we report that in addition to Ku70/80 and DNA-PK (Labhart, 1999; Postow et al., 2008; Di Virgilio and Gautier, 2005), end joining in X. laevis egg extract requires the NHEJ factors XLF and XRCC4-LIG4. These results further validate egg extracts as a physiologically realistic in vitro system for studying NHEJ. Using single-molecule fluorescence imaging in extract, we have monitored a complete double-strand break repair reaction at nanometer resolution in real time.
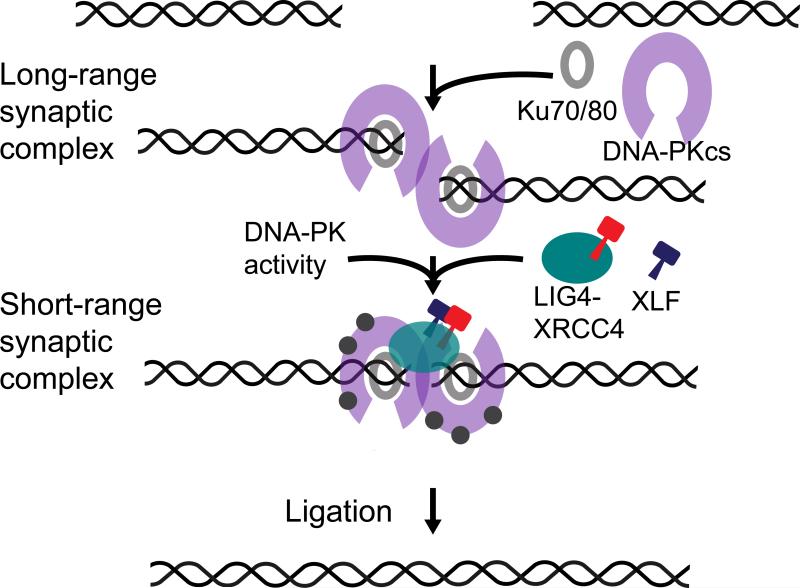
Our results identify two stages of DNA end synapsis during NHEJ (Fig. 6). Ku70/80 and DNA-PKcs form an initial long-range complex in which DNA ends are held sufficiently far apart that no FRET is detected between the Cy3 and Cy5 labels. The absence of FRET in this initial complex is not surprising given the large dimensions of DNA-PKcs (ca. 160 Å × 120 Å × 100 Å for a monomer) (Sibanda et al., 2010). Conversion of the long-range synaptic complex to the short-range complex, in which the DNA ends are closely aligned, requires DNA-PK catalytic activity, XLF, and LIG4:XRCC4, but not LIG4 catalytic activity.
Figure 6. Two-Stage Model of NHEJ Synaptic Complex Assembly.
DNA ends are initially tethered in a long-range complex and then brought together after several seconds into a short-range complex. Formation of the initial long-range complex requires Ku70/80 and DNA-PKcs, whereas formation of the short-range complex requires DNA-PK catalytic activity, XLF, and LIG4:XRCC4, but not LIG4 catalytic activity. Close alignment of the ends in the short-range complex positions them to be ligated.
Given that our extract system contains the complete soluble proteome, and that end joining depends on key NHEJ factors, it is reasonable to expect that the stepwise process of synapsis that we observe in vitro also occurs in intact cells. However, it is likely that the kinetics of the process are different between extract and intact cell nuclei, given differences in the overall concentration of DNA and repair factors. We also cannot exclude the possibility that alternative mechanisms of synapsis might occur in intact cells. For instance, DNA bridging interactions within chromatin domains larger than our in vitro substrates (Bassing and Alt, 2004) might increase the effective local concentration of DNA ends, facilitating subsequent long- and short-range complex formation.
Recently, Reid and colleagues proposed a model in which XLF, XRCC4, and LIG4 filaments initially bridge DNA ends in a side-by-side orientation. The ends subsequently slide past one another, aligning them for ligation (Reid et al., 2015). This model is based primarily on their observation that in reactions containing Cy3- and Cy5-labeled DNA duplexes and purified Ku70/80, XLF, XRCC4, and LIG4, FRET efficiency fluctuated in individual traces, resulting in a broad distribution of values. However, it is unclear whether DNA bridging complexes formed under these conditions were physiological, given that DNA-PKcs was not present in the mixture and that ligation was not shown to depend on Ku70/80 or XLF. In contrast, our extract system ligates DNA ends in a manner that requires key NHEJ factors, including DNA-PKcs. A central role for DNA-PK in synaptic complex formation is consistent with its importance for NHEJ in vivo (Dobbs et al., 2010; Jette and Lees-Miller, 2015; Jiang et al., 2015; Lees-Miller et al., 1995; Peterson et al., 1995). Notably, the model of Reid et al would predict that in our experiments, formation of the short-range complex is preceded by a transient high-FRET intermediate as donor and acceptor dyes slide past each other, yet this is not observed (Fig. 3B and S3H-O). Instead, our FRET efficiency measurements exhibit a bimodal distribution indicating that alignment of DNA ends involves a transition between discrete structural intermediates. Reid et al. also based their model on super-resolution images of fixed cells in which a fraction of immunostained XRCC4, XLF, and LIG4 foci colocalized with Ku or TUNEL staining and appeared filamentous (Reid et al., 2015). However, these experiments lack the resolution to determine the location of DNA ends relative to each other and therefore cannot distinguish between different models of synaptic complex assembly. Thus, we believe that our cell extract-based approach currently provides the highest resolution view of physiological end joining.
Our result that LIG4 is required for synapsis independent of its catalytic activity is in line with previous results from pull-down experiments in human cell extracts (Cottarel et al., 2013). While Cottarel et al. (2013) observed an overall decrease in DNA bridging in the absence of LIG4, we find that LIG4 is required for short-range but not long-range synapsis. Our observations and those of Cottarel et al. (2013) are reconciled by the fact that the short-range complex is much longer-lived than the long-range complex (Fig. 2D and 5B). Although an ensemble pull-down assay could potentially detect both complexes, our results suggest it would preferentially detect the short-range complex, which would dissociate more slowly than the long-range complex during wash steps. This interpretation also explains why depletion of Ku70/80 or DNA-PKcs, which disrupt both long- and short-range complex formation in our assay, caused a more complete DNA bridging defect than LIG4 depletion in ensemble experiments (Cottarel et al., 2013).
An important question is how end processing is coordinated with the two stages of end synapsis. Previous work suggested that autophosphorylation induces a conformational change in DNA-PKcs that makes bound DNA ends accessible to processing enzymes (Calsou et al., 1999; Ding et al., 2003; Dobbs et al., 2010; Hammel et al., 2010; Weterings et al., 2003). The DNA-PK-dependent conformational transition that we observe in the synaptic complex may therefore regulate not only ligation, but also processing of DNA ends. Consistent with this idea, XRCC4-LIG4 and XLF, which we have shown are also required for this conformational transition, are necessary for DNA-PKcs autophosphorylation and for some types of end processing (Akopiants et al., 2009; Cottarel et al., 2013; Lee et al., 2003). Coupling LIG4 recruitment with the switch to a processing-competent state would position LIG4 to ligate broken ends as soon as they are compatible (Waters et al., 2014b), helping to minimize genetic alterations during NHEJ.
EXPERIMENTAL PROCEDURES
Egg Extract Preparation
High speed supernatant (HSS) of egg cytosol was prepared as described previously (Lebofsky et al., 2009).
Cloning and Protein Purification
Bulk End Joining Assays
Extract was supplemented with nocodazole to a final concentration of 7.5 ng/μl if nocodazole had not already been added prior to immunodepletion. For the experiments shown in Fig. 1, 10 μl extract was mixed with 0.5 μl of ~20 ng/μl radiolabeled linear substrate DNA (see Supplemental Experimental Procedures for description of substrate preparation), 1 μl of 1 μg/μl closed-circular pBluescript II DNA, and 0.3 μl of an ATP regeneration mixture containing 65 mM ATP, 650 mM phosphocreatine, and 160 ng/μl creatine phosphokinase (Sigma, Cat. # C-3755; Type I from rabbit muscle). The addition of closed-circular “carrier” DNA to the reaction was necessary for efficient end joining of dilute linear substrate, similar to the dependence on total DNA concentration seen for DNA replication in extract (Lebofsky et al., 2011). An initial 2 μl sample (“0 min”) was withdrawn while the reactions were on ice and mixed with 5 μl stop solution/loading dye (80 mM Tris, pH 8, 8 mM EDTA, 0.13% phosphoric acid, 10% Ficoll, 5% SDS, 0.2% bromophenol blue). Reactions were transferred to room temperature, and additional 2 μl samples were withdrawn at the indicated times and mixed with 5 μl stop solution/loading dye. Samples were digested at room temperature for at least 1 h with 1 μg proteinase K per sample, and products were separated by electrophoresis on a 1x Tris-borate-EDTA (TBE), 0.8% agarose gel. Gels were sandwiched between filter paper and a HyBond-XL nylon membrane (GE Healthcare), dried on a gel dryer, and exposed to a storage phosphorscreen, which was imaged using a Personal Molecular Imager (BioRad) or Typhoon FLA 7000 imager (GE Healthcare Life Sciences).
Antibody Generation
Antibodies were raised in rabbits by New England Peptide against synthetic peptides corresponding to C-terminal sequences from X. laevis XLF [Ac-CGASKPKKKAKGLFM-OH], XRCC4 (Ac-CKNTPDPDDLFSDI-OH), and Ku80 (Ac-CMEDEGDVDDLLDMM-OH). Antibodies were affinity purified by the supplier using resin coupled to the corresponding peptide through its N-terminal cysteine. Anti-DNA-PKcs antibody was raised by Pocono Rabbit Farm and Laboratory against an insoluble fragment of X. laevis DNA-PKcs spanning the PIKK and FATC domains (see Supplemental Experimental Procedures). For affinity purification of antibody, antigen dissolved in 1x PBS with 6 M urea and 5 mM β-mercaptoethanol was coupled to AminoLink Coupling Resin (Thermo Fisher Scientific) following the manufacturer's instructions. Rabbit serum was passed over antibody-coupled resin by gravity flow, and the resin was washed with 1x PBS. Bound antibody was eluted with 200 mM glycine, pH 2.6, and elution fractions were rapidly neutralized with 0.14 volumes of 1 M Tris-HCl, pH 8.8. Mock IgG for the experiment in Fig. 1B was purified by protein A sepharose affinity chromatography from pre-immune serum of the rabbit used to produce anti-DNA-PKcs antibody. Antibody was eluted from the resin as described above. Details about immunodepletion and rescue experiments can be found in Supplemental Experimental Procedures.
Western Blotting
Extract samples for western blotting were diluted with four volumes of SDS-PAGE sample buffer, and additional serial dilutions were made of undepleted extract in SDS-PAGE sample buffer. Samples were separated by SDS-PAGE, transferred to PVDF membranes, and probed with the indicated antibodies. Details can be found in Supplemental Experimental Procedures.
Single-Molecule Tethering Assay
Biotin-100 bp-Cy3 duplexes (see Supplemental Experimental Procedures) were tethered at a 1:1000 dilution on a streptavidin-coated coverslip in degassed egg lysis buffer (ELB; 10 mM HEPES, pH 7.7, 50 mM KCl, 2.5 mM MgCl2) for 3-5 min, after which the flowcell was washed with 200 μl ELB (see Supplemental Experimental Procedures for details about microscope and flowcell construction). 25 μl HSS was mixed with 2.5 μl of 1 mg/ml closed-circular pBluescript II DNA, 0.8 μl of ATP regeneration mix (see “Bulk End Joining Assays”), 0.6 μl of 250 mM protocatechuic acid (PCA) in ELB (adjusted to pH 7.7), 0.6 μl of 5 μM protocatechuate 3,4-dioxygenase (PCD; storage buffer 10 mM HEPES, pH 7.5, 50 mM KCl, 1.25 mM MgCl2, 50% glycerol), 0.6 μl of 50 mM Trolox (6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid) in DMSO, and 1 μl of 100 nM Cy5-labeled 100 bp duplex, in that order. PCA and PCD form an oxygen-scavenging system, while Trolox serves as a triplet-state quencher. The extract was drawn into the channel, and images were acquired continuously at a rate of 1 s/frame with alternating 532 nm and 641 nm laser excitation. The surface power density was 4 W/cm2 for the 532 nm laser and 0.9 W/cm2 for the 641 nm laser. To determine the nonspecific background rate of tethering (Fig. 2E), Biotin-100 bp-Cy3 duplexes were omitted, and the correct plane of focus was maintained by imaging 605 nm quantum dots (Life Technologies) nonspecifically adsorbed to the surface. For Cy5 photostability measurements (Fig. 2D), a biotinylated, Cy5-labeled PCR product was imaged in egg extract under the same imaging conditions. Data were analyzed using custom MATLAB scripts (see Supplemental Experimental Procedures).
Single-Molecule Circularization Assay
The intramolecular end joining substrate shown in Fig. 3A (see Supplemental Experimental Procedures) was tethered to a streptavidin-coated coverslip within a flowcell channel. An extract mixture was prepared as described above for single-molecule tethering experiments and drawn into the flowcell. For the experiments shown in Fig. 3C, 4, and 5A, 100 ms exposures were taken stroboscopically every 1 s, alternating between two frames of 532 nm excitation (surface power density 16 W/cm2) and one frame of 641 nm excitation (surface power density 7 W/cm2). A different field of view was typically imaged every 18 frames. To obtain the kymographs shown in Fig. 4A-B and the kinetic traces shown in Fig. 4C-F and 5A, FRET efficiency data from all replicates were pooled and binned in 36 s windows. For long timecourse imaging (Fig. 3B, 3D, 5B, and S3), images were taken continuously at a rate of 1 frame/s, alternating between 2 frames of 532 nm excitation (surface power density 4 W/cm2) and 1 frame of 641 nm excitation (surface power density 0.9 W/cm2). Data analysis is described in Supplemental Experimental Procedures.
Supplementary Material
Non-homologous end joining (NHEJ) was monitored on single DNA molecules
Synapsis of DNA ends during NHEJ proceeds through two distinct stages.
Different NHEJ factors are required at different stages of synapsis.
DNA-PK activity is required to transition between the two synaptic complexes.
ACKNOWLEDGMENTS
We thank members of the Walter and Loparo laboratories for helpful discussions, James Kath for homemade Pfu polymerase, Hyeongjun Kim and Jacob Sargent for help with microscope construction and advice on FRET experiments, Dan Floyd for calibration grid fabrication, Ravi Amunugama for advice on protein expression in Sf9 cells, Hironori Funabiki for plasmids and a sample of his lab's anti-Ku80 antibody, Martin Wühr for help searching X. laevis sequence databases, Ross Tamaino at the Taplin Mass Spectrometry Facility for assistance with mass spectrometry, and Katheryn Meek for anti-DNA-PKcs mouse monoclonal antibody used for western blotting. We would also like to thank Jennifer Waters and Talley Lambert at the Harvard Medical School Nikon Imaging Facility for assistance with preliminary time course FRET experiments. This work was funded by a National Science Foundation Graduate Research Fellowship (to T.G.W.G.), a National Institutes of Health grant R01GM115487 (to J.J.L), the Stewart Trust Fellows Award (to J.J.L.), and the Howard Hughes Medical Institute (J.C.W.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
All authors designed experiments and wrote the manuscript. T.G.W.G. performed experiments and data analysis.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, 3 Supplemental Text items, 5 figures, and 2 tables.
REFERENCES
- Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 2006;124:301–313. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- Akopiants K, Zhou R-Z, Mohapatra S, Valerie K, Lees-Miller SP, Lee K-J, Chen DJ, Revy P, de Villartay J-P, Povirk LF. Requirement for XLF/Cernunnos in alignment-based gap filling by DNA polymerases lambda and mu for nonhomologous end joining in human whole-cell extracts. Nucleic Acids Res. 2009;37:4055–4062. doi: 10.1093/nar/gkp283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassing CH, Alt FW. H2AX may function as an anchor to hold broken chromosomal DNA ends in close proximity. Cell Cycle. 2004;3:149–153. doi: 10.4161/cc.3.2.689. [DOI] [PubMed] [Google Scholar]
- Bryans M, Valenzano MC, Stamato TD. Absence of DNA ligase IV protein in XR-1 cells: Evidence for stabilization by XRCC4. Mutat. Res. - DNA Repair. 1999;433:53–58. doi: 10.1016/s0921-8777(98)00063-9. [DOI] [PubMed] [Google Scholar]
- Buck D, Malivert L, de Chasseval R, Barraud A, Fondanèche M-C, Sanal O, Plebani A, Stéphan J-L, Hufnagel M, le Deist F, et al. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- Calsou P, Frit P, Humbert O, Muller C, Chen DJ, Salles B. The DNA-dependent protein kinase catalytic activity regulates DNA end processing by means of Ku entry into DNA. J. Biol. Chem. 1999;274:7848–7856. doi: 10.1074/jbc.274.12.7848. [DOI] [PubMed] [Google Scholar]
- Carter T, Vancurová I, Sun I, Lou W, DeLeon S. A DNA-activated protein kinase from HeLa cell nuclei. Mol. Cell. Biol. 1990;10:6460–6471. doi: 10.1128/mcb.10.12.6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary RB, Peterson SR, Wang J, Bear DG, Bradbury EM, Chen DJ. DNA looping by Ku and the DNA-dependent protein kinase. Proc. Natl. Acad. Sci. U. S. A. 1997;94:4267–4272. doi: 10.1073/pnas.94.9.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiruvella KK, Liang Z, Wilson TE. Repair of double-strand breaks by end joining. Cold Spring Harb. Perspect. Biol. 2013;5 doi: 10.1101/cshperspect.a012757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottarel J, Frit P, Bombarde O, Salles B, Négrel A, Bernard S, Jeggo PA, Lieber MR, Modesti M, Calsou P. A noncatalytic function of the ligation complex during nonhomologous end joining. J. Cell Biol. 2013;200:173–186. doi: 10.1083/jcb.201203128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craxton A, Somers J, Munnur D, Jukes-Jones R, Cain K, Malewicz M. XLS (c9orf142) is a new component of mammalian DNA double-stranded break repair. Cell Death Differ. 2015;22:890–897. doi: 10.1038/cdd.2015.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Critchlow SE, Bowater RP, Jackson SP. Mammalian DNA double- strand break repair protein XRCC4 interacts with DNA ligase IV. Curr. Biol. 1997;7:588–598. doi: 10.1016/s0960-9822(06)00258-2. [DOI] [PubMed] [Google Scholar]
- DeFazio LG, Stansel RM, Griffith JD, Chu G. Synapsis of DNA ends by DNA-dependent protein kinase. EMBO J. 2002;21:3192–3200. doi: 10.1093/emboj/cdf299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Reddy YVR, Wang W, Woods T, Douglas P, Ramsden DA, Lees- Miller SP, Meek K. Autophosphorylation of the catalytic subunit of the DNA-dependent protein kinase is required for efficient end processing during DNA double-strand break repair. Mol. Cell. Biol. 2003;23:5836–5848. doi: 10.1128/MCB.23.16.5836-5848.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbs TA, Tainer JA, Lees-Miller SP. A structural model for regulation of NHEJ by DNA-PKcs autophosphorylation. DNA Repair (Amst) 2010;9:1307–1314. doi: 10.1016/j.dnarep.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvir A, Peterson SR, Knuth MW, Lu H, Dynan WS. Ku autoantigen is the regulatory component of a template-associated protein kinase that phosphorylates RNA polymerase II. Proc. Natl. Acad. Sci. U. S. A. 1992;89:11920–11924. doi: 10.1073/pnas.89.24.11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvir A, Stein LY, Calore BL, Dynan WS. Purification and characterization of a template-associated protein kinase that phosphorylates RNA polymerase II. J. Biol. Chem. 1993;268:10440–10447. [PubMed] [Google Scholar]
- Gottlieb TM, Jackson SP. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell. 1993;72:131–142. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- Grawunder U, Wilm M, Wu X, Kulesza P, Wilson TE, Mann M, Lieber MR. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature. 1997;388:492–495. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- Gu J, Lu H, Tsai AG, Schwarz K, Lieber MR. Single-stranded DNA ligation and XLF-stimulated incompatible DNA end ligation by the XRCC4-DNA ligase IV complex: influence of terminal DNA sequence. Nucleic Acids Res. 2007;35:5755–5762. doi: 10.1093/nar/gkm579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammel M, Yu Y, Mahaney BL, Cai B, Ye R, Phipps BM, Rambo RP, Hura GL, Pelikan M, So S, et al. Ku and DNA-dependent protein kinase dynamic conformations and assembly regulate DNA binding and the initial non-homologous end joining complex. J. Biol. Chem. 2010;285:1414–1423. doi: 10.1074/jbc.M109.065615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob B, Splinter J, Durante M, Taucher-Scholz G. Live cell microscopy analysis of radiation-induced DNA double-strand break motion. Proc. Natl. Acad. Sci. U. S. A. 2009;106:3172–3177. doi: 10.1073/pnas.0810987106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jette N, Lees-Miller SP. The DNA-dependent protein kinase: A multifunctional protein kinase with roles in DNA double strand break repair and mitosis. Prog Biophys Mol Biol. 2015;117:194–205. doi: 10.1016/j.pbiomolbio.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Crowe JL, Liu X, Nakajima S, Wang Y, Li C, Lee BJ, Dubois RL, Liu C, Yu X, et al. Differential phosphorylation of DNA-PKcs regulates the interplay between end-processing and end-ligation during nonhomologous end-joining. Mol. Cell. 2015;58:172–185. doi: 10.1016/j.molcel.2015.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruhlak MJ, Celeste A, Dellaire G, Fernandez-Capetillo O, Müller WG, McNally JG, Bazett-Jones DP, Nussenzweig A. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J. Cell Biol. 2006;172:823–834. doi: 10.1083/jcb.200510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labhart P. Ku-dependent nonhomologous DNA end joining in Xenopus egg extracts. Mol. Cell. Biol. 1999;19:2585–2593. doi: 10.1128/mcb.19.4.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskey RA, Mills AD, Morris NR. Assembly of SV40 chromatin in a cell-free system from Xenopus eggs. Cell. 1977;10:237–243. doi: 10.1016/0092-8674(77)90217-3. [DOI] [PubMed] [Google Scholar]
- Lebofsky R, Takahashi T, Walter JC. DNA replication in nucleus-free Xenopus egg extracts. Methods Mol. Biol. 2009;521:229–252. doi: 10.1007/978-1-60327-815-7_13. [DOI] [PubMed] [Google Scholar]
- Lebofsky R, van Oijen AM, Walter JC. DNA is a co-factor for its own replication in Xenopus egg extracts. Nucleic Acids Res. 2011;39:545–555. doi: 10.1093/nar/gkq739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Yannone SM, Chen DJ, Povirk LF. Requirement for XRCC4 and DNA ligase IV in alignment-based gap filling for nonhomologous DNA end joining in vitro. Cancer Res. 2003;63:22–24. [PubMed] [Google Scholar]
- Lees-Miller SP, Chen YR, Anderson CW. Human cells contain a DNA- activated protein kinase that phosphorylates simian virus 40 T antigen, mouse p53, and the human Ku autoantigen. Mol. Cell. Biol. 1990;10:6472–6481. doi: 10.1128/mcb.10.12.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees-Miller SP, Godbout R, Chan DW, Weinfeld M, Day RS, Barron GM, Allalunis-Turner J. Absence of p350 subunit of DNA-activated protein kinase from a radiosensitive human cell line. Science. 1995;267:1183–1185. doi: 10.1126/science.7855602. [DOI] [PubMed] [Google Scholar]
- Lu H, Pannicke U, Schwarz K, Lieber MR. Length-dependent binding of human XLF to DNA and stimulation of XRCC4.DNA ligase IV activity. J. Biol. Chem. 2007;282:11155–11162. doi: 10.1074/jbc.M609904200. [DOI] [PubMed] [Google Scholar]
- Lucas JS, Zhang Y, Dudko OK, Murre C. 3D trajectories adopted by coding and regulatory DNA elements: First-passage times for genomic interactions. Cell. 2014;158:339–352. doi: 10.1016/j.cell.2014.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Lu H, Schwarz K, Lieber MR. Repair of double-strand DNA breaks by the human non-homologous DNA end joining pathway: The iterative processing model. Cell Cycle. 2005;4:1193–1200. doi: 10.4161/cc.4.9.1977. [DOI] [PubMed] [Google Scholar]
- Mahaney BL, Hammel M, Meek K, Tainer JA, Lees-Miller SP. XRCC4 and XLF form long helical protein filaments suitable for DNA end protection and alignment to facilitate DNA double strand break repair. Biochem. Cell Biol. 2013;91:31–41. doi: 10.1139/bcb-2012-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochi T, Blackford AN, Coates J, Jhujh S, Mehmood S, Tamura N, Travers J, Wu Q, Draviam VM, Robinson CV, et al. DNA repair. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair. Science. 2015 doi: 10.1126/science.1261971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson SR, Kurimasa A, Oshimura M, Dynan WS, Bradbury EM, Chen DJ. Loss of the catalytic subunit of the DNA-dependent protein kinase in DNA double-strand-break-repair mutant mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 1995;92:3171–3174. doi: 10.1073/pnas.92.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postow L, Ghenoiu C, Woo EM, Krutchinsky AN, Chait BT, Funabiki H. Ku80 removal from DNA through double strand break-induced ubiquitylation. J. Cell Biol. 2008;182:467–479. doi: 10.1083/jcb.200802146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan SK, Jette N, Lees-Miller SP. Non-homologous end joining: Emerging themes and unanswered questions. DNA Repair (Amst) 2014;17:2–8. doi: 10.1016/j.dnarep.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsden DA, Gellert M. Ku protein stimulates DNA end joining by mammalian DNA ligases: a direct role for Ku in repair of DNA double-strand breaks. EMBO J. 1998;17:609–614. doi: 10.1093/emboj/17.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid DA, Keegan S, Leo-Macias A, Watanabe G, Strande NT, Chang HH, Oksuz BA, Fenyo D, Lieber MR, Ramsden DA, et al. Organization and dynamics of the nonhomologous end-joining machinery during DNA double-strand break repair. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E2575–E2584. doi: 10.1073/pnas.1420115112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, de Melo AJ, Xu Y, Tadi SK, Négrel A, Hendrickson E, Modesti M, Meek K. XRCC4/XLF interaction is variably required for DNA repair, and is not required for Ligase IV stimulation. Mol. Cell. Biol. 2015 doi: 10.1128/MCB.01503-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibanda BL, Chirgadze DY, Blundell TL. Crystal structure of DNA PKcs reveals a large open-ring cradle comprised of HEAT repeats. Nature. 2010;463:118–121. doi: 10.1038/nature08648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutoglou E, Dorn JF, Sengupta K, Jasin M, Nussenzweig A, Ried T, Danuser G, Misteli T. Positional stability of single double-strand breaks in mammalian cells. Nat. Cell Biol. 2007;9:675–682. doi: 10.1038/ncb1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spagnolo L, Rivera-Calzada A, Pearl LH, Llorca O. Three-Dimensional Structure of the Human DNA-PKcs/Ku70/Ku80 Complex Assembled on DNA and Its Implications for DNA DSB Repair. Mol. Cell. 2006;22:511–519. doi: 10.1016/j.molcel.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Thode S, Schäfer A, Pfeiffer P, Vielmetter W. A novel pathway of DNA end-to-end joining. Cell. 1990;60:921–928. doi: 10.1016/0092-8674(90)90340-k. [DOI] [PubMed] [Google Scholar]
- Tsai CJ, Kim SA, Chu G. Cernunnos/XLF promotes the ligation of mismatched and noncohesive DNA ends. Proc. Natl. Acad. Sci. U. S. A. 2007;104:7851–7856. doi: 10.1073/pnas.0702620104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Virgilio M, Gautier J. Repair of double-strand breaks by nonhomologous end joining in the absence of Mre11. J. Cell Biol. 2005;171:765–771. doi: 10.1083/jcb.200506029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters CA, Strande NT, Wyatt DW, Pryor JM, Ramsden DA. Nonhomologous end joining: A good solution for bad ends. DNA Repair (Amst) 2014a;17:39–51. doi: 10.1016/j.dnarep.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters CA, Strande NT, Pryor JM, Strom CN, Mieczkowski P, Burkhalter MD, Oh S, Qaqish BF, Moore DT, Hendrickson EA, et al. The fidelity of the ligation step determines how ends are resolved during nonhomologous end joining. Nat. Commun. 2014b;5:4286. doi: 10.1038/ncomms5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weterings E, Verkaik NS, Brüggenwirth HT, Hoeijmakers JHJ, van Gent DC. The role of DNA dependent protein kinase in synapsis of DNA ends. Nucleic Acids Res. 2003;31:7238–7246. doi: 10.1093/nar/gkg889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing M, Yang M, Huo W, Feng F, Wei L, Jiang W, Ning S, Yan Z, Li W, Wang Q, et al. Interactome analysis identifies a new paralogue of XRCC4 in non-homologous end joining DNA repair pathway. Nat. Commun. 2015;6:6233. doi: 10.1038/ncomms7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha S, Alt FW, Cheng H-L, Brush JW, Li G. Defective DNA repair and increased genomic instability in Cernunnos-XLF-deficient murine ES cells. Proc. Natl. Acad. Sci. U. S. A. 2007;104:4518–4523. doi: 10.1073/pnas.0611734104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.