Abstract
Background
Microbes have been extensively explored for production of environment-friendly fuels and chemicals. The microbial fermentation pathways leading to these commodities usually involve many redox reactions. This makes the fermentative production of highly reduced products challenging, because there is a limited NADH output from glucose catabolism. Microbial production of n-butanol apparently represents one typical example.
Results
In this study, we addressed the issue by adjustment of the intracellular redox state in Escherichia coli. This was initiated with strain BuT-8 which carries the clostridial CoA-dependent synthetic pathway. Three metabolite nodes in the central metabolism of the strain were targeted for engineering. First, the pyruvate node was manipulated by enhancement of pyruvate decarboxylation in the oxidative pathway. Subsequently, the pentose phosphate (PP) pathway was amplified at the glucose-6-phosphate (G6P) node. The pathway for G6P isomerization was further blocked to force the glycolytic flux through the PP pathway. It resulted in a growth defect, and the cell growth was later recovered by limiting the tricarboxylic acid cycle at the acetyl-CoA node. Finally, the resulting strain exhibited a high NADH level and enabled production of 6.1 g/L n-butanol with a yield of 0.31 g/g-glucose and a productivity of 0.21 g/L/h.
Conclusions
The production efficiency of fermentative products in microbes strongly depends on the intracellular redox state. This work illustrates the flexibility of pyruvate, G6P, and acetyl-CoA nodes at the junction of the central metabolism for engineering. In principle, high production of reduced products of interest can be achieved by individual or coordinated modulation of these metabolite nodes.
Electronic supplementary material
The online version of this article (doi:10.1186/s13068-016-0467-4) contains supplementary material, which is available to authorized users.
Keywords: n-Butanol, The redox state, Metabolic engineering, Escherichia coli
Background
Our daily life is tightly linked to the petroleum-based industries. However, the rising price, the insecure supply, and the environmental concern of fossil fuels have currently overshadowed these industries. Consequently, it provokes the demand for renewable and environment-friendly fuels and chemicals [1]. The bioprocess production of these chemical commodities appears to be appealing [2, 3]. Bio-based fuels and chemicals of interest are generally fermentative products of living microbes. The microbial fermentation pathways involve many redox reactions, which usually require NADH and NAD+ as cofactors. With NAD+ as an electron acceptor, the oxidation of sugars produces NADH. NAD+ is regenerated when intermediate metabolites in the sugar catabolism are subsequently reduced at the expense of NADH. The result of the reductive reactions usually leads to production of ethanol, lactate, and succinate as exemplified in fermentative Escherichia coli [4]. Therefore, maintaining the redox balance of NADH and NAD+ is a key to ensure the continued operation of cellular metabolism under the fermentative condition.
Fermentative production of n-butanol in Clostridium species is a well-known bioprocess [5]. The fermentation process mainly consists of the acidogenesis and the solventogenesis phase [6]. In the acidogenesis stage, the growing Clostridium species ferment glucose to mainly produce acetate and butyrate. Upon reaching the stationary growth phase, the bacteria re-assimilate these organic acids, while acetone, n-butanol, and ethanol are produced as end products. In view of the clostridial synthetic pathway, the direct synthesis of n-butanol from glucose can cause the NADH/NAD+ redox imbalance because more NADH is required in the synthetic pathway than that generated in the glycolytic pathway. This may explain why the synthesis of n-butanol in Clostridium species proceeds in two stages.
n-Butanol is an alternative fuel of potential because its property is superior to ethanol in terms of the energy density, the vapor pressure, and hygroscopicity [7]. Moreover, n-butanol can be used for the transportation fuel after blended with gasoline at any concentrations and transported with the existing pipeline infrastructure [8]. These merits make microbial production of n-butanol industrially attractive. Many approaches have been proposed for production of n-butanol in a variety of surrogate strains [9–12]. However, these attempts are generally discouraged by their low n-butanol titer. Recognized as the biotechnology workhorse, E. coli has been commonly employed to produce value-added chemicals and biofuels [13, 14]. Production of n-butanol in E. coli is proven feasible after introduction of the clostridial synthetic pathway into the strain [9, 15, 16]. In addition, the production titer is improved by manipulating pyruvate dehydrogenase (PDH) and formate dehydrogenase (FDH) in glycolysis to increase the NADH availability in the cell [15, 17, 18]. Recently, we have proposed a new production platform based on two strains: a butyrate-conversion strain and a butyrate-producing strain [19]. The dual-culture system rebuilds a redox-balanced synthetic pathway, which enables effective production of n-butanol.
Microbial production of a highly reduced fermentation product such as n-butanol remains challenging because there is a limited NADH output from glucose catabolism. This issue was addressed by rerouting the central metabolic pathways in E. coli. The NADH availability is manifested by the interplay of glycolysis, the pentose phosphate (PP) pathway, and the tricarboxylic acid (TCA) cycle involved in central metabolism (Fig. 1). In addition to PDH and FDH, the enzymes that regulate the metabolite pools at the junction of central metabolism were systematically manipulated to modulate the intracellular NADH. As a result, the engineered strain with the remodeled pathways enabled effective production of n-butanol.
Fig. 1.
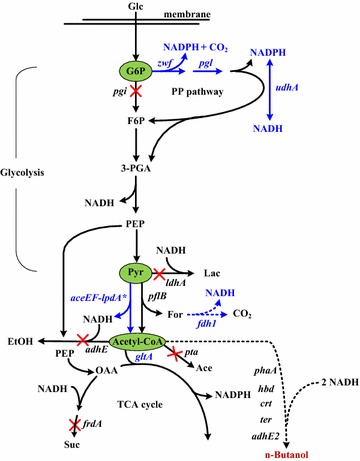
The central metabolic pathways leading to n-butanol in E. coli. The dotted lines denote the heterologous pathways. The CoA-dependent synthetic pathway of n-butanol is composed of heterologous phaA, hbd, crt, ter, and adhE2 genes as shown. Three metabolite nodes including G6P, pyruvate, and acetyl-CoA are targeted for engineering and marked. The genes involved in the metabolic pathways: aceEF-lpdA*, pyruvate dehydrogenase complex; adhE, aldehyde-alcohol dehydrogenase; adhE2, butyraldehyde-butanol dehydrogenase; crt, crotonese; gltA, citrate synthase; hbd, 3-hydroxybutyryl-CoA dehydrogenase; ldhA, lactate dehydrogenase; fdh1, formate dehydrogenase; frdA, subunit of fumarate reductase; pflB, pyruvate formate-lyase; pgi, phosphoglucose isomerase; pgl, lactonase; phaA, acetoacetyl-CoA thiolase; pta, phosphate acetyltransferase; ter, trans-enoyl-CoA reductase; udhA, transhydrogenase; zwf, glucose-6-phosphate dehydrogenase. The deleted genes are indicated by “X”. Abbreviations: Ace acetate; EtOH ethanol; F6P fructose-6-phosphate; Lac lactate; For formate; G6P glucose-6-phosphate; Glc glucose; OAA oxaloacetate; PEP phosphoenolpyruvate; 3-PGA 3-phosphoglyceraldehyde; Pyr pyruvate; Suc succinate
Results and discussion
Amplification of the pyruvate oxidation pathway
As shown in Fig. 1, the reductive synthesis of one n-butanol from one glucose requires more NADH than that provided in glycolysis. Therefore, the approach of NADH replenishment is expected to favor the fermentative production of n-butanol. In this context, the pyruvate node connecting glycolysis and the TCA cycle appears to be a potential target for manipulation. In E. coli, pyruvate is oxidized to acetyl-CoA by a reaction mediated by PDH under the aerobic growth and by pyruvate formate-lyase (PFL) under the fermentative growth [4]. Formate is the reduced product of the PFL reaction. Found in other microbes, FDH such as Candida boidinii fdh and Saccharomyces cerevisiaefdh1 catalyzes oxidation of formate to CO2 associated with NADH generation [20]. These two genes have been employed in E. coli to elevate intracellular NADH, resulting in an increase in n-butanol production [17, 21]. Accordingly, S. cerevisiaefdh1 under the control of the trc promoter (Ptrc) without lacO was integrated into strain BuT-8. Strain BuT-8 was previously constructed with a CoA-dependent pathway of n-butanol consisting of the constitutive λPL promoter (PλPL)-driven hbd, crt, and adhE2 of Clostridium acetobutylicum, phaA of Cupriavidus necator, and ter of Treponema denticola [19]. In addition, the undesired pathways involving endogenous adhE, ldhA, pta, and frdA were removed from this strain to curtail carbon waste and conserve NADH. Equipped with S. cerevisiaefdh1, the resulting strain BuT-8-Fdh1 produced 3.1 g/L n-butanol at 24 h (Fig. 2a). This production titer accounts for a 25 % increase over that for strain BuT-8 (Table 1).
Fig. 2.
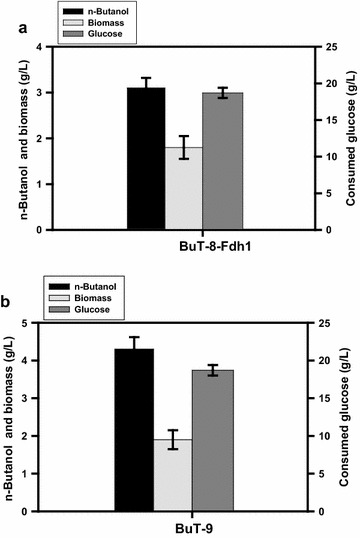
Production of n-butanol in strains with the amplification of the pyruvate oxidation pathway. The engineered E. coli strains were grown in M9Y medium containing 20 g/L glucose. The fermentations were conducted under the oxygen-limited condition for 24 h. The experiments were conducted in triplicate. Keys: a the fermentation performance for strain BuT-8-Fdh1; b the fermentation performance for strain BuT-9
Table 1.
Summary of the fermentation kinetics for main engineered strains
| Strain | YB (g/L) | PB (g/L/h) | YB/G (g/g) | NADH (μmol/g cell) | Specific enzyme activity (U/mg protein) | |||
|---|---|---|---|---|---|---|---|---|
| PDH | Zwf | Pgl | GltA | |||||
| BuT-8 | 2.7a | 0.11a | 0.14a | 42.2 | 0.7 | ND | ND | ND |
| BuT-9 | 4.3 | 0.18 | 0.22 | 60.9 | 1.6 | 6.1 | ND | ND |
| BuT-10 | 4.9 | 0.20 | 0.25 | 75.8 | 18.3 | 0.5 | ND | ND |
| BuT-12 | 5.4 | 0.23 | 0.27 | 82.6 | ND | ND | 6.2 | 2.2 |
| BuT-14 | 6.1 | 0.21 | 0.31 | 96.1 | ND | ND | ND | 1.5 |
In contrast to PFL, the PDH reaction generates NADH as the reduced product. Therefore, manipulation of the PDH level is expected to alter intracellular NADH. This was conducted by fusion of PλPL with aceEF operon to enhance the gene expression in strain BuT-8-Fdh1. To render PDH less sensitive to NADH inhibition, the endogenous lpdA (encoding dihydrolipoamide dehydrogenase) was deleted and a mutation site (E354K) in lpdA was additionally created [22]. The mutant lpdA* under the control of PλPL was then inserted into strain BuT-8-Fdh1 to obtain strain BuT-9. Consequently, strain BuT-9 exhibited 1.3-fold higher PDH activity and 45 % higher NADH level as compared to strain BuT-8 (Table 1). The accumulated pyruvate in strain BuT-8 was greatly reduced and carbon flux was diverted from the synthetic pathways of byproducts (Table 2). Strain BuT-9 finally produced 4.3 g/L n-butanol (Fig. 2b), which accounts for a 60 % increase over that in strain BuT-8 (Table 1).
Table 2.
Carbon recovery of fermentation products for engineered strains during the oxygen-limited growth on glucose
| Strain | Pyruvate | Succinate | Ethanol | Lactate | Acetate | Butyrate | Butanol | Total (%) |
|---|---|---|---|---|---|---|---|---|
| BuT-8 | 20.7 | 3.5 | 7.8 | 2.7 | 1.5 | 4.1 | 21.9 | 62.2 |
| BuT-9 | 2.6 | 2.1 | 5.2 | 1.8 | 1.2 | 2.7 | 34.8 | 50.4 |
| BuT-10 | 0.01 | 1.1 | 5.2 | 1.5 | 0.8 | 2.0 | 39.6 | 50.2 |
| BuT-12 | Nil | 0.8 | 5.2 | 1.4 | 0.7 | 1.4 | 43.7 | 53.2 |
| BuT-14 | Nil | 0.6 | 2.6 | 1.3 | 0.5 | 1.4 | 49.4 | 55.8 |
Carbon recovery was calculated as the molar percent of carbon in products per carbon in consumed glucose. Nil, carbon recovery less than 0.01
The similar approach has been previously applied for n-butanol production in E. coli that carries the CoA-dependent synthetic pathway. By using C. boidinii fdh, the best strain in their study showed a 1.3-fold increase in n-butanol production [17]. In another work, a 1.6-fold improvement in the production yield was reported for a strain with enhanced PDH [15]. In addition, it was reported that a strain with the optimal activation of PDH exhibited a 12 % improvement in n-butanol production [18]. Further improvement of the strain by optimization of S. cerevisiae fdh1 expression led to a 35.4 % increase in the production titer [18]. These studies were conducted using super-rich TB medium, in contrast to ours which employed M9Y medium. Although displaying a various degree of improvement in the n-butanol production, aforementioned studies and ours illustrate the feasibility in manipulating the pyruvate node to modulate intracellular NADH.
Amplification of the pentose phosphate pathway
The glycolytic pathway bifurcates at the glucose-6-phosphate (G6P) node (Fig. 1). With G6P as a starting metabolite, the PP pathway produces precursors for the synthesis of nucleic acids and aromatic amino acids and also provides a major source of NADPH for biosynthesis which is involved in most of the reductive pathways [4]. It is possible to increase NADH availability by manipulation of the G6P node. G6P dehydrogenase (encoded by zwf) catalyzes the first step in the PP pathway. Therefore, zwf of strain BuT-9 was fused to PλPL. In E. coli, pyridine nucleotide transhydrogenase (encoded by udhA) functions to interconvert NADPH and NADH [23]. Therefore, strain BuT-10 was obtained by further fusion udhA of strain BuT-9 with PλPL to enhance its expression. As compared to strain BuT-9, strain BuT-10 displayed twofold higher Zwf activity and a 10 % increase in the n-butanol production (4.9 g/L) (Table 1; Fig. 3a).
Fig. 3.
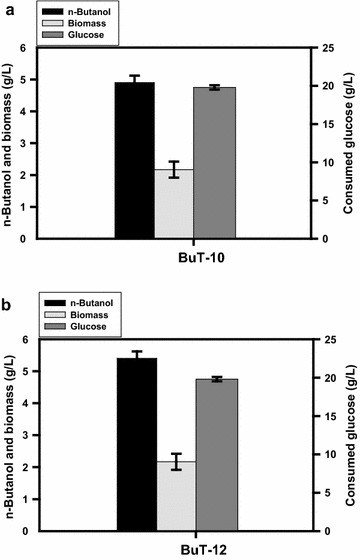
Production of n-butanol in strains with the amplification of the PP pathway. The engineered E. coli strains were grown in M9Y medium containing 20 g/L glucose. The fermentations were conducted under the oxygen-limited condition for 24 h. The experiments were conducted in triplicate. Keys: a the fermentation performance for strain BuT-10; b the fermentation performance for strain BuT-12
The developed strain is derived from strain BL21 which lacks pgl [24], a gene encoding lactonase that is responsible for the reaction following Zwf in the PP pathway. It is likely that yieK with an annotated function of Pgl is functioning in E. coli B strain but less active. Therefore, the carbon flux channeled into the PP pathway by elevated Zwf may be limited at the Pgl-mediated reaction step. To address this issue, the PλPL-driven pgl from E. coli K-12 strain was re-introduced into strain BuT-10. Finally, the resulting strain (BuT-12) enabled production of 5.4 g/L n-butanol (Fig. 3b). As compared to strain BuT-10, strain BuT-12 displayed a tenfold higher Pgl activity, a 36 % more NADH, and a 25.6 % improvement in the n-butanol production (Table 1). The approach by enhancing the PP pathway results in more NADH production, which drives more acetyl-CoA into the synthetic pathway of n-butanol. This is supported by the observed decrease in pyruvate and succinate (Table 2).
It is apparent that redistribution of carbon flux in glycolysis and the PP pathway can greatly affect intracellular NADH level. Notice that entry of one glucose into the oxidative PP pathway generates two reducing equivalents but wastes one CO2. Nevertheless, strain BuT-12 which is manipulated at the pyruvate and G6P nodes displays a 96 % increase in the NADH level and doubles the n-butanol production as compared to strain BuT-8 (Table 1).
Rerouting catabolic pathways of glucose
According to the central metabolism of E. coli (Fig. 1), glucose catabolism proceeding via the PP pathway generates 85 % more reducing power per gram mole of glucose than that via glycolysis. It seems useful to increase intracellular NADH by diverting the glycolytic flux to the PP pathway. Phosphoglucose isomerase (encoded by pgi) is responsible for isomerization of G6P, and its inactivation makes the PP pathway the primary route of glucose catabolism [25]. Therefore, strain BuT-13 was obtained by deletion of pgi in strain BuT-12. In comparison with strain BuT-12, strain BuT-13 grew poorly (0.31/h vs. 0.5/h), while it exhibited a 32 % and 30 % decrease in biomass yield and glucose utilization, respectively (Fig. 4). After fermentation for 30 h, strain BuT-13 was unable to consume all glucose and produced less n-butanol (4.6 g/L).
Fig. 4.
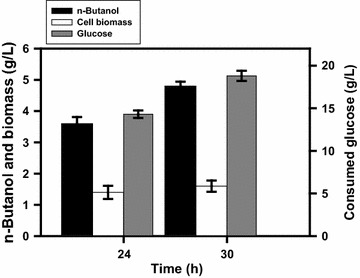
Production of n-butanol in the strain with the glucose catabolism via the PP pathway. Strain BuT-13 grown in M9Y medium containing 20 g/L glucose. The fermentations were conducted under the oxygen-limited condition for 30 h. The experiments were conducted in triplicate
Improving NADPH availability was realized by a strain deprived of pgi, whereas the strain showed a 47 % decrease in the specific growth rate [26]. The severe growth defect resulting from pgi knockout is attributed to a surplus of NADPH, which in turn perturbs the physiological state of cells [27]. Interestingly, an elevated level of either UdhA or Zwf can lead to growth recovery of the pgi-deficient strain by 25 and 68 %, respectively [27, 28]. Strain BuT-13 is still afflicted by the growth defect although it exhibits higher UdhA and Zwf activities and harbors the n-butanol synthetic pathway that consumes the reducing equivalent. The result implies the presence of an imbalanced redox state in the strain.
In response to the oxygen tension, the TCA cycle operates as either an oxidative pathway or a reductive pathway associated with production of various levels of reducing equivalents [4]. Citrate synthase (encoded by gltA) catalyzes the first committed step in the TCA cycle (Fig. 1). The approach to divert carbon flux from the TCA cycle by lowering the GltA activity is expected to conserve acetyl-CoA (the precursor of n-butanol ) and modulate production of reducing equivalents. It may be helpful to ameliorate the negative impact on the strain imposed by null pgi. This was carried out by replacement of the gltA cognate promoter P2 with lacO site in strain BuT-13. The resulting strain BuT-14 was then cultured and examined for its fermentation performance. Consequently, strain BuT-14 grew almost normally (ca. 0.46/h), and its biomass yield was comparable to strain BuT-12. All fermentation byproducts were significantly reduced in strain BuT-14 which consequently produced 6.1 g/L n-butanol at 29 h (Table 2; Fig. 5). As expected, strain BuT-14 exhibited 32 % less GltA activity and 16 % more NADH as compared to strain BuT-12. Direction of carbon flux through the PP pathway by pgi deletion increases the reducing equivalent, whereas prevention of carbon flux from entering the TCA cycle by lowering GltA reduces the reducing equivalent. It leads to a net outcome of a moderate increase in NADH, which suffices the need for the synthesis of n-butanol. As a result, a redox-balanced state is established in strain BuT-14 that recovers from the null pgi-induced growth defect. It was intriguing to learn the response of this strain to the act of further lowering the GltA activity. Strain BuT-14-A was thus obtained by integration of lacIQ into strain BuT-14. Consequently, strain BuT-14-A displayed 50 % less GltA activity than strain BuT-12 (Table 1). The strain exhibited a poor growth and consumed only 40 % glucose associated with the n-butanol production of 1.8 g/L at 30 h (data not shown). It was reported that the growth of E. coli on glucose remains unaffected by a 90 % decrease in the GltA activity [29]. In contrast, the growth of the n-butanol-producing strain that lacks pgi on glucose is closely linked to the GltA activity. By modulation of the GltA activity, the producer strain enables recovery from the growth defect caused by null pgi. It is likely that the alteration of the GltA activity perturbs the intracellular redox state of the strain. Apparently, the engineered strain displays high susceptibility to the intracellular redox state and optimal adjustment of GltA activity is necessary to ensure the superior performance of the strain.
Fig. 5.
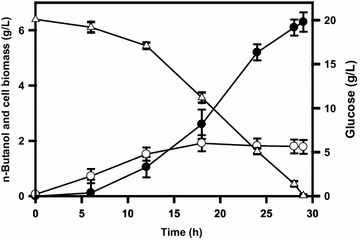
Time course of n-butanol production in the strain with the limited TCA cycle. Strain BuT-14 grown in M9Y medium containing 20 g/L glucose. The fermentations were conducted under the oxygen-limited condition. The experiments were conducted in triplicate
Recruitment of FDH and enhanced expression of PDH have been proposed to increase NADH availability favoring the n-butanol production in E. coli. In one study, the approach by recruiting FDH achieved the n-butanol productivity (PB) of 0.2 g/L/h and the conversion yield on glucose (YB/G) of 0.36 g/g [17]. An alternative study by manipulating PDH reported PB of 0.065 g/L/h and YB/G of 0.19 g/g [15]. In addition, PB of 0.26 g/L/h and YB/G of 0.27 g/g were obtained by optimization of both FDH and PDH activity in a recent work [18]. All these studies were conducted with super-rich TB medium, and plasmids were employed for the episomal expression of multiple target genes to increase their expression levels, which is important to obtain the observed effect. Therefore, the discrepancy in the fermentation production by these reports is likely attributed to various expression levels of the cloned genes. However, plasmid maintenance is known to impose a metabolic burden on cells, thus resulting in a reduced growth rate and perturbation of the cell physiology [30]. A metabolic load is additionally imposed on E. coli after the forced expression of the plasmid-borne genes, which suppresses the primary carbon and energy metabolism of cells [31]. The use of TB medium (12 g/L tryptone, 24 g/L yeast extract, 2.31 g/L KH2PO4, 12.54 g/L KHPO4, 4 mL/L glycerol) is expected to improve the cell growth under the anaerobic condition and helps to ameliorate the plasmid-induced stress on cells. It should be noted that components other than glucose in the TB medium could contribute 15 % of the n-butanol production according to the previous study [17]. Therefore, these reported yields based on glucose may be re-estimated. In contrast, the engineered strains were free of plasmids and grown on the M9Y medium with glucose in this study. First, the starter strain BuT-8 displayed PB of 0.11 g/L/h and YB/G of 0.14 g/g (Table 1). With both enhanced FDH and PDH, strain BuT-9 achieved PB of 0.18 g/L/h and YB/G of 0.22 g/g. Finally, strain BuT-14 was developed by rerouting of the glycolytic flux through the PP pathway and further reducing GltA activity in the TCA cycle. It exhibited 2.3-fold higher NADH and n-butanol production titer than the starter strain BuT-8. Notice that n-butanol was not produced for strain BuT-14 grown on M9Y medium without glucose (data not shown). Therefore, the n-butanol production in the strain comes merely from glucose. As a result, strain BuT-14 displayed PB of 0.21 g/L/h and YB/G of 0.31 g/g. In theory, 0.85 mol instead of 1 mol n-butanol per mole glucose is produced in the glucose catabolism via the PP pathway due to CO2 loss, which leads to the theoretical yield of n-butanol as 0.35 g/g. Accordingly, n-butanol remains the main product for strain BuT-14 (Table 2) with YB/G reaching 89 % of the theoretical. It is feasible to recycle CO2 in E. coli with the expression of Synechococcus ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) and phosphoribulokinase [32]. The issue of increasing YB/G may be addressed by recruitment of the Rubisco-based CO2 recycling system in strain BuT-14. Overall, it indicates that our proposed approach is effective for production of the highly reduced product without the need for a super-rich medium.
Conclusions
The intracellular redox state in microbes is recognized as a key factor determining the production efficiency of fermentation products. The current work illustrates that pyruvate, G6P, and acetyl-CoA at the junction of the central catabolism are flexible for engineering. It is known that the enzymes, such as PDH and GltA, involved in the use of these metabolite nodes are subjected to the physiological control mediated by the effector metabolites [4]. Nevertheless, genetic manipulation of these metabolite nodes can lead to the redistribution of carbon flux, which in turn alters the cellular redox state. In principle, these metabolite nodes can be individually or coordinately modulated to fulfill the intracellular need for high production of reduced products of interest.
Methods
Bacterial culturing
The method for bacterial culturing under the oxygen-limited condition essentially followed the previous report [19]. The seeding cultures were prepared by growing E. coli strains on Luria–Bertani medium [33] with 2 g/L glucose overnight. The cell density was measured turbidimetrically at 550 nm (OD550). The overnight culture was inoculated into capped Erlenmeyer flasks (125 mL) containing 50 mL M9Y medium (6 g/L Na2HPO4, 3 g/L KH2PO4, 0.5 g/L NaCl, 1 g/L NH4Cl, 1 mM MgSO4, 0.1 mM CaCl2, 10 mg/L vitamin B1, 5 g/L yeast extract) with 20 g/L glucose to attain the initial cell density at OD550 of 0.2. The shake`flask cultures were maintained in an orbital shaker set at 100 rpm.
Strain construction
The strains and primers applied in this study are listed in Table 3. Genomic insertion of fdh1 into strain BuT-8 was constructed previously [34]. In brief, Ptrc-driven fdh1 of S. cerevisiae was amplified from plasmid pTrc-Fdh1 [21] by PCR with primers RC12171/RC12314. After digestion with BamHI, the PCR DNA was spliced into BamHI-NruI of plasmid pP21-Km to obtain plasmid pP21-Fdh1. Based on our reported methods, the DNA containing Ptrc-fdh1 was then integrated into E. coli using plasmid pP21-Fdh1, and the inserted kanamycin-resistant marker in the strain was later removed [35]. Strain BuT-8 with fdh1 was renamed BuT-8-Fdh1.
Table 3.
The strains and primers applied in this study
| Characteristics | Source | |
|---|---|---|
| Strains | ||
| BuT-8 | ΔfrdA ɸ80attB:: PλPL-crt | |
| ΔadhE::ɸ80attB::PλPL-pha-hbd | ||
| ΔldhA::λattB::PλPL-adhE2 | 19 | |
| BuT-8-Fdh1 | as P21attB:: Ptac-fdh1 | 34 |
| BuT-9 | as But-8-Fdh ∆lpdA λattB::PλPL-lpdA * PλPL-aceEF | This study |
| BuT-10 | as But-9 PλPL-zwf PλPL-UdhA | This study |
| BuT-12 | as But-10 ΔatoD::PλPL-pgl | This study |
| BuT-13 | as But-12 ∆pgi | This study |
| BuT-14 | as But-13 lacO-gltA | This study |
| Primers | ||
| RC10178 | ATAAGGATCCATATCTAACACCGTGCGTG | |
| RC11210 | CACACCATATGTTAGAATTCATTACCTTCG | |
| RC11403 | TTTGCGGTACCAAGCCCTTTGCAAATTGC | |
| RC11404 | CAGCAGAGCTCGAATGGATCGCGTTATC | |
| RC11405 | AGAATCATATGGCGGTAACGCAAACAG | |
| RC11406 | CTTAAGGATCCTAACCCGGTACTTAAGCCAG | |
| RC11407 | CGTAAGGTACCTGACGCATGCGCGTTTG | |
| RC11408 | ACTTAGAGCTCTAAATGCGGCTTCCACCAG | |
| RC11409 | GCCCTCATATGCCACATTCCTACGATTAC | |
| RC11410 | TGTTCGGATCCATAAAAGCAACAGAATGGTAAC | |
| RC11417 | CCAAGCCCTTTGCAAATTGC | |
| RC11418 | CTCGAATGGATCGCGTTATC | |
| RC11419 | CCTGACGCATGCGCGTTTG | |
| RC11420 | CTAAATGCGGCTTCCACCAG | |
| RC12058 | AATAACATATGTCAGAACGTTTCCCAAATG | |
| RC12059 | CTATCTCTAGACGTTGAGTTTTCTGGAACC | |
| RC12060 | CCAGTTCGAGGTCTTTTTTCG | |
| RC12085 | TATGGGGTACCAGTTCGAGGTCTTTTTTCG | |
| RC12086 | CAATGGAGCTCTGCTTCATCTGCTAAGG | |
| RC12154 | GCGATATCGTCGGTCAACC | |
| RC12155 | TGAGAAGCTTCAGTCCGCATCACCAGAG | |
| RC12171 | GCAAGCTTATTTCTTCTGTCCATAAGC | |
| RC12215 | GTCCATCGCCTATACCAAACCAGAAGTTGCATG | |
| RC12216 | CATGCAACTTCTGGTTTGGTATAGGCGATGGAC | |
| RC12288 | AACTGCTCGAGTTACTTCTTCTTCGCTTTCG | |
| RC12289 | AAGTGGATCCATACCCGTCGTCTTTCAGG | |
| RC12290 | CCATGAGCTCGGCTTTTTTCTGGTAATCTC | |
| RC12314 | TCTGGGGATCCTTCTGAAATGAGCTGTTGAC | |
| RC12331 | ACTCTCGAATTCTGGTCGTCCTATCGCTTC | |
| RC13001 | TTGAATTCCGCCTTTAAAGATCGCCATG | |
| RC13034 | CATCTCACCAGATATCATGC | |
| RC13035 | AATCGGAGCTCGAAAGTGAACTGTTTGG | |
| RC13195 | ATCTTCCCGGGCGGAATTCATTACCGTTC | |
| RC13196 | GAAATTGTTATCCGCTCACAATTCCGGGTACCCAATTC | |
| RC13197 | CAGCAAAATACCTTCATCACC | |
| RC13198 | TTCAGGGGAAGAGAGGCTG | |
| RC13199 | TCAATGGGCCCACACTGTTACATAAGTTAATC | |
| RC13200 | TTAATGTCGACGATTGCTAAGTACTTGATTCG | |
| RC13201 | GGTACCCAGAAGCCACAG | |
| RC13292 | ATCCCGGGAAGCAAACAGTTTATATCGC | |
| RC13293 | ATCTCGAGTTAGTGTGCGTTAACCACCAC | |
| RC14025 | GAGGAATTCTGTAGGCTGGAGCTGCTTC | |
| RC14026 | AACGGTCGACATGGGAATTAGCCATGG | |
Plasmid pMCS-lpdA was obtained by PCR-amplification of lpdA from strain BL21 with primers RC12154/RC12155 and subsequent incorporation into NdeI-XhoI of plasmid pMCS-5. The E354K mutation was introduced into lpdA on plasmid pMSC-lpdA by the site-directed mutagenesis using primers RC12215/RC12216. The mutant lpdA (lpdA*) was confirmed by DNA sequencing and removed from plasmid pMCS-lpdA by NdeI-XhoI. The recovered lpdA* was subcloned into plasmid pLoxKm-PR [36] which carries a cassette of PλPL fused with LE*-kan-RE* (LE*-kan-RE*-PλPL). The resulting plasmid pLoxKm-lpdA* contains the LE*-kan-RE*-PλPL-regulated lpdA* (LE*-kan-RE*-PλPL-lpdA*). Meanwhile, the upstream region of lpdA was amplified by PCR with primers RC12289/RC12290 and spliced into BamHI-SacI of plasmid pBluescript to produce plasmid pBlue-ac. The DNA containing LE*-kan-RE*-PλPL-lpdA* was recovered from plasmid pLoxKm-lpdA* by BamHI-XhoI and then incorporated into plasmid pBlue-ac to give plasmid pBlue-ac/lpdA*. In addition, PCR was conducted on plasmid pBlue-ac/lpdA* with primers RC11210/RC12331. The PCR DNA was digested with EcoRI and self-ligated to give plasmid pBlue-Ac-lpd, which carries lpdA interrupted with LE*-kan-RE*. To knockout lpdA, the truncated lpdA was amplified from plasmid pBlue-Ac-lpd by PCR with primers RC12288/RC12290 and electroporated into E. coli following our protocol. Finally, the DNA containing PλPL-lpdA* was amplified from plasmid pBlue-ac/lpdA* by PCR with primers RC10178/RC12288 and then restricted by BamHI. Plasmid pLam-LpdA* was obtained by incorporation of the PCR DNA into BamHI-EcoRV of plasmid pLam-Crt [36]. Similarly, the DNA containing PλPL-lpdA* was integrated into E. coli followed by removal of the inserted marker [21].
To enhance the expression of endogenous genes, PλPL was placed in front of the structural genes with their cognate promoters intact. This was carried out as follows: First, the upstream region and 5′-end structural regions of zwf, udh, and aceE were amplified from strain BL21 by PCR with primers RC11403/RC11404, RC11407/RC11408, and RC12085/RC12086, respectively. Each PCR DNA was digested by KpnI-SacI and incorporated into plasmid pBluescript to obtain plasmid pBlue-zwf, pBlue-udhA, and pBlue-aceE. Secondly, the NdeI-BamHI site was introduced into plasmid pBlue-zwf and pBlue-udhA by PCR with primers RC11405/RC11406 and RC11409/RC11410 while the NdeI-XbaI site into plasmid pBlue-aceE with primers RC12058/RC12059. The LE*-kan-RE*-PλPL cassette was recovered from plasmid pLoxKm-PR by NdeI-BamHI or NdeI-XbaI digestion and then incorporated into plasmid pBlue-zwf, pBlue-udhA, and pBlue-aceE to obtain plasmid pPR-zwf, pPR-udhA, and pPR-aceE. Finally, the PCR DNAs were amplified from plasmid pPR-zwf, pPR-udhA, and pPR-aceE with primers RC11417/RC11418, RC11419/RC11420, and RC12060/RC12086, respectively. These passenger DNAs were individually integrated into the strain by electroporation according to the reported method [21]. The associated marker was finally rescued.
To obtain pgl, the gene was amplified from strain MG1655 with primers RC13292/RC13293. After cleavage by EcoRV-SacI, the PCR DNA and plasmid pBluescript were spliced together to give plasmid pBlue-pgl. The pgl-containing DNA was recovered by SmaI-XhoI and incorporated into plasmid pLoxKm-PL. The construction resulted in plasmid pSPL-pgl, which fuses LE*-kan-RE*-PλPL with pgl. The LE*-kan-RE*-PλPL-pgl DNA was amplified by PCR with primers RC13001/RC13293. Plasmid pAto-pgl was obtained by incorporation of the PCR DNA into EcoRI-NruI of plasmid pSPL-atoD [36]. Similarly, the passenger DNA was amplified from plasmid pSPL-atoD with primers RC13034/RC13035 and then electroporated to the strain. Later removal of the inserted marker was carried out.
To modulate the gltA expression, its P2 promoter was replaced with lacO. This was done in several steps. First, lacO was created in plasmid pLoxKm-PR by PCR with primers RC13195/RC13196. After cleavage by SmaI, the PCR DNA was self-ligated to produce plasmid pLoxCm-LacO, which carries the fusion of LE*-kan-RE*-lacO. Secondly, the DNA containing the upstream region and 5′-end structural sequence of gltA was amplified from strain BL21 by PCR with primers RC13197/RC13198. Plasmid pBlue-GltA was generated by incorporation of the PCR DNA into KpnI-SmaI of plasmid pBluescript. Moreover, the ApaI-SalI site was introduced into plasmid pBlue-GltA by PCR with primers RC13199/RC13200. The LE*-kan-RE*-lacO cassette was recovered from plasmid pLoxCm-LacO by ApaI-SalI and incorporated into plasmid pBlue-GltA to give plasmid pBlue-GltO. Finally, the FRT-Cm-FRT cassette was amplified from plasmid pKD3 by PCR with primers RC14025/RC14026. The LE*-kan-RE* cassette was replaced by FRT-Cm-FRT by incorporation of the PCR DNA into EcoRI-SalI of plasmid pBlue-gltO, leading to plasmid pB-gltO-Cm. The passenger DNA was amplified from plasmid pB-gltO-Cm with primers RC13197/RC13201 and then electroporated to the strain in a similar way.
Analytical method
The analytical method essentially followed our reported protocol [19]. Glucose was measured by high-performance liquid chromatography (HPLC) with Reflective Index RID-10A (Shimadzu, Japan). n-Butanol was determined by Gas Chromatograph Trace 1300 (Thermo Scientific, USA).
The intracellular NADH level was measured by using the fluorescent NAD/NADH detection kit (Cell Technology, USA). The assay procedure exactly followed the manufacturer’s instruction. In brief, bacterial cultures were harvested by centrifugation and the cell pellets were resuspended in 200 µL NADH extraction buffer plus 200 µL lysis buffer. The mixture was kept at 60 °C for 20 min. After centrifugation, the supernatant was recovered and mixed with the reaction reagent for the measurement. The reaction was kept in dark at room temperature for 1 h. The NADH level was then measured with the excitation at 530–570 nm and emission at 590–600 nm.
Enzyme activity assay
Bacterial cultures were harvested by centrifugation and the cell pellets were resuspended in 1 mL solution buffer. Cells were disrupted by sonication after centrifugation. The supernatant was saved as the cell-free extract (CFX). The total protein content in CFX was determined using Bio-Rad protein assay kit. The activity of pyruvate dehydrogenase was determined by monitoring the reduction of NAD+ at 340 nm at room temperature according to the previous report [15]. The reaction solution (1 mL) contains 50 mM potassium phosphate (pH 7.9), 5 mM sodium pyruvate, 1.3 mM CoA, 2 mM NAD+, 0.5 mM thiamine pyrophosphate, and 5 mM MgCl2. To start the reaction, 100 µL CFX was added to the solution. The activity of glucose-6-phosphate dehydrogenase was determined by monitoring the reduction of NADP+ at 340 nm following the reported protocol [37]. The reaction solution (1 mL) is composed of 2 mM glucose-6-phosphate, 0.67 mM NADP+, 10 mM MgCl2, and 50 mM Tris–HCl (pH 7.5). The reaction was initiated by adding 100 µL CFX to the solution at 30 °C. The method for measuring the activity of lactonase was similar to the glucose-6-phosphate dehydrogenase activity assay [38]. The reaction solution comprises 50 μM glucose-6-phosphate, 0.5 mM NADP+, 50 mM Tris–HCl, 10 mM MgCl2, and 50 mM Tris–HCl (pH 7.5). In addition, the citrate synthase activity was measured as reported previously [29]. The composition of the assay solution includes 0.1 mM acetyl-CoA, 0.5 mM oxaloacetate, 0.2 mM 5′5-dithiobis-(2-nitrobenzoic acid), and 50 mM Tris–HCl (pH 7.5).
Authors’ contributions
MS constructed bacterial strains and performed fermentation; SYL analyzed data and drafted the manuscript; ZWW helped gene cloning; CJC and YPC assisted in the design of the study and revised the manuscript. All authors read and approved the final manuscript.
Acknowledgements
This work is supported by Taiwan Ministry of Science and Technology (MOST 102-2221-E-039-014-MY3, 104-2221-E-035-072, 104-2623-E-035-004-ET, and 104-2622-E-035-011-CC1).
Competing interests
The authors declare that they have no competing interests.
Abbreviations
- PP
pentose phosphate
- G6P
glucose-6-phosphate
- PDH
pyruvate dehydrogenase
- FDH
formate dehydrogenase
- TCA
tricarboxylic acid
- PFL
pyruvate formate-lyase
- HPLC
high-performance liquid chromatography
- CFX
cell-free extract
- Ace
acetate
- EtOH
ethanol
- F6P
fructose-6-phosphate
- Lac
lactate
- For
formate
- Glc
glucose
- OAA
oxaloacetate
- PEP
phosphoenolpyruvate
- 3-PGA
3-phosphoglyceraldehyde
- Pyr
pyruvate
- Suc
succinate
Additional file
10.1186/s13068-016-0467-4 The development course of E. coli strains for the fermentative production of n-butanol.
Contributor Information
Mukesh Saini, Email: mksaini1@gmail.com.
Si-Yu Li, Email: syli@dragon.nchu.edu.tw.
Ze Win Wang, Email: t7gene1@gmail.com.
Chung-Jen Chiang, Phone: 886-4-22053366, Email: oleosin91@yahoo.com.tw.
Yun-Peng Chao, Phone: 886-4-24517250, Email: ypchao@fcu.edu.tw.
References
- 1.Schubert C. Can biofuels finally take center stage? Nat Biotechnol. 2006;24:777–784. doi: 10.1038/nbt0706-777. [DOI] [PubMed] [Google Scholar]
- 2.Hermann BG, Blok K, Patel MK. Producing bio-based bulk chemicals using industrial biotechnology saves energy and combats climate change. Environ Sci Technol. 2007;41:7915–7921. doi: 10.1021/es062559q. [DOI] [PubMed] [Google Scholar]
- 3.Stephanopoulos G. Challenges in engineering microbes for biofuels production. Science. 2007;315:801–804. doi: 10.1126/science.1139612. [DOI] [PubMed] [Google Scholar]
- 4.White D. The physiology and biochemistry of prokaryotes. 3. New York: Oxford University Press; 2007. [Google Scholar]
- 5.Jones DT, Woods DR. Acetone-butanol fermentation revisited. Microbiol Rev. 1986;50:484–524. doi: 10.1128/mr.50.4.484-524.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee SY, Park JH, Jang SH, Nielsen LK, Kim J, Jung KS. Fermentative butanol production by Clostridia. Biotechnol Bioeng. 2008;101:209–228. doi: 10.1002/bit.22003. [DOI] [PubMed] [Google Scholar]
- 7.Mussatto S, Dragone G, Guimarães PM, Silva JP, Carneiro LM, Roberto IC, et al. Technological trends, global market, and challenges of bio-ethanol production. Biotechnol Adv. 2010;28:817–830. doi: 10.1016/j.biotechadv.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 8.Dürre P. Biobutanol: an attractive biofuel. Biotechnol J. 2007;2:1525–1534. doi: 10.1002/biot.200700168. [DOI] [PubMed] [Google Scholar]
- 9.Atsumi S, Cann AF, Connor MR, Shen CR, Smith KM, Brynildsen MP, et al. Metabolic engineering of Escherichia coli for 1-butanol production. Metab Eng. 2008;10:305–311. doi: 10.1016/j.ymben.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 10.Berezina OV, Zakharova NV, Brandt A, Yarotsky SV, Schwarz WH, Zverlov V. Reconstructing the clostridial n-butanol metabolic pathway in Lactobacillus brevis. Appl Microbiol Biotechnol. 2010;87:635–646. doi: 10.1007/s00253-010-2480-z. [DOI] [PubMed] [Google Scholar]
- 11.Nielsen DR, Leonard E, Yoon SH, Tseng HC, Yuan C, Prather KL. Engineering alternative butanol production platforms in heterologous bacteria. Metab Eng. 2009;11:262–273. doi: 10.1016/j.ymben.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 12.Steen EJ, Chan R, Prasad N, Mayers S, Petzold CJ, Redding A, et al. Metabolic engineering of Saccharomyces cerevisiae for the production of n-butanol. Microb Cell Fact. 2008;7:36. doi: 10.1186/1475-2859-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clomburg JM, Gonzalez R. Biofuel production in Escherichia coli: the role of metabolic engineering and synthetic biology. Appl Biochem Biotechnol. 2010;86:419–434. doi: 10.1007/s00253-010-2446-1. [DOI] [PubMed] [Google Scholar]
- 14.Yu C, Cao Y, Zou H, Xian M. Metabolic engineering of Escherichia coli for biotechnological production of high-value organic acids and alcohols. Appl Biochem Biotechnol. 2011;89:573–583. doi: 10.1007/s00253-010-2970-z. [DOI] [PubMed] [Google Scholar]
- 15.Bond-Watts BB, Bellerose RJ, Chang MCY. Enzyme mechanism as a kinetic control element for designing synthetic biofuel pathways. Nat Chem Biol. 2011;7:222–227. doi: 10.1038/nchembio.537. [DOI] [PubMed] [Google Scholar]
- 16.Lim HG, Lim JH, Jung GY. Modular design of metabolic network for robust production of n-butanol from galactose–glucose mixtures. Biotechnol Biofuels. 2015;8:137. doi: 10.1186/s13068-015-0327-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen CR, Lan EI, Dekishima Y, Baez A, Cho KM, Liao JC. Driving forces enable high-titer anaerobic 1-butanol synthesis in Escherichia coli. Appl Environ Microbial. 2011;77:2905–2915. doi: 10.1128/AEM.03034-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lim JH, Seo SW, Kim SY, Jung GY. Model-driven rebalancing of the intracellular redox state for optimization of a heterologous n-butanol pathway in Escherichia coli. Metab Eng. 2013;20:56–62. doi: 10.1016/j.ymben.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 19.Saini M, Chen MH, Chiang CJ, Chao YP. Potential production platform of n-butanol in Escherichia coli. Metab Eng. 2015;27:76–82. doi: 10.1016/j.ymben.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 20.Berríos-Rivera SJ, Bennett GN, San KY. The effect of increasing NADH availability on the redistribution of metabolic fluxes in Escherichia coli chemostat cultures. Metab Eng. 2002;4:230–237. doi: 10.1006/mben.2002.0228. [DOI] [PubMed] [Google Scholar]
- 21.Chiang CJ, Saini M, Lee HM, Wang ZW, Chen PT, Chao YP. Genomic engineering of Escherichia coli by the phage attachment site-based integration system with mutant loxP sites. Proc Biochem. 2012;47:2246–2254. doi: 10.1016/j.procbio.2012.08.022. [DOI] [Google Scholar]
- 22.Kim Y, Ingram LO, Shanmugam KT. Dihydrolipoamide dehydrogenase mutation alters the NADH sensitivity of pyruvate dehydrogenase complex of Escherichia coli K-12. J Bacteriol. 2008;190:3851–3858. doi: 10.1128/JB.00104-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Canonaco F, Hess AT, Heri S, Wang T, Szyperski T, Sauer U. Metabolic flux response to phosphoglucose isomerase knock-out in Escherichia coli and impact of overexpression of the soluble transhydrogenase UdhA. FEMS Microbiol Lett. 2001;204:247–252. doi: 10.1111/j.1574-6968.2001.tb10892.x. [DOI] [PubMed] [Google Scholar]
- 24.Meier S, Jensen PR, Duus JØ. Direct observation of metabolic differences in living Escherichia coli strains K-12 and BL21. ChemBioChem. 2012;13:308–310. doi: 10.1002/cbic.201100654. [DOI] [PubMed] [Google Scholar]
- 25.Hua Q, Yang C, Baba T, Mori H, Shimizu K. Responses of the central metabolism in Escherichia coli to phosphoglucose isomerase and glucose-6-phosphate dehydrogenase knockouts. J Bacteriol. 2003;185:7053–7067. doi: 10.1128/JB.185.24.7053-7067.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chemler JA, Fowler ZL, McHugh KP, Koffas MA. Improving NADPH availability for natural product biosynthesis in Escherichia coli by metabolic engineering. Metab Eng. 2010;12:96–104. doi: 10.1016/j.ymben.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 27.Canonaco F, Hess TA, Heri S, Wang T, Szyperski T, Sauer U. Metabolic flux response to phosphoglucose isomerase knock-out in Escherichia coli and impact of overexpression of the soluble transhydrogenase UdhA. FEMS Microbiol Lett. 2001;204:247–252. doi: 10.1111/j.1574-6968.2001.tb10892.x. [DOI] [PubMed] [Google Scholar]
- 28.Flores S, de Anda-Herrera R, Gosset G, Bolívar FG. Growth-rate recovery of Escherichia coli cultures carrying a multicopy plasmid, by engineering of the pentose-phosphate pathway. Biotechnol Bioeng. 2004;87:485–494. doi: 10.1002/bit.20137. [DOI] [PubMed] [Google Scholar]
- 29.Walsh K, Koshland DE., Jr Characterization of rate-controlling steps in vivo by use of an adjustable expression vector. Proc Natl Acad Sci USA. 1985;82:3577–3581. doi: 10.1073/pnas.82.11.3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ow DSW, Nissom PM, Philp R, Oh SKW, Yap GS. Global transcriptional analysis of metabolic burden due to plasmid maintenance in Escherichia coli DH5α during batch fermentation. Enzyme Microb Technol. 2006;39:391–398. doi: 10.1016/j.enzmictec.2005.11.048. [DOI] [Google Scholar]
- 31.Neubauer P, Lin HY, Mathiszik B. Metabolic load of recombinant protein production: inhibition of cellular capacities for glucose uptake and respiration after induction of a heterologous gene in Escherichia coli. Biotechnol Bioeng. 2003;83:5364. doi: 10.1002/bit.10645. [DOI] [PubMed] [Google Scholar]
- 32.Zhuang ZY, Li SY. Rubisco-based engineered Escherichia coli for in situ carbon dioxide recycling. Bioresour Technol. 2013;150:79–88. doi: 10.1016/j.biortech.2013.09.116. [DOI] [PubMed] [Google Scholar]
- 33.Miller JH. Experiments in molecular genetics. New York: Cold Spring Harbor; 1972. [Google Scholar]
- 34.Saini M, Chiang CJ, Li SY, Chao YP. Production of biobutanol from cellulose hydrolysate by the Escherichia coli coculture system. FEMS Microbiol Lett. 2016 doi: 10.1093/femsle/fnw008. [DOI] [PubMed] [Google Scholar]
- 35.Chiang CJ, Chen PT, Chao YP. Replicon-free and markerless methods for genomic insertion of DNAs in phage attachment sites and controlled expression of chromosomal genes in Escherichia coli. Biotechnol Bioeng. 2008;101:985–995. doi: 10.1002/bit.21976. [DOI] [PubMed] [Google Scholar]
- 36.Saini M, Wang ZW, Chiang CJ, Chao YP. Metabolic engineering of Escherichia coli for production of butyric acid. J Agri Food Chem. 2014;62:4342–4348. doi: 10.1021/jf500355p. [DOI] [PubMed] [Google Scholar]
- 37.Snoep JL, Arfman N, Yomano LP, Westerhoff HV, Conway T, Ingram LO. Control of glycolytic flux in Zymomonas mobilis by glucose-6-phosphate dehydrogenase activity. Biotechnol Bioeng. 1996;15:191–197. doi: 10.1002/(SICI)1097-0290(19960720)51:2<190::AID-BIT8>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 38.Sinha A, Maitra PK. Induction of specific enzymes of the oxidative pentose pathway by glucono-delta-lactone in Saccharomyces cerevisiae. J Gen Microbiol. 1992;138:1865–1873. doi: 10.1099/00221287-138-9-1865. [DOI] [PubMed] [Google Scholar]


