Abstract
The sarcomere of muscle is composed of tens of thousands of myosin motors that self-assemble into thick filaments and interact with surrounding actin-based thin filaments in a dense, near-crystalline hexagonal lattice1. Together, these actin–myosin interactions enable large-scale movement and force generation, two primary attributes of muscle. Research on isolated fibres has provided considerable insight into the collective properties of muscle, but how actin–myosin interactions are coordinated in an ensemble remains poorly understood2. Here, we show that artificial myosin filaments, engineered using a DNA nanotube scaffold, provide precise control over motor number, type and spacing. Using both dimeric myosin V- and myosin VI-labelled nanotubes, we find that neither myosin density nor spacing has a significant effect on the gliding speed of actin filaments. This observation supports a simple model of myosin ensembles as energy reservoirs that buffer individual stochastic events to bring about smooth, continuous motion. Furthermore, gliding speed increases with cross-bridge compliance, but is limited by Brownian effects. As a first step to reconstituting muscle motility, we demonstrate human β-cardiac myosin-driven gliding of actin filaments on DNA nanotubes.
In a large motor ensemble, such as in muscle, the percentage of motors that simultaneously interact with an actin filament is determined by the duty ratio, which is the fraction of time myosin spends bound to actin during its kinetic cycle. In the case of muscle myosin, the low duty ratio (0.02–0.05)3–6, among other factors, primarily allows the higher velocities needed in muscle contraction (compared to the low velocities than can be achieved with processive motors). Furthermore, the low duty ratio may also constitute a mechanism to avoid interference between the numerous lever arm strokes that together drive muscle contraction. Nevertheless, given the large number of actin–myosin interactions in a single muscle fibre, the discrete stroke of a single myosin lever will still experience resistance from the many myosin cross-bridges that are bound at any given time7,8. Despite the fact that these myosin cross-bridges can be widely spaced, they are still mechanically linked through the sarcomeric lattice, so mechanical coordination between these events will influence collective myosin function.
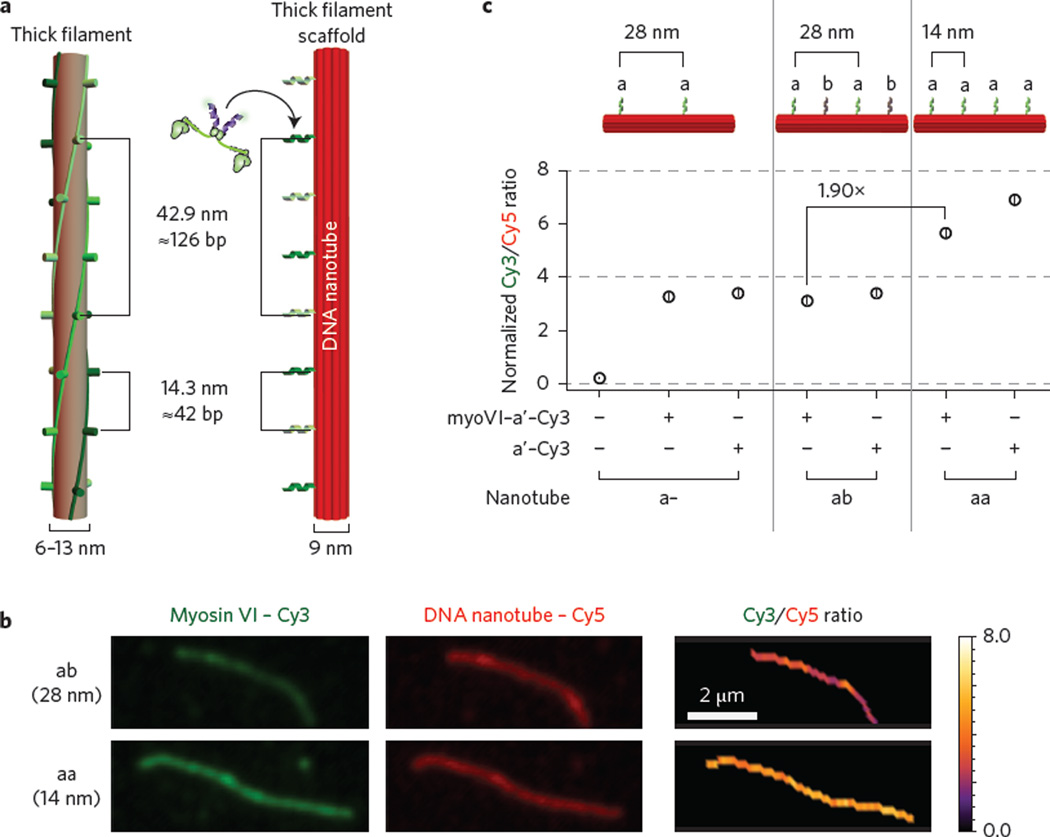
A range of experimental and theoretical approaches have demonstrated that molecular motors behave differently in isolation than in an ensemble2,9–12, stressing the importance of studying multi-motor behaviour. The most widely used of these is the in vitro gliding assay, which provides a good measure of motor directionality, average speed and processivity13. However, an accurate and precise comparison between motors, especially the effects of mutations that influence the catalytic cycle, is limited by the variability in measured speeds, which is generally attributed to heterogeneity in surface preparation and motor density. Here, we overcome this limitation with DNA nanotechnology, which enables the spatial organization of macromolecules, such as molecular motors, with nanometre precision. This positional control allows the measurement of collective transport by defined ensembles of motor proteins10,11,14. The DNA structures used thus far, however, are discrete units with a limited number of motor binding sites (1–15 sites)10,11. To study larger ensembles of motors, as in muscle, we engineered ten-helix DNA nanotubes15 with 14, 28 or 42 nm spacing between protein attachment points. The 42 nm pattern of attachment points on the DNA nanotube models the interactions of one actin-based thin filament with one myosin-based thick filament. The 14 nm pattern models the actin–myosin interactions occurring in a sarcomere, where a thin filament is surrounded by three interacting thick filaments1,8 (Fig. 1a). DNA strands with specific sequence extensions or chemical modifications were incorporated such that each nanotube unit included a modified strand for surface attachment (biotin), imaging (Cy5) and sequence-specific protein attachment (oligo-a or b) (Supplementary Fig. 1 and Supplementary Table 1). Unlike their previous DNA counterparts, these nanotubes self-assemble into long crystalline one-dimensional tracks with an average length on the order of 5 µm (ref. 15). This long contour length is not only desirable for gliding assays, but probes interactions on the length scale of a muscle sarcomere (1.5–2.5 µm; refs 1, 8). Furthermore, these polymerizable nanotubes require significantly fewer strands than previous DNA-motor scaffolds (approximately threefold less; Supplementary Fig. 1 and Supplementary Table 1).
Figure 1. Formation of synthetic myosin filaments using DNA nanotubes.
a, Schematics of a DNA nanotube platform (right) and a natural myosin thick filament (left). Attachment oligos (a or b) are placed 42 base pairs apart (or ~14 nm), matching the 14.3 nm vertical spacing observed between myosin heads in a muscle thick filament1. b, Example of nanotube labelling ratio determination. Cy5-nanotubes with 14 nm (aa) and 28 nm (a– and ab) oligo-a spacing were labelled with myosin VI-Cy3-a′ and the ratio of Cy3 (green) to Cy5 (red) intensities was determined. c, Summary of normalized labelling ratios for a–, aa and ab nanotubes (N ≥ 140). Ratios were normalized with the myosin Cy3-conjugation efficiency (Supplementary Fig. 2) and nanotubes were either annealed with Cy3-a′ or incubated with myosin VI-Cy3-a′ at room temperature. Nanotubes without Cy3-a′ labelling have negligible Cy3/Cy5 ratios, as shown with a– labelling ratio data. For myosin V labelling, see Supplementary Fig. 3. Error bars indicate standard error of the mean (s.e.m.).
In lieu of the difficulty in expressing recombinant muscle myosin, we initially utilized two different processive motors, dimeric myosin V and VI. High duty-ratio motors like myosin V and VI also increase the density of actin–myosin interactions over a finite stretch of an actin filament, enabling us to examine the mechanical interactions between a range of myosin group sizes (10–140 myosins). To assess the labelling efficiency of Cy5-labelled nanotubes, SNAP-tagged dimeric myosin V and VI were labelled with Cy3-ssDNA complementary to oligo-a attachment strands (Cy3-a′; Supplementary Fig. 2) and the ratios of Cy3 to Cy5 were determined (Fig. 1b,c and Supplementary Fig. 3). Nanotubes labelled with myosin VI-Cy3-a′ have normalized Cy3/Cy5 ratios that are similar to those of control-labelled nanotubes and have a 1.9-fold increase for aa nanotubes compared to ab and a– nanotubes (Fig. 1c). The doubling of the Cy3 to Cy5 ratios is also reproducible when running the myosin–nanotube structures in an agarose gel (Supplementary Fig. 4). Overall, this labelling assessment serves as an in situ quality control to ensure homogeneity between measurements of different nanotubes and between experiments.
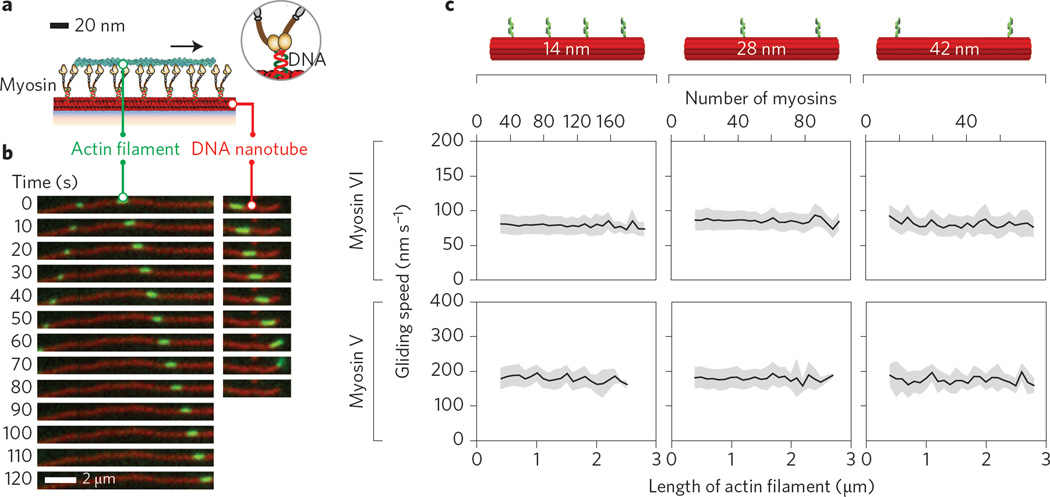
For nanotubes with 14, 28 or 42 nm spacing, actin filaments move along the artificial myosin V and myosin VI filaments with high processivity and run length (Fig. 2a,b and Supplementary Movie 1), consistent with the engagement of each actin filament with a large number of myosins. The mean actin gliding speeds over myosin V- or myosin VI-labelled nanotubes (Fig. 2c) are significantly lower than the published single myosin V and VI speeds on actin filaments at 2 mM ATP16. This speed decrease is similar to that found in previous reports for two myosin V motors on a single actin filament14 and one to six myosin V or myosin VI motors on two-dimensional actin networks11. For both myosin V- and myosin VI-labelled nanotubes, neither myosin density nor total myosin number had an effect on the mean actin gliding speed (Fig. 2c). The length-independent speed for large motor numbers is similar to previous observations from standard in vitro gliding assays with non-processive muscle myosins4,17.
Figure 2. Density and number of myosins on synthetic filaments have negligible effect on gliding speeds.
a, Schematic of the myosin nanotube filament motility assay set-up. Biotinylated Cy5-DNA nanotubes (red) are attached to a glass coverslip through a biotin–BSA–neutravidin interface and uniquely labelled with either myosin V or VI Cy3-a′ (brown) at defined positions (Supplementary Table 1). b, Time-lapse images of three phalloidin-labelled actin filaments (green) moving along a myosin VI-Cy3-labelled 14 nm nanotube (red). Actin filaments move along the nanotube until reaching the end, then detach. c, Mean velocities (black) of actin along myosin V- and VI-labelled 14, 28 and 42 nm nanotubes as a function of actin length (or myosin number). Mean gliding speeds for myosin VI-labelled 14, 28 and 42 nm nanotubes are 80 ± 14 nm s−1 (N = 6,637 actin filaments), 82 ± 14 nms−1 (N=8,091) and 81 ± 15 nm s−1 (N = 696), respectively. For myosin V-labelled nanotubes, the mean speeds are 183 ± 29 nm s−1 (N=658 actin filaments), 179 ± 30 nm s−1 (N = 1,446) and 175 ± 33 nm s−1 (N = 222) for 14, 28 and 42 nm nanotubes, respectively. Error bars (grey shaded regions) indicate s.d.
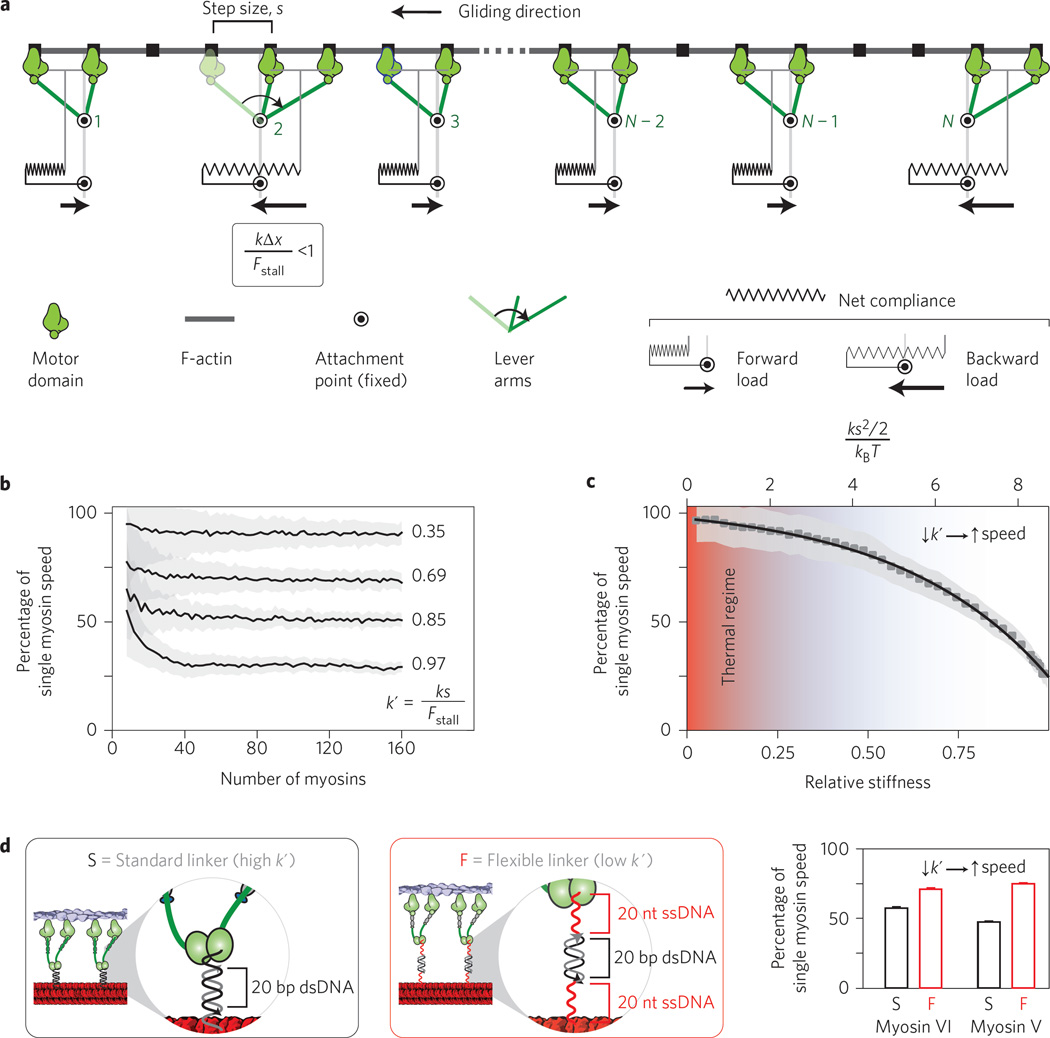
To gain insights into the molecular basis for this non-trivial collective behaviour, we designed a minimal stochastic simulation that models the net stiffness of each cross-bridge as a linear spring (Fig. 3a). Each myosin releases from the actin filament and undergoes a processive step, the direction of which is determined by the myosin type (V, towards the plus end; VI, towards the minus end). Because of the high motor densities (≥1 motor per 42 nm) and high processivities of myosin V and VI18,19, all motor dimers are assumed to always have at least one head bound to the gliding actin filament. In the model, a successful step or stroke is only possible if the resisting load is less than the stall force, Fstall. The load experienced by each myosin is proportional to the net stiffness of each cross-bridge (k) and its displacement from the resting position. The higher this load (high k), the slower the load-dependent individual actomyosin kinetics (increased cycle time) and hence the greater the reduction in ensemble speed compared to the speed of an individual myosin. In contrast, flexible cross-bridges (low k) result in higher ensemble speeds, ultimately approaching single myosin speed (Fig. 3b,c). However, as cross-bridge stiffness decreases, the potential energy stored in each cross-bridge connection approaches the thermal regime (kBT) such that smooth continuous gliding of the actin filament is no longer possible in the face of increasing thermal noise. Comparison of the experimental data and stochastic simulations shows that the relative stiffness of our model myosin filament system is estimated to be ~0.7, which translates to a mechanical energy per step of ~7 kBT (top horizontal axis; Fig. 3c). This stiffness reflects the most compliant element in the ensemble, namely the myosin protein. Thus, despite obvious differences between native myosin filaments and our synthetic scaffold, our measurements suggest that the myosin cross-bridge has evolved to operate sufficiently outside the thermal regime. The combination of experiment and simulation reveals that the actomyosin system balances the requirements of smooth motion and maximal speed.
Figure 3. Stochastic simulation of actin gliding along myosin filaments predicts actin length-independent gliding speed for sufficiently high motor number.
a, A periodic ensemble of N motors (green), attached to a rigid DNA nanotube scaffold (circles), interacts with a rigid actin filament (grey). Actin and interacting myosins are coupled viscoelastically, and each myosin experiences either a forward or a backward load. If tension T is less than the stall force (Fstall), a myosin is able to take a forward step of size s, as illustrated with the pre- (light green) and post- (dark green) stroke states of motor 2. A step will result in a net displacement of the actin filament of s/N (see Supplementary Information). See Supplementary Movie 2 for an animation of the model. b, Ensemble gliding velocity of dimeric myosin motors for various values of k′ (relative motor stiffness) as a function of motor number N. The mean velocity generated by the model is designated by the black line, with the s.d. in grey. c, Ensemble gliding speed and kinetic cycle time for the myosin motor as a function of k′. Based on the nanotube experiments, the elastic energy per motor cycle of myosin V and VI (~7 kBT) is well outside the thermal regime (red shaded area). Error bars (grey shaded regions) indicate s.d. d, Increasing linker flexibility through extension of the attachment strands with two single-stranded DNA segments (Supplementary Table 1) results in increased actin gliding speed for both myosin V and VI.
As mentioned above, our model predicts that the net stiffness of the myosin cross-bridge tunes the length-independent gliding speed. To test this model, we decreased the motor attachment stiffness (k′) by extending the attachment oligos with two 20-nucleotidelong ssDNA regions14 (Fig. 3d and Supplementary Table 1). For both myosin V and VI, this decreased relative stiffness results in a significant increase in gliding speeds along 28 nm DNA nanotubes (Fig. 3d). The speed increase is in agreement with previous reports for collective movements of two myosin V motors with different linker stiffnesses14. This speed increase supports our model that gliding movement emerges from the collective inter-motor tensions between elastically-coupled myosin motors.
The communication between motors is incorporated through the known load-dependent lengthening of the dwell time of the myosin on the actin filament20,21. This parameter captures the decrease in speed relative to single processive myosins (Fig. 3b,c) and, more importantly, predicts the length-independent gliding speed for sufficiently high motor number (N > 40; Figs 2c and 3b,c). What, then, is the advantage of larger assemblies of myosins? In muscle, of course, larger ensembles of myosins are needed to obtain the requisite force output of that particular muscle. Furthermore, our model predicts that external opposing loads will be distributed evenly over a larger collective of myosins, thereby lowering the local resisting force and allowing the overall ensemble to successfully function in opposition to a greater range of loads. We therefore propose that myosin ensembles function as energy reservoirs, where the individual steps collectively feed into the potential energy of the system, which is then released at a steady rate as the individual motors release and rebind to the actin filaments.
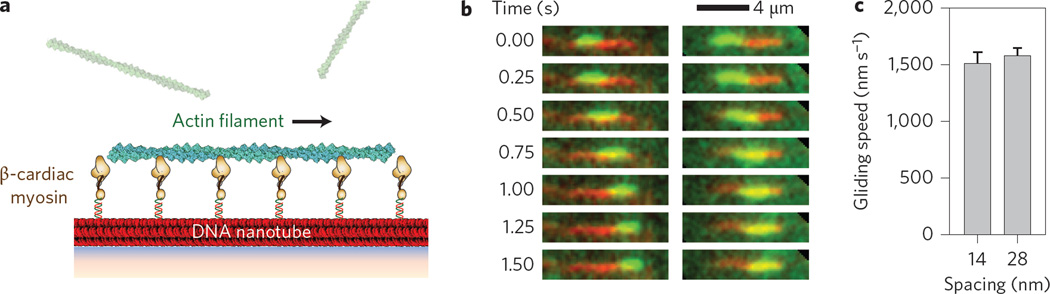
Finally, to test our system with a non-processive muscle myosin, we used truncated, single-headed, human β-cardiac myosin bound by its essential light chain5 (Fig. 4 and Supplementary Movie 3). Overall, the landing rate was lower with this monomeric β-cardiac myosin than with dimeric myosin V and VI (Fig. 2), as expected from the substantially lower duty ratio of muscle myosin (~0.05 for β-cardiac myosin and >0.8 for myosin V and VI3,5). Mean gliding speeds on β-cardiac myosin nanotubes with spacings of 14 and 28 nm are not significantly different, supporting the model (Fig. 3) based on similar observations for myosin V and VI. Interestingly, the actin gliding speed on DNA nanotubes for β-cardiac myosin, but not myosin V or VI, was significantly higher than that measured using the standard motility assay setup (850 ± 30 nm s−1 versus 1,580 ± 70 nm s−1 for β-cardiac myosin; 73 ± 4 nm s−1 versus 80 ± 14 nm s−1 for myosin VI; Fig. 4c). We estimate that, unlike with myosin V and VI (Fig. 3b), β-cardiac myosin-driven actin gliding on DNA nanotubes approaches the unloaded limit, with at most four myosin heads bound to the actin filament at any given time (based on a mean actin filament length of 1 µm, a 14 nm spacing and a 0.05 duty ratio). Hence, one possible explanation for the observed differences with β-cardiac myosin is that, in the standard gliding assay, myosins are able to bind off-axis and interact with actin at higher densities4. This probably contributes to the observed higher landing rate in the standard gliding assay, but at the expense of reduced speed from inter-motor interference.
Figure 4. Actin gliding on synthetic human β-cardiac myosin filaments.
a, Schematic of the nanotube filament motility assay with single-headed β-cardiac muscle myosin (brown). b, Time-lapse images of two phalloidin-labelled actin filaments (green) moving along human β-cardiac myosin-Cy3-labelled 14 nm nanotubes (red). c, Comparison of mean gliding speeds for actin filaments by human β-cardiac myosin using either 14 or 28 nm nanotubes. Mean gliding speeds for the 14 and 28 nm nanotubes are 1,580 ± 70 nm s−1 (N = 259 actin filaments) and 1,510 ± 100 nm s−1 (N = 125), respectively. Error bars indicate s.e.m.
In conclusion, the use of DNA nanotubes, combined with systematic and quantitative myosin labelling, provides a reliable, programmable and inexpensive alternative to the traditional gliding assay, and with controlled motor organization. Given the widespread interest in mapping the effects of cardiomyopathy-causing mutations (>200 mutations in β-cardiac myosin alone22), at the level of a single molecule, sarcomere, myocyte and whole heart, we propose that this technology is a timely addition to bridge single-molecule and myocyte biology. In addition to β-cardiac myosin, the artificial filaments can also incorporate the actin/myosin binding domains from myosin binding protein C (MyBP-C), a key regulator of cardiac muscle23. The engineered myosin filaments, combined with recent advances in generating recombinant skeletal and cardiac myosins5,24, are an essential first step to reconstituting the muscle thick/thin filament interface.
Methods
Nanotube assembly
DNA strands were ordered, unpurified, from Integrated DNA Technologies (IDT). DNA nanotubes were prepared in 1× TAE/Mg2+ buffer (40 mM Tris, 20 mM acetic acid, 2 mM EDTA, 12.5 mM MgCl2, pH 8.0). For 14 and 28 nm nanotubes, strands UM-01 to UM-40 were used. For 42 nm nanotubes, strands UM-01 to UM-30 and strands UM42-31 to UM42-60 were used (Supplementary Table 1). Each strand was combined to a final concentration of 2 µM, except strands 11, 18 and 31, which were each added in excess at a final concentration of 3.25 µM (Supplementary Table 1 and Supplementary Fig. 1). Strands 11 and 31 contained the sequence for myosin attachment (oligo-a, Flex oligo-a, or oligo-b). For the positive labelling control, an excess of Cy3-labelled strand complementary to oligo-a (Cy3-a′) was added in a twofold excess of attachment strands (14 µM). The anneal protocol was performed using a BioRad MyCycler (Supplementary Table 2) by decreasing the temperature from 90 to 70 °C over 20–200 min, from 70 to 40 °C over 900 min, from 40 to 20 °C over 200 min and finally holding at 4 °C.
Myosin expression, purification and labelling
Myosin V and VI were constructed, expressed in Sf9 insect cells, purified through FLAG affinity, and Cy3-a′, labelled as previously described11. Myosin V and VI constructs contained (from the N- to C-terminus) myosin (myosin VI, residues 1–992 from Sus scrofa, containing both the IQ and SAH domains; myosin VA, residues 1–1,103 from Gallus gallus, containing the IQ domains), a GCN4 leucine zipper (for dimerization25), a SNAP tag (for DNA oligo attachment), a FLAG tag (for purification) and finally a 6×His tag (for alternative purification). Myosin VI was cloned in pBiex-1 (Novagen) and expressed through transient transfection using the Escort VI system (Sigma). Myosin V was cloned in pFastBac for dual calmodulin co-expression and expressed through baculovirus infection. Human β-cardiac myosin II containing a SNAP tag was expressed as described previously5. β-Cardiac myosin constructs contained (from the N- to C-terminus) MHY7 residues 1–808, a SNAP tag and a C-tag (for affinity-based attachment using a PDZ-based system26). Replication-deficient recombinant adenoviruses were produced and amplified in HEK293 cells for both myosin and the essential light chain (MYL3) containing an N-terminal FLAG tag (for purification). Murine C2C12 myoblasts were differentiated and infected with myosin and light chain viruses for protein expression. Motor protein was used in motility experiments within one day of preparation.
Cells were lysed, incubated with Anti-FLAG resin and washed according to the method of Hariadi and co-authors11. Myosin bound to Anti-FLAG resin was incubated with excess (>5 µM) BG-Cy3-a′ at room temperature (~25 °C) for 30 min followed by overnight incubation on ice. Resin was washed three times with wash buffer (20 mM imidazole, 150 mM KCl, 5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 1 µg ml−1 phenylmethylsulphonyl fluoride, 10 µg ml−1 aproprotin, 10 µg ml−1 leupeptin, pH 7.4). Resin was then washed twice with wash buffer + 55% (vol/vol) glycerol. Finally, labelled myosin was incubated with 0.2 mg ml−1 FLAG-peptide (Sigma). Calmodulin was added to a concentration of 5–10 µM for stability and protein was stored at −20 °C. Labelling efficiency was assessed with a 10% SDS-PAGE gel on a Typhoon gel imager (GE Healthcare) for Cy3 followed by Coomassie staining (Supplementary Fig. 2). Myosin labelled with BG-Cy3-a′ showed a distinct gel-shift.
Motility assay
For gliding assays, the final wash buffer contained Alexa-488 phalloidin-labelled F-actin (Invitrogen), 2 mM ATP, 1 mM phosphocreatine, 0.1 mg ml−1 creatine-phosphokinase, 45 µg ml−1 catalase, 25 µg ml−1 glucose oxidase, 0.3–0.6% glucose and, in the case of myosin V and VI, 8 µM calmodulin. To obtain short actin filaments, a dilute solution of actin filaments was sheared by passing through a high-performance liquid chromatography syringe (National Scientific, NS600502, 22s gauge) one to five times. For each nanotube-based surface the relative Cy3 and Cy5 intensities were measured to determine the labelling efficiency. Time-lapse imaging was acquired at room temperature. Only fields of view with high labelling efficiency were analysed (Supplementary Methods). For myosin V and VI, movies were taken at a frame rate of 2 Hz and for β-cardiac myosin, the frame rate was 4 Hz.
Supplementary Material
Acknowledgments
The authors thank M. Westfall, D. Smith and L. Hilbert for useful discussions. Research was funded by the American Heart Association Scientist Development Grant (13SDG14270009), National Institutes of Health (NIH) grants 1DP2 CA186752-01 and 1-R01-GM-105646-01-A1 to S.S. and NIH grants GM33289 and HL117138 to J.A.S. R.F.S. is a Life Sciences Research Foundation Fellow. R.E.T. is supported by the NIH (F32 HL123247-02) and A.S.A. is supported by a Lucile Packard CHRI Postdoctoral Award.
Footnotes
Author contributions
R.F.H., R.F.S., A.S.A., R.E.T., J.A.S. and S.S. planned and designed experiments. R.F.H., R.F.S., A.S.A., R.E.T. and S.Su. performed experiments and analysed the results. R.F.H. and S.S. performed the mathematical modelling. R.F.H., R.F.S., J.A.S. and S.S. wrote the manuscript.
Additional information
Supplementary information is available in the online version of the paper.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Huxley HE. The mechanism of muscular contraction. Science. 1969;164:1356–1365. doi: 10.1126/science.164.3886.1356. [DOI] [PubMed] [Google Scholar]
- 2.Guérin T, Prost J, Martin P, Joanny JF. Coordination and collective properties of molecular motors: theory. Curr. Opin. Cell Biol. 2010;22:14–20. doi: 10.1016/j.ceb.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 3.De La Cruz EM, Ostap EM. Relating biochemistry and function in the myosin superfamily. Curr. Opin. Cell Biol. 2004;16:61–67. doi: 10.1016/j.ceb.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 4.Uyeda TQ, Kron SJ, Spudich JA. Myosin step size. Estimation from slow sliding movement of actin over low densities of heavy meromyosin. J. Mol. Biol. 1990;214:699–710. doi: 10.1016/0022-2836(90)90287-V. [DOI] [PubMed] [Google Scholar]
- 5.Sommese RF, et al. Molecular consequences of the R453C hypertrophic cardiomyopathy mutation on human β-cardiac myosin motor function. Proc. Natl Acad. Sci. USA. 2013;110:12607–12612. doi: 10.1073/pnas.1309493110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harris DE, Warshaw DM. Smooth and skeletal muscle myosin both exhibit low duty cycles at zero load in vitro. J. Biol. Chem. 1993;268:14764–14768. [PubMed] [Google Scholar]
- 7.Piazzesi G, et al. Skeletal muscle performance determined by modulation of number of myosin motors rather than motor force or stroke size. Cell. 2007;131:784–795. doi: 10.1016/j.cell.2007.09.045. [DOI] [PubMed] [Google Scholar]
- 8.Spudich JA. Hypertrophic and dilated cardiomyopathy: four decades of basic research on muscle lead to potential therapeutic approaches to these devastating genetic diseases. Biophys. J. 2014;106:1236–1249. doi: 10.1016/j.bpj.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baker JE, Brosseau C, Joel PB, Warshaw DM. The biochemical kinetics underlying actin movement generated by one and many skeletal muscle myosin molecules. Biophys. J. 2002;82:2134–2147. doi: 10.1016/S0006-3495(02)75560-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derr ND, et al. Tug-of-war in motor protein ensembles revealed with a programmable DNA origami scaffold. Science. 2012;338:662–665. doi: 10.1126/science.1226734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hariadi RF, Cale M, Sivaramakrishnan S. Myosin lever arm directs collective motion on cellular actin network. Proc. Natl Acad. Sci. USA. 2014;111:4091–4096. doi: 10.1073/pnas.1315923111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walcott S, Warshaw DM, Debold EP. Mechanical coupling between myosin molecules causes differences between ensemble and single-molecule measurements. Biophys. J. 2012;103:501–510. doi: 10.1016/j.bpj.2012.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rock RS, Rief M, Mehta AD, Spudich J. A. In vitro assays of processive myosin motors. Methods. 2000;22:373–381. doi: 10.1006/meth.2000.1089. [DOI] [PubMed] [Google Scholar]
- 14.Lu H, et al. Collective dynamics of elastically coupled myosin V motors. J. Biol. Chem. 2012;287:27753–27761. doi: 10.1074/jbc.M112.371393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yin P, et al. Programming DNA tube circumferences. Science. 2008;321:824–826. doi: 10.1126/science.1157312. [DOI] [PubMed] [Google Scholar]
- 16.Ali MY, et al. Myosin Va and myosin VI coordinate their steps while engaged in an in vitro tug of war during cargo transport. Proc. Natl Acad. Sci. USA. 2011;108:E535–E541. doi: 10.1073/pnas.1104298108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hilbert L, Cumarasamy S, Zitouni NB, Mackey MC, Lauzon AM. The kinetics of mechanically coupled myosins exhibit group size-dependent regimes. Biophys. J. 2013;105:1466–1474. doi: 10.1016/j.bpj.2013.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rief M, et al. Myosin-V stepping kinetics: a molecular model for processivity. Proc. Natl Acad. Sci. USA. 2000;97:9482–9486. doi: 10.1073/pnas.97.17.9482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rock RS, et al. Myosin VI is a processive motor with a large step size. Proc. Natl Acad. Sci. USA. 2001;98:13655–13659. doi: 10.1073/pnas.191512398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gebhardt JCM, Clemen AEM, Jaud J, Rief M. Myosin-V is a mechanical ratchet. Proc. Natl Acad. Sci. USA. 2006;103:8680–8685. doi: 10.1073/pnas.0510191103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Veigel C, Molloy JE, Schmitz S, Kendrick-Jones J. Load-dependent kinetics of force production by smooth muscle myosin measured with optical tweezers. Nature Cell Biol. 2003;5:980–986. doi: 10.1038/ncb1060. [DOI] [PubMed] [Google Scholar]
- 22.Walsh R, Rutland C, Thomas R, Loughna S. Cardiomyopathy: a systematic review of disease-causing mutations in myosin heavy chain 7 and their phenotypic manifestations. Cardiology. 2010;115:49–60. doi: 10.1159/000252808. [DOI] [PubMed] [Google Scholar]
- 23.Weith A, et al. Unique single molecule binding of cardiac myosin binding protein-C to actin and phosphorylation-dependent inhibition of actomyosin motility requires 17 amino acids of the motif domain. J. Mol. Cell. Cardiol. 2012;52:219–227. doi: 10.1016/j.yjmcc.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Resnicow DI, Deacon JC, Warrick HM, Spudich JA, Leinwand LA. Functional diversity among a family of human skeletal muscle myosin motors. Proc. Natl Acad. Sci. USA. 2010;107:1053–1058. doi: 10.1073/pnas.0913527107. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 25.Trybus KM, Freyzon Y, Faust LZ, Sweeney HL. Spare the rod, spoil the regulation: the necessity for a myosin rod. Proc. Natl Acad. Sci. USA. 1997;94:48–52. doi: 10.1073/pnas.94.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang J, Nagy SS, Koide A, Rock RS, Koide S. A peptide tag system for facile purification and single-molecule immobilization. Biochemistry. 2009;48:11834–11836. doi: 10.1021/bi901756n. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.