Among the most fascinating enzymes are the molecular motors, which exquisitely couple adenosine triphosphate (ATP) hydrolysis to directional mechanical motion. They power the movement of intracellular vesicles, chromosomes, and messenger RNA–protein complexes through the cytoplasm of nearly all eukaryotic cells, using actin filaments and microtubules as their tracks. A prominent theme in these motors is allostery, or communications that occur across the enzyme at several nanometer distances. Chemical events occurring in the motor’s active site, for instance, are coordinated with tight binding of the motor to the track along which it moves, and then its subsequent release, and with mechanical elements that amplify small movements occurring near the active site. In the 1980s, researchers used quantitative in vitro motility assays, sensitive to single molecules, to study two of the three major classes of motor enzymes: the microtubule-based kinesin family and the actin-based myosin family. In the 1990s, investigators solved the crystal structures of kinesin 1 (1) and muscle myosin II (2). These complementary approaches ushered in a new era of understanding the mechanisms of these molecular machines. Dynein, the third important class of molecular motor, is a complex that processively moves along microtubules in the opposite direction to kinesin 1. Although single molecule assays have been applied to dynein, detailed structural information on this mammoth machine has remained elusive, until now. On page 1159 of this issue, Carter et al. (3) report a crystal structure for a 610-kD homodimer of yeast cytoplasmic dynein. The structure reveals surprises about how this massive molecular motor might work.
Dynein was first discovered by Gibbons in 1965 (4) as the adenosine triphosphatase (ATPase) that drives the beating of cilia and flagella. Subsequent studies showed that dyneins play diverse motility roles in eukaryotic cells. Paschal et al. (5) identified a cytoplasmic version of dynein, which subsequent studies showed powers the movement of many cargoes. A final type of dynein (cytoplasmic dynein 2) powers the movement of protein building blocks in cilia and flagella.
Given how much we know about kinesins and myosins from structural and single-molecule studies, one might think that we could make a reasonable guess about how dynein works. However, dynein emerged from an evolutionary lineage that is separate from kinesin and myosin (which share an ancient evolutionary origin) and seems to be a completely different type of molecular machine. Phylogenetic sequence analysis (6) showed that dynein is a member of the AAA family (ATPases associated with diverse cellular activities), but it is an odd uncle. Many AAA ATPases are protein unfoldases, which target proteins for degradation or break apart protein complexes (7). These AAA ATPases work by self-assembling into hexameric rings and then use ATP energy to “stuff ” the polypeptide chain into the central pore. Some AAA proteins also act as DNA and RNA helicases, again by feeding the nucleic acid polymer through the central pore. Dynein, however, has evolved a way of using the same basic ATPase module to walk along a microtubule track.
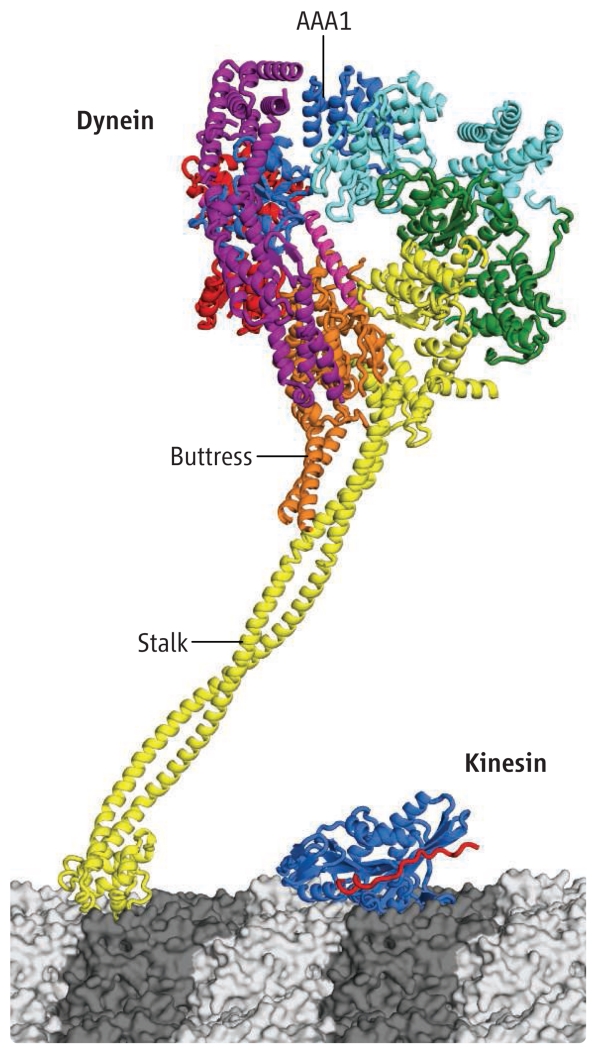
Dynein also is more complicated than most AAA ATPases. It has integrated six AAA domains (four are functional for ATP binding and two are nonfunctional) into one polypeptide chain that folds into a ringlike structure (see the figure). Dynein also has added various bells and whistles to this basic ring-shaped scaffold, which allow it to function as a cytoskeletal motor. One such additional feature is a mechanical element, called the linker, which sits on top of the ring and might swing from one position to another, perhaps similar to the lever arm of myosins (8). A second feature is a long (~15 nm) coiled coil, called the stalk, which emerges from the ring and has a microtubule binding domain perched on its end. Remarkably, the affinity of the microtubule binding domain is modulated by transitions in the ATPase cycle (the binding of ATP, hydrolysis and product release), primarily by one of the four AAA ATPase domains (AAA1). However, this ATPase site resides on the opposite side of the ring from where the stalk and microtubule binding domain protrude out of the ring. How an allosteric conformational change propagates from AAA1, around the ring, and then through a long coiled coil to the microtubule binding domain remains a mystery.
Figure. Structure of the dynein motor.
Dynein is depicted bound to a microtubule next to the motor domain of kinesin 1. The six AAA domains (dark blue, light blue, green, yellow, orange, and red), linker (purple), buttress, and stalk are indicated. The affinity of the microtubule binding domain is modulated by transitions in the ATPase cycle, primarily by the AAA1 ATPase domain (dark blue). In Carter et al.’s crystal structure, the stalk was truncated just below the point at which the buttress meets the stalk. The structure of the distal microtubule binding domain is from (12), and an intervening coiled coil of the proper length is introduced in this figure.
Past studies using EM were instrumental in providing the first structural insights into dynein, revealing the approximate locations of the AAA domains, the linker, and the stalk, and showing the movement of the linker in different nucleotide states (9, 10). Carter et al. have taken the problem an important step further with x-ray crystallography, solving a 6 Å structure of the cytoplasmic dynein motor domain. The structure is not of high enough resolution to resolve the amino acid side chains, but it shows virtually all of the helices and β sheets. For a complex machine like dynein, however, such information is important because it reveals the organization and interactions of the ATPase domains, how the stalk integrates into the ATPase ring, and the secondary structure of the linker.
The structure uncovered several surprises. First, the microtubule binding stalk appears to be supported by a second coiled coil that emerges from the ring. The authors call this second and shorter coiled coil the “buttress.” However, it may not just prop up the stalk; it may also play an important role in changing the structure of the stalk’s coiled coil during dynein’s ATPase cycle in a way that alters microtubule binding affinity. Second, the linker, which is composed of helical bundles, does not sit flat on the ring but rather arches over it, looking a bit like the handle of a basket. The contact of the linker with one end of the ring looks tenuous, however, suggesting that it may break and come loose at some stage of dynein’s ATPase cycle. Third, the ATPase domains do not form a symmetric hexameric ring, as predicted (11), but instead are arranged in a very irregular pattern. Particularly intriguing is a large gap between AAA1 (dynein’s main hydrolytic site) and AAA2. This is surprising, because hydrolysis of ATP requires a neighboring subunit in close proximity to provide key residues that promote γ-phosphate bond cleavage. The authors crystallized dynein without nucleotide and speculate that if ATP bound to AAA1, it would draw AAA2 closer, thus decreasing the gap and allowing hydrolysis to occur. The movement of AAA2 toward AAA1 might start a domino effect of movements of other AAA domains around the ring. Such a domino effect could explain how binding of ATP to AAA1 could affect the stalk/microtubule binding domain, the buttress, and the linker, even though they are on the opposite side of the ring.
Many questions remain unanswered. Carter et al. do not address the role of nucleotide in other AAA domains (AAA2, AAA3, and AAA4), and it is unclear at this resolution whether these sites might contain tightly bound nucleotide. We also are left with the question of why it was advantageous for cells to have evolved such a mammoth motor, when a much smaller microtubule motor, kinesin, is available. Perhaps the answer lies in the specific mechanisms that regulate dynein motor function. Co-crystals of the dynein motor domain with some of its regulatory proteins (e.g., lissencephaly 1) might provide answers.
Finally, this is only one snapshot of the motor in action. Obtaining views in two or more nucleotide states is essential. To ultimately understand how dynein works, researchers will need crystal structures of dynein with better than 3 Å resolution, and in different nucleotide states. This first structure, however, has provided a wealth of information, new hypotheses that can be tested, and optimism that crystals of different nucleotide states may be obtained in the near future.
Footnotes
X-ray crystallography provides some surprising insights into the dynein class of molecular motors.
References
- 1.Kull FJ, et al. Nature. 1996;380:6574. doi: 10.1038/380555a0. [DOI] [PubMed] [Google Scholar]
- 2.Rayment I, et al. Science. 1993;261:5117. doi: 10.1126/science.8316858. [DOI] [PubMed] [Google Scholar]
- 3.Carter AP, et al. Science. 2011;331:1159. doi: 10.1126/science.1202393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gibbons IR, Rowe AJ. Science. 1965;149:424. doi: 10.1126/science.149.3682.424. [DOI] [PubMed] [Google Scholar]
- 5.Paschal BM, Shpetner HS, Vallee RB. J. Cell Biol. 1987;105:1273. doi: 10.1083/jcb.105.3.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neuwald AF, Aravind L, Spouge JL, Koonin EV. Genome Res. 1999;9:27. [PubMed] [Google Scholar]
- 7.Hanson PI, Whiteheart SW. Nat. Rev. Mol. Cell Biol. 2005;6:519. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 8.Spudich JA, Sivaramakrishnan S. Nat. Rev. Mol. Cell Biol. 2010;11:128. doi: 10.1038/nrm2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burgess SA, Walker ML, Sakakibara H, Knight PJ, Oiwa K. Nature. 2003;421:615. doi: 10.1038/nature01377. [DOI] [PubMed] [Google Scholar]
- 10.Roberts AJ, et al. Cell. 2009;136:485. doi: 10.1016/j.cell.2008.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mocz G, Gibbons IR. Structure. 2001;9:93. doi: 10.1016/s0969-2126(00)00557-8. [DOI] [PubMed] [Google Scholar]
- 12.Carter AP, et al. Science. 2008;322:1691. doi: 10.1126/science.1164424. [DOI] [PMC free article] [PubMed] [Google Scholar]