Who is not fascinated by the myriad biological movements that define life? From cell migration, cell division and a network of translocation activities within cells to highly specialized muscle contraction, molecular motors operate by burning ATP as fuel. Three types of molecular motors—myosin, kinesin and dynein—and nearly 100 different subtypes transduce that chemical energy into mechanical movements to carry out a wide variety of cellular tasks. Understanding the molecular basis of energy transduction by these motors has taken decades.
Our understanding of molecular motors could be viewed as beginning with the two 1954 papers in Nature by Hugh Huxley and Jean Hanson and Andrew Huxley and Rolf Niedergerke, respectively, where the authors proposed the sliding-filament theory of muscle contraction. But a good place to start my story is 1969, when Hugh Huxley, on the basis of his remarkable X-ray diffraction experiments on live muscle coupled with electron microscopy, postulated the swinging cross-bridge hypothesis of muscle contraction1. Thus, more than 40 years ago, he proposed the basic concepts of how the myosin molecule produces the sliding of actin filaments to produce contraction. Hugh Huxley laid the foundation for the molecular motor field, and we are all indebted to him.
My beginnings in myosin research began as a postdoctoral fellow in Hugh’s laboratory at the Medical Research Council Laboratory of Molecular Biology in Cambridge, England, coincidentally in 1969. But my fascination with science began much earlier.
Recently at a biophysics meeting in Cavtat, Croatia, I pondered the path that brought me, the grandson of a Croatian immigrant, to experimental science. My grandfather George Spudich and his family left Zagreb at the turn of the twentieth century. They settled in Benld, a small coal-mining town in Illinois with a population of about 1,500, both then and now. My father worked the ‘number 2’ mine, sometimes for two eight-hour shifts when a family member was injured. Neither of my parents was college educated, but they both had keen intellects, positive and enthusiastic outlooks and profound work ethics. My father was intrigued by how things work and shared that interest with my brother John and me. After the coal mines closed, my father taught himself electrical engineering, founded the Spudich Electric Company and patented one of his inventions. He often told John and me, “do whatever excites you, but do it well and be respectful of people you interact with.”
I was captivated with chemistry from a young age. Beginning at the age of six, I mastered every chemistry set I could get. The myriad chemical reactions that could be created using everyday materials, sometimes with marvelously explosive results, fed my excitement for chemistry. It was a world unfamiliar to my parents, but they respected my preoccupations and cleared the pantry of our modest home for me to set up a lab with discarded equipment given to me by my high school chemistry teacher Robert Brandsmark. My brother John has also followed the allure of science into an exciting and distinguished career in basic research. His work has established the molecular basis of signaling in an important class of rhodopsins that he discovered in 1982 (ref. 2). John was my first collaborator. We once had three fire engines responding to a substantial explosion we set off in a drainage ditch using chemicals we could easily get at that time from a company in St. Louis.
My public high school education in an industrial town in southern Illinois prepared me well for the rigorous chemistry curriculum at the University of Illinois–Urbana-Champaign, and a chance encounter with Woody Hastings at the University of Illinois launched my experimental-science career. Throughout my undergraduate years, I worked with Woody on bioluminescence in Vibrio fischeri3. I was inspired by his high-spirited fascination with biology and was fortunate to be invited to help him teach in the physiology course at the Marine Biological Laboratory (MBL) in Woods Hole (Fig. 1). At the MBL, I was introduced to the breadth and potential of many biological systems, including muscle contraction.
Figure 1.
Woods Hole Physiology Course, 1963. Woody Hastings is in the top row, eighth from left.
In 1963 I joined the PhD program in the new Department of Biochemistry at Stanford University, founded by Arthur Kornberg. One of the many remarkable aspects of the biochemistry department was that, although Arthur was my thesis advisor, all the faculty members were my mentors. This unique environment shaped the way I do research and taught me how to be a responsible colleague and a mentor to others (Fig. 2). I learned how important it is to reduce complex biological systems to their essential components and create quantitative in vitro assays for the function of interest. Those years also made it clear to me that interdisciplinary approaches would be key to understanding complex biological processes. So I decided to do postdoctoral work in both genetics and structural biology. I spent one year at Stanford with another influential role model, Charley Yanofsky, working on the genetics of the Escherichia coli tryptophan operon. I then joined Hugh Huxley’s laboratory in Cambridge. My postdoctoral work with Hugh defined the structure of the actin-tropomyosin-troponin complex and led us to propose the steric blocking mechanism of regulation of muscle contraction4.
Figure 2.
Many of the contributors to the discoveries regarding energy transduction by myosins and the roles of myosins in nonmuscle cells. This 2007 photo includes students, postdoctoral fellows, sabbatical visitors and two of my former mentors, Paul Berg and Charley Yanofsky, at a Spudich Symposium at Stanford that was organized by former members of my laboratory.
When I set up my own laboratory at the University of California–San Francisco, I chose to apply a combination of biochemistry, genetics and structural biology to study fundamental unanswered questions in cell biology at the time: how the chemical energy of ATP hydrolysis brings about mechanical movement and what roles a myosin-like motor might have in nonmuscle cells. I was then following the advice I now give my students: get as much interdisciplinary training as you can early in your career, work on what you are captivated by and stay focused on the essential issues.
The essential first steps were to develop a quantitative in vitro motility assay for myosin movement on actin, which is crucial for understanding the molecular mechanism of energy transduction by this system, and to develop a model organism to unravel the molecular basis of the myriad nonmuscle-cell movements that are apparent by light microscopy. We explored Neurospora crassa, Saccharomyces cerevisiae, Physarum polycephalum, Dictyostelium discoideum, Nitella axillaris and other organisms, all unfamiliar to me at the time. The giant cells of the alga Nitella were particularly intriguing because of their striking intracellular cytoplasmic streaming that was visible under a simple light microscope. Although not suitable for biochemistry or genetics, Nitella would assume an important role in my lab a decade later, after Yolande Kersey in Norm Wessells’s laboratory in the Department of Biological Sciences at Stanford showed oriented actin cables lying along chloroplast rows in these cells5.
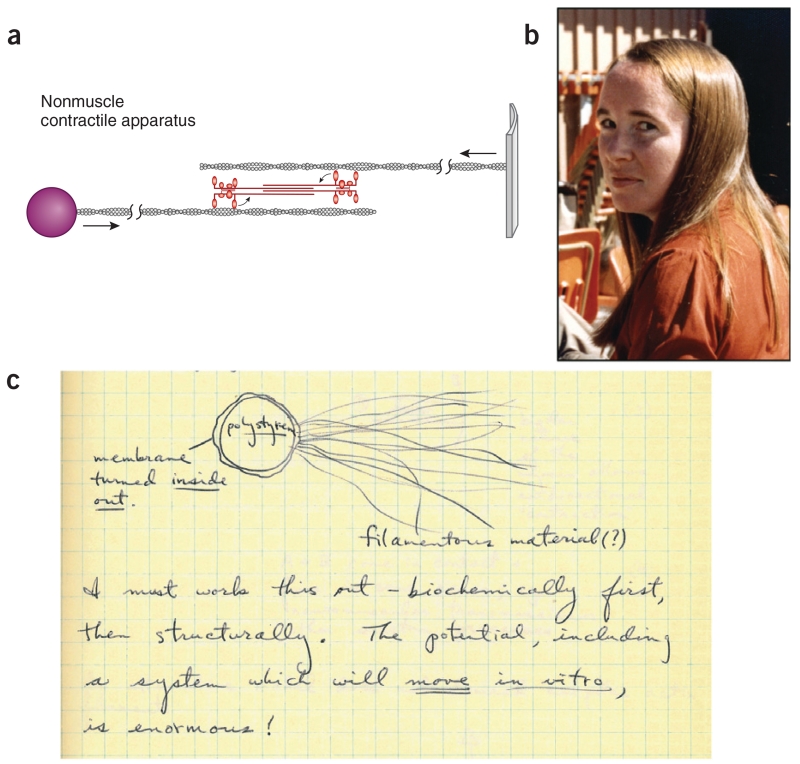
The slime mold Dictyostelium proved best for our initial biochemical approach6. Margaret Clarke, my postdoctoral fellow, identified a myosin in Dictyostelium. We also showed that actin is associated with the cell membrane in this organism, and we isolated membrane-coated polystyrene beads with actin filaments emanating from them. We were tremendously excited about the possibilities these results presented as a small step along the way to an in vitro motility assay where these actin-coated particles could move along a myosin-coated surface (Fig. 3). And in the late 1970s we succeeded in growing purified actin filaments with the correct polarity off polystyrene beads, which fueled our optimism toward reaching the same goal.
Figure 3.
Dictyostelium has a muscle-like myosin and membrane-associated actin. (a) A possible scheme for pulling two membranes together (redrawn from ref. 6). (b) Margaret Clarke discovered myosin II in Dictyostelium and showed that it forms bipolar thick filaments, similar to muscle myosin. (c) Phagocytized polystyrene beads offered an opportunity to explore one version of an in vitro motility assay where the beads may be pulled along by myosin. Taken from my laboratory notebook, 21 January 1973.
In 1977 I joined the Department of Structural Biology at Stanford. In the next years we extensively characterized the actin-myosin system in Dictyostelium. My student Arturo De Lozanne made the chance discovery that genes in Dictyostelium can be knocked out by homologous recombination and provided the first genetic proof that myosin II is essential for cytokinesis but is not required for cell migration7. The latter was a surprising and important observation because it was assumed up to that time that myosin II drove the forward movement of cells. Dietmar Manstein, Meg Titus and Arturo then extended these experiments to create a myosin-null cell8, which was crucial to our later work using mutational analysis to define the structure-function relationships of the myosin molecule and for important experiments in support of the swinging cross-bridge hypothesis9.
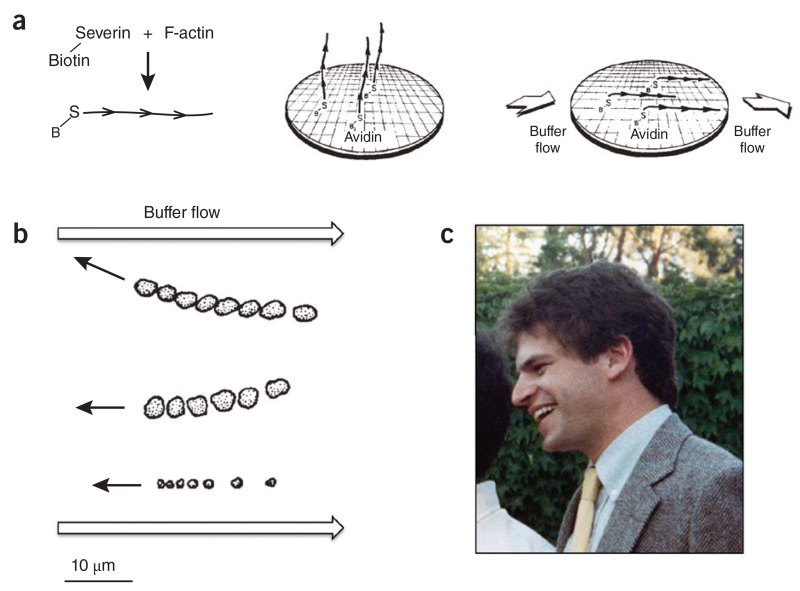
Interestingly, reports from a number of laboratories between 1969 and 1980 did not support the swinging cross-bridge model, and it was more imperative than ever to develop a quantitative in vitro motility system to test the various models under consideration. In 1981 we identified and purified Dictyostelium severin, a protein that tightly binds the ‘barbed ends’ of actin filaments. This provided an opportunity to try another version of an in vitro motility assay. Using biotinylated severin, we attached the actin filament barbed ends to an avidin-coated slide and flowed aqueous solution over them. Long filaments attached to the surface at one end would be expected to orient in the direction of the flowing solution (Fig. 4a). We placed myosin-coated beads on these actin-coated slides and added ATP but saw only sporadic movements. In retrospect, we probably did not have sufficient alignment of filaments; we were not monitoring filament alignment at that time by electron microscopy, as we did later.
Figure 4.
One approach to an in vitro motility assay from a totally defined system. (a) The concept was to observe myosin-coated beads moving along fixed actin filaments oriented by buffer flow. The actin filaments had biotinylated severin bound to their barbed ends; the barbed ends were attached to an avidin-coated surface by way of the tight avidin-biotin link. The filaments were oriented by buffer flow. B, biotin; S, severin. (b) Myosin-coated beads were observed by light microscopy to move upstream toward the barbed end of the surface-attached actin filaments. The position of each of the three bead aggregates is shown as a function of time. This was the first demonstration of quantitative, directed movement of myosin along actin with a totally defined system (taken from ref. 11). (c) Graduate student Steve Kron at a colleague’s wedding.
A key breakthrough occurred in 1982 when Mike Sheetz came to my laboratory on sabbatical. Not certain what component of our system might be limiting our approach, we took advantage of the known orientation of actin filaments in Nitella5 to overcome the actin filament alignment problem. Peter Sargent, a neurobiologist in the Structural Biology Department at that time, helped us cut open a Nitella cell, and we attached it to a surface to expose the actin fibers. We added myosin-coated beads and eureka! We saw robust ATP-dependent unidirectional movement along chloroplast rows, which mark the actin fibers10.
Armed with the Nitella results, Mike left my lab and went to the MBL to explore whether myosin-coated vesicles may account for the particle movements observed in squid axons. Ron Vale, then a graduate student at Stanford with Eric Shooter, was fascinated by the movement of organelles in nerve axons and joined Mike at the MBL. To their great surprise, they found that movement in axons is not myosin driven. Instead, they discovered the new molecular motor kinesin, a discovery that completely energized the field and opened up years of exciting work from their laboratories and many others.
Meanwhile, back at Stanford, although our Nitella results were exciting, we were working to eliminate the vagaries of the complex Nitella substratum and establish a totally defined in vitro motility assay for myosin moving on actin. We returned to the biotinylated-severin actin-filament approach but now used electron microscope grids attached to the glass slides to monitor the orientation of the actin filaments under various buffer flow conditions (Fig. 4a). With well-oriented actin filaments, my graduate student Steve Kron and I achieved the long-term goal of observing ATP-dependent directed movement of purified myosin along purified actin filaments at rates that were consistent with the speeds of muscle contraction and other forms of cell motility11 (Fig. 4b).
Thanks to a report by Toshio Yanagida et al.12, we returned to the myosin-coated surface concept that we had considered earlier. In the 1970s, we could only consider monitoring movement by observing the translocation of beads to which the actin filaments were attached; there was no way to visualize the 9-nm-wide actin filaments directly. In 1984, Yanagida et al.12 showed that individual actin filaments labeled with rhodamine-phalloidin could be observed by fluorescence microscopy. Using Toshio’s observation, Steve Kron achieved robust ATP-dependent movement of fluorescent actin filaments on glass slides coated with purified myosin13. Using this ‘Kron assay’, postdoctoral fellow Yoko Toyoshima and others in my laboratory showed that the globular head, or subfragment 1 (S1), of myosin is the motor domain14. This observation eliminated competing theories to the swinging cross-bridge hypothesis and focused research on the S1 head to understand how the myosin family of molecular motors works. Almost two decades of hard work led us to the early goals we had set for ourselves.
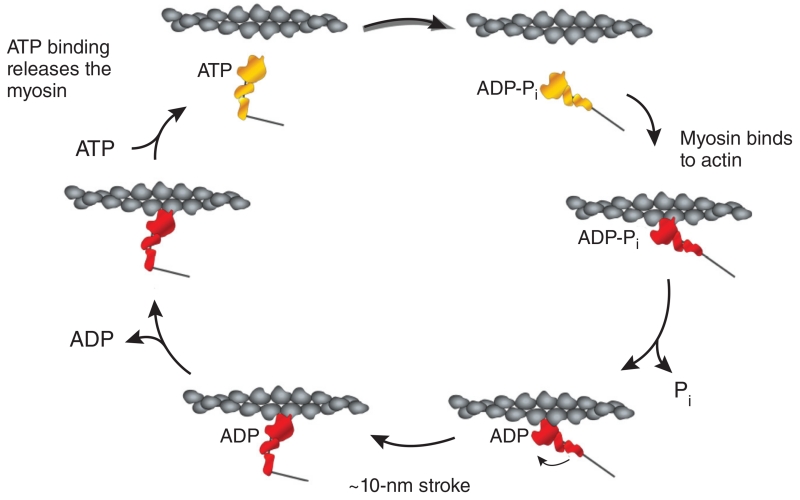
The combination of the in vitro motility assay and the Dictyostelium myosin-null cell provided powerful tools for Kathy Ruppel, Taro Uyeda, Dietmar Manstein, William Shih, Coleen Murphy, Meg Titus, Tom Egelhoff and others in my lab to use mutations along myosin to define the biochemical, biophysical and assembly properties of the molecule. Our results were consistent with the proposed actin-activated myosin chemomechanical cycle derived largely from the elegant biochemical kinetic studies from Edward Taylor’s laboratory in the early 1970s (ref. 15) (Fig. 5). Then, in 1993, Ivan Rayment and his colleagues16 obtained a high-resolution crystal structure of myosin S1. Ivan’s pivotal work allowed us to place our mutational analyses in a myosin structure-function context.
Figure 5.
The actin-activated myosin chemomechanical cycle. This cycle, extensively studied by many researchers over several decades, was derived from kinetic studies of Lymn and Taylor15. A mechanical stroke only occurs when the myosin is strongly bound to actin. Our mutational analyses of Dictyostelium myosin II probed each of the steps shown and provided structure-function analyses that helped define how the myosin motor works. ADP-Pi, ADP and inorganic phosphate, the products of ATP hydrolysis, remain bound to the active site until actin binds to the myosin.
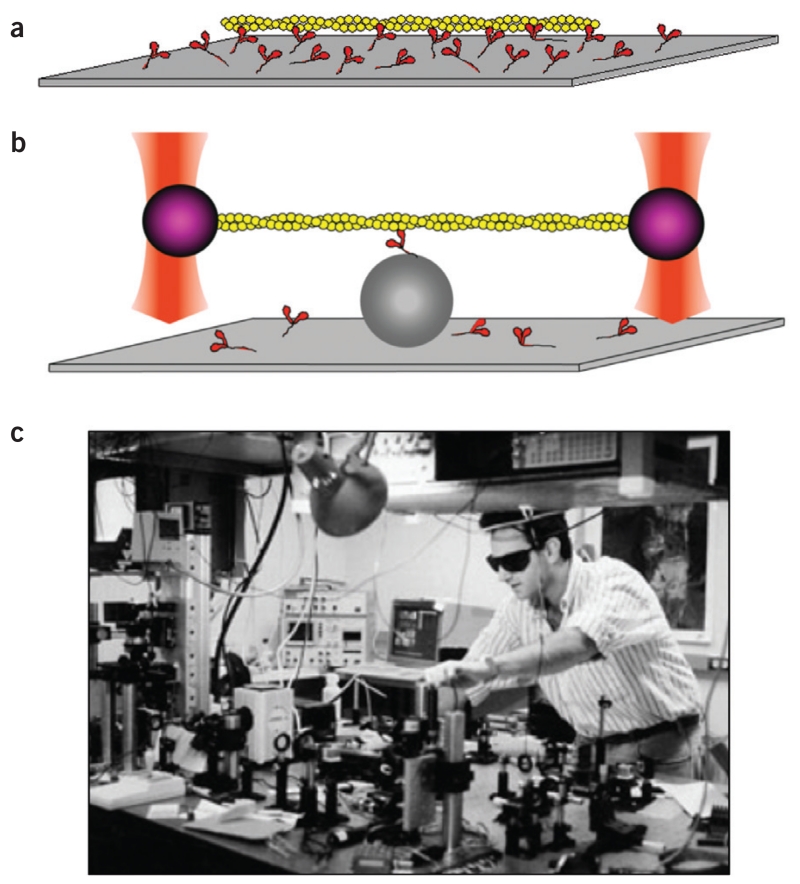
Fundamental issues still remained—primarily to establish the step size that the myosin takes for each ATP hydrolysis, which was under considerable debate. A step size larger than ~10 nm would be inconsistent with Hugh Huxley’s swinging cross-bridge model, and one would be forced to consider alternative molecular mechanisms. It was therefore crucial to observe a single myosin molecule go through one cycle of ATP hydrolysis and measure the step size directly. This was achieved when my graduate student Jeff Finer and sabbatical visitor Robert Simmons modified the Kron assay by building a dual-beam laser trap for single-molecule analysis (Fig. 6). Using the dual-beam laser trap, we lowered a single actin filament onto a single myosin molecule and were able to measure the step size as ~10 nm and the force produced at ~5 pN (ref. 17).
Figure 6.
In vitro motility taken to the single-molecule level using the physics of laser trapping. (a) The Kron in vitro motility assay observing fluorescent actin filaments (yellow) moving on a myosin-coated (red) surface. (b) Two polystyrene beads attached to the ends of a single actin filament are trapped in space by laser beams. The filament is lowered onto a single myosin molecule on a bump on the surface (gray sphere). (c) Jeff Finer building the dual-beam laser trap in around 1990.
Building the laser trap involved a collaboration between our group and the physicist (now US Secretary of Energy) Steve Chu. Our collaboration was highly unusual in that members of our labs physically moved to each other’s laboratory environments for extended periods of time. This was so productive for both labs that we convinced then provost Condoleezza Rice and our deans that such interdisciplinary exchange of students should occur broadly—between biology, physics, chemistry, computational sciences, engineering and clinical sciences. This led to our cofounding the global interdisciplinary Bio-X program at Stanford.
The dual-beam laser trap experiments led to a host of studies on nonmuscle myosins in my laboratory and others. Strong additional evidence in support of Hugh Huxley’s swinging cross-bridge hypothesis came from analyses of nonmuscle myosins. Of particular interest was myosin VI, which first seemed as though it may be the myosin motor that would disprove the swinging cross-bridge hypothesis but turned out to strongly support the hypothesis, with a full ~180° swing of its lever arm18. This work by my postdoctoral fellow Zev Bryant on myosin VI was a vivid demonstration of how in vitro motility and single-molecule laser trap assays can reveal functional structural transitions in molecular motors and, potentially, other enzymes.
One of my great satisfactions is that the more detailed understanding of energy transduction by myosin has led to potential clinical therapies. A small molecule that binds and activates β-cardiac myosin is now in clinical trials for the treatment of heart failure, and another small molecule currently in clinical trials activates skeletal muscle contraction and may aid patients with amyotropic lateral sclerosis and other diseases.
I am privileged to have had many opportunities in my career. I am grateful to the many teachers and people committed to the superb public education system that helped me achieve my aspirations. In turn, it is my ardent hope that my work on understanding the complexities of molecular motors will bring benefit to society at large. Although the potential therapeutic benefits are exciting, one of the greatest pleasures of my career has been to see members of my extended scientific family thriving in their chosen careers.
Throughout my career, my wife Anna, a scientist and scholar in her own right, has worked closely with me, having fruitful science conversations with me both in the lab and at home. In addition to all that we share in our family life, we are two scientific colleagues who constantly bounce ideas off each other, and she therefore contributes immensely to all that I do.
ACKNOWLEDGMENTS
Discoveries in science are a community enterprise involving scores of investigators making pivotal contributions along the way. In this short essay, I have necessarily left out the names of many people who contributed to the breakthroughs we made on the workings of cellular motors. I thank all the members of my laboratory over these many years, with whom I have shared the joys of discovery. My mentors have my gratitude for their support and encouragement and for sharing with me their own ways of creative research. It has also been my privilege to share the excitement of discoveries with my fellow faculty colleagues first at the University of California–San Francisco and for the last 35 years at Stanford University. I also thank K. VijayRaghavan, S. Mayor and my colleagues at the National Center for Biological Sciences, Bangalore for incorporating me into their inspiring and innovative scientific activities for the last ten years. And my immense gratitude goes to my family, first and foremost to my wife Anna for bringing her energy, keen intellect and unwavering support into my life. My daughters Rani and Serena carry on the values my parents imparted to me, and they, together with my erudite sons-in-law Dan and Dave and my ‘cool’ grandchildren Indira, Hana, Anjali, Alexander and Nathaniel, are the joys of my life. Our work would not have been possible without the generous financial support from the US National Institutes of Health and grants from the Human Frontiers Science Program.
Footnotes
COMPETING FINANCIAL INTERESTS
The author declares no competing financial interests.
References
- 1.Huxley HE. The mechanism of muscular contraction. Science. 1969;164:1356–1365. doi: 10.1126/science.164.3886.1356. [DOI] [PubMed] [Google Scholar]
- 2.Spudich JL. The multitalented microbial sensory rhodopsins. Trends Microbiol. 2006;14:480–487. doi: 10.1016/j.tim.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 3.Spudich JA, Hastings JW. Inhibition of the bioluminescent oxidation of reduced flavin mononucleotide by 2-decenal. J. Biol. Chem. 1963;238:3106–3108. [PubMed] [Google Scholar]
- 4.Spudich JA, Huxley HE, Finch J. Regulation of skeletal muscle contraction. II. Structural studies of the interaction of the tropomyosin-troponin complex with actin. J. Mol. Biol. 1972;72:619–632. doi: 10.1016/0022-2836(72)90180-5. [DOI] [PubMed] [Google Scholar]
- 5.Kersey YM, Hepler PK, Palevitz BA, Wessells NK. Polarity of actin filaments in Characean algae. Proc. Natl. Acad. Sci. USA. 1976;73:165–167. doi: 10.1073/pnas.73.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spudich JA. Biochemical and structural studies of actomyosin-like proteins from nonmuscle cells. II. Purification, properties, and membrane association of actin from amoebae of Dictyostelium discoideum. J. Biol. Chem. 1974;249:6013–6020. [PubMed] [Google Scholar]
- 7.De Lozanne A, Spudich JA. Disruption of the Dictyostelium myosin heavy chain gene by homologous recombination. Science. 1987;236:1086–1091. doi: 10.1126/science.3576222. [DOI] [PubMed] [Google Scholar]
- 8.Manstein DJ, Titus MA, De Lozanne A, Spudich JA. Gene replacement in Dictyostelium: generation of myosin null mutants. EMBO J. 1989;8:923–932. doi: 10.1002/j.1460-2075.1989.tb03453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uyeda TQP, Abramson PD, Spudich JA. The neck region of the myosin motor domain acts as a lever arm to generate movement. Proc. Natl. Acad. Sci. USA. 1996;93:4459–4464. doi: 10.1073/pnas.93.9.4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheetz MP, Spudich JA. Movement of myosin-coated fluorescent beads on actin cables in vitro. Nature. 1983;303:31–35. doi: 10.1038/303031a0. [DOI] [PubMed] [Google Scholar]
- 11.Spudich JA, Kron SJ, Sheetz MP. Movement of myosin-coated beads on oriented filaments reconstituted from purified actin. Nature. 1985;315:584–586. doi: 10.1038/315584a0. [DOI] [PubMed] [Google Scholar]
- 12.Yanagida T, Nakase M, Nishiyama K, Oosawa F. Direct observation of motion of single F-actin filaments in the presence of myosin. Nature. 1984;307:58–60. doi: 10.1038/307058a0. [DOI] [PubMed] [Google Scholar]
- 13.Kron SJ, Spudich JA. Fluorescent actin filaments move on myosin fixed to a glass surface. Proc. Natl. Acad. Sci. USA. 1986;83:6272–6276. doi: 10.1073/pnas.83.17.6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toyoshima YY, et al. Myosin subfragment-1 is sufficient to move actin filaments in vitro. Nature. 1987;328:536–539. doi: 10.1038/328536a0. [DOI] [PubMed] [Google Scholar]
- 15.Lymn RW, Taylor EW. Mechanism of adenosine triphosphate hydrolysis by actomyosin. Biochemistry. 1971;10:4617–4624. doi: 10.1021/bi00801a004. [DOI] [PubMed] [Google Scholar]
- 16.Rayment I, et al. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 17.Finer JT, Simmons RM, Spudich JA. Single myosin molecule mechanics: piconewton forces and nanometre steps. Nature. 1994;368:113–119. doi: 10.1038/368113a0. [DOI] [PubMed] [Google Scholar]
- 18.Bryant Z, Altman D, Spudich JA. The power stroke of myosin VI and the basis of reverse directionality. Proc. Natl. Acad. Sci. USA. 2007;104:772–777. doi: 10.1073/pnas.0610144104. [DOI] [PMC free article] [PubMed] [Google Scholar]