Abstract
Colocalization of fluorescent probes is commonly used in cell biology to discern the proximity of two proteins in the cell. Considering that the resolution limit of optical microscopy is on the order of 250 nm, there has not been a need for high-resolution colocalization techniques. However, with the advent of higher resolution techniques for cell biology and single-molecule biophysics, colocalization must also improve. For diffraction-limited applications, a geometric transformation (i.e., translation, scaling, and rotation) is typically applied to one color channel to align it with the other; however, to achieve high-resolution colocalization, this is not sufficient. Single-molecule high-resolution colocalization (SHREC) of single probes uses the local weighted mean transformation to achieve a colocalization resolution of at least 10 nm. This article describes the process of collecting a calibration data set of fiducials and the appropriate analysis to determine the transformation for colocalization.
INTRODUCTION
How disparate parts of biological systems (e.g., single proteins and whole cells) relate to one another is often inferred by their physical proximity, as observed by microscopy. The distances between proteins, whether in vivo or in vitro, are used to determine whether they interact. As new technologies increase the positional resolution of each individual protein (Gustafsson 1999, 2005; Betzig et al. 2006; Donnert et al. 2006; Hess et al. 2006; Rust et al. 2006), their colocalization resolution will also need to improve. In the field of single-molecule biophysics, the study of enzymatic mechanisms can also benefit from precise colocalization of separate protein domains. Visualizing the structural dynamics of an individual enzyme sheds light on the conformational changes involved in its mechanics. Both cellular biology and enzymatic biology have benefited from the use of Förster resonance energy transfer (FRET), a technique to detect proximity. However, FRET does not easily measure absolute distances or distances greater than ~10 nm. Accurate and precise colocalization of chromatically differing fluorescent probes is a technique that can be tremendously useful in the study of biological problems. It picks up approximately where FRET drops off and results in dynamic distance measurements of the absolute distance between probes.
A variety of strategies, which differ in the resolution obtained and ease of implementation, have been used for colocalization of fluorescent probes in the 10–100 nm range (van Oijen et al. 1998; Lacoste et al. 2000; Schmidt et al. 2000; Michalet et al. 2001; Koyama-Honda et al. 2005). This article describes one such technique: single-molecule high-resolution colocalization (SHREC) of single probes (Churchman et al. 2005). Of all the colocalization techniques, SHREC has the highest reported colocalization resolution and is among the easiest to apply. This article describes the instrumentation needed and the analysis required to obtain a high-resolution mapping between imaging channels. A protocol for acquiring the registration data can be found in Single-Molecule High-Resolution Colocalization of Single Probes (Churchman and Spudich 2012).
THEORY
Typically, a sample is labeled differentially with probes of distinct emission frequencies. For dual-color far-field microscopy, an image produced in the sample plane is split into two images by wavelength and recorded separately. The different channels will not map directly onto each other because of their separate imaging paths. Aberrations arise because of the different lenses and mirrors used in the two optical paths and because of chromatic effects. In theory, one channel’s image would simply need to be transformed with respect to the other in a linear conformal manner including only a rotation, scaling, and translation. However, the imperfect nature of the optical elements induces aberrations that must be mapped in a local nonlinear manner if colocalization resolution in the nanometer range is desired. In this article, we describe mapping that can correct for these aberrations using the local weighted mean (LWM) method described by Goshtasby (1988). Although this may not be the only mapping method that can produce such high-resolution results, an advantage to implementing the LWM method is that it is simple to do so using the software program MATLAB (The MathWorks, Natick, MA) and its image processing toolbox.
A mapping is best calculated from a large data set of control points that consists of pairs of positions that correspond to each other. In practice, this is done with what is called a fiducial marker: an object that, when put in the sample plane, can be seen in each of the imaging channels. Fluorescent beads are an excellent choice because they are bright and relatively photostable. Additionally, if the fluorescent bead is diffraction limited, then the image it produces in each imaging channel is the point-spread function of the microscope, and the position of the bead can be found to high precision by a fit with a two-dimensional Gaussian function. By imaging the fiducial bead over the entire field of view, a group of control points can be determined and a mapping can be calculated. This mapping can be applied to future data sets to perform transformations from one space to another. A position measured in one imaging channel can be placed accurately in the context of the other imaging channel.
An important point to note is that there is an error associated with the measurement of the control points. A successful mapping can be performed only when the control point error is smaller than that sought for a colocalization error. Many fluorescent beads are bright enough to be localized with high resolution. However, it is necessary to verify that such is the case in each channel.
INSTRUMENTATION
To observe single fluorophores, it is helpful to illuminate the sample using a total internal reflection fluorescence microscope (TIRFM), as it eliminates most of the fluorescent background (Axelrod et al. 1984). The following are two ways that TIRFM is implemented in single-molecule experiments: (1) Using a quartz prism coupled to a quartz slide with the sample attached on the opposite side, and then collecting the signal through a glass coverslip placed after an ~100-μm layer of imaging buffer and (2) using an objective with a numerical aperture (NA) >1.4, which bends the excitation light to the critical angle. The objective also serves to collect the emitted light. Both TIRFM methods are compatible with the SHREC technique, although the through-the-objective approach of TIRFM was used in development. The microscope details provided here can be altered to suit individual purposes, but they are described here to give an idea of one way to build the instrument. It should be noted that SHREC does not rely on TIRFM. For applications with probes brighter than single fluorophores, other illumination techniques can be used with the SHREC analysis.
Excitation Path
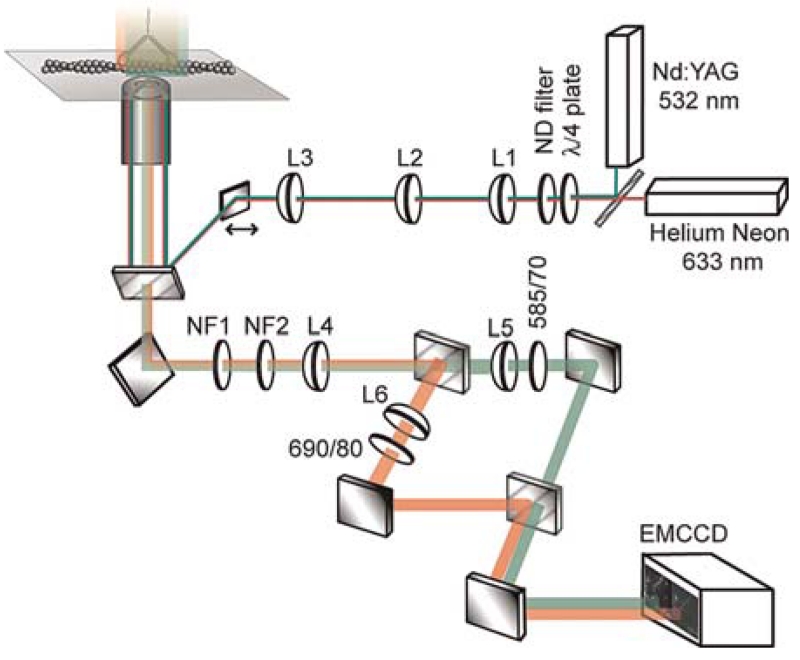
The excitation illumination uses laser lines displaced from the excitation peak of the fluorescent probes so that the probes’ emission can be fully filtered from the incident laser light. For a system designed to excite and image the fluorescent probes Cy3 and Cy5 (Fig. 1), an Nd:Yag laser (Compass 215 M, 15 mW, 532 nm; Coherent Inc., Santa Clara, CA) is combined with a Helium Neon laser (25 mW, 633 nm; JDS Uniphase, San Jose, CA) using a dichroic mirror (595 LP beam splitter; Chroma Technology Corp., Rockingham, VT). It is useful to be able to control the excitation intensity of each laser separately, and thus an OD filter wheel in front of each laser is encouraged or, alternatively, an OD filter wheel can be placed after the dichroic mirror along with a filter wheel between the dichroic mirror and the brighter of the two lasers. A quarter wave plate (quartz multiorder λ/4 plate; Newport Corporation, Irvine, CA) placed after the dichroic circularizes the polarization of the excitation light to ensure that all orientations of the fluorescent probes are equally excited. To fill the field of view and to make the illumination intensity as uniform as possible, two lenses make a telescope to increase the diameter of the combined beams from 2 to 10 mm. In the setup shown in Figure 1, the telescope is made from L1 (f = 2.5 mm) and L2 (f = 95 mm) lenses. An adjustable iris following the telescope cuts out regions of the beam that are not needed. A third lens (L3; f = 500 mm) placed following the telescope focuses the beams onto the back aperture of the objective (Olympus 1.65 NA 100× TIRFM). Because the light leaving the telescope is collimated, the third lens should have a focal length equal to the distance of this lens to the objective’s back aperture. A mirror placed on a translation stage reflects the beam toward a dichroic mirror that reflects both 532 and 633-nm light but not the Cy3 and Cy5 emitted light (beam splitter 532–632 nm; Chroma Technology Corp.). The translation stage allows the incident beam to be targeted to the center or to the edge of the objective. When it is sent through the center of the objective, the microscope is operating in an epifluorescent manner and when it is sent through the side of the objective, it is bent so that at the glass/water interface of the sample, it is totally internally reflected. Placed above the TIRFM objective is a closed-loop, two-axis, piezo nano-translation stage equipped with capacitive sensors for position measurement (Physik Instrumente, Auburn, MA). The piezo stage is used in our implementation of the SHREC technique to ease the recording of fiducial data. However, it is not necessary. In Single-Molecule High-Resolution Colocalization of Single Probes (Churchman and Spudich 2012), we also describe methods for applying the SHREC technique without the use of a piezo nanostage.
FIGURE 1.
Schematic of the microscope used for SHREC. Light leaving the two lasers is combined by a dichroic mirror, circularized by a λ/4 wave plate, attenuated by a neutral density (ND) filter and expanded by the telescope formed by L1 and L2. L3 focuses the light onto the back aperture of the objective so that the light rays are parallel leaving the top of the objective. All of the light is then totally internally reflected off the coverslip/water interface and a thin layer of the sample is exposed to the resulting evanescent wave. The emitted light from the sample is collected by the same objective and transmitted into the emission path. Notch filters (NF1 and NF2) block any laser light reaching the emission path. The emitted light is focused by L4 to an image plane where it is then separated by wavelength via a dichroic mirror. Each path is positioned by a mirror and sent through a lens (L5, L6) before another identical dichroic mirror combines them. The separated image from the emitted light is then sent to the EMCCD camera (see main text for the technical specifications of the individual components).
Emission Path
The probes’ emitted light is collected by the objective and transmitted through the dichroic mirror. A mirror below the dichroic mirror sends the light through two notch filters (NF1 and NF2) (StopLine U-grade [OD 6] 532/17 and 633/25 nm, respectively; Semrock Corp., Rochester, NY) to block any transmitted laser light. A tube lens (L4 = 120 mm in Figs. 1 and 2) focuses the emitted light onto two razor blades separated by about a 1-mm slit, thus blocking any light that is not to be imaged by the camera. This ensures that light from the two channels does not mix on the camera’s CCD chip. A dichroic mirror (625 LP beam splitter; Chroma Technology Corp.) separates the emission from the two probes and sends them down different paths. Each path has a lens (L5 and L6, f = 50 mm) focusing the light onto the electron-multiplying charge-coupled device (EMCCD) camera (iXon DV 887 EMCCD; Andor Technology, Belfast, Ireland), allowing the focus of each path to be adjusted separately. An emission filter is placed following the lens in each path that transmits as much of the probe’s emission peak as possible. In the case of Cy3 and Cy5, 585/70 and 690/40 filters were used, respectively. A second dichroic mirror (625 LP beam splitter; Chroma Technology Corp.) combines the emission light from each path such that they are placed side by side when shone onto the EMCCD camera. An EMCCD camera is very powerful for single-molecule imaging because of its low readout noise and its high quantum efficiency.
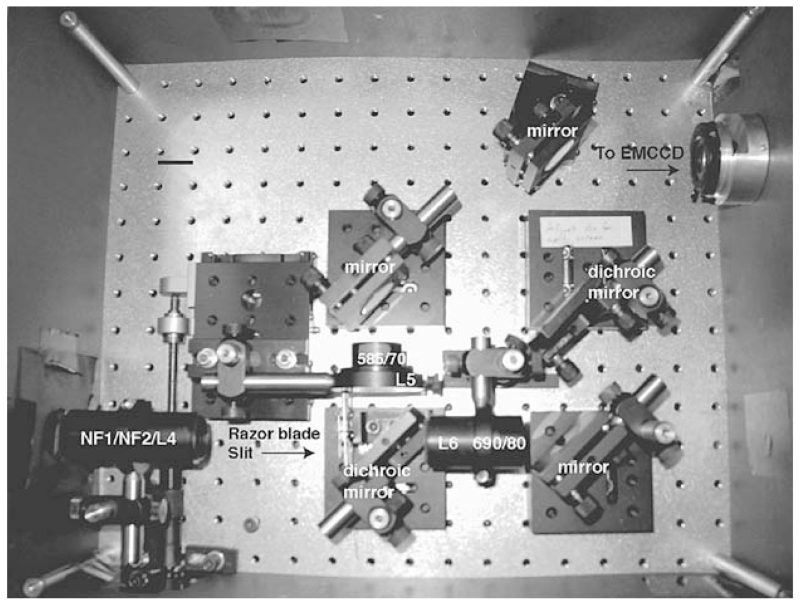
FIGURE 2.
Photo of emission path used for SHREC. The notch filters along with lens 4 (NF1/NF2/L4) are all mounted in a single tube. The razor blades are each mounted on a very small translation stage, which is mounted just before the first dichroic mirror. Lens 6 and the emission filter for Cy5 (L6, 690/80) are mounted in the same tube following the transmission side of the dichroic mirror. Lens 5 (L5) is mounted on a translation stage, which can be adjusted outside the box for the emission path. This allows separate focusing of each channel, which is occasionally necessary because of drift. An emission filter for Cy3 (585/70) is mounted directly following L5. Regular optical mirrors redirect each path to a second identical dichroic mirror, which combines the paths. A final mirror directs the split image onto an EMCCD camera. The dichroic mirrors and the regular mirrors are mounted using magnetic mounts, allowing for easy rearrangement of the optical path when imaging one color over a larger field of view or when using different dyes.
An emission path equipped with the lenses displayed in Figures 1 and 2 results in a pixel size of 27.7 nm. Localization of diffraction-limited spots gives better resolution when the pixel size is much larger than 27.7 nm (Thompson et al. 2002). In practice, the camera was run in a 4 × 4 binning mode so that the effective pixel size was 110.7 nm and the field of view was 14 × 7 μm. A different choice for lenses in the emission path could provide a smaller magnification. This would render the binning mode unnecessary, and a larger field of view could be imaged. However, when using a larger area for imaging, more registration data and analysis are needed. We found it simplest to register the smallest area possible. Regardless, because the mapping is local, the registration of a larger area should result in a similar colocalization resolution.
OBTAINING FIDUCIAL DATA
A fiducial marker should be chosen so that it can be localized to high enough precision that its own localization error can be disregarded. A good choice is a fluorescent bead that emits brightly in each color channel and does not photobleach substantially within the time it takes to acquire the registration data. A large bead will emit more brightly than a smaller one. However, if the bead is not diffraction limited, it will then be localized with greater error due to the slight asymmetry of fluorescent beads. A diffraction-limited bead will be better localized because a two-dimensional Gaussian function can be fit well to its image. In the development of the SHREC technique for Cy3 and Cy5, 100-nm TransFluoSphere beads (543/620; Molecular Probes/Invitrogen) were used.
Fiducial data are taken by moving and imaging an individual bead stuck to a coverslip through the field of view in a grid pattern using a piezo nanostage. The smaller the spacing of the grid, the better the resolution of the colocalization. However, the trade-off is a larger registration data set and more computer cycles needed to analyze them. In theory, the limit of the grid spacing depends on the localization resolution of the fiducials, the resolution of the piezo nanostage, and the photostability of the fiducial marker. A grid spacing of 0.5 μm over a 14 × 7 μm field of view yielded a colocalization resolution of 3.3 nm using 30 Mb of registration data and a few minutes of computer analysis done on a standard laptop computer.
The fiducial distribution criterion for a high-resolution mapping is simply that there is even sampling of the field of view. The pattern in which the fiducial markers (beads) lie is irrelevant, but making a grid pattern with a piezo nanostage is a simple way to ensure that the criterion is met. However, a field of nonoverlapping beads can be imaged and then moved with a traditional microscope stage to adequately sample the entire field of view. The piezo nanostage simply allows for the automation of the collection of fiducial data and ensures that all parts of the field of view are represented. It is a costly addition, however, especially if it is not used for another application.
ANALYZING FIDUCIAL DATA
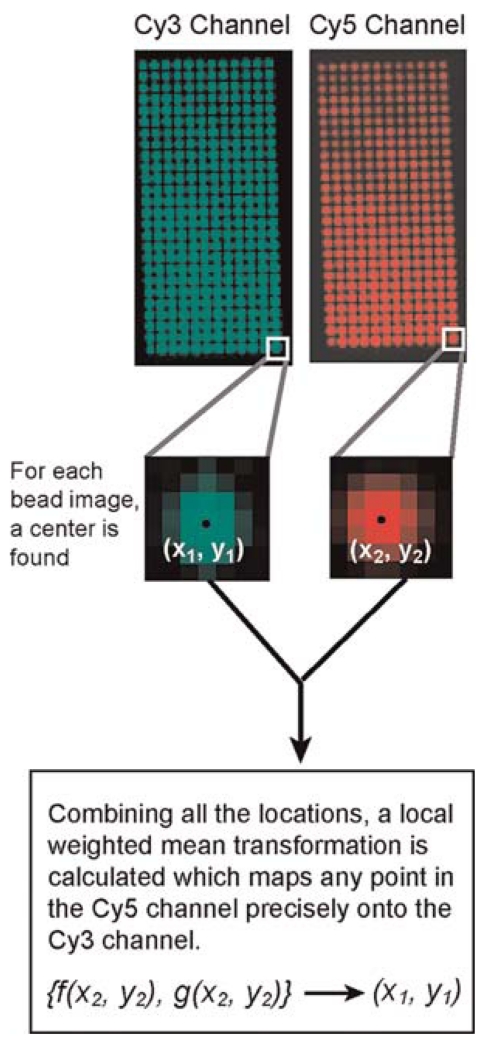
The fiducial data set consists of a stack of N images, each with an image of a fluorescent bead in each channel at a particular position on the stage (Fig. 3). To calculate the mapping from one channel to the next, the position of the bead in each channel must first be determined. If the fiducial data have been taken without a piezo nanostage, there may be fewer than N images, with each image containing many bead images. In this case, the position of every bead in both imaging channels will need to be determined. For diffraction-limited beads, this is done most accurately by a least-squares fit with a two-dimensional Gaussian function:
FIGURE 3.
Explanation of the data collected and analysis that determines the transformation from one channel to the other. A bead that emits broadly so that it can be seen in each imaging channel is moved in a grid-like manner through the field of view. (Top panels) Resulting grid. Each bead is localized in both channels, which produces a pair of control points, {(x1, y1), (x2, y2)} (center). All of the pairs of control points contribute to the calculation of an LWM transformation from one imaging channel to the other (bottom).
In the shot-noise limited case, the error affiliated with this localization is:
where Nγ is the number of photons. This error is also the fiducial localization error. For more details on this type of localization, see Cheezum et al. (2001), Thompson et al. (2002), and Yildiz et al. (2003). Again, the fiducial localization error should be equal to, or less than, the desired final registration error.2
Once fits have been performed for each image, a pair of locations is known for each of the N positions the bead had in the field of view. These pairs of locations are called control points. The set of control points can be used to calculate an LWM mapping that can be applied to any future data point. First, for each control point, at least six (or more, if desired) of the control points closest to the selected control point in channel 1 are chosen. Using these control points from channel 1 along with their counterparts in channel 2, an affiliated second-order polynomial can be found by the least squares method for each coordinate.
By measuring the distance from the selected control point to the furthest of the six surrounding control points in channel 1, one can determine the radius of influence that this polynomial will have in the final transformation. Finally, all the polynomials are combined in a weighted sum to create the final transformation (Goshtasby 1988):
where
and
This is performed for each coordinate.
The “cp2tform” command of MATLAB does all of this at once and yields a transformation structure that can be used in other MATLAB functions to perform additional transformations.
There are two metrics to estimate the error associated with this transformation (Fitzpatrick et al. 2000). The easiest to calculate is the fiducial registration error (FRE). Using the found an LWM mapping function, fLWM (x), one transforms the control points from channel 2, {xi,2}, onto channel 1’s space. FRE is then calculated as follows:
A more accurate metric is the target registration error (TRE). This is found by sequentially leaving out a pair of control points and calculating an LWM mapping with the remaining pairs of control points. The mapping is then applied to the pair that was left out, and the distance between the registered control points is the error. This is done for each control point, and the TRE is calculated using the following function:
Once the transformation has been determined, it can be applied to a variety of analyses. For example, for single-molecule data, it can be applied to locations found from single diffractionlimited probes in channel 2. These locations can then be appropriately compared to probes localized in channel 1. In addition, when colocalizing images of objects larger then single probes (such as cells), the investigator may wish to transform the entire image in channel 2 and overlap it with the image in channel 1 to see colocalization by eye. We have not explored this image registration application of SHREC, but MATLAB makes this procedure straightforward with the function, “imtransform.”
A major goal of data registration and calibration is to look at enzymatic conformational changes or intra/intermolecular distances. In such studies, a distance is often measured after data have been registered. The noise due to the probes’ localization and the channel registration will make the distance probability distribution asymmetric (i.e., non-Gaussian) as follows (Fig. 4):
where I0 is the modified Bessel function of integer order zero, x is the distance measured, σ is the localization error of the points between which the distance was measured, and μ is the true distance between the probes (Churchman et al. 2006).
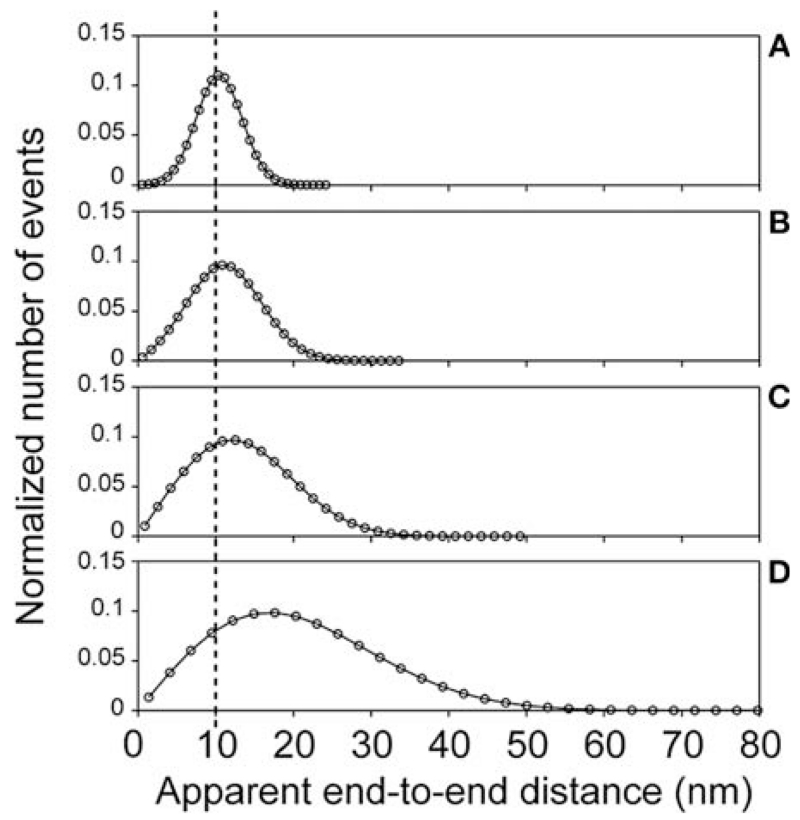
FIGURE 4.
Asymmetric distribution for distance measurements in two dimensions. The distance measurement distributions for a 10-nm separation (dotted line) with four signal-to-noise ratios show how the skewness increases with signal to noise. The signal-to-noise ratios μ/σ in (A–D) are 3.33, 2, 1.25, and 0.667, respectively. A Gaussian analysis of these distributions would result in values that differ from the simulated value, μ, by 5% (A), 13% (B), 32% (C), and 112% (D).
Fitting the above equation to a data set of measured distances yields an estimate for the true distance (μ), with higher accuracy than calculating the mean value of the data set. The true distance (μ), deviates from the distribution’s maximum, 〈x〉, as a function of measurement error, σ:
For any amount of error, the distribution is asymmetric, and the mean value of the distribution will be systematically biased toward larger numbers.
FUTURE DIRECTIONS
Improving the colocalization resolution using SHREC beyond what has already been shown is certainly possible. A significant improvement would come from decreasing the localization error of the fiducial markers by increasing the signal-to-noise ratio when imaging them. Scattering light off a gold, silver, or plastic bead would provide many more photons than the photons resulting from a fluorescent bead without any photostability problems. Techniques for scattering and imaging light off gold beads are described in Single-Molecule Gold-Nanoparticle Tracking (Dunn and Spudich 2011). Additionally, the technology involved in making CCD cameras will continue to decrease the noise associated with taking the fiducial data. As the signal-to-noise ratio increases, it will be important to move beyond localizing by a least-squares fit with a two-dimensional Gaussian function, as eventually the two-dimensional Gaussian will no longer serve as the best approximation for the point spread function. With higher resolution of fiducial locations, fiducial markers can sample more points in the field of view.
REFERENCES
- Axelrod D, Burghardt TP, Thompson NL. Total internal reflection fluorescence. Annu Rev Biophys Bioeng. 1984;13:247–268. doi: 10.1146/annurev.bb.13.060184.001335. [DOI] [PubMed] [Google Scholar]
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging intracellular fluorescent-proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- Cheezum MK, Walker WF, Guilford WH. Quantitative comparison of algorithms for tracking single fluorescent particles. Biophys J. 2001;81:2378–2388. doi: 10.1016/S0006-3495(01)75884-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman LS, Spudich JA. Single-molecule high-resolution colocalization of single probes. Cold Spring Harb Protoc. 2012 doi: 10.1101/pdb.prot067926. doi: 10.1101/pdb. prot067926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman LS, Okten Z, Rock RS, Dawson JF, Spudich JA. Single molecule high-resolution colocalization of Cy3 and Cy5 attached to macromolecules measures intramolecular distances through time. Proc Natl Acad Sci. 2005;102:1419–1423. doi: 10.1073/pnas.0409487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman LS, Flyvbjerg H, Spudich JA. A non-Gaussian distribution quantifies distances measured with fluorescence localization techniques. Biophys J. 2006;90:668–671. doi: 10.1529/biophysj.105.065599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnert G, Keller J, Medda R, Andrei MA, Rizzoli SO, Luhrmann R, Jahn R, Eggeling C, Hell SW. Macromolecular-scale resolution in biological fluorescence microscopy. Proc Natl Acad Sci. 2006;103:11440–11445. doi: 10.1073/pnas.0604965103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn AR, Spudich JA. Single-molecule gold-nanoparticle tracking. Cold Spring Harb Protoc. 2011 doi: 10.1101/pdb.prot066977. doi: 10.1101/pdb.prot066977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick JM, Hill DLG, Maurer CR., Jr . Image registration. In: Fitzpatrick JM, Sonka M, editors. Handbook of medical imaging, vol 2. Medical image processing and analysis. SPIE; Bellingham, Washington: 2000. pp. 447–513. [Google Scholar]
- Goshtasby A. Image registration by local approximation methods. Image Vision Comput. 1988;6:255–261. [Google Scholar]
- Gustafsson MG. Extended resolution fluorescence microscopy. Curr Opin Struct Biol. 1999;9:627–634. doi: 10.1016/s0959-440x(99)00016-0. [DOI] [PubMed] [Google Scholar]
- Gustafsson MG. Nonlinear structured-illumination microscopy: Wide-field fluorescence imaging with theoretically unlimited resolution. Proc Natl Acad Sci. 2005;102:13081–13086. doi: 10.1073/pnas.0406877102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess ST, Girirajan TP, Mason MD. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy (FPALM) Biophys J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama-Honda I, Ritchie K, Fujiwara T, Iino R, Murakoshi H, Kasai RS, Kusumi A. Fluorescence imaging for monitoring the colocalization of two single molecules in living cells. Biophys J. 2005;88:2126–2136. doi: 10.1529/biophysj.104.048967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacoste TD, Michalet X, Pinaud F, Chemla DS, Alivisatos AP, Weiss S. Ultrahigh-resolution multicolor colocalization of single fluorescent probes. Proc Natl Acad Sci. 2000;97:9461–9466. doi: 10.1073/pnas.170286097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalet X, Lacoste TD, Weiss S. Ultrahigh-resolution colocalization of spectrally separable point-like fluorescent probes. Methods. 2001;25:87–102. doi: 10.1006/meth.2001.1218. [DOI] [PubMed] [Google Scholar]
- Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Nagomi M, Hell S. Subresolution axial distance measurements in far-field fluorescence microscopy with precision of 1 nanometer. Rev Sci Instrum. 2000;71:2742–2745. [Google Scholar]
- Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophys J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oijen A, Kohler J, Schmidt J, Muller M, Brakenhoff G. 3-dimensional super-resolution by spectrally selective imaging. Chem Phys Lett. 1998;292:183–187. [Google Scholar]
- Yildiz A, Forkey JN, McKinney SA, Ha T, Goldman YE, Selvin PR. Myosin V walks hand-over-hand: Single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]