Abstract
Background. Uropathogenic Escherichia coli (UPEC), a leading cause of urinary tract and invasive infections worldwide, is rapidly acquiring multidrug resistance, hastening the need for selective new anti-infective agents. Here we demonstrate the molecular target of DU011, our previously discovered potent, nontoxic, small-molecule inhibitor of UPEC polysaccharide capsule biogenesis and virulence.
Methods. Real-time polymerase chain reaction analysis and a target-overexpression drug-suppressor screen were used to localize the putative inhibitor target. A thermal shift assay quantified interactions between the target protein and the inhibitor, and a novel DNase protection assay measured chemical inhibition of protein-DNA interactions. Virulence of a regulatory target mutant was assessed in a murine sepsis model.
Results. MprA, a MarR family transcriptional repressor, was identified as the putative target of the DU011 inhibitor. Thermal shift measurements indicated the formation of a stable DU011-MprA complex, and DU011 abrogated MprA binding to its DNA promoter site. Knockout of mprA had effects similar to that of DU011 treatment of wild-type bacteria: a loss of encapsulation and complete attenuation in a murine sepsis model, without any negative change in antibiotic resistance.
Conclusions. MprA regulates UPEC polysaccharide encapsulation, is essential for UPEC virulence, and can be targeted without inducing antibiotic resistance.
Keywords: E. coli, polysaccharide capsule, multidrug efflux pumps, small-molecule capsule inhibitor
The global rise in antibiotic resistance threatens the effectiveness of treatment for even common community-acquired infections. Urinary tract infection (UTI) is among the most common infections and accounts for about 10.5 million ambulatory visits annually [1], leading to frequent and recurrent antibiotic use at an annual treatment cost approaching $3.5 billion [2]. Uropathogenic Escherichia coli (UPEC) produces 75%–90% of community-acquired UTIs [3, 4] and is becoming increasingly resistant to antibiotics, with universal resistance to amoxicillin in >50% of isolates and regional resistance to trimethoprim-sulfamethoxazole (TMP-SMX) and ciprofloxacin among >20% [5–9]. Further complicating UTI therapy, E. coli ST-131 clonal type is a global threat, with multidrug resistance against extended-spectrum β-lactams, aminoglycosides, and carbapenems [10]. The redundant use of the same antibiotics for multiple types of infections, including common infections like UTI, may be a driver for the emergence and persistence of multidrug-resistant E. coli, which may be addressed through the development of specific therapies for UTI due to E. coli. We and others have proposed targeted inhibition of virulence-associated factors such as polysaccharide capsule for this approach [11–13].
Prior research has demonstrated a role for the ubiquitous polysaccharide encapsulation of UPEC in UTI and for more-invasive diseases, such as sepsis and neonatal meningitis [14–17]. We identified small molecules that inhibit the biogenesis of UPEC group 2 polysaccharide capsule, regardless of serotype, and facilitate immune clearance of invasive E. coli with sufficient potency to abrogate an otherwise lethal bloodstream infection in a murine infection model [13]. However, the mechanism of action has remained unknown.
In this current work, we investigated the mechanism through which the small-molecule DU011 (3-[2,6-difluorobenzamido]-5-[4-ethoxyphenyl] thiophene-2-carboxylic acid; Molecular Libraries Program [MLP] probe number ML317) inhibits E. coli group 2 capsule production [13]. DU011 and other capsule inhibitors were identified in phenotypic screens [12, 13]. Yet, the targets of small molecules identified from such phenotypic screens often remain unidentified [18]. However, using a combination of genetics and biochemical assays, we demonstrate that DU011 mediates inhibition of capsule expression through a direct interaction with the highly conserved multidrug efflux pump transcriptional regulator MprA (previously referred to as EmrR) without altering antibiotic susceptibility. We demonstrate that mutation of mprA abrogates capsule expression and fully attenuates E. coli in a murine sepsis model. This study also provides a novel link between multidrug efflux pump regulation and polysaccharide capsule expression, while, of importance, identifying small molecules that separate the virulence regulatory effects from the drug efflux effects, yielding potential antiinfective agents that do not have the negative consequence of increased antibiotic resistance. This work is further illustration of the power of chemical genetics to define bacterial molecular virulence.
MATERIALS AND METHODS
Bacterial Strains, Plasmids, Phage, and Growth Conditions
All E. coli strains, plasmids, and phages used in the present study are listed in Table 1. Bacteria were grown in Luria-Bertani medium (LB) with shaking at 250 rpm at 37°C. LB was supplemented with 1% dimethyl sulfoxide (DMSO; Acros), with or without the addition of small molecules of interest. Phage lysates were prepared from 50-mL cultures of E. coli strains UTI89 (for K1F phage) or MG1655 (for T7 phage) and stored at 4°C over drops of chloroform as described previously [24].
Table 1.
Strains, Plasmids, and Bacteriophage
| Strain/Phage | Description or Relevant Genotype | Reference |
|---|---|---|
| Bacterial strains | ||
| UTI89 | K1 Escherichia coli cystitis isolate | [19] |
| UTI89 Δneu | Region II K1 capsule synthesis mutant | [14] |
| UTI89 ΔmprA | mprA deletion mutant | NA (laboratory collection) |
| EV36 | K-12/K1 hybrid produced by conjugation with an Hfr kps+ strain; K1 encapsulated and susceptible to K1-specific phage | [20] |
| Plasmid | ||
| pASKA-mprA | mprA deletion mutant complemented with pASKA containing mprA | [21] |
| Phage | ||
| T7 phage (T7φ) | Inhibited by K1 capsule | [22] |
| K1F phage (K1Fφ) | K1 capsule specific | [23] |
K1F Phage Validation of Capsule Expression
K1F, a K1 polysialic acid capsule–dependent lytic phage [23], was used to quantify the presence of encapsulation, as previously described [12]. Overnight cultures of pathogenic cystitis E. coli K1 strain UTI89 and isogenic mutant strains were diluted at a ratio of 1:100 into LB, and compounds were added in the appropriate concentrations. The plates were shaken vigorously for 1.5 hours. Then, K1F phage (5 µL of a high-titer phage lysate [>109 plaque-forming units/mL]) was added, and the plates were returned to the shaker. The OD600 was measured after 3 hours to determine the extent of phage-mediated lysis. Plates were read at ambient temperature in a BioTek µQuant plate reader, and the degree of phage-mediated lysis was determined based on the absorbance.
T7 Phage–Mediated Lysis Assay
T7 entry into E. coli is inhibited by cell-associated polysaccharide encapsulation and was used as a reciprocal assay to the K1F assay [22].
Overexpression Screen to Identify Capsule Inhibitor Targets
This screen was initially conducted using DU003 (MLP number ML333). However, once the target was identified, it was validated for DU011, as well. The ASKA library, a complete set of E. coli K-12 open reading frames carrying a His-tag [21], was pooled and transformed into EV36 (a K1:K12 hybrid) [20]. Cells were resuspended in LB with 0.1 mM ITPG at an OD600 of 2.3 and included in the T7 phage assay, using 50 µM DU003. At an OD600 of 0.1–0.2, phage was added, and cells were incubated for 3 hours. Clones of EV36 that were resistant to lysis by T7 phage would likely have a capsule present and be resistant to DU003 because of overexpression of its target. The aforementioned screen was repeated twice (total number of screens, 3), to maximize the recovery of DU003-resistant clones. Plasmids were isolated from individual colonies (Thermo Scientific) and sequenced using previously published primers (Supplementary Table 1) [21].
Thermal Shift Assay (TSA)
TSA (also known as differential scanning fluorimetry; [25]) was performed using SYPRO Orange (Life Sciences) as the shift reporter dye. Briefly, 12.5 µg of protein was incubated with the indicated concentration of compound for 20 minutes, dye was added, and the reactions were monitored in real time (Bio-Rad MiniOpticon; excitation, 490; emission, 575 nm) from 29°C to 95°C with a rate of change of 1°C/minute. The melt curve is represented as normalized data and calculated as d(fluorescence)/d(temperature), consistent with prior publications [25].
Morphological Chemical-Similarity Analysis
Morphological similarity analysis was performed using Surflex-Sim's molecular similarity technique called “morphological similarity,” which bases similarity on a molecule's shape, hydrogen bonding, and electrostatic properties [26–28]. The similarity analysis was performed by Surflex-Sim from SYBYL Certara. Surflex-Sim optimizes the pose of a molecule to maximize 3-dimensional similarity to the target molecule. Morphological similarity bases similarity on a molecule's shape (with a focus on surfaces, not volumes), hydrogen bonding, and electrostatic properties. In this analysis, the minimum root-mean-square deviation between final poses are 0.05.
DNase Protection Assay
A real-time DNase protection assay was developed to assess the effect of chemical ligands on MprA binding to target DNA consensus sequences. The binding site of MprA was amplified by polymerase chain reaction (PCR), using UTI89 genomic DNA as the template, based on primers (Supplementary Table 1) flanking a known MprA consensus element immediately upstream of the start of the mprA gene [29], which provided a 229-nucleotide target DNA. MprA (2 µg) was incubated with the indicated concentration of compound in 20 mM HEPES buffer at pH 7.4 for 20 minutes, followed by the addition of the DNA probe or control for 20 minutes. DNAse (0.5 µg; Promega) was added to this mixture and incubated at 37°C for 10 minutes, followed by addition of 20 mM ethylenediaminetetraacetic acid to stop the reaction. EvaGreen dye (1 µL of 20x, Biotium) was added, and DNA melt curves were recorded (Bio-Rad MiniOpticon; excitation, 490 mm; emission, 520 mm). A. The area under curve for the melting curve was calculated using GraphPad Prism software, version 4.
Murine Sepsis Model
All animal experiments were conducted with prior approval from the Institutional Animal Care and Use Committee of Duke University. Groups of five 5–6-week-old C57BL/6NCr female mice (Frederick National Laboratory for Cancer Research) were injected subcutaneously twice daily, starting 12 hours prior to infection, with 100 µL of 1 mg/mL of DU011 or 1% DMSO (vehicle control). At the time of infection, a dose of DU011 or vehicle control was also given intraperitoneally along with 50 µL of 108 UTI89 or UTI89ΔmprA in phosphate-buffered saline (PBS). Overnight cultures were pelleted and resuspended in 1 mL of PBS. Absorbance was adjusted to an OD600 of 0.8, and the cultures were then diluted 1:10 in PBS. Animals were monitored every 6 hours from the time of infection until conclusion for severe morbidity. Surviving mice received another dose, based on treatment group, and underwent continued monitoring. The experiment was concluded 48 hours after infection. Experiments were performed in 2 independent trials. When a moribund state was suspected or anticipated, animals were immediately euthanized to minimize potential pain and/or suffering.
RESULTS
Small-Molecule Inhibitors Decreased Capsule Gene Transcription
We previously identified multiple small molecules that inhibit the production of E. coli group II capsules, including serotypes K1, K5, and K15 [13]. We hypothesized that one mechanism of the inhibitors is through modulation of a transcription factor. Transcriptional analysis of the capsule genes was performed by quantitative reverse-transcription PCR (qRT-PCR). Chemical treatment with DU003 and DU011 resulted in decreased levels of kpsD and kpsM transcripts (representing region 1 and region 3 polycistronic transcription, respectively; Supplementary Figure 1). Similar results were obtained following treatment with additional, previously described [13] small-molecule inhibitors of polysaccharide capsule, DU001, DU007, and DU008 (data not shown). These data indicated that DU003 and DU011 inhibition of capsule production is at the level of transcription or transcript stability.
Identification of MprA as the Putative Capsule Inhibitor Target
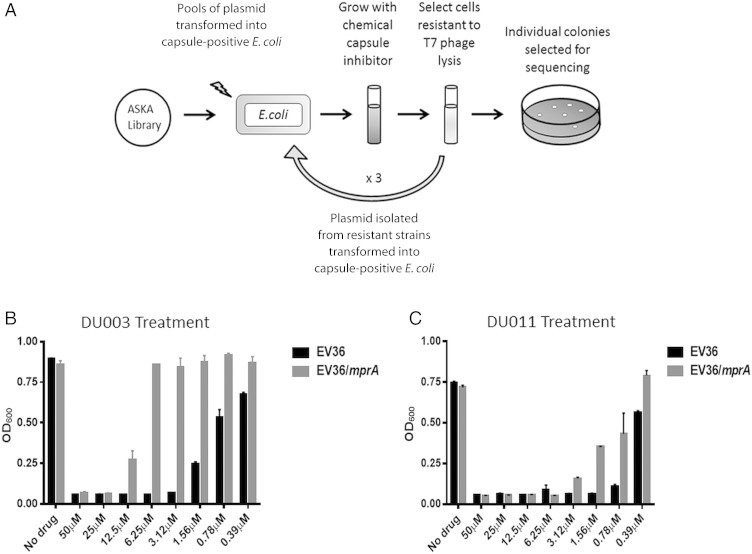
We predicted that overexpression of the DU003 and DU011 target would reduce the relative amount of capsule inhibition by the inhibitors and thus decrease the susceptibility to T7 phage entry and lysis, which is inhibited by capsule (Figure 1A). Through this approach, we expected to discern the identity of the inhibitor target. Sequences in all of the pASKA-based plasmids selected from the screen corresponded to mprA, a known transcriptional repressor of an operon encoding the EmrAB efflux pump [30]. pASKA-mprA was independently transformed into E. coli EV36, and increased resistance to compounds DU003 and DU011 was confirmed (Figure 1B and 1C).
Figure 1.
Schematic diagram showing the approach to identify genetic suppressors of capsule chemical inhibitors. A, An overview of the approach to identify chemical inhibitor suppressors. B and C, Measurement of lysis by T7 phage with and without mprA expression and treatment with DU003 and DU011, respectively. Abbreviations: E. coli, Escherichia coli; OD600, optical density.
Inhibition of K1 Capsule Expression by High-Concentration Salicylate and 2,4-Dinitrophenol (DNP) Is Similar to That by DU003 and DU011
Salicylate and DNP have been described as ligands of MprA [30, 31], and thus we predicted that they would also inhibit group 2 capsule production. Treatment of UTI89 with ≥500 µM salicylate or ≥62.5 µM DNP resulted in inhibition of cell lysis in the presence of the K1 phage, consistent with loss of encapsulation (Supplementary Figure 2).
Genetic Knockout of mprA Results in the Loss of Capsule
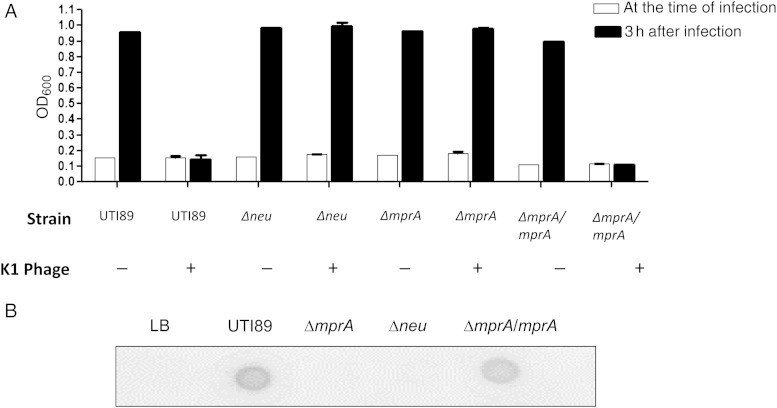
With multiple results implicating MprA in the regulation of group 2 capsules, we predicted that knockout of mprA would produce an unencapsulated mutant strain. Similar to the capsule synthesis mutant UTI89Δneu, UTI89ΔmprA was resistant to lysis by K1 phage, suggesting the loss of K1 encapsulation (Figure 2A). Complementation of the ΔmprA mutant with mprA in trans resulted in restoration of capsule, as measured through K1 phage sensitivity. To further confirm the loss of encapsulation in the mprA mutant, a K1 antigen immunodot blot was performed (Figure 2B). Anti-K1 antibody was reactive with polysaccharide extracts from wild-type UTI89 but not the Δneu or ΔmprA mutant strain extracts. K1 capsular antigen was detected upon complementation of ΔmprA with mprA in trans.
Figure 2.
Determination of the UTI89ΔmprA capsule phenotype. A, Measurement of susceptibility to capsule-dependent K1 phage lysis among isogenic capsule and mprA mutants of Escherichia coli UTI89. Optical density (OD600) is shown at the time of phage infection and 3 hours after infection. B, K1 polysaccharide antigen immunodot blot analysis using anti-K1 antibody.
DU003 and DU011 Do Not Increase Antibiotic Resistance at Concentrations in Excess of Capsule-Inhibition Concentrations
MprA has previously been shown to repress the expression of emrAB, which encodes for a multidrug efflux pump [29]. qRT-PCR was used to measure emrA transcripts in UTI89ΔmprA. The level of emrA transcript was 2.9-fold higher in UTI89ΔmprA, compared with wild-type UTI89 (Supplementary Figure 3). Treatment of wild-type UTI89 with DU011 (200 µM) or DNP (200 µM) produced 2.6-fold and 2.4-fold changes, respectively, in emrA transcript levels, compared with no treatment. There was no significant difference in the fold change between UTI89ΔmprA and UTI89 treated with DU011 (P = .16) or between UTI89ΔmprA treated with DNP (P = .13). DU003 (200 µM) and salicylate (200 µM) only produced 1.3-fold and 1.5-fold changes in emrA transcript levels, respectively (Supplementary Figure 3). To determine whether the increased transcript level of emrAB was equated with increased antibiotic resistance, the minimum inhibitory concentration (MIC) of different classes of antibiotics was tested in the presence and absence of the inhibitors and in comparison with UTI89ΔmprA. Notably, neither the capsule inhibitors nor mutation of mprA produced a clinically significant increase in the MICs of key representatives from multiple classes of antibiotics that are routinely used for the treatment of UTI or E. coli infections (Supplementary Table 2). These data suggest that each ligand at these concentrations insufficiently upregulates the EmrAB efflux pump to result in clinically important antibiotic resistance.
DU011 but Not DU003 Interacts Directly With MprA
To identify small-molecule/MprA interactions, we performed a TSA. Incubation of MprA with salicylate or DNP produced a reproducible shift in the MprA melting curve and peak melting temperature (Tm), as shown by a normalized representation of the melt curve in Figure 3A. MprA and nalidixic acid did not produce a shift in Tm, suggesting that, within the conditions of this assay, nalidixic acid is not a ligand of MprA, which is different from a prior report [29]. DU011 but not DU003 produced a concentration-dependent shift in the MprA Tm (Figure 3A and 3B). These data suggest that DU011 binds directly to MprA but that DU003 does not. Interestingly, the Tm for DU011-MprA was higher than it was for the other tested ligands, suggesting that DU011 not only interacts with MprA, but also yields a more stable configuration of the protein.
Figure 3.
Thermal shift assay to measure MprA conformational change in response to DU011 and prototypic ligands. A, Comparison of the thermal profiles of MprA with and without DU011, DU003, salicylate, 2,4-dinitrophenol (DNP), or nalidixic acid. B, Thermal shift of MprA with increasing concentrations of DU011. The melt curve is represented as normalized data calculated as follows: d(fluorescence)/d(temperature).
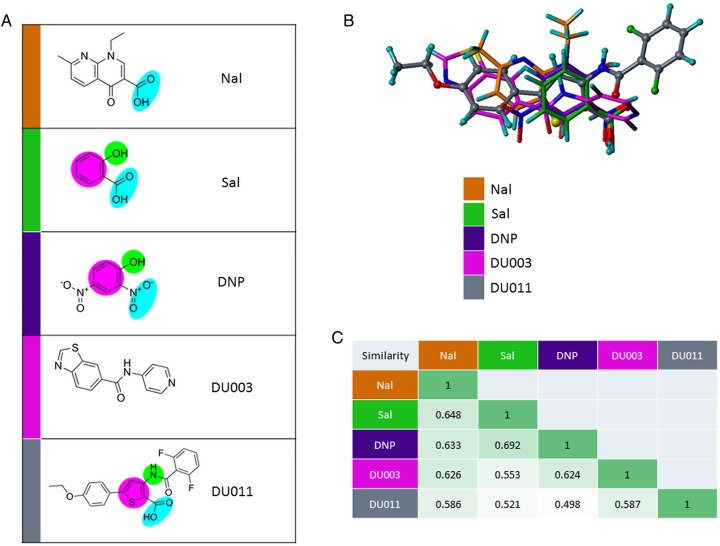
Analysis of Structural Similarity Between Salicylate, DNP, and DU011
Each compound was examined for related and recurrent bioisosteric functional groups (Figure 4A). Nalidixic acid and DU003 appeared to share few or none of the proposed related groups found among the other compounds. Three-dimensional positioning of the molecules highlighted the spatial dissimilarity of DU003 to the more conserved planar relationship of salicylate, DNP, and DU011 (Figure 4B). The fluorinated phenyl and phenol ether rings of DU011 were noted to occupy a unique space as compared to the other molecules. Interestingly, morphological-based similarity analysis (Figure 4C) suggested that DU011 is the least similar among all of the analyzed structures, with its dissimilarity likely being driven by the unique fluorinated phenyl and phenol ether rings. This analysis cannot distinguish between the structural components required for potential interactions with MprA or differences in the conformation MprA may assume following binding. However, it does provides the basis for interpreting the ligand-MprA interaction studies, suggesting that DU011 shares functional biosteric groups with salicylate and DNP, perhaps providing a basis for why they all bind to MprA. The unique space occupancy of the DU011 fluorinated phenyl and phenol ether rings may provide some basis for the potentially different and more stable conformation it produces in MprA.
Figure 4.
Comparative structural analysis capsule inhibitors DU003, DU011, and prototypic MprA ligands. A, Structures for nalidixic acid (Nal), salicylate (Sal), 2,4-dinitrophenol (DNP), DU003, and DU011, with the proposed recurrent bioisosteric functional groups highlighted in blue (carboxylic acid and nitro), green (phenol and aminothiophene/amide nitrogen), and purple (phenyl and thiophene rings). B, Graphic structure overlay results from morphological similarity analysis on Nal, Sal, DNP, DU003, and DU011. C, Morphologic similarity values between Nal, Sal, DNP, DU003, and DU011. Darker green indicates higher similarity.
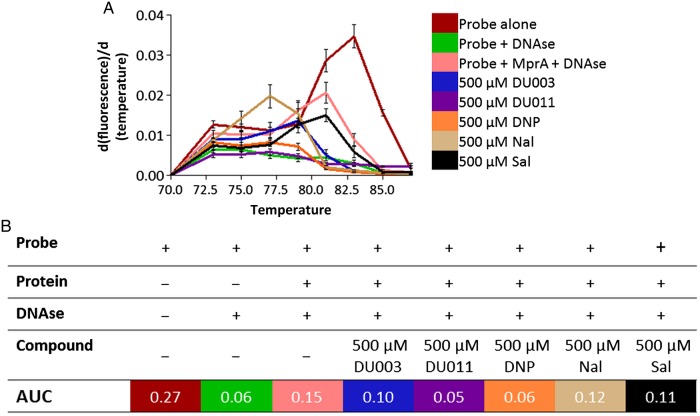
DU011 and DNP Increase DNase Sensitivity
We predicted that ligands interacting with MprA would relieve MprA of binding to consensus-containing target DNA. This hypothesis was tested in a modified real-time DNase protection assay. Purified MprA was coincubated with DU011 or DNP and the mprA promoter region [29]. Inclusion of either compound in the assay resulted in a loss of fluorescence relative to the no-treatment control, as measured by the area under the curve (Figure 5). Gel electrophoresis of the reaction products confirmed the DNase-dependent degradation of the probe when coincubated with MprA and DU011 or DNP but not with MprA alone (data not shown). In contrast, DU003, salicylate, and nalidixic acid did not produce a change in fluorescence relative to the no-treatment control and consistent with their lack of significant interaction with MprA and thus persistent protection of the DNA probe from DNase by bound MprA. We further demonstrated that DU011 produced a concentration-dependent change in DNase sensitivity of the probe in the presence of MprA (Supplementary Figure 4).
Figure 5.
Measurement of MprA real-time DNase protection in the presence of DU011 and other ligands. A, Melting curves of MprA operator DNA probe in the presence of MprA with and without DU003, DU011, 2,4-dinitrophenol (DNP), nalidixic acid (Nal), and salicylate (Sal). Concentrations of each ligand in the binding reactions are indicated in the legend. Error bars indicated standard deviations of the mean from 3 independent experiments. The melt curve is represented as normalized data, calculated as follows: d(fluorescence)/d(temperature). B, A table showing reaction conditions for each assay and the mean area under the curve (AUC).
The mprA Knockout Mutant Is Completely Attenuated in a Murine Lethal Sepsis Model
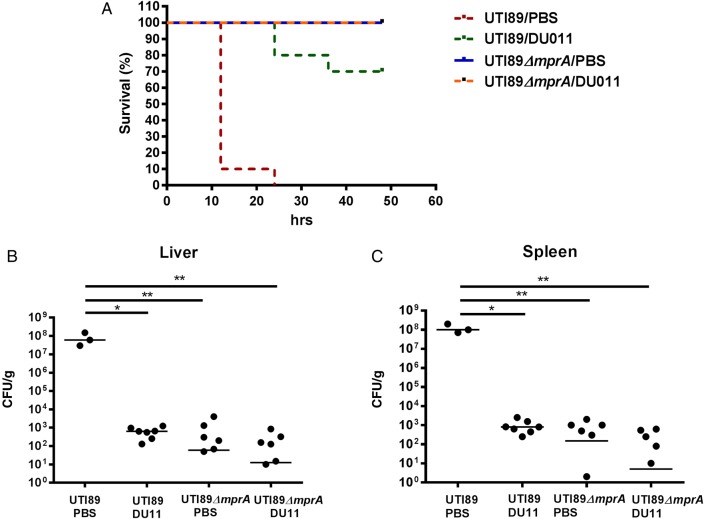
We have previously described the near-complete attenuation of a systemic lethal infection by DU011 in a murine sepsis model, using E. coli UTI89 [13]. We predicted that the mprA knockout strain would have a similar fitness defect, mimicking the phenotype of DU011 binding to MprA in vivo, with the bacteria becoming unencapsulated and thus being exposed to innate immune clearance. Mice infected with wild-type UTI89 had 100% mortality, whereas those infected with the mprA knockout strain had no mortality or evident morbidity. DU011 administration to mice infected with wild-type mprA also attenuated infection and mortality (Figure 6A). Furthermore, animals infected with the mprA knockout mutant or with wild-type mprA and treated with DU011 had a significantly lower bacterial load in both the liver and the spleen (Figure 6B and 6C), suggesting that the unencapsulated bacteria are cleared more efficiently by the host immune system than the wild-type bacteria.
Figure 6.
Comparison of DU011 treatment and mutation of mprA in a murine disseminated Escherichia coli infection model. A, Female mice (n = 10 in 2 separate experiments) with and without DU011 pretreatment were infected with wild-type or mprA mutant E. coli UTI89 at a dose previously shown to be lethal for the wild-type E. coli infection. B and C, The number of colony-forming units (CFU) per gram of tissue from the liver (left) and spleen (right) at the time mice were euthanized owing to reaching a moribund state or the end of the experiment. Horizontal lines indicate median values. *P = .01 and **P = .007, by the Mann–Whitney U test. Abbreviation: PBS, phosphate-buffered saline.
DISCUSSION
Chemical inhibition of virulence factors represents a therapeutic approach that is gaining attention for the treatment of infectious diseases, particularly as rising antibiotic resistance threatens the effectiveness of conventional broad-spectrum therapeutics. Multiple potential benefits of targeting virulence mechanisms have been proposed, including less development of antibiotic resistance and reduced collateral effects on the commensal microbiota. In this study, we identified MprA as the target of the polysaccharide capsule inhibitor DU011, and our work interconnects the regulation of multidrug efflux with the regulation of E. coli group 2 capsule, a widely recognized preeminent virulence factor of extraintestinal pathogenic E. coli responsible for UTI, bloodstream infections, meningitis, pneumonia, and osteomyelitis [14, 32–34]. Perhaps the most important outcome from this study is the demonstration that specific ligands of the MarR family of regulators, in this case MprA, can limit virulence without the expense of inducing antibiotic resistance.
MprA is a member of the MarR family of winged helix transcriptional regulators that have known roles in controlling the expression of multidrug efflux pumps and virulence-associated factors in a range of pathogenic bacteria, including Pseudomonas aeruginosa, Salmonella typhimurium, and Bordetella pertussis [35–37]. Prior studies suggest that the efflux pumps may have roles in pathogenicity beyond providing xenobiotic resistance; they may be required for resistance against the host inflammatory response, as well. For instance, Salmonella typhimurium lacking all efflux pumps is rendered avirulent [38], perhaps due to its inability to effectively expel host immune factors.
Here, we demonstrate that virulence may be selectively and potently altered by small molecules without the negative consequence of induced antibiotic resistance. Our initial objective was to identify uropathogenic E. coli–specific virulence inhibitors. However, the presence and conservation of MprA among many gram-negative bacteria, including commensal E. coli, suggests that DU011 and other capsule inhibitors may affect other members of the human microbiota. Future studies will be required to determine whether this is consequential or benign. Certainly the lack of overt inhibition of E. coli growth in the presence of the capsule inhibitors portends far fewer effects on the microbiota than conventional antibiotics.
While salicylate, DNP, and DU011 have similar proposed bioisosteric functional groups, DU011 is distinguished by unique space occupancy due to the fluorinated phenyl and phenol ether rings. This may be one reason why the MprA-DU011 complex may assume a more stable conformation relative to DNP-MprA and salicylate-MprA complexes. The differences in MprA-ligand conformations may be responsible for the distinct regulatory activities of the MprA-ligand complexes. We were able to use the mprA operator as a probe for MprA-ligand interactions [29] and demonstrate that MprA binding to this operator is decreased with the addition of DU011 and DNP. Using the same approach, we were not able to demonstrate direct binding of MprA to the capsule promoter regions, and bioinformatic searches failed to identify MprA operator sequences (Arshad and Seed, unpublished data). Thus, MprA is likely regulating polysaccharide capsule expression through intermediate regulators that may be part of a broader coordinate regulatory network. RNA sequencing and genome-wide chromatin immunoprecipitation sequencing studies will be critical to define the regulatory network and determine whether as we anticipate, the different MprA ligands stimulate different global transcriptional states as the result of the unique conformations each MprA-ligand complex assumes. Where the target of DU003 is within this regulatory network remains to be determined. It is interesting that MprA overexpression rendered K1 E. coli less susceptible to DU003 capsule inhibition, yet our data indicate that DU003 does not directly interact with MprA. Although the target for DU003 currently remains unknown to us, we suspect that DU003 interacts with an intermediate regulator between MprA and capsule transcription and that high levels of MprA are able to suppress the DU003 effect.
We show that mprA is a potent regulator of virulence. Knockout of mprA resulted in full attenuation in a murine sepsis model; DU011 treatment significantly reduced mortality, from 100% to 30%. The difference in survival in the chemical-treated mice as compared to mice infected with the mprA mutant may be due to lack of optimization of DU011 pharmacokinetics or may suggest that the targeting of MprA can be optimized. A better understanding of the binding pocket of MprA, combined with structural modeling, may lead to development of DU011 derivatives or as of yet not envisioned molecules with high affinity and conformational-exacting effects on MprA. Together with rigorous pharmacokinetic studies, we foresee the opportunity to develop a novel class of antiinfective agents that have lesser consequences on the normal commensal microbiota than conventional antibiotics, including induction of drug resistance while attenuating uropathogenic E. coli in the act of causing invasive disease.
Supplementary Data
Supplementary materials are available at http://jid.oxfordjournals.org. Consisting of data provided by the author to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the author, so questions or comments should be addressed to the author.
Notes
Acknowledgments. We thank Richard Silver, for kindly providing the horse group B meningococcal antiserum (H46), K1F bacteriophage, and useful E. coli strains; and Dr Jenna Wang, for performing the analysis of morphological chemical-similarity analysis.
Financial support. This work was supported by the Eunice Kennedy Shriver National Institute of Child Health (K12HD000850 and K12-HD043494 to M. A.), the Department of Defense (W81XWH-13-1-0450 to P. C. S.), the National Institutes of Health and the US Department of Health and Human Services (NIGMS 1R01GM108494-01 to P. C. S., NHGRI 5U54HG005031 to Jeffrey Aubé, PI), the March of Dimes (6-FY12-277 to P. C. S.), the Christopher and Dana Reeve Foundation (to P. C. S.), and the Paralyzed Veterans of America (to P. C. S.).
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Foxman B. Urinary tract infection syndromes: occurrence, recurrence, bacteriology, risk factors, and disease burden. Infect Dis Clin North Am 2014; 28:1–13. [DOI] [PubMed] [Google Scholar]
- 2.Litwin MS, Saigal CS. Urologic diseases in America. NIH publication 07-5512:3–7. Bethesda: National Institute of Diabetes and Digestive and Kidney Diseases, 2007. [Google Scholar]
- 3.Czaja CA, Scholes D, Hooton TM, Stamm WE. Population-based epidemiologic analysis of acute pyelonephritis. Clin Infect Dis 2007; 45:273–80. [DOI] [PubMed] [Google Scholar]
- 4.Hayami H, Takahashi S, Ishikawa K et al. Nationwide surveillance of bacterial pathogens from patients with acute uncomplicated cystitis conducted by the Japanese surveillance committee during 2009 and 2010: antimicrobial susceptibility of Escherichia coli and Staphylococcus saprophyticus. J Infect Chemother 2013; 19:393–403. [DOI] [PubMed] [Google Scholar]
- 5.Gupta K, Sahm D, Mayfield D, Stamm WE. Antimicrobial resistance among uropathogens that cause community-acquired urinary tract infections in women: a nationwide analysis. Clin Infect Dis 2001; 33:89–94. [DOI] [PubMed] [Google Scholar]
- 6.Yolbaş I, Tekin R, Kelekci S et al. Community-acquired urinary tract infections in children: pathogens, antibiotic susceptibility and seasonal changes. Eur Rev Med Pharmacol Sci 2013; 17:971–6. [PubMed] [Google Scholar]
- 7.Edlin RS, Shapiro DJ, Hersh AL, Copp HL. Antibiotic resistance patterns of outpatient pediatric urinary tract infections. J Urol 2013; 190:222–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanchez GV, Master RN, Karlowsky JA, Bordon JM. In vitro antimicrobial resistance of urinary Escherichia coli isolates among U.S. outpatients from 2000 to 2010. Antimicrob Agents Chemother 2012; 56:2181–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karlowsky JA, Adam HJ, Desjardins M et al. Changes in fluoroquinolone resistance over 5 years (CANWARD 2007–11) in bacterial pathogens isolated in Canadian hospitals. J Antimicrob Chemother 2013; 68:39–46. [DOI] [PubMed] [Google Scholar]
- 10.Accogli M, Giani T, Monaco M et al. Emergence of Escherichia coli ST131 sub-clone H30 producing VIM-1 and KPC-3 carbapenemases, Italy. J Antimicrob Chemother 2014; 69:2293–6. [DOI] [PubMed] [Google Scholar]
- 11.Mushtaq N, Redpath MB, Luzio JP, Taylor PW. Treatment of experimental Escherichia coli infection with recombinant bacteriophage-derived capsule depolymerase. J Antimicrob Chemoth 2005; 56:160–5. [DOI] [PubMed] [Google Scholar]
- 12.Goller CC, Seed PC. High-throughput identification of chemical inhibitors of E. coli Group 2 capsule biogenesis as anti-virulence agents. PLoS One 2010; 5:e11642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goller CC, Arshad M, Noah JW et al. Lifting the Mask: Identification of new small molecule inhibitors of uropathogenic Escherichia coli group 2 capsule biogenesis. PLoS One 2014; 9:e96054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson GG, Goller CC, Justice S, Hultgren SJ, Seed PC. Polysaccharide capsule and sialic acid-mediated regulation promote biofilm-like intracellular bacterial communities during cystitis. Infect Immun 2010; 78:963–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bahrani-Mougeot FK, Buckles EL, Lockatell CV et al. Type 1 fimbriae and extracellular polysaccharides are preeminent uropathogenic Escherichia coli virulence determinants in the murine urinary tract. Mol Microbiol 2002; 45:1079–93. [DOI] [PubMed] [Google Scholar]
- 16.Smith SN, Hagan EC, Lane MC, Mobley HL. Dissemination and systemic colonization of uropathogenic Escherichia coli in a murine model of bacteremia. MBio 2010; 1:pii:e00262–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim KS, Itabashi H, Gemski P, Sadoff J, Warren RL, Cross AS. The K1 capsule is the critical determinant in the development of Escherichia coli meningitis in the rat. J Clin Invest 1992; 90:897–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swinney DC, Anthony J. How were new medicines discovered? Nat Rev Drug Discov 2011; 10:507–19. [DOI] [PubMed] [Google Scholar]
- 19.Mulvey MA, Lopez-Boado YS, Wilson CL et al. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science 1998; 282:1494–7. [DOI] [PubMed] [Google Scholar]
- 20.Vimr ER, Troy FA. Regulation of sialic acid metabolism in Escherichia coli: role of N-acylneuraminate pyruvate-lyase. J Bacteriol 1985; 164:854–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kitagawa M, Ara T, Arifuzzaman M et al. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res 2005; 12:291–9. [DOI] [PubMed] [Google Scholar]
- 22.Scholl D, Adhya S, Merril C. Escherichia coli K1's Capsule Is a Barrier to Bacteriophage T7. Appl Environ Microbiol 2005; 71:4872–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vimr ER, McCoy RD, Vollger HF, Wilkison NC, Troy FA. Use of prokaryotic-derived probes to identify poly(sialic acid) in neonatal neuronal membranes. Proc Natl Acad Sci U S A 1984; 81:1971–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory, 1989. [Google Scholar]
- 25.Schmidt J, Klingler FM, Proschak E, Steinhilber D, Schubert-Zsilavecz M, Merk D. NSAIDs ibuprofen, indometacin, and diclofenac do not interact with farnesoid X receptor. Sci Rep 2015; 5:14782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jain AN. Morphological similarity: a 3D molecular similarity method correlated with protein-ligand recognition. J Comput Aided Mol Des 2000; 14:199–213. [DOI] [PubMed] [Google Scholar]
- 27.Jain AN. Surflex: fully automatic flexible molecular docking using a molecular similarity-based search engine. J Med Chem 2003; 46:499–511. [DOI] [PubMed] [Google Scholar]
- 28.Jain AN. Ligand-based structural hypotheses for virtual screening. J Med Chem 2004; 47:947–61. [DOI] [PubMed] [Google Scholar]
- 29.Xiong A, Gottman A, Park C, Baetens M, Pandza S, Matin A. The EmrR protein represses the Escherichia coli emrRAB multidrug resistance operon by directly binding to its promoter region. Antimicrob Agents Chemother 2000; 44:2905–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lomovskaya O, Lewis K, Matin A. EmrR is a negative regulator of the Escherichia coli multidrug resistance pump EmrAB. J Bacteriol 1995; 177:2328–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brooun A, Tomashek JJ, Lewis K. Purification and ligand binding of EmrR, a regulator of a multidrug transporter. J Bacteriol 1999; 181:5131–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamzaoui A, Salem R, Koubaa M et al. Escherichia coli osteomyelitis of the ischium in an adult. Orthop Traumatol Surg Res 2009; 95:636–8. [DOI] [PubMed] [Google Scholar]
- 33.Sader HS, Farrell DJ, Flamm RK, Jones RN. Antimicrobial susceptibility of Gram-negative organisms isolated from patients hospitalised with pneumonia in US and European hospitals: results from the SENTRY Antimicrobial Surveillance Program, 2009–2012. Int J Antimicrob Agents 2014; 43:328–34. [DOI] [PubMed] [Google Scholar]
- 34.Gaschignard J, Levy C, Romain O et al. Neonatal Bacterial Meningitis: 444 Cases in 7 Years. Pediatr Infect Dis J 2011; 30:212–7. [DOI] [PubMed] [Google Scholar]
- 35.Grkovic S, Brown MH, Skurray RA. Regulation of bacterial drug export systems. Microbiol Mol Biol Rev 2002; 66:671–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stover CK, Pham XQ, Erwin AL et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 2000; 406:959–64. [DOI] [PubMed] [Google Scholar]
- 37.Conover MS, Redfern CJ, Ganguly T et al. BpsR modulates Bordetella biofilm formation by negatively regulating the expression of the Bps polysaccharide. J Bacteriol 2012; 194:233–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishino K, Latifi T, Groisman EA. Virulence and drug resistance roles of multidrug efflux systems of Salmonella enterica serovar Typhimurium. Mol Microbiol 2006; 59:126–41. [DOI] [PubMed] [Google Scholar]
- 39.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 2000; 6:6640–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.