Abstract
Cryptosporidiosis is a common cause of diarrhea morbidity and mortality worldwide. Research progress on this infection has been slowed by lack of methods to genetically manipulate Cryptosporidium parasites. Small interfering RNA (siRNA) is widely used to study gene function, but Cryptosporidium species lack the enzymes necessary to process siRNA. By preassembling complexes with the human enzyme Argonaute 2 (hAgo2) and Cryptosporidium single-stranded RNA (ssRNA), we induced specific slicing in Cryptosporidium RNA targets. We demonstrated the reduction in expression of target genes at the mRNA and protein levels by transfecting live parasites with ssRNA-hAgo2 complexes. Furthermore we used this method to confirm the role of selected molecules during host cell invasion. This novel method provides a novel means of silencing Cryptosporidium genes to study their role in host-parasite interactions and as potential targets for chemotherapy.
Keywords: Argonaute, Cryptosporidium, Cryptosporidium parvum, cryptosporidiosis, silencing, siRNA
The parasites of the genus Cryptosporidium are important causes of diarrheal diseases worldwide. A recent multicenter study in sub-Saharan Africa and south Asia identified Cryptosporidium as second to rotavirus as a cause of childhood morbidity and mortality due to diarrhea [1]. Similarly, studies from throughout the resource-poor world are demonstrating that Cryptosporidium is a major contributor to childhood malnutrition [2]. Despite the morbidity and mortality from this pathogen, there is currently no reliable drug treatment for cryptosporidiosis in compromised hosts. The lack of methods to manipulate gene expression has been identified as a major obstacle in the drug development for cryptosporidiosis [2].
RNA interference is a process that most eukaryotic cells use to silence genes. When exogenous double-stranded RNA (dsRNA) is detected in the cytoplasm, it is processed into small interfering RNAs (siRNAs) by the RNase III–like enzyme dicer. Transactivation response RNA-binding protein, Argonaute (Ago), and the siRNA bind to dicer to form the RNA-inducing silencing complex (RISC). Target RNAs are then cleaved by the Ago guided by the single-stranded RNA (ssRNA) to produce silencing (Figure 1A). Many protozoan parasites, including Cryptosporidium, lack RISC components and cannot process siRNA [3–6]. Previous studies have demonstrated that human Argonaute 2 (hAgo2) produces the final cleavage in the RNA target [7–10]. We hypothesized that protein hAgo2 preloaded with heterologous ssRNA can be transferred into live organisms that lack the RISC pathway to induce gene silencing (Figure 1A). Here we tested this hypothesis by using preassembled Cryptosporidium-ssRNA-hAgo2 complexes to transfect live Cryptosporidium and to study molecules involved in host cell invasion.
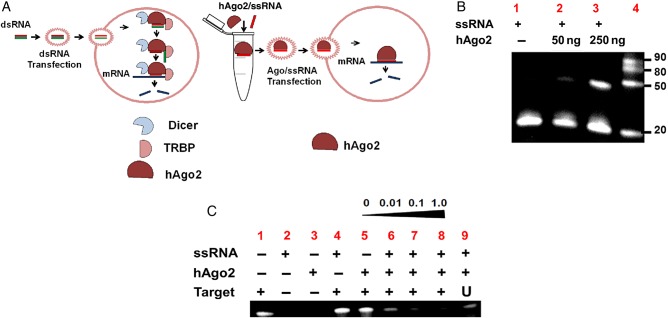
Figure 1.
Complex assembling and human Argonaute 2 (hAgo2) activity. A, In human cells, dicer enzyme processes double-stranded RNA (dsRNA; left). The resulting small interfering RNA (siRNA) is loaded by dicer and transactivation response RNA-binding protein onto hAgo2 (RISC complex) to producing target degradation. hAgo2 and single-stranded RNA (ssRNA) are preassembled in vitro, and the complex is introduced into organisms without RISC enzymes, such as Cryptosporidium, to produce silencing (right). B, Complex assembly was confirmed by the electrophoretic mobility-shift assay assay, using ssRNA labeled with FAM. Lanes 1–3, Samples incubated with hAgo2 (0, 50, 250 ng). Lane 4, Molecular markers. C, ssRNA-hAgo2 complex activity is demonstrated by denaturing gel electrophoresis, using an RNA target (50 nucleotides) labeled with carboxyfluorescein. 1, Cp15 target; 2, Cp15-ssRNA with no target; 3, Cp15-ssRNA with hAgo2 alone; 4, Cp15 target incubated with ssRNA alone; 5, Cp15 target incubated with hAgo2 alone; 6, Cp15 target loaded with 0.01 µM of Cp15-ssRNA; 7, Cp15 target loaded with 0.1 µM of Cp15-ssRNA; 8, Cp15 target loaded with 1 µM of Cp15-ssRNA; 9, unrelated target (60 nucleotides) with 1 µM of Cp15-ssRNA. Abbreviation: mRNA, messenger RNA.
MATERIAL AND METHODS
ssRNA-hAgo2 Complex Assembly
To form complexes, we used hAgo2 (Sino-Biological, Beijing, China) and ssRNA designed from Cryptosporidium targets with defined functions. Cp15 (L34568.1) is a 15-kDa surface antigen implicated in attachment [11], glycoprotein (Gp900; AF068065) is a 900-kDa mucin-like protein implicated in adhesion to host cells [12], calcium-dependent kinase 1 (CDPK; XM_001388059) is implicated in parasite motility and invasion in apicomplexans [13], inosine-5-monophosphate dehydrogenase (Ino-5mod; XM_625342) is part of the guanine biosynthesis pathway [14], and Cp17 is an immunodominant 17-kDa antigen (AF1141660) [15]. We designed ssRNAs by using RNA design tool (Integrated DNA technologies, Coralville, Iowa). We conducted in silico analysis on RNA and ssRNA by using RNAfold (Institute for Theoretical Chemistry, University of Vienna). To reduce off-target effects, we conducted nucleotide-blast analysis of ssRNAs. The selected ssRNAs (Supplementary Table 1) were synthetized with the modifications reported by Rivas et al [8], which includes 5′P-uN-(21–23)-dTT-3′. We assembled complexes by using the method described by Miyoshi et al [9] with modifications. Briefly, for assembling, we used 50–250 ng of hAgo2 and 1 µM of Cp15 ssRNA (21 nucleotides) in 14 µL of assembly buffer (30 mM HEPES, pH 7.4, 150 mM KOAc, and 2 mM MgCl2). We incubated the sample for 90 minutes at room temperature. The formation of complexes was evaluated by an electrophoretic mobility-shift assay (EMSA) as described previously [16]. For EMSA experiments, the 3′ end of the Cp15 ssRNA was labeled with carboxyfluorescein (FAM) for the visualization of RNA in agarose gels 4%.
ssRNA-hAgo2 Activity
To evaluate the activity of the complexes, we used the slicing assay described by Miyoshi et al [9], with modifications. Briefly, we assembled the complexes as described above, but we used a fixed level of hAgo2 (250 ng) with different concentrations of ssRNA (0, 10 nM, 100 nM, and 1 µM). After incubation (for 90 minutes at room temperature), we added 1 µL (100 nM) of target Cp15 RNA (50 nucleotides) or unrelated RNA sequence (60 nucleotides) as a negative control. We stopped the reaction by heating (for 5 minutes at 95°C). The RNA was separated by Tris/borate/ethylenediaminetetraacetic acid–urea electrophoresis, using 10% agarose gels (Bio-Rad, Hercules, California). To measure the degradation of target, RNA was visualized with SYBR Gold (Life Technologies, Carlsbad, California).
Transfection of Parasites
Cryptosporidium parvum oocysts were purchased from the University of Arizona. To show protein transfection of the sporozoites within oocysts, we used human immunoglobulin G (IgG) protein labeled with phycoerythrin (New England Biolabs, Massachusetts) and hAgo2 labeled with NHS-rhodamine (Thermo-Fisher, Rockford, Illinois). For transfection experiments, we used fluorochrome-labeled anti-Cryptosporidium antibodies to detect the oocyst wall of Cryptosporidium (Merifluor, Meridian Life Science, Memphis, Tennessee). For transfections, the protein transfection reagent (PTR) Pro-Jet (Thermo-Fisher, Rockford, Illinois) was resuspended in 1 mL of HEPES (100 mM). PTR (35 µL) was added to the sample (20 µL) containing IgG-FITC, hAgo2-NHS-Rhodamine, or ssRNA-hAgo2. The samples were incubated 5 minutes at room temperature to allow encapsulation of the complexes within the PTR. Then, 106 oocysts were added to the sample and incubated at 37°C for 2 hours to increase permeability and to improve transfection into sporozoites. For silencing, the oocysts were incubated for 4 hours at 37° with the preassembled complexes. After incubation, samples were concentrated by centrifugation (at 15 000 × g for 30 seconds) and washed with phosphate-buffered saline (PBS). To induce excystation, the pellets were resuspended in 25 µL of acidic water (pH 2–3) and incubated for 10 minutes on ice. We then added 225 µL of Roswell Park Memorial Institute (RPMI) 1640–taurocholate 0.8% medium and incubated at 37°C for 30 minutes. The presence of motile sporozoites after the excystation was evaluated by microscopy. The parasites were collected by centrifugation (at 15 000g) and then frozen for RNA extraction or fixed in formalin.
Viability Assays
A total of 1 × 106 oocysts were transfected with ssRNA-hAgo2 but followed by staining with the vital dye carboxyfluorescein succinimidyl ester (CFSE; Life Technologies, Grand Island, New York) as described before [17] and then analyzed by fluorescent microscopy. The cytotoxicity of transfection was quantified by flow cytometry. For these experiments, 1 × 106 oocysts were transfected and then stained using the Baclight Viability Kit, following the instructions of the vendor (Life Technologies, Carlsbad, California). Stained oocysts were incubated (at room temperature for 20 minutes) with 0.5 mL of fixation buffer (2% paraformaldehyde) and washed 3 times with PBS, and the proportion of viable organisms was quantitated by flow cytometry, using the Stratedigm SE500 Analyzer (Stratedigm, San Jose, California).
Quantitative Polymerase Chain Reaction (qPCR) Assays
We evaluated silencing by measuring the reduction of messenger RNA (mRNA) by qPCR. The RNA was isolated from the transfected parasites using a commercial kit (RNeasy Plus kit, Qiagen, Valencia, California). The concentration of the RNA was evaluated by spectrophotometry (NanoDrop 1000, Thermo-Scientific, Wilmington, Delaware). For amplification, we used 100 ng of total RNA as template and the SuperScript III Platinum SYBR Green One-Step qRT-PCR Kit (Life Technologies, Grand Island, New York). For the reaction, we used the following conditions: 55°C for 20 minutes, 95°C for 5 minutes, and 40 cycles at 95°C for 15 seconds and 65°C for 1 minute, using the AB7500 Fast RT-PCR System (Life Technologies, Grand Island, New York). The primers used are described in table 2 (Supplementary data). We evaluated the relative fold change by the delta-delta method (ratio = 2ΔCt sample−ΔCt control), which compares the cycle threshold (Ct) values of treated samples and untransfected parasites. For controls and reference genes we used the untargeted mRNA surface protein Cp23 (DQ389174) and 18S ribosomal RNA (AB089290.1). All samples were analyzed in triplicate in at least 2 independent experiments.
Western Blot Assays
To demonstrate correlation between silencing and reduction at protein levels, we analyzed expression of Gp900 by Western blot. Protein crude extracts were obtained from transfected oocysts (2 × 106) with hAgo2-Gp900 ssRNA (or not). After transfection with preassembled complexes, the parasites were concentrated by centrifugation (at 16 000 × g for 1 minute), resuspended in Laemmli buffer with 1% sodium dodecyl sulfate (SDS), and lysed by heating (95°C for 15 minutes). Proteins were resolved by SDS polyacrylamide gel electrophoresis, using 4%–20% gradient polyacrylamide gels (Bio-Rad, Hercules, California). Proteins were transferred onto a nitrocellulose membrane and blocked for 1 hour at room temperature with 5% bovine serum albumin–TBST (Tris/Tris-HCl 25 mM, NaCl 0.13 M, KCl 0.0027 M, and 0.05% Tween 20). The membrane was incubated with 1 µg/mL of primary monoclonal antibody 4E9 [12] (Genway biotech, San Diego, California) diluted (1:1000) in TBST. The membrane was blotted with an horseradish peroxidase–conjugated secondary antibody diluted in TBST (1:5000). After 3 washes with PBS, the proteins were visualized using the Supersignal West Pico Chemiluminescent Substrate according to manufacturer specifications (Life Technologies, Grand Island, New York).
RNA Ligase–Mediated Rapid Amplification of Complementary DNA Ends (RLM-RACE)
We used the RLM-RACE technique to verify the cleavage site of the hAgo2-ssRNA [18]. For these experiments, total RNA was purified with the RNeasy Plus kit (Qiagen) from C. parvum oocysts previously transfected with hAgo2-CDPK ssRNA complex. Purified total RNA was then ligated to a 45-base RNA (5′-GCU GAU GGC GAU GAA UGA ACA CUG CGU UUG CUG GCU UUG AUG AAA-3) not present in the C. parvum transcriptome (adapter sequence), using T4 RNA ligase (New England Biolabs, Ipswich, Massachusetts). For ligation experiments, total RNA (approximately 50 ng in 4 µL) was mixed with 1 µL of adapter sequence (1 µg), 2 µL of T4 RNA ligase buffer, 2 µL of 10 µM ATP, 10 µL of polyethylene glycol 1000, and 1 µL of T4 RNA ligase. After 16 hours at room temperature, the enzyme was inactivated (at 75°C for 15 minutes). The reaction mixture (2 µL) was amplified by nested qPCR, using the outer primers RACE-outer 5′-GCT GAT GGC GAT GAA TGA ACA CTG 3′ and CDPK-R420 5′ AAT GTA GAA GCT AGA TGA ATC-3′ and the inner primers RACE-inner 5′ GAA CAC TGC GTT TGC TGG CTT TGA 3′ and CDPK-R 260 5′-AAA AGA ACC TTT GCC AAG CA-3′. The amplified RACE products were cloned using TOPO-TA cloning (Life Technologies, Grand Island, New York), according to the manufacturer's instructions, and sequenced using purified plasmid as template.
Invasion Experiments
We evaluated the effect of silencing, using an invasion model for Cryptosporidium [19]. Briefly, we infected human intestinal cells (HCT-8; ATCC, Manassas, Virginia) with sporozoites transfected with PTR alone or PTR with hAgo2-Cp 15 ssRNA, hAgo2-Gp900 ssRNA, or hAgo2-CDPK ssRNA. We incubated the epithelial cells for 2 hours at 37°C to allow sporozoite invasion. After incubation, the medium was discarded, and cells were washed 3 times with 500 µL of RPMI 1640 medium to remove supernatant with dead sporozoites. After washing, the cells were lysed by adding 350 µL of RNA extraction buffer (Qiagen) and frozen at −80°C until use. For RNA purification, the samples were processed with the RNeasy plus extraction kit (Qiagen). The number of parasites in infected cells was quantified by qPCR, using a standard curve for RNA extracted from a known number of parasites (ranging from 1 × 106 to 0.1 parasites). The reduction was calculated from the number of parasites in cells infected with parasites treated with PTR alone, compared with those treated with each complex. All qPCR experiments were conducted in triplicate in at least 2 independent experiments.
RESULTS
ssRNA-hAgo2 Complexes Have Slicer Activity
We demonstrated formation of complexes, by EMSA (Figure 1C). In this assay, the interaction between the RNA and protein is stabilized, and the ratio of bound to unbound nucleic acid on the gel reflects the fraction of free and bound probe molecules as the binding reaction enters the gel. Only samples incubated with hAgo2 produced retardation of ssRNA-FAM, RNA alone, or unbounded RNA migrate faster (Figure 1B). Therefore, complex formation was observed by the appearance of approximately 50-nucleotide bands in lanes 2 and 3 (Figure 1C), compared with naked ssRNA (Figure 1B). The bands of approximately 21 nucleotides in lanes 2 and 3 correspond to the unbound ssRNA. Differences in intensity observed in lanes 1, 2, and 3 correlated with the amount of protein. To test for slicer activity of the complex, synthetic Cp15 ssRNA (50 nucleotides) was incubated with hAgo2, ssRNA, or both. In these activity assays (Figure 1C), intense bands were observed in untreated target, ssRNA alone and 0 µM (lanes 1 and 5), ssRNA and hAgo2 alone did not show fluorescence (lanes 2 and 3). Partial degradation was observed with 0.01 and 0.1 µM (lanes 6 and 7) and complete target degradation with 1 µM of ssRNA (lane 8). However, ssRNA (1 µM) plus hAgo2 had no effect on an unrelated RNA target (lane 9). The degradation varied with the concentration of ssRNA, and the greatest degradation was observed when using higher levels of protein and ssRNA (250 ng and 1 µM, respectively). No degradation was observed with unrelated RNA target.
Protein Transfection in Cryptosporidium
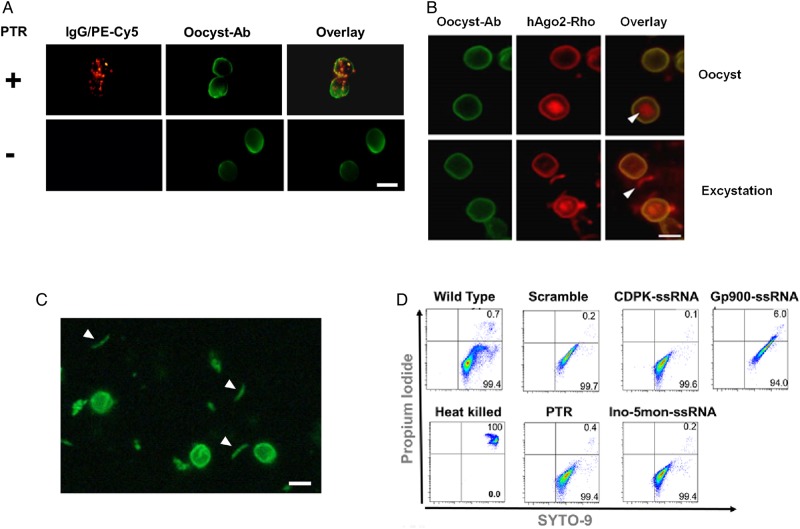
We hypothesized that protein transfection reagents could introduce proteins into Cryptosporidium as previously done with other cells [20–22]. We initially transfected oocysts using PTR loaded with fluorescent IgG (Mw approximately 150 kDa, Figure 2A). We detected the labeled protein only in parasites treated with PTR complexes (Figure 2A). No nonspecific binding was observed, since no signal was detected in samples incubated with the IgG alone (Figure 2A). To demonstrate sporozoite transfection, we mixed PTR with rhodamine labeled–hAgo2, transfected it into oocysts, and induced excystation of motile sporozoites. Both oocysts (Figure 2B) and sporozoites (Figure 2B) incorporated fluorescent hAgo2. Some labeled-hAgo2 was observed in the oocyst shell, which may reflect protein in transit. Microscopic analysis using PTR with fluorescent proteins indicated that 50%–80% of oocysts were transfected. The viability of transfected sporozoites was evaluated by fluorescent microscopy after inducing excystation and staining oocysts with the vital dye CFSE (Figure 2C) as previously described [17, 23]. We detected the vital dye CFSE in sporozoites, but we also observed some nonspecific fluorescence in the walls of some oocysts. However, we confirmed the viability by inducing excystation of stained motile sporozoites as shown in Figure 2C. We quantified the viability of parasites treated with different ssRNA-hAgo2 complexes by flow cytometry, using propium iodide, which stains live parasites. We confirmed that the majority of parasites remained alive after transfection, with no significant differences compared with hAgo2-scramble ssRNA or PTR alone or untransfected parasites (Figure 2D).
Figure 2.
Cryptosporidium transfection and viability assays. A, Fluorescent immunoglobulin G (IgG) labeled with phycoerythrin (red) was encapsulated within protein transfection reagent (PTR) and then used to transfect Cryptosporidium sporozoites within oocysts. Oocysts were incubated with anti-Cryptosporidium oocyst antibody-FITC (Oocyst Ab-FITC; green). IgG was only introduced into samples treated with PTR (top). No signal was detected in the parasites with samples incubated only with IgG (bottom). B, PTR was loaded with labeled human Argonaute 2 (hAgo2)–NHS-rhodamine (red) and then used to transfect oocysts. Cyst wall was stained with oocyst antibody (green). hAgo2 was detected in oocysts (white triangle; top) or in released sporozoites after excystation (white triangle; bottom). C, We stained oocysts with the vital dye carboxyfluorescein succinimidyl ester (CFSE; green). Stained and motile sporozoites were observed (white triangles). White bar, 5 µm. D, Viability was quantified by flow cytometry for untransfected parasites (wild type), heat-killed parasites, parasites treated with empty PTR, or parasites transfected with complexes of hAgo2 preloaded with scramble single-stranded RNA (ssRNA), calcium-dependent kinase 1 (CDPK)–ssRNA, inosine-5-monophosphate dehydrogenase (Ino-5mon)–ssRNA, and glycoprotein (Gp900)–ssRNA.
ssRNA-hAgo2 Produce Specific mRNA Cleavage in Live Parasites
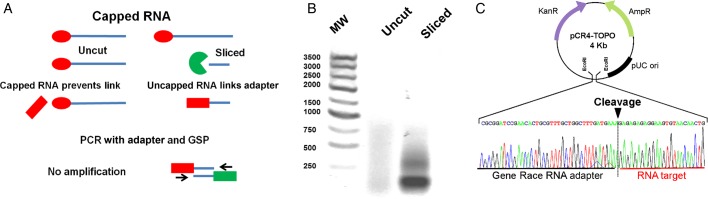
We used RLM-RACE to demonstrate the specificity of slicing produced by ssRNA-hAgo2 complexes (Figure 3A). In these assays, sliced mRNA was ligated to an adapter, and this ligated molecule was amplified with nested qPCR, cloned, and sequenced. The sequenced fragment showed where the adapter sequence ligated sliced mRNA, which corresponded to the place where ssRNA-hAgo2 sliced the mRNA target. For these experiments, CDPK expression was silenced by using the ssRNA-hAgo2 CDPK complex, and then the purified total RNA was ligated to an adaptor sequence. The molecule resulting from the ligation of the adaptor to the cleaved mRNA was amplified with a forward adaptor primer and a reverse gene-specific primer. We obtained a PCR product only in the sample sliced with the complex (Figure 3B). Sequencing results of the PCR product (Supplementary Figure 2) confirmed the cleavage of CDPK mRNA at a site in the middle of the sequence complementary to the CDPK ssRNA (Figure 3C).
Figure 3.
Specific RNA slicing in Cryptosporidium was demonstrated by rapid amplification of 5′ complementary DNA ends (RACE). A, Capped RNA (red circle) does not link the primer adaptor (red rectangle), the adaptor is linked only if the RNA target is sliced by human Argonaute 2 (hAgo2)—single-stranded RNA (ssRNA) complex (green), and a RACE–polymerase chain reaction (PCR) product is obtained only from sliced products by using an adaptor and gene-specific primer (GSP; green rectangle) but not with capped RNA. B, Electrophoresis of PCR-RACE products of parasites transfected only with protein transfection reagent (uncut) or with specific hAgo2-ssRNA (sliced). C, The PCR-RACE product was cloned and sequenced. The sequence of the sliced target (underlined in red) is located immediately after the adapter (underlined black), where the cleavage site is indicated (black triangle). Abbreviation: MW, molecular weight. This figure is available in black and white in print and in color online.
ssRNA-hAgo2 Silences Genes in Transfected Parasites
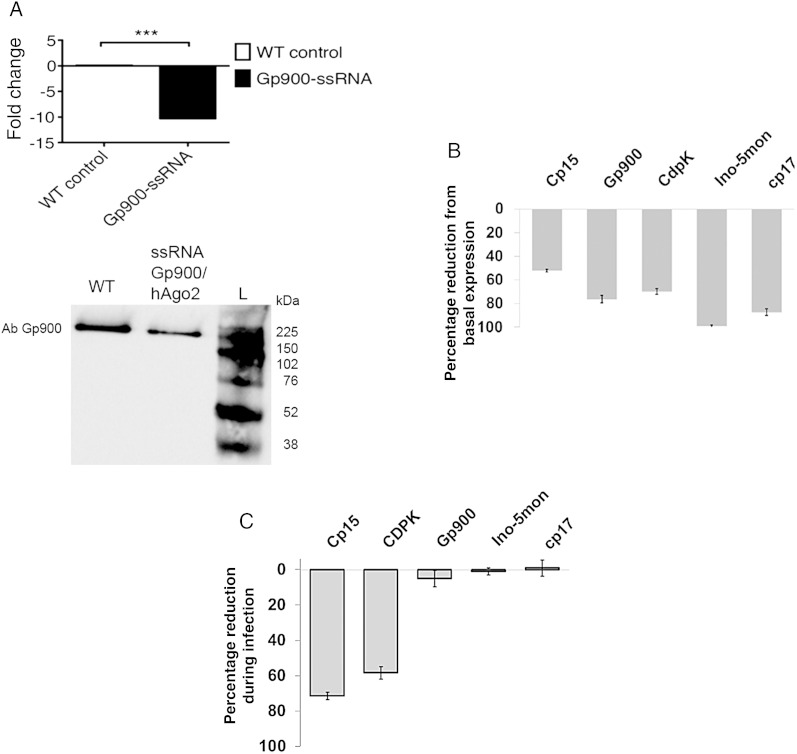
The majority of Cryptosporidium proteins are expressed during excystation [24]. We incubated the parasites for 4 hours to allow silencing and protein turnover, and then we evaluated the abundance of RNA after excystation to determine the silencing rate. qPCR results confirmed that mRNA was silenced by 10-fold, compared with basal mRNA levels of untransfected parasites (Figure 4A). To test the effects of silencing at the protein level, we evaluated expression of Gp900, compared with expression in untransfected parasites, by Western blot, using a commercially available antibody. There was a significant reduction in the expression of Gp900 after silencing (Figure 4A), and moderate levels of protein were detected in parasites treated with Gp900 ssRNA-hAgo2; the protein levels were most likely due the presence of residual preformed protein. To confirm the reproducibility of the method, we silenced Cp15, CDPK, Cp17, and Ino-5mon. All of these targets were reduced by 50%–95%, compared with the expression in parasites transfected only with PTR alone (Figure 4B). By contrast, expression of untargeted genes 18S and Cp23 remained unaltered after transfection. In addition, we demonstrated that silencing is dependent on hAgo2 by assembling reactions with different amounts of hAgo2 but a fixed concentration of CDPK ssRNA (Supplementary Figure 1). Induction of silencing was dose dependent for the amount of hAgo2, and no effect was observed when naked ssRNA was used. Overall, these results suggest that reduction in mRNA and protein levels are attributed to cleavage and blocking (by binding target) produced by ssRNA-Ago complexes.
Figure 4.
Silenced genes in Cryptosporidium. A, Quantitative polymerase chain reaction (qPCR; top) shows a fold reduction in messenger RNA target from transfected parasites with human Argonaute 2 (hAgo2) loaded with single-stranded RNA (ssRNA) specific for glycoprotein (Gp900; black bar). Control parasites were treated only with transfection reagent (wild type [WT]). Western blot analysis of Gp900 (bottom) in parasites transfected (ssRNA Gp900) or not transfected (WT control). B, qPCR analysis was used to calculate the percentage reduction in expression, relative to basal expression, for the Cryptosporidium targets Cp15, glycoprotein (Gp900), calcium-dependent kinase 1 (CDPK), inosine-5-monophosphate dehydrogenase (Ino-5mon), and Cp17 (gray bars). C, Transfected parasites were used to evaluate the role of silenced genes during infection. Percentages are relative to control (parasites were transfected only with protein transfection reagent [PTR]). In panels B and C, data are mean ± SD of qPCR triplicates and are representative of at least 3 individual experiments with similar results.
Silencing Decreased Parasite Invasion
After transfection with Cp15, CDPK, Gp900, Cp17, and Ino-5mon ssRNA complexes, oocysts were hatched and used to invade HCT-8 cells. Two hours after invasion, the number of intracellular trophozoites was evaluated by qPCR, which measured parasite RNA in the infected cells. There was a significant reduction of parasite RNA when we tested sporozoites silenced with Cp15 and CDPK ssRNA. By contrast, silencing of Gp900, cp17, and Ino-5mon did not significantly reduce the parasite burden (Figure 4C). Thus, we showed a differential effects of silencing during the invasion process.
DISCUSSION
We have developed a novel and rapid strategy to silence genes in Cryptosporidium. Gene silencing by siRNA is a potent tool to study gene function, but this tool has not been available for Cryptosporidium. Human RISC enzymes expressed in other organisms are functional [25]. Thus, we reasoned that RISC components could be used to reconstitute the siRNA pathway in Cryptosporidium. Since ssRNA-Ago2 complexes can be assembled in vitro and retain slicer activity [7–9, 26], we hypothesized that preassembled complexes could be introduced directly to Cryptosporidium by using protein transfection methods. Sporozoites are fragile and only live for a few hours, while oocysts live for longer periods. We demonstrated that a cationic lipid mixture efficiently introduces ssRNA-hAgo2 complexes into sporozoites within the oocysts, which overcomes the problem of the limited viability of sporozoites.
For other apicomplexans, proteins expressed before and during excystation have a key role in invasion of host cells, and many of them have been used as targets for chemotherapy and vaccination [27, 28]. Genes expressed by sporozoites are also potential targets for vaccination [29]. Thus, to demonstrate the silencing by our method, we targeted genes expressed during the sporozoite stage. We demonstrated silencing of all the selected targets, with up to 95% reduction in mRNA levels. Indeed, knocking down sporozoite Cp15 and CDPK reduced infection in HCT-8 cells. By contrast, knocking down expression of Gp900, Cp17, and Ino-5mon did not interfere with the infection process. Thus, we have demonstrated variable effects on parasite invasion by using this novel method.
The genome of Cryptosporidium has been sequenced [3, 6], but the biological function of many genes has not been identified [24]. Therefore, it is unclear which genes are essential for the parasite to complete its life cycle. Thus, the development of a fast and scalable method to determine biological function would allow a quick comparison of the role of genes during key steps of the infection (eg, invasion, proliferation, and egress). Vinayak et al recently reported a method of knocking out Cryptosporidium genes [30]. However, the method is complex and limited because it requires transfection of the fragile sporozoite stage, which must be immediately surgically implanted in the intestines of immunodeficient mice to amplify and select for mutants. The parasites can only be studied following recovery of oocysts from feces [30, 31]. Thus, this strategy is not amenable to the scale-up needed for drug screening. The major advantage of our method over other technologies is the simplicity and speed of analysis of the silencing. The use of preloaded ssRNA-Ago complexes ready to silence bypasses the transcription-translation process associated with gene expression, reducing the time until the parasites can be assayed from days to hours. This may allow the scale-up of the method for high-throughput analysis.
In summary, we have developed a novel method to silence genes expression in Cryptosporidium, using preassembled complexes of hAgo2 and ssRNA. It might also be useful for other organisms with a deficient or absent siRNA pathway [4, 5]. This method may prove to be a new tool to identify drug targets for cryptosporidiosis and for attenuation of parasites for live vaccines.
Supplementary Data
Supplementary materials are available at http://jid.oxfordjournals.org. Consisting of data provided by the author to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the author, so questions or comments should be addressed to the author.
Notes
Financial support. This work was supported by the Institute for Translational Sciences at the University of Texas Medical Branch at Galveston; the National Center for Advancing Translational Sciences, National Institutes of Health (Clinical and Translational Science Award UL1TR000071); and the University of Texas Medical Branch at Galveston (John Sealy Memorial Endowment Fund); S. N. was supported by a fellowship from the Sealy Center for Vaccine Development at UTMB.
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Kotloff KL, Nataro JP, Blackwelder WC et al. . Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 2013; 382:209–22. [DOI] [PubMed] [Google Scholar]
- 2.Checkley W, White AC Jr, Jaganath D et al. . A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for cryptosporidium. Lancet Infect Dis 2015; 15:85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu P, Widmer G, Wang Y et al. . The genome of Cryptosporidium hominis. Nature 2004; 431:1107–12. [DOI] [PubMed] [Google Scholar]
- 4.Baum J, Papenfuss AT, Mair GR et al. . Molecular genetics and comparative genomics reveal RNAi is not functional in malaria parasites. Nucleic Acids Res 2009; 37:3788–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lye LF, Owens K, Shi H et al. . Retention and loss of RNA interference pathways in trypanosomatid protozoans. PLoS Pathog 2010; 6:e1001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abrahamsen MS, Templeton TJ, Enomoto S et al. . Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 2004; 304:441–5. [DOI] [PubMed] [Google Scholar]
- 7.Deerberg A, Willkomm S, Restle T. Minimal mechanistic model of siRNA-dependent target RNA slicing by recombinant human Argonaute 2 protein. Proc Natl Acad Sci U S A 2013; 110:17850–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rivas FV, Tolia NH, Song JJ et al. . Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol 2005; 12:340–9. [DOI] [PubMed] [Google Scholar]
- 9.Miyoshi K, Uejima H, Nagami-Okada T, Siomi H, Siomi MC. In vitro RNA cleavage assay for Argonaute-family proteins. Methods Mol Biol 2008; 442:29–43. [DOI] [PubMed] [Google Scholar]
- 10.Ye X, Huang N, Liu Y et al. . Structure of C3PO and mechanism of human RISC activation. Nat Struct Mol Biol 2011; 18:650–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jenkins MC, Fayer R. Cloning and expression of cDNA encoding an antigenic Cryptosporidium parvum protein. Mol Biochem Parasitol 1995; 71:149–52. [DOI] [PubMed] [Google Scholar]
- 12.Cevallos AM, Bhat N, Verdon R et al. . Mediation of Cryptosporidium parvum infection in vitro by mucin-like glycoproteins defined by a neutralizing monoclonal antibody. Infect Immun 2000; 68:5167–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Billker O, Lourido S, Sibley LD. Calcium-dependent signaling and kinases in apicomplexan parasites. Cell Host Microbe 2009; 5:612–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorla SK, McNair NN, Yang G et al. . Validation of IMP dehydrogenase inhibitors in a mouse model of cryptosporidiosis. Antimicrob Agents Chemother 2014; 58:1603–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jakobi V, Petry F. Differential expression of Cryptosporidium parvum genes encoding sporozoite surface antigens in infected HCT-8 host cells. Microbes Infect 2006; 8:2186–94. [DOI] [PubMed] [Google Scholar]
- 16.Hellman LM, Fried MG. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat Protoc 2007; 2:1849–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng H, Nie W, Bonilla R, Widmer G, Sheoran A, Tzipori S. Quantitative tracking of Cryptosporidium infection in cell culture with CFSE. J Parasitol 2006; 92:1350–4. [DOI] [PubMed] [Google Scholar]
- 18.Lasham A, Herbert M, Coppieters 't Wallant N et al. . A rapid and sensitive method to detect siRNA-mediated mRNA cleavage in vivo using 5′ RACE and a molecular beacon probe. Nucleic Acids Res 2010; 38:e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Z, Ojo KK, Johnson SM et al. . Benzoylbenzimidazole-based selective inhibitors targeting Cryptosporidium parvum and Toxoplasma gondii calcium-dependent protein kinase-1. Bioorg Med Chem Lett 2012; 22:5264–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiang WC, Geel TM, Altintas MM, Sever S, Ruiters MH, Reiser J. Establishment of protein delivery systems targeting podocytes. PLoS One 2010; 5:e11837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weill CO, Biri S, Adib A, Erbacher P. A practical approach for intracellular protein delivery. Cytotechnology 2008; 56:41–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nossa CW, Jain P, Tamilselvam B et al. . Activation of the abundant nuclear factor poly(ADP-ribose) polymerase-1 by Helicobacter pylori. Proc Natl Acad Sci U S A 2009; 106:19998–20003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castellanos-Gonzalez A, Yancey LS, Wang HC et al. . Cryptosporidium infection of human intestinal epithelial cells increases expression of osteoprotegerin: a novel mechanism for evasion of host defenses. J Infect Dis 2008; 197:916–23. [DOI] [PubMed] [Google Scholar]
- 24.Sanderson SJ, Xia D, Prieto H et al. . Determining the protein repertoire of Cryptosporidium parvum sporozoites. Proteomics 2008; 8:1398–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi H, Tschudi C, Ullu E. Functional replacement of Trypanosoma brucei Argonaute by the human slicer Argonaute2. RNA 2006; 12:943–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, Ye X, Jiang F et al. . C3PO, an endoribonuclease that promotes RNAi by facilitating RISC activation. Science 2009; 325:750–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ojo KK, Larson ET, Keyloun KR et al. . Toxoplasma gondii calcium-dependent protein kinase 1 is a target for selective kinase inhibitors. Nat Struct Mol Biol 2010; 17:602–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Panchal D, Bhanot P. Activity of a trisubstituted pyrrole in inhibiting sporozoite invasion and blocking malaria infection. Antimicrob Agents Chemother 2010; 54:4269–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perryman LE, Jasmer DP, Riggs MW, Bohnet SG, McGuire TC, Arrowood MJ. A cloned gene of Cryptosporidium parvum encodes neutralization-sensitive epitopes. Mol Biochem Parasitol 1996; 80:137–47. [DOI] [PubMed] [Google Scholar]
- 30.Vinayak S, Pawlowic MC, Sateriale A et al. . Genetic modification of the diarrhoeal pathogen Cryptosporidium parvum. Nature 2015; 523:477–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beverley SM. Parasitology: CRISPR for Cryptosporidium. Nature 2015; 523:413–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.