Abstract
Array Comparative Genomic Hybridization (aCGH) is a rapid screening technique to detect gene deletions and duplications, providing an overview of chromosomal aberrations throughout the entire genome of a tumor, without the need for cell culturing. However, the heterogeneity of aCGH data obfuscates existing methods of data analysis. Analysis of aCGH data from a systems biology perspective or in the context of total aberrations is largely absent in the published literature. We present here a novel alternative to the functional analysis of aCGH data using the phylogenetic paradigm that is well-suited to high dimensional datasets of heterogeneous nature, but has not been widely adapted to aCGH data. Maximum parsimony phylogenetic analysis sorts out genetic data through the simplest presentation of the data on a cladogram, a graphical evolutionary tree, thus providing a powerful and efficient method for aCGH data analysis. For example, the cladogram models the multiphasic changes in the cancer genome and identifies shared early mutations in the disease progression, providing a simple yet powerful means of aCGH data interpretation. As such, applying maximum parsimony phylogenetic analysis to aCGH results allows for the differentiation between drivers and passenger genes aberrations in cancer specimens. In addition to offering a novel methodology to analyze aCGH results, we present here a crucial software suite that we wrote to carry out the analysis. In a broader context, we wish to underscore that phylogenetic analysis of aCGH data is a non-parametric method that circumvents the pitfalls and frustrations of standard analytical techniques that rely on parametric statistics. Organizing the data in a cladogram as explained in this research article provides insights into the disease common aberrations, as well as the disease subtypes and their shared aberrations (the synapomorphies) of each subtype. Hence, we report the method and make the software suite publicly and freely available at http://software.phylomcs.com so that researchers can test alternative and innovative approaches to the analysis of aCGH data.
Introduction
Array Comparative Genomic Hybridization (aCGH) is a rapid screening technique to detect gene deletions and duplications and to provide an overview of chromosomal aberrations throughout the entire genome of a tumor without the need for cell culturing (Oostlander et al., 2004; Weiss et al., 1999). Although data acquisition software and imaging technology have improved chip logistics, sample measurement, and data collection, the heterogeneity of aCGH data obfuscates attempts to identify meaningful conclusions—even in instances of small sample number (Brim et al., 2012; 2014).
There are different methods to analyze aCGH data (Lai et al., 2005). One common method employs statistical analysis or algorithms to determine duplications and deletions of aberrant genes from copy number variations (CNVs), producing a text-based aCGH data (Picard et al., 2005; van de Wiel et al., 2011; Van Wieringen et al., 2008). Dimension reduction analysis may be applied for the detection of smaller copy aberrations that might otherwise be missed. Statistical methods can be applied to organize the identified aberrant genes into weighted clusters, which is useful for subtype discovery (Van Wieringen et al., 2008). Furthermore, some software packages such as Asterias offer statistical tools for aCGH data analysis (Diaz-Uriarte et al., 2007).
Software packages that parse aCGH data produce results in a variety of formats, ultimately generating a list of aberrant chromosomal regions, information on whether the region was amplified or deleted, and a list of the genes affected. A common next step is to conduct a functional analysis on the affected genes in an attempt to determine relevant pathways and employ biological context to identify significant genes or biomarkers for further study. The nature of aCGH results as predominantly non-numerical and highly dimensional featuring large numbers of affected genes presents obstacles to applying traditional interpretations such as functional analysis and visualizations. Phylogenetic analyses, by applying Neighbor-Joining (NJ), parsimony, and Bayesian algorithms, have been carried out previously for systematics' studies (Edwards-Ingram et al., 2004; Gilbert et al., 2011a; 2011b; Renn et al., 2010). However, our data preparation (namely the polarity assessment step in our method) differs drastically from theirs and none of these were applied to cancer specimens.
Confronted with these limitations, we utilized a novel alternative to statistical analysis of aCGH results using the systems biology paradigm of phylogenetics (Wiley and Lieberman, 2011). Phylogenetics aims to classify objects (taxa, specimens, etc.) according to their shared derived variables (synapomorphies) into a hierarchical scheme represented by a graphical dichotomous tree (cladogram). Except for our two previous publications (Brim et al., 2012; 2014), analysis of aCGH cancer data from a systems biology perspective or in the context of total aberrations is largely absent from published literature.
Phylogenetic algorithms are well-suited to big sets of heterogeneous biological data (Albert, 2005), but have not been widely adapted to aCGH cancer data. Maximum parsimony phylogenetic analysis attempts to sort genetic information through the simplest presentation of the data on a cladogram, a graphical evolutionary tree, thus providing a powerful and efficient method for aCGH data analysis (Brim et al., 2012; Salazar et al., 2015). The cladogram is a dichotomous tree diagram that models the multiphasic spectrum of changes in the cancer genome and helps identify shared early mutations in the disease progression. Maximum parsimony phylogenetic analysis eschews the aforementioned limitations of other analysis techniques and presents a simple, robust, and rapid technique for analyzing highly dimensional and heterogeneous data and producing easily interpretable results (Abu-Asab et al., 2012; Salazar et al., 2015).
This article provides a step by step explanation of applying maximum parsimony phylogenetic analysis to aCGH data, including instructions on using the software programs that we wrote for this purpose. We are publishing this technique that we have already successfully utilized in the analysis of aCGH data of colorectal cancer patients (Brim et al., 2012; 2014) because it holds promise for improving the interpretation of aCGH data.
Materials and Methods
The aCGH data was collected from 27 colorectal cancer patients (for details on patients' specimens, see Brim et al., 2014) and first analyzed through Agilent Technologies Standard Software Packages (Agilent Feature Extraction software 9) and Agilent Genomic Workbench 5.0 software, followed by extraction of gained or lost genes in each tumor specimens (for details, see Brim et al., 2014). The output of the last step was a spreadsheet file as shown in Table 1 (see the full spreadsheet data in Supplementary Data 1; supplementary material is available online at www.liebertpub.com/omi). While the specific formatting of the initial aCGH data may differ between software packages used in the initial analysis, the basic features required for the maximum parsimony phylogenetic method are the specimens' identification and the aberrant genes names. These two components are all that is necessary to generate the binary matrix format by using our program CGHExtractor that was written specifically for this purpose (see below). CGHExtractor produces the necessary files for the processing of the data in the parsimony programs MIX and TNT to produce a phylogenetic cladogram.
Table 1.
Partial Listing of aCGH Data Used in Parsimony Phylogenetics Analysis of Shiraz-3T Subset*
| Aberration Number1 | Chromosome2 | Cytoband2 | Start2 | Stop2 | # Probes3 | Amplification4 | Deletion4 | P value5 | Gene names6 |
|---|---|---|---|---|---|---|---|---|---|
| 1 | chr1 | p36.33–p36.32 | 892526 | 3753050 | 135 | 0.707248 | 0 | 2.74E-183 | FLJ39609, SAMD11, NOC2L, KLHL17, PLEKHN1, HES4, HES4 … etc. |
| 2 | chr1 | p36.11–p33 | 26169808 | 46735833 | 889 | 0.12146 | 0 | 1.43E-38 | GRRP1, ZNF593, CNKSR1, CNKSR1, CATSPER4, CCDC21, SH3BGRL3, UBXN11 … etc. |
| 3 | chr1 | p33–p32.3 | 48075091 | 51080935 | 106 | 0 | −0.34596 | 1.34E-37 | SKINT1, SLC5A9, SPATA6, AGBL4, BEND5, ELAVL4, ELAVL4, ELAVL4, ELAVL4, DMRTA2, FAF1 … etc. |
See Supplementary Data 1 for whole data table.
Aberration Number = identifier for all aberrations found (883 in total), organized by sample identification values.
Chr, Cytoband, Start, and Stop = chromosomal positions of the aberrations. “Chr” is an abbreviation for Chromosome, and the other headings have the standard definitions for chromosome mappings.
Probe = number of probes used to identify the aberrations.
Amplification and Deletion = amplification or deletion of chromosomal.
Significance of findings is indicated by P value.
Gene Names = names of all afflicted genes in that location and specific aberration grouping.
Computer programs used in the analysis
An analysis that produces 1) a phylogenetic cladogram, 2) lists of synapomorphies for the cladogram's nodes (points of bifurcations) and specimens, and 3) lists of chromosomes carrying the mutations, requires the tandem application of the following programs: a) CGHExtractor, b) MIX or TNT, c) SynapExtractor, and d) ChromExtractor.
CGHExtractor and ChromExtractor are two new programs that we created and have not been published before; therefore, we are describing below their user and program procedures. CGHExtractor and ChromExtractor were tested separately by each of the authors, and their results were compared with that of spreadsheet calculations and verified that the results were accurate. SynapExtractor has been described by Salazar et al. (2015), MIX by Felsenstein (1989), and TNT by Goboloff (1999). The last two are off-the-shelf parsimony programs that are widely used to generate cladograms.
Downloading the programs
CGHExtractor, SynapExtractor, and ChromExtractor are freely available to the public for academic research and teaching only, and not for commercial use or resale. To download the programs, visit: http://software.phylomics.com/.
Preparing aCGH data for parsimony phylogenetic analysis using CGHExtractor
To achieve our goal of carrying out maximum parsimony phylogenetic analysis on the aCGH data, we wrote a software program, CGHExtractor (Fig. 1 shows the interface window), that used the spreadsheet data (Table 1, complete dataset at Supplementary Data 1) to create a new data matrix which transformed the aCGH data into qualitative values of zeros (0s), ones (1s), and twos (2s) if the user will use TNT to create the cladogram, or into 0s and 1s only when using MIX, which handles only binary values.
FIG. 1.
CGHExtractor interface.
CGHExtractor performs the following tasks:
1. builds a complete list of all the genes that appear as aberrations in the last column of Table 1 of all specimens;
2. lists aberrant genes per specimen;
3. scores each gene in the complete list as either 1 or 2 when the gene is present in the aberration list of the specimen (1 for deletion or 2 for duplication). When the gene is not present in the aberration list of the specimen, 0 is recorded;
4. generates a new matrix containing only 0s, 1s, and 2s (Table 2 shows a partial view of the new matrix. Full matrix in Supplementary Data 2).
Table 2.
Partial View of Polarized Values of aCGH Data as Produced by CGHExtractor*
| Gene name | Shiraz-9T | Shiraz-12T | Shiraz-18T | Shiraz-22T |
|---|---|---|---|---|
| GNB1 | 2 | 0 | 1 | 2 |
| CALML6 | 2 | 0 | 1 | 2 |
| TMEM52 | 2 | 0 | 1 | 2 |
| C1orf222 | 2 | 0 | 1 | 2 |
| KIAA1751 | 2 | 0 | 1 | 2 |
| GABRD | 2 | 0 | 1 | 2 |
| PRKCZ | 2 | 0 | 1 | 2 |
| LOC100128003 | 2 | 0 | 1 | 2 |
| C1orf86 | 2 | 0 | 1 | 2 |
| SKI | 2 | 0 | 1 | 2 |
| MORN1 | 2 | 0 | 1 | 2 |
| LOC100129534 | 2 | 0 | 1 | 2 |
| RER1 | 2 | 0 | 1 | 2 |
See Supplementary Data 2 for whole dataset.
Gene names are row headers in the first column, whereas specimens' names are column headers. A value of 1 and 2 indicates that the aCGH results from the specimen named at the top of the column contain an aberration in the gene named in the corresponding row. A value of 0 indicates that there is no such aberration in the specimen. The table is an abbreviated example of the matrix used to generate the input files of MIX and TNT to produce the maximum parsimony phylogenetic cladogram of Figure 3.
To verify the accuracy of CGHExtractor, we carried out the calculations manually using the MATCH function of Microsoft Excel, which returned the same results as CGHExtractor.
This conversion to binary or ternary values, also termed polarity assessment in phylogenetic terminology, characterizes each gene mutation as present or absent for each specimen. Based on this new matrix, two additional files were also generated by CGHExtractor and used by the parsimony program MIX of the PHYLIP package or TNT to produce a maximum-parsimony phylogenetic cladogram (Fig. 2).
FIG. 2.
Partial view of MIX and TNT input files that were produced by CGHExtractor. These filese were processed with MIX and TNT to produce the cladogram of Figure 3. (A) Partial view of the MIX input file. (B) Partial view of the TNT input file. The view shows the embedded commands before and after the data matrix that control TNT settings; these can be modified as needed.
MIX is a parsimony program that allows the selection between Wagner parsimony (allows reversals), Camin-Sokal parsimony (does not allow reversals), or a mix of the two methods. The methods consider changes from an ancestral state (0) to a derived (1 for deletion or duplication) state, conducting a heuristic search to find cladogram(s) that require the fewest of such changes—the simplest. MIX outputs all the equally most parsimonious cladograms. Several options within the program allow the user to modify the cladogram search. Once the cladograms are saved in an output file, the parsimony analysis is complete and the results are available for interpretation. For further information on the methodology and instructions for using MIX of the PHYLIP package, see the documentation available at the PHYLIP website (http://evolution.genetics.washington.edu/phylip/doc/main.html).
The TNT program is similar to MIX but with a graphical user interface. It handles multistate input, in this case it is ternary (0, 1, 2), and runs faster than MIX.
CGHExtractor: User Procedure
CGHExtractor utilizes the data table from aCGH datasets; header and footer information is unneeded and must be removed from the file prior to using CGHExtractor.
Upon running CGHExtractor and pressing the Select File button, the user is prompted to browse for a CSV file to open; after that, the user names the output CSV file. If the CSV file is in the format of aCGH data table with header and footer information removed, CGHExtractor will then process and output the extracted table of polarized values as the specified file.
CGHExtractor: Program procedure
Building a master list of all the genes of the dataset
CGHExtractor iterates over the dataset, checking every row that begins with an aberration number. In these rows, the tenth column contains a list of aberrant genes; CGHExtractor parses these gene names to build a complete list of the genes, the master list, that appear as aberrations in the dataset. Duplicate names are not added.
Polarity assessment of genes
The second pass through the dataset generates the polarized data values (0,1,2) and saves them in a matrix where one dimension is indexed by specimens' identifiers and the other dimension is indexed by genes. To fill this table, CGHExtractor iterates through the dataset, passing over each specimen's data in succession. Each specimen's data corresponds to a column in the matrix, so CGHExtractor fills an entire column before concatenating it to the matrix. For each deletion under a specimen, 1 is written to the column for every gene listed as a deletion aberration. Likewise, 2 is written for every gene listed under an amplification aberration. In the case of a gene appearing in multiple aberrations for the same specimen, the final polarity value is the one from the latest aberration in the dataset.
Finally, CGHExtractor writes the data from the polarity-value matrix to a CSV table (Supplementary Data 2). This cannot be done at the same time as the previous pass because here the gene names are in a set order; since the table has gene rows and specimen columns, the polarity value of a gene must be known for every specimen before that row may be written to a file. After this table is written to the CSV file, CGHExtractor uses it to generate MIX and TNT input files (Fig. 2). Once the three files have been generated, CGHExtractor is finished.
CGHExtractor: Program abstraction
Let P(G, S) be the polarity value for a given gene G and specimen S. CGHExtractor uses aberration data to define P over the domain of the specimens and genes present in the dataset.
The dataset defines a list of involved genes La for each aberration a.
P(G, S) = 1 if there exists for S some aberration d such that d is a deletion aberration and Ld contains G.
P(G, S) = 2 if there exists for S some aberration a such that a is an amplification aberration and La contains G.
Otherwise, P(G, S) = 0.
If there exist both amplification and deletion aberrations for S and G, P(G, S) takes its value based on the last aberration involving S and G (i.e., the aberration with the highest ID number).
Generating parsimony cladograms
Two input files generated by CGHExtractor were used to generate the parsimony cladograms by processing the files with MIX and TNT. This process has been described previously (Salazar et al., 2015).
Extracting Synapomorphies of Clades Using SynapExtractor
Synapomorphies are the chromosomal aberrations whose character states can be used to define a set of specimens as a clade (i.e., a set of specimens that share aberrations). MIX and TNT produce lists of synapomorphies of the cladogram nodes in their output files. However, we use the SynapExtractor to organize the synapomorphies in a comma-delimited text file (CSV) that is easier to view with a text editor or spreadsheet program (such as Microsoft Excel). Supplementary Data 3 lists all the node of the cladogram (Fig. 3) and their synapomorphies as produced by SynapExtractor from the output files of MIX and TNT. The synapomorphies of each node can be further analyzed with ChromExtractor to reveal which chromosomes contributed the mutations. A description of SynapExtractor is detailed in Salazar et al. (2015). SynapExtractor requires the master list of mutated genes (from the output of CGHExtractor) in a CSV file and the output file of MIX or TNT to generate a CSV file containing the synapomorphies of each node of the cladogram.
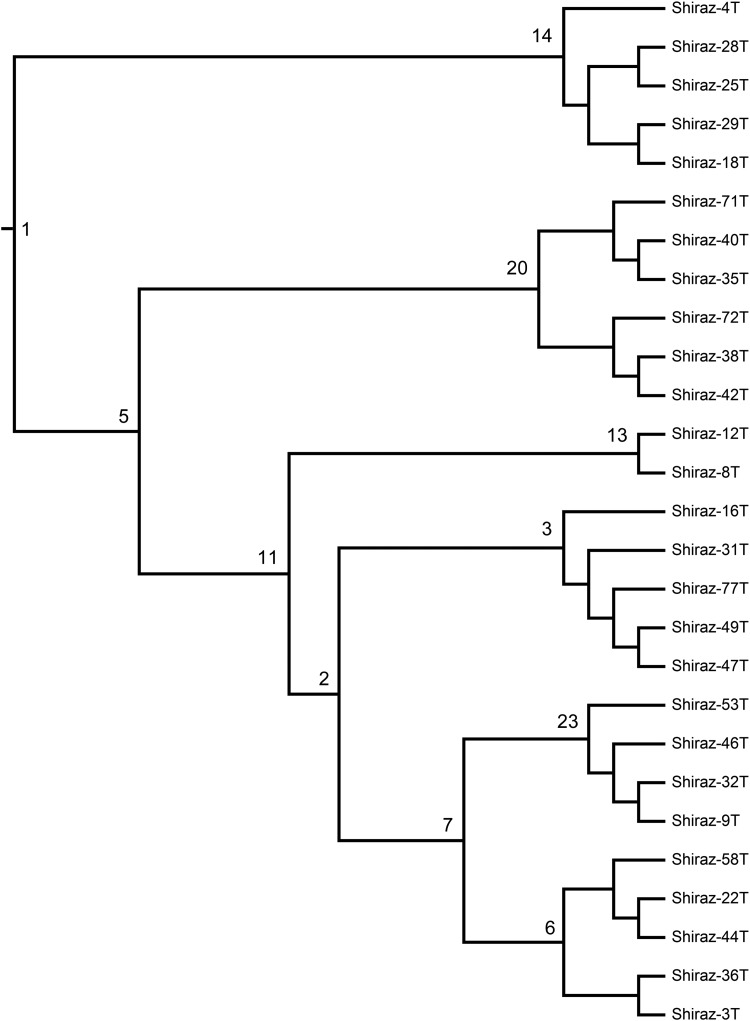
FIG. 3.
A most parsimonious cladogram based on aCGH dataset of the 27 colorectal cancer specimens produced by MIX's Camin-Sokal parsimony. Nodes are numbered for easy references, and each node is defined by a number of synapomorphies that were identified by MIX (see Supplementary Data 3 for listing of the nodes' synapomorphies).
Generating Table of Aberrant Chromosomes Using ChromExtractor
The cladogram's clades and their synapomorphies will also be used to find out which of the chromosomes have the clonal aberrations and thus contributed to carcinogenesis. For this purpose, we have written a second program, ChromExtractor (Fig. 4 shows the interface window), that extracts the synapomorphies from MIX's output file (usually called outfile), or TNT's output file, and match them with their chromosomes, thus producing a file containing this data (Table 3).
FIG. 4.
ChromExtractor interface.
Table 3.
Partial Output of ChromExtractor Program Showing Results for Gene AAA1 at Node 14 (1 to 14) from Cladogram (Fig. 3)*
| Gene name | Chromosome | Specimen | Aberration # | Amplification | Deletion |
|---|---|---|---|---|---|
| AAA1 | chr7 | Shiraz-3T | 87 | 0.198593 | 0 |
| Shiraz-4T | 252 | 0.192083 | 0 | ||
| Shiraz-36T | 376 | 0.159952 | 0 | ||
| Shiraz-44T | 631 | 0 | −0.18031 | ||
| Shiraz-46T | 942 | 0.069143 | 0 | ||
| Shiraz-18T | 1433 | 0.250973 | 0 | ||
| Shiraz-22T | 1544 | 0 | −0.19358 | ||
| Shiraz-25T | 1697 | 0.21945 | 0 | ||
| Shiraz-28T | 1734 | 0.362882 | 0 | ||
| Shiraz-29T | 1777 | 0.156747 | 0 | ||
| Shiraz-38T | 1950 | 0.265916 | 0 | ||
| Shiraz-71T | 2236 | 0.320072 | 0 | ||
| Shiraz-3T | 87 | 0.198593 | 0 | ||
| Shiraz-4T | 252 | 0.192083 | 0 | ||
| Shiraz-36T | 376 | 0.159952 | 0 | ||
| Shiraz-44T | 631 | 0 | −0.18031 | ||
| Shiraz-46T | 942 | 0.069143 | 0 | ||
| Shiraz-18T | 1433 | 0.250973 | 0 | ||
| Shiraz-22T | 1544 | 0 | −0.19358 | ||
| Shiraz-25T | 1697 | 0.21945 | 0 | ||
| Shiraz-28T | 1734 | 0.362882 | 0 | ||
| Shiraz-29T | 1777 | 0.156747 | 0 |
See Supplementary Data 4 for full table.
General description of ChromExtractor
This program processes lists of gene names such as the lists of synapomorphies generated by SynapExtractor. ChromExtractor uses these lists to uncover the mutated chromosomes for each node of the cladogram (i.e., for each clade of specimens). Thus, it makes possible the discovery of the chromosomes contributing the shared aberrations for each clade. This chromosome information is mined from the original CGH dataset used in CGHExtractor (Supplementary Data 1) based on the synapomorphies list generated by SynapExtractor (Supplementary Data 3).
ChromExtractor: User procedure
Upon running ChromExtractor and pressing the Select Files button, the user selects a dataset CSV file (the original dataset as in Supplementary Data 1) as well as any number of CSV gene lists (the nodes' synapomorphies: lists of columns generated by SynapExtractor, each column should be separated into its own CSV file). The gene list in the CSV file has one column containing, after the header (usually an identifier for a cladogram node), a list of gene names. ChromExtractor searches the dataset file and creates a CSV output file for each gene list, where each gene name is now a header for aberration data relevant to that gene, followed by the following columns: Chromosome, Specimen, Aberration #, Amplification, and Deletion. The aberration number can be used for reference back to the original dataset.
The user will find the ChromExtractor output CSV files in the same directory as their respective input files, with the same name plus the suffix of “_output”.
ChromExtractor: Program procedure
After checking file names to determine which input files are the gene lists and which is the dataset, ChromExtractor creates a dictionary mapping genes to tables of text; ChromExtractor iterates through each gene list file in order to define the keys for the dictionary.
At first, each key maps to an empty table. ChromExtractor iterates through the dataset to populate the tables. Each aberration lists affected genes in the tenth column, so that a row of data can be written to the tables for those genes. The specific data values copied over are the chromosome identifier in the second column; the aberration number in the first column; and the amplification and deletion values in the seventh and eighth columns. The specimen identifier is also written, but from memory: ChromExtractor keeps track of the last specimen identifier it passes, since these labels occur between sections of data rather than on each aberration entry.
Once the iteration is complete, every gene present in the input gene lists maps to aberration data for every aberration in the dataset that lists that gene. ChromExtractor then writes an output CSV file for each input gene list; this output list is in the same format as the input list, but after each gene name are the rows of data extracted from the chromosome aberration file. After every output file is saved (with the same name as the corresponding input with the suffix “_output”), ChromExtractor is finished.
ChromExtractor: Program abstraction
The dataset can be characterized as a function D(x) that gives aberration data (chromosome and specimen identifiers, amplification/deletion values, etc.) for every aberration number x, ChromExtractor provides the information in an inverse format. For every gene G in an input gene list, ChromExtractor writes x as well as part of D(x) for all x where D(x) contains G.
Results
Files produced by CGHExtractor
CGHExtractor produced three files. The first is a CSV file of the polarized data matrix (Table 2; partial listing of the polarized data as 0s, 1s, and 2s; full results in Supplementary Data 2), and the second and the third are text files based on the first and used as input files for either MIX or TNT (Fig. 2A and B show partial view). In the MIX input file, the first line specifies the dimensions of the matrix, and then subsequent lines provide data values for each specimen. In the TNT input file, in addition to specifying the data matrix, the embedded commands before and after the data matrix control TNT settings; these can be modified to control the execution settings of TNT.
The cladogram
The input files generated by CGHExtractor (Fig. 2) were processed with MIX and TNT and both produced only one most parsimonious cladogram (Fig. 3). The cladogram had six major clades on the following nodes numbered 14, 20, 13, 3, 23, and 6. The cladogram arrangement is hierarchical, and the upper clades (6 and 23) share some synapomorphies with the lower clades at nodes 5, 11, and 2. In the context of aCGH data, specimens are grouped into clades according to the shared chromosomal aberrations, the synapomorphies (i.e., each node separated sister clades according to the synapomorphies of the clades).
Each of the nodes in Figure 3 had a list of synapomorphies. The bifurcation of the cladogram allows for subtyping of specimens according to their shared genetic aberrations. The cladogram and its synapomorphies are the end product of the parsimony phylogenetic analysis of MIX or TNT, and the cladogram is open for interpretation and application depending on the desired outcome.
Extracting clades' synapomorphies using SynapExtractor
The synapomorphies of each node were extracted with SynapExtractor (Salazar et al., 2015). SynapExtractor produced a CSV file listing all the nodes of the cladogram and their synapomorphies (see Supplementary Data 3).
Extracting the list of aberrant chromosomes for each node/clade using ChromExtractor
ChromExtractor used the original data file (Table 1, Supplementary Data 1) and the CSV file of SynapExtractor (Supplementary Data 3) and produced a CSV file that listed the gene name, chromosome, specimen, aberration identification number, amplification, and deletion for the selected node of the cladogram. Table 3 shows partial view for node 14 (for full file, see Supplementary Data 4). For example, the analysis showed that the synapomorphies of node 14 were generated by mutations on chromosomes 1, 4, 5, 7, 22, X, and Y, with bulk of mutations on 7, X, and 5 respectively; the other chromosomes each contributed only one synapomorphy.
Discussion
Data produced by aCGH is particularly challenging to analyze due to its volume and text format. Maximum parsimony phylogenetics is a powerful and efficient analytical technique capable of mining complex datasets that are heterogeneous and highly dimensional. The method produces a parsimonious cladogram, which is the cladogram with the minimal number of steps to construct. Maximum parsimony has been applied to a variety of biomedical omics data such as gene-expression microarray (Abu-Asab et al., 2013a; Salazar et al., 2015), mass spectrometry proteomics (Abu-Asab et al., 2006), and aCGH data (Brim et al., 2012; 2014).
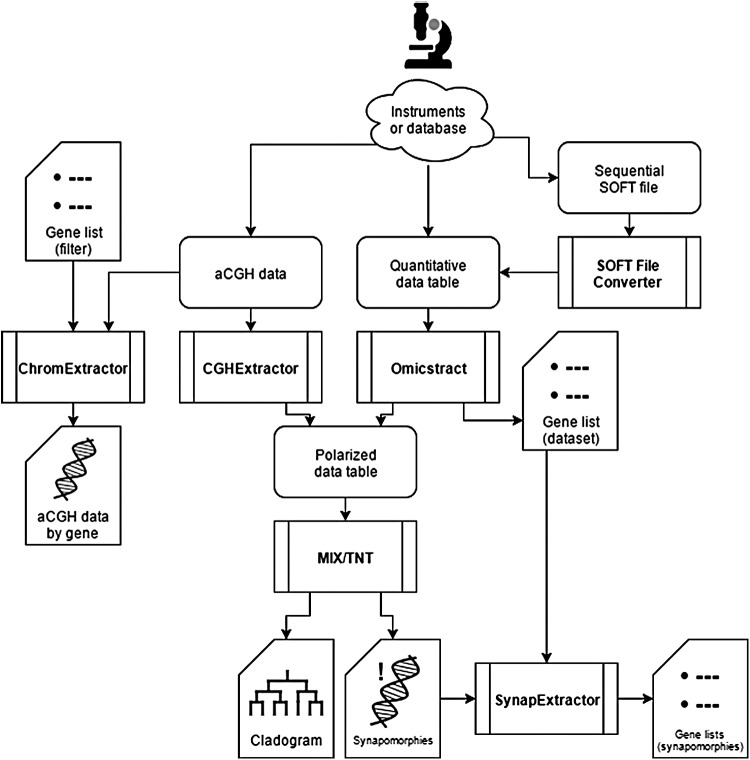
The construction of a parsimonious cladogram of aCGH data allows for the easy stratification of specimens (e.g., subtyping of a cancer type) and the identification of shared chromosomal aberrations among a group of patients, thus producing a better understanding of the disease initiation, progression, and subtypes (see Fig. 5 and Table 4 for a summary of the analytical process).
FIG. 5.
Schematic relational diagram showing our various programs used in the analytical process of aCGH and other omics data (metabolomics, proteomics, and microarrays).
Table 4.
Summary of Analytical Process of aCGH Dataset and Programs Used
| Task | Program | Output file | Files' content |
|---|---|---|---|
| 1. List and polarize aberrations of each specimen compared to total aberrations of all specimens | CGHExtractor | CSV file listing the aberrations of each specimens in a matrix format as: 1 = deletion 2 = duplication 0 = not present |
Ternary coding of aberrations in relation to complete list of aberrations in all specimens. |
| 2. Generation of input files for parsimony programs MIX and TNT | CGHExtractor | Text files in input format of MIX and TNT. | The polarized values of aberrations. Binary for MIX, and ternary for TNT |
| 3. Generation parsimonious cladograms | MIX and TNT | • MIX produces two files named “output” and “treefile.” • TNT saves its analysis in file that has .tnt extension (saving should be done by the user) |
Cladograms, and steps used to generate them |
| 4. Extraction list of synapomorphies of each node of cladogram | SynapExtractor | • CSV file with lists of synapomorphies of eachnode. • The program processes together the gene list in a CSV file and output file from MIX or TNT. |
Lists the headers of nodes from cladogram, and below each node's number a list of its synapomorphies |
| 5. Extraction of a list of chromosomes that have aberrations for each node | ChromExtractor | • CSV file with four headers per nodes. • The program processes together the CSV file from SynapExtractor and the original aCGH data file. |
Lists the following attributes: • gene name • chromosome • specimen(s) • aberration id • amplification • deletion |
Our programs, CGHExtractor, SynapExtractor, and ChromExtractor, are publicly available online for downloading at http://software.phylomics.com.
One possible use for the maximum parsimony phylogenetic analysis and its cladogram is the further narrowing of candidate driver genes for a particular disease or condition. Stratifying patients into clades of a cladogram opens new opportunities of study in functional analysis for the impact of chromosomal aberrations in driving disease progression in its various subtypes (Abu-Asab et al., 2011). The directionality of the cladogram offers insight into colorectal cancer progression with accumulating shared chromosomal aberrations (Abu-Asab et al., 2008a).
Additionally, when the cladogram topology (shape as defined by the bifurcations) and segmentation of specimens is viewed in the context of disease progression for each specimen, it is possible to separate driver genes from passenger genes in the different cancers. This is especially valuable due to the heterogeneous nature of variation in cancer specimens and saves valuable time by focusing functional studies and pathway studies on the driver genes (clonal) rather than the passenger genes (nonexpanding) (Fox et al., 2013).
Parsimony phylogenetic analysis of aCGH data may produce one best parsimonious cladogram as in our example (Fig. 3), or possibly multiple equally parsimonious cladograms. The number of equally parsimonious cladograms is dependent on the heterogeneity of the data, which could affect the robustness of the analysis since MIX and TNT may not be able to resolve some of the character states distribution among the specimens. A smaller number of equally parsimonious cladograms suggests that the genetic variants and chromosomal aberrations from the aCGH results can be organized easily amongst the different specimens. In other words, a smaller number of equally parsimonious cladograms is indicative of clearer segmentation of specimens by synapomorphies (Abu-Asab et al., 2013b).
The otherwise daunting task of organizing aCGH data into a usable format for phylogenetic analysis is simplified through the use of our novel program CGHExtractor, which converts raw chromosomal aberration data into a series of polarized values (0/1 or 0/1/2) that can be easily processed by phylogenetic programs such as MIX and TNT.
Phylogenetic analysis of aCGH data is a non-parametric method that circumvents the pitfalls and frustrations of standard analytical techniques that rely on parametric statistics. As can be seen from our data analysis, attempting to analyze such a large body of non-numerical text data statistically would have been exceptionally challenging, whereas organizing the data in a cladogram provided insights into the disease common aberrations, as well as the disease subtypes and their shared aberrations (the synapomorphies) of each subtype.
We are publishing the method and making the software suite publicly and freely available in order to make it possible for researchers to test alternative approaches to the analysis of aCGH data.
Supplementary Material
Acknowledgment
The authors thank Dr. Hassan Brim of Howard University College of Medicine for the opportunity to analyze the data. The research presented here was partially supported by the intramural program of the National Institutes of Health.
Author Disclosure Statement
The authors declare they have no conflicting financial interests.
References
- Abu-Asab M, Chaouchi M, and Amri H. (2006). Phyloproteomics: What phylogenetic analysis reveals about serum proteomics. J Proteome Res 5, 2236–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Asab M, Chaouchi M, and Amri H. (2008a). Evolutionary medicine: A meaningful connection between omics, disease, and treatment. Proteomics Clin Appl 2, 122–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Asab M, Koithan M, Shaver J, and Amri H. (2012). Analyzing heterogeneous complexity in complementary and alternative medicine research: A systems biology solution via parsimony phylogenetics. Forsch Komplementarmed 19, 42–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Asab MS, Abu-Asab N, Loffredo CA, Clarke R, and Amri H. (2013a). Identifying early events of gene expression in breast cancer with systems biology phylogenetics. Cytogen Genome Res 139, 206–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Asab MS, Chaouchi M, Alesci S, et al. (2011). Biomarkers in the age of omics: Time for a systems biology approach. Omics 15, 105–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Asab MS, Salazar J, Tuo J, and Chan CC. (2013b). Systems biology profiling of AMD on the basis of gene expression. J Ophthalmol 2013, 453934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert VA. (2005). Parsimony, Phylogeny, and Genomics. Oxford University Press, Oxford, New York [Google Scholar]
- Brim H, Abu-Asab MS, Nouraie M, et al. (2014). An integrative CGH, MSI and candidate genes methylation analysis of colorectal tumors. PloS One 9, e82185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brim H, Lee E, Abu-Asab MS, et al. (2012). Genomic aberrations in an African American colorectal cancer cohort reveals a MSI-specific profile and chromosome X amplification in male patients. PloS One 7, e40392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Uriarte R, Alibes A, Morrissey ER, Canada A, Rueda OM, and Neves ML. (2007). Asterias: Integrated analysis of expression and aCGH data using an open-source, web-based, parallelized software suite. Nucleic Acids Res 35, W75–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards-Ingram LC, Gent ME, Hoyle DC, Hayes A, Stateva LI, and Oliver SG. (2004). Comparative genomic hybridization provides new insights into the molecular taxonomy of the Saccharomyces sensu stricto complex. Genome Res 14, 1043–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. (1989). PHYLIP: Phylogeny Inference Package (Version 3.2). Cladistics 5, 164–166 [Google Scholar]
- Fox EJ, Prindle MJ, and Loeb LA. (2013). Do mutator mutations fuel tumorigenesis? Cancer Metastasis Rev 32, 353–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LB, Chae L, Kasuga T, and Taylor JW. (2011a). Array Comparative Genomic Hybridizations: Assessing the ability to recapture evolutionary relationships using an in silico approach. BMC Genomics 12, 456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LB, Kasuga T, Glass NL, and Taylor JW. (2011b). Array CGH phylogeny: How accurate are comparative genomic hybridization-based trees? BMC Genomics 12, 487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goloboff PA. (1999). Analyzing large data sets in reasonable times: Solutions for composite optima. Cladistics 15, 415–428 [DOI] [PubMed] [Google Scholar]
- Lai WR, Johnson MD, Kucherlapati R, and Park PJ. (2005). Comparative analysis of algorithms for identifying amplifications and deletions in array CGH data. Bioinformatics 21, 3763–3770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostlander AE, Meijer GA, and Ylstra B. (2004). Microarray-based comparative genomic hybridization and its applications in human genetics. Clin Genet 66, 488–495 [DOI] [PubMed] [Google Scholar]
- Picard F, Robin S, Lavielle M, Vaisse C, and Daudin JJ. (2005). A statistical approach for array CGH data analysis. BMC Bioinformatics 6, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renn SC, Machado HE, Jones A, Soneji K, Kulathinal RJ, and Hofmann HA. (2010). Using comparative genomic hybridization to survey genomic sequence divergence across species: A proof-of-concept from Drosophila. BMC Genomics 11, 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar J, Amri H, Noursi D, and Abu-Asab M. (2015). Computational tools for parsimony phylogenetic analysis of Omics data. Omics 19, 471–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van De Wiel MA, Picard F, Van Wieringen WN, and Ylstra B. (2011). Preprocessing and downstream analysis of microarray DNA copy number profiles. Brief Bioinf 12, 10–21 [DOI] [PubMed] [Google Scholar]
- Van Wieringen WN, Van De Wiel MA, and Ylstra B. (2008). Weighted clustering of called array CGH data. Biostatistics 9, 484–500 [DOI] [PubMed] [Google Scholar]
- Weiss MM, Hermsen MA, Meijer GA, et al. (1999). Comparative genomic hybridisation. Mol Path MP 52, 243–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley EO, and Lieberman BS. (2011). Phylogenetics: Theory and Practice of Phylogenetics Systematics. Wiley-Blackwell, Hoboken, N.J [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.