Abstract
Manually curating chemicals, diseases and their relationships is significantly important to biomedical research, but it is plagued by its high cost and the rapid growth of the biomedical literature. In recent years, there has been a growing interest in developing computational approaches for automatic chemical-disease relation (CDR) extraction. Despite these attempts, the lack of a comprehensive benchmarking dataset has limited the comparison of different techniques in order to assess and advance the current state-of-the-art. To this end, we organized a challenge task through BioCreative V to automatically extract CDRs from the literature. We designed two challenge tasks: disease named entity recognition (DNER) and chemical-induced disease (CID) relation extraction. To assist system development and assessment, we created a large annotated text corpus that consisted of human annotations of chemicals, diseases and their interactions from 1500 PubMed articles. 34 teams worldwide participated in the CDR task: 16 (DNER) and 18 (CID). The best systems achieved an F-score of 86.46% for the DNER task—a result that approaches the human inter-annotator agreement (0.8875)—and an F-score of 57.03% for the CID task, the highest results ever reported for such tasks. When combining team results via machine learning, the ensemble system was able to further improve over the best team results by achieving 88.89% and 62.80% in F-score for the DNER and CID task, respectively. Additionally, another novel aspect of our evaluation is to test each participating system’s ability to return real-time results: the average response time for each team’s DNER and CID web service systems were 5.6 and 9.3 s, respectively. Most teams used hybrid systems for their submissions based on machining learning. Given the level of participation and results, we found our task to be successful in engaging the text-mining research community, producing a large annotated corpus and improving the results of automatic disease recognition and CDR extraction.
Database URL: http://www.biocreative.org/tasks/biocreative-v/track-3-cdr/
Introduction and motivation
Chemicals, diseases and their relations are among the most searched topics by PubMed users worldwide (1, 2), reflecting their central roles in many areas of biomedical research and healthcare such as drug discovery and safety surveillance. Developing a drug takes time and money: on average, around 14 years and $2 billion or more (3). Greater than 95% of potential drugs fail during development for reasons such as undesired side effects due to either off-target binding or unanticipated physiologic roles of the intended target (4). Although the ultimate goal in drug discovery is to develop chemicals for therapeutics, recognition of adverse drug reactions (ADRs) between chemicals and diseases is important for improving chemical safety and toxicity studies and facilitating new screening assays for pharmaceutical compound survival. In addition, ADRs are an integral part of drug post-marketing surveillance. Identification of chemicals as biomarkers can also be helpful in informing potential relationships between chemicals and pathologies. Hence, manual annotation of such mechanistic and biomarker/correlative chemical-disease relations (CDR) from unstructured free text into structured knowledge to facilitate identification of potential toxicity has been an important theme for several bioinformatics databases, such as the Comparative Toxicogenomics Database (CTD; http://ctdbase.org/) (5). NOTE: We consider the words ‘drug’ and ‘chemical’ to be interchangeable in this document.
Manual curation of CDRs from the literature is costly and insufficient to keep up with the rapid literature growth (6). In response, there have been many attempts to extract such relations by automated natural language processing (NLP) methods. Over the years, a wide variety of relation extraction approaches have been proposed, such as simple co-occurrence, pattern matching, machine learning and knowledge-driven methods (7–9). A small number of test corpora were also developed, but they are limited in size and annotation scope (10, 11). More recently, a similar set of computational methods has been applied to a number of diverse datasets such as the FDA’s Adverse Event Reporting System (FAERS) (12), electronic medical records (13), tweets, and user comments in social media (14). In comparison, the scholarly publications contain richer information about drug-induced phenomena in a variety of settings such as in vitro and in vivo systems and across species for approved indications, off-label uses, and for drugs in development.
Despite these previous attempts and other closely related studies [e.g. PPI (15)], automatic biomedical relation detection from free text remains challenging, from identifying relevant concepts [e.g. diseases (16–19)], to extracting relations. The lack of a comprehensive benchmarking dataset has limited the comparison of different computational techniques in order to assess and improve the current state of the art. In addition, few software tools for relation extraction have been made freely available and, to the best of our knowledge, been incorporated into practical applications such as biocuration.
Materials and methods
Through BioCreative V, one of the major formal evaluation events (20) for BioNLP research, we organized a challenge task of automatic extraction of mechanistic and biomarker CDRs from the biomedical literature with the goal of supporting biocuration, new drug discovery and drug safety surveillance. More specifically, we designed two subtasks:
Disease named entity recognition (DNER). An intermediate step for automatic CDR extraction is DNER and normalization, which was found to be highly difficult on its own based on previous BioCreative CTD tasks (21, 22) and other studies (19). For the subtask, participating systems were given PubMed titles and abstracts and asked to return normalized disease concept identifiers.
Chemical-induced disease (CID) relation extraction. Participating systems were provided with titles and abstracts from PubMed articles as input (same as DNER input) and asked to return a ranked list of <chemical, disease> pairs with normalized concept identifiers for which drug-induced diseases are associated in the abstract.
Note that both chemical and diseases were described using the National Library of Medicine’s Medical Subject Headings (MeSH) controlled vocabulary (23). Systems were required to return entity pairs; both entities needed to be normalized into MeSH identifiers, along with their text spans in the article.
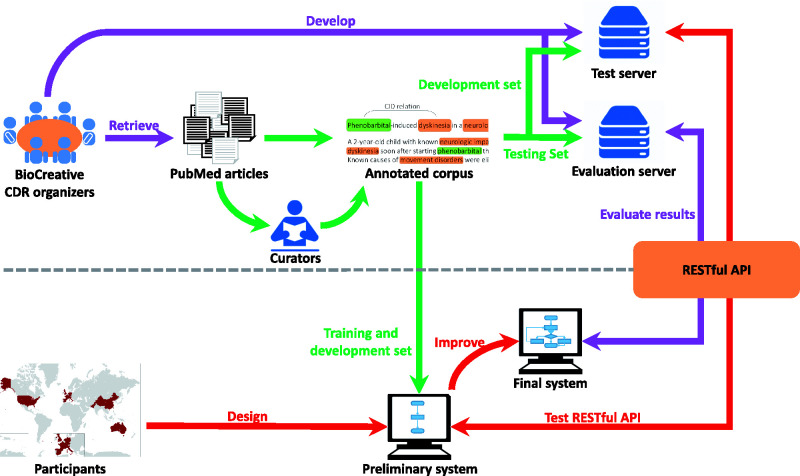
Figure 1 illustrates our task pipeline. As the task organizers, we prepared the corpus and developed an evaluation server that uses a Representational State Transfer (REST) API to submit test data to participating systems and immediately collect their results for real-time evaluation. On the participant side, teams could use any techniques to design their CDR system and were provided a test server and sample data so that they were able to iteratively improve their system before the final test.
Figure 1.
The pipeline of the task workflow. The task organization is shown in purple; corpus development is shown in green; and team participation is shown in red.
Task data
For our task, we prepared a total of 1500 PubMed articles: 500 each for the training, development and test set. Of all 1500 articles, most (1400) were selected from an existing CTD-Pfizer collaboration-related dataset (see details below). The remaining 100 articles represented completely new curation and were incorporated into the test set (The dataset can be found at http://www.biocreative.org/resources/corpora/biocreative-v-cdr-corpus/.).
For both tasks, we prepared manual annotations. For the DNER task, a number of NCBI-based MeSH annotators were recruited to annotate every disease and chemical occurrence in the abstract with both text spans and concept identifiers. We refer readers to (24) for more details regarding this annotation.
During a previous collaboration with Pfizer (6), CTD curated over 150 000 chemical-disease interactions. CTD biocurators followed CTD’s rigorous curation process and curated interactions from just the abstract whenever possible, except in cases where referencing the full text was necessary to resolve relevant issues mentioned in the abstract. For the CDR task, we mostly leveraged existing curated data from the 1400 articles. The relation data for the additional 100 articles was generated during the CDR challenge by CTD staff, and this curation was not made public until the challenge was complete.
Table 1 describes the chemical, disease and relation annotations for the three data sets. The chemical and disease mention columns are non-distinct mentions per abstract. The ID and CID relations columns are distinct per abstract.
Table 1.
Statistics of the CDR data sets
| Task dataset | Articles | Chemical |
Disease |
CID relation | ||
|---|---|---|---|---|---|---|
| Mention | ID | Mention | ID | |||
| Training | 500 | 5,203 | 1,467 | 4,182 | 1,965 | 1,038 |
| Development | 500 | 5,347 | 1,507 | 4,244 | 1,865 | 1,012 |
| Test | 500 | 5,385 | 1,435 | 4,424 | 1,988 | 1,066 |
Task evaluation
For final evaluation of the participant systems, text-mined entities (diseases) and relations (<chemical, disease> pairs) were compared to manually annotated data using standard precision, recall and F-score metrics. More specifically, the DNER results are evaluated by comparing the set of disease concepts annotated within the document with the set of disease concepts predicted by the participant system. Similarly, the CID results are evaluated by comparing the set of chemical-disease relationships annotated within the document with the set of chemical-disease relationships predicted by the system.
For results submission, participants followed the procedure implemented for the previous BioCreative-CTD task (21) where teams submitted their results through web services (We allowed offline submissions for manual runs.). In particular RESTful was selected as the architectural style for the participant web services. To assist participants, the organizers provided executable files together with a step-by-step installation guide. Also, a testing web site was provided to the teams in order to simulate the exact system-testing environment that would be used later to test the participant systems. Indeed, this testing facility was heavily used by teams during the system development phase. Because of the use of REST, we are able to report the response time of each system in addition to their accuracy.
Benchmarking systems
For comparison, we benchmarked several systems for the DNER and CID tasks.
For DNER, we first developed a straightforward dictionary look-up baseline approach that relied on disease names from CTD. We also retrained models using the out-of-box DNorm, NCBI’s previous work for DNER and normalization (16). DNorm combines an approach based on rich features and conditional random fields for named entity recognition [using BANNER (25)] with a novel machine learning method for normalization based on pairwise learning to rank (26). DNorm is a competitive system which achieved the highest performance in a previous disease challenge (17, 27); its performance therefore provides a very strong benchmark.
For the CID task, we established a baseline using a simple co-occurrence method with two variants: co-occurrence of chemicals and diseases at the abstract-level, and at the sentence-level. The chemical and disease entities were automatically recognized using NCBI’s in-house tools, DNorm (16) and tmChem (28), respectively.
Post-challenge ensemble system
To benefit from all participated systems and further improve the results, we combined the team results using an ensemble approach based on machine learning. We used one binary feature to represent each participant system output. For the DNER task, each feature represents whether the respective system returned the disease concept. For the CID task, each feature represents whether the individual system returned a <Chemical ID, Disease ID > pair. Our implementation used Support Vector Machine. We performed a 5-fold cross validation on the test set to evaluate our ensemble method.
Results
A total of 25 teams submitted 34 systems for testing in the CDR task: 16 systems were tested in conjunction with the DNER task, and 18 for the CID task. Each team was allowed to submit up to three runs (i.e. three different versions of their tool) for each task; a total of 86 runs were submitted. The 25 teams represented 12 different countries in four continents: Australia (1), Asia (12), Europe (9) and North America (3).
DNER results
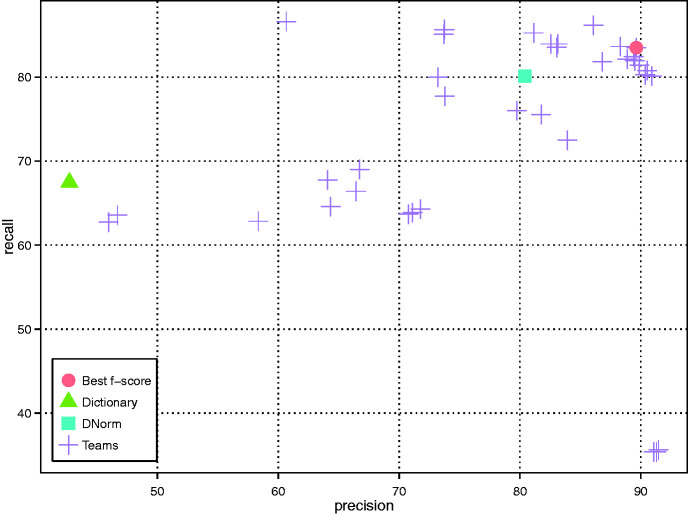
A total of 16 teams successfully submitted DNER results in 40 runs. As shown in Figure 2 (only the best run of each team is included), multiple teams achieved an F-score higher than 85% with the highest being 86.46% (Team 314), a result that approaches the inter-annotator agreement of the human annotators (0.8875) (24). The average precision, recall and F-score were 78.99%, 74.81% and 76.03%, respectively. The best precision result was obtained by using CRF model together with word2vec models trained over Wikipedia and Medline (Team 296); the best recall result was obtained by using rules and dictionary with a low level of spelling correction (Team 304); and the best F-score result was obtained by using CRF model with post-processing (Team 314). Detailed results are shown in Appendix Tables A1 and A2.
Figure 2.
DNER results of all teams as well as the baseline (dictionary look up) and DNorm systems.
All teams but one achieved a higher F-score than our baseline dictionary method, which obtained an F-score of 52.30%. While we did not perform any additional development on DNorm to adapt it to this dataset, it sets a significantly stronger benchmark with a performance of 80.64% F-score. A total of seven teams achieved performance higher than DNorm.
CID results
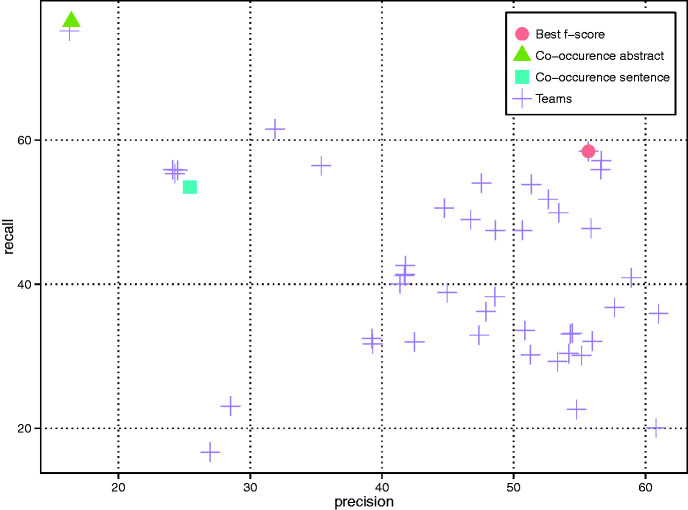
A total of 18 teams successfully submitted CID results in 46 runs. As shown in Figure 3 (only the best run of each team is included), the F-score ranges from 32.01% to 57.03% (Team 288) with an average of 43.37%. All teams outperformed the baseline results by the simple abstract-level co-occurrence method (16.43% in precision, 76.45% in recall and 27.05% in F-score). The best results were obtained by combining two Support Vector Machine (SVM) classifiers (29), which were trained on sentence and document level respectively (Team 288). Detailed results are shown in Appendix Tables A3 and A4.
Figure 3.
CID results of all teams as well as two variants of the co-occurrence baseline method (i.e. abstract- and sentence-level).
Response time results
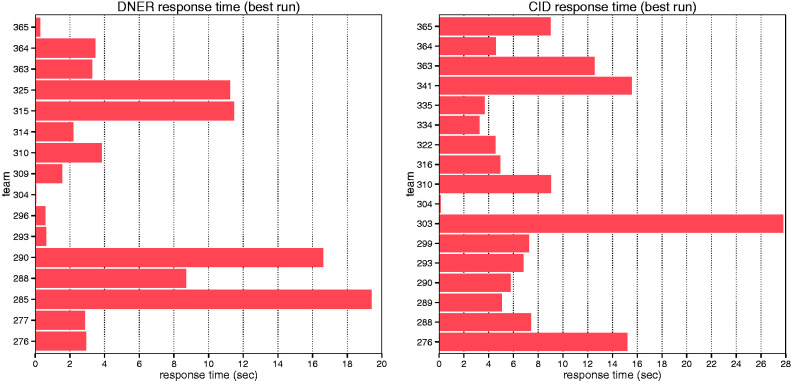
The average response time for DNER teams was 5.57 s, with a standard deviation of 6.1 (Figure 4), ranging from 0.053 to 19.4 s per request. The average response time for CID teams was 8.38 s, with a standard deviation of 6.5, ranging from 0.119 to 27.8 s. The quickest response time was obtained by Team 304, which used a rule-based system for both DNER and CID tasks.
Figure 4.
Average response time of each individual team for DNER and CID tasks.
Ensemble approach results
Tables 2 and 3 show the evaluation results of the combined DNER and CID results, respectively. The first row shows the best result of individual teams, and the second row shows an upper bound performance score by comparing independent annotations carried manually by either human curators or crowds. From the results in the third row, we observed that using the ensemble approach we were able to achieve a 2.8% and 10.1% improvement in F-score over the best individual system for DNER and CID, respectively.
Table 2.
DNER results using a combination method
| DNER | P | R | F |
|---|---|---|---|
| Best team result | 89.63 | 83.50 | 86.46 |
| Inter-annotator agreement | – | – | 88.75 |
| All teams combined (16 teams) | 93.21 | 84.96 | 88.89 |
Performance is shown in Precision (P), Recall (R) and F-score (F).
Table 3.
CID results using a combination method
| CID | P | R | F |
|---|---|---|---|
| Best team result | 55.67 | 58.44 | 57.03 |
| Crowdsourcinga | 56.10 | 76.50 | 64.70 |
| All teams combined (18 teams) | 76.45 | 53.28 | 62.80 |
Performance is shown in Precision (P), Recall (R) and F-score (F).
100 abstracts from training set. (Li T, Bravo A, Furlong LI, Good BM, Su AI, Extracting structured CID relations from free text via crowdsourcing. BioCreative V, 2015)
Discussion
The DNER task of BioCreative V showed that the automatic recognition of disease entities from PubMed abstracts is a feasible task by automated named entity recognition. To determine the difficulty of DNER and CID tasks, we examined how many teams correctly identified each of the gold standard DNER concepts and CID relations in the test set. As shown in Table 4, only ∼5% of the 1988 DNER concepts in the test set were not found by any of the teams (i.e. 95% of the concepts were retrieved by at least one team, the complete list is shown in Appendix Table A5). The 103 unrecognized mentions represented 83 distinct DNER concepts; of those 83 concepts, 26 (31.3%) did not appear in either the training or development sets.
Table 4.
Overview of how many teams correctly identified DNER concepts and CID relations
| No. of teams | DNER concepts |
CID relations |
||
|---|---|---|---|---|
| No. | % | No. | % | |
| 0 | 103 | 5.18 | 128 | 12.01 |
| 1 | 44 | 2.21 | 71 | 6.66 |
| 2 | 39 | 1.96 | 74 | 6.94 |
| 3 | 31 | 1.56 | 69 | 6.47 |
| 4 | 35 | 1.76 | 63 | 5.91 |
| 5 | 25 | 1.26 | 69 | 6.47 |
| 6 | 25 | 1.26 | 55 | 5.16 |
| 7 | 35 | 1.76 | 46 | 4.32 |
| 8 | 31 | 1.56 | 45 | 4.22 |
| 9 | 59 | 2.97 | 42 | 3.94 |
| 10 | 87 | 4.38 | 40 | 3.75 |
| 11 | 111 | 5.58 | 34 | 3.19 |
| 12 | 137 | 6.89 | 53 | 4.97 |
| 13 | 135 | 6.79 | 42 | 3.94 |
| 14 | 194 | 9.76 | 44 | 4.13 |
| 15 | 416 | 20.93 | 40 | 3.75 |
| 16 | 481 | 24.20 | 39 | 3.66 |
| 17 | — | — | 50 | 4.69 |
| 18 | — | — | 62 | 5.82 |
| Sum | 1988 | 100.00 | 1066 | 100.00 |
For the CID task, 128 of the 1066 CID pairs (12%) were not detected by any of the teams. This means that only 88% CID relations could be retrieved by at least one team, demonstrating the difficulty of the CID task. For those 128 pairs, most (92.2%) were not present in the training and development sets.
We carried out a post-challenge survey in order to better understand the commonly used participating techniques, tools and resources. Results are presented in the Appendix Table A7. Overall, most of the teams developed their hybrid systems using machine-learning techniques: primarily CRF for the DNER task and SVM for CID (30). The other two general approaches are pattern matching (CID) and dictionary look-up (DNER). Only a few teams explored the use of other machine learning techniques such as maximum entropy and logistic regression. Although machine learning-based approaches attained the highest scores in general, it’s worth noting that some rule-based systems were also highly competitive, as demonstrated by one team which ranked second and third in the DNER and CID tasks, respectively.
When developing their own systems, many teams adapted existing packages that implement machine learning algorithms (e.g. LibSVM or CRFsuite) or general NLP software (e.g. Stanford CoreNLP or OpenNLP). BioNLP software tools such as tmChem and DNorm for chemical and disease entity recognition were heavily used in the relation extraction task as a pre-step.
In terms of resources, many used external databases or terminologies with UMLS and CTD being the most commonly used resources.
Conclusions
Given the level of participation and team results, we conclude that the CDR challenge task was run successfully and is expected to make significant contributions to both the text-mining and biocuration research communities. To the best of our knowledge, the constructed corpus is the largest of its kind for both disease annotations and disease-chemical relations. In addition, our corpus includes both the text spans and normalized concept identifiers of entity annotations, as well as relation annotations, in the same abstract. We believe this data set will be invaluable in advancing text-mining techniques for relation extraction tasks. Furthermore, our annotated data includes ∼30% of the CDR relations that are asserted across sentence boundaries (i.e. not in the same sentences).
Unlike most challenge tasks in BioNLP (20), our task was designed to provide practical benefits to assist literature-based biocuration through two distinct requests: (i) all text-mined entities and relations were to be normalized to database identifiers so that they could be readily used for data curation and (ii) through web services, biocuration groups can remotely request text-mined results in real-time without additional investment in text-mining tool adoption and technical infrastructure. By doing so, we hope that the state-of-the-art will be advanced for BioNLP systems toward higher standards for interoperability and scalability in future development efforts.
Supplementary Material
Funding
This work was supported by the National Institutes of Health Intramural Research Program; National Library of Medicine and the National Institute of Environmental Health Sciences [ES014065 and ES019604].
Conflict of interest. None declared.
References
- 1. Doğan R.I., Murray G.C., Névéol A. et al. (2009) Understanding PubMed user search behavior through log analysis. Database, 2009, bap018, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Névéol A., Doğan R.I., Lu Z. (2011) Semi-automatic semantic annotation of PubMed queries: a study on quality, efficiency, satisfaction. J. Biomed. Inform., 44, 310–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li J., Zheng S., Chen B. et al. (2016) A survey of current trends in computational drug repositioning. Brief. Bioinform., 1, 2–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hurle M.R., Yang L., Xie Q. et al. (2013) Computational drug repositioning: from data to therapeutics. Clin. Pharmacol. Ther., 93, 335–341. [DOI] [PubMed] [Google Scholar]
- 5. Davis A.P., Grondin C.J., Lennon-Hopkins K. et al. (2015) The Comparative Toxicogenomics Database's 10th year anniversary: update 2015. Nucleic Acids Res., 43, D914–D920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Davis A.P., Wiegers T.C., Roberts P.M. et al. (2013) A CTD-Pfizer collaboration: manual curation of 88,000 scientific articles text mined for drug-disease and drug-phenotype interactions. Database, 2013, bat080, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kang N., Singh B., Bui C. et al. (2014) Knowledge-based extraction of adverse drug events from biomedical text. BMC Bioinform., 15(64), 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xua R., Wang Q. (2014) Automatic construction of a large-scale and accurate drug-side-effect association knowledge base from biomedical literature. J. Biomed. Inform., 51, 191–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gurulingappa H., Mateen-Rajput A., Toldo L. (2012) Extraction of potential adverse drug events from medical case reports. J. Biomed. Semant., 3(15), 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mulligen E.M., Fourrier-Reglat A., Gurwitz D. et al. (2012) The EU-ADR corpus: annotated drugs, diseases, targets, and their relationships. J. Biomed. Inform., 45, 879–884. [DOI] [PubMed] [Google Scholar]
- 11. Gurulingappa H., Rajput A.M., Roberts A. et al. (2012) Development of a benchmark corpus to support the automatic extraction of drug-related adverse effects from medical case reports. J. Biomed. Inform., 45, 885–892. [DOI] [PubMed] [Google Scholar]
- 12. Harpaz R., Vilar S., DuMouchel W. et al. (2013) Combing signals from spontaneous reports and electronic health records for detection of adverse drug reactions. J. Am. Med. Inform. Assoc., 20, 413–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Iyer S.V., Harpaz R., LePendu P. et al. (2014) Mining clinical text for signals of adverse drug-drug interactions. J. Am. Med. Inform. Assoc., 21, 353–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Leaman R., Wojtulewicz L., Sullivan R. et al. (2010) Towards internet-age pharmacovigilance: extracting adverse drug reactions from user posts to health-related social networks. In: Proceedings of the 2010 Workshop on Biomedical Natural Language Processing. Association for Computational Linguistics, Uppsala, Sweden. pp. 117–125.
- 15. Krallinger M., Vazquez M., Leitner F. et al. (2011) The protein-protein interaction tasks of BioCreative III: classification/ranking of articles and linking bio-ontology concepts to full text. BMC Bioinform., 12(Suppl 8):S3, 1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Leaman R., Doğan R.I., Lu Z. (2013) DNorm: disease name normalization with pairwise learning to rank. Bioinformatics, 29, 2909–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Leaman R., Khare R., Lu Z. (2013) NCBI at 2013 ShARe/CLEF shared task: disorder normalization in clinical notes with DNorm. In: Proceedings of the CLEF 2013 Evaluation Labs and Workshop, Valencia, Spain.
- 18. Doğan R.I., Leaman R., Lu Z. (2014) NCBI disease corpus: a resource for disease name recognition and concept normalization. J. Biomed. Inform., 47, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Leaman R., Khare R., Lu Z. (2015) Challenges in clinical natural language processing for automated disorder normalization. J. Biomed. Inform., 57, 28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang C.C., Lu Z. (2015) Community challenges in biomedical text mining over 10 years: success, failure and the future. Brief. Bioinform., 17(1), 132–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wiegers T.C., Davis A.P., Mattingly C.J. (2014) Web services-based text-mining demonstrates broad impacts for interoperability and process simplification. Database, 2014, bau050, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wiegers T.C., Davis A.P., Mattingly C.J. (2012) Collaborative biocuration-text-mining development task for document prioritization for curation. Database, 2012, bas037, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Coletti M.H., Bleich H.L. (2001) Medical subject headings used to search the biomedical literature. J. Am. Med. Inform. Assoc., 8, 317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li J., Sun Y., Johnson R.J. et al. (2015) Annotating chemicals, diseases and their interactions in biomedical literature. In: Proceedings of the Fifth BioCreative Challenge Evaluation Workshop, Sevilla, Spain. pp. 173–182.
- 25. Leaman R., Gonzalez G. (2008) BANNER: an executable survey of advances in biomedical named entity recognition. In: Pacific Symposium on Biocomputing, Fairmont Orchid, Hawaii, USA. pp. 652–663. [PubMed]
- 26. Burges C., Shaked T., Renshaw E. et al. (2005) Learning to rank using gradient descent. In: International Conference on Machine Learning. ACM, Bonn, Germany. pp. 89–96.
- 27. Pradhan S., Elhadad N., South B.R. et al. (2014) Evaluating the state of the art in disorder recognition and normalization of the clinical narrative. J. Am. Med. Inform. Assoc., 22(1), 143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Leaman R., Wei C.H., Lu Z. (2015) tmChem: a high performance approach for chemical named entity recognition and normalization. J. Cheminform., 7(Suppl 3):S3, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cortes C., Vapnik V. (1995) Support-vector networks. Mach. Learn., 20, 273–297. [Google Scholar]
- 30. Lafferty J., McCallum A., Pereira F. (2001) Conditional random fields: probabilistic models for segmenting and labeling sequence data. In: Proceedings of the International Conference on Machine Learning (ICML 01), Bellevue, Washington, USA. pp. 282–289.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.