Abstract
In the process of seeking novel lung host defense regulators by analyzing genome-wide RNA sequence data from normal human airway epithelium, we detected expression of POU2AF1, a known transcription co-factor previously thought to be expressed only in lymphocytes. Lymphocyte contamination of human airway epithelial samples obtained by bronchoscopy and brushing was excluded by immunohistochemistry staining, the observation of up-regulation of POU2AF1 in purified airway basal stem/progenitor cells undergoing differentiation and analysis of differentiating single basal cell clones. Lentivirus-mediated up-regulation of POU2AF1 in airway basal cells induced up-regulation of host defense genes, including MX1, IFIT3, IFITM and known POU2AF1 downstream genes HLA-DRA, ID2, ID3, IL6, BCL6. Interestingly, expression of these genes paralleled changes of POU2AF1 expression during airway epithelium differentiation in vitro, suggesting POU2AF1 helps to maintain a “host defense tone” even in pathogen-free condition. Cigarette smoke, a known risk factor for airway infection, suppressed POU2AF1 expression both in vivo in humans and in vitro in human airway epithelial cultures, accompanied by deregulation of POU2AF1 downstream genes. Finally, enhancing POU2AF1 expression in human airway epithelium attenuated the suppression of host defense genes by smoking. Together, these findings suggest a novel function of POU2AF1 as a potential regulator of host defense genes in the human airway epithelium.
Introduction
The airway epithelium functions as a complex barrier that serves as the front line of the host defense system of the lung1. In response to inhaled pathogens, airway epithelial cells generate two major types of host defense molecules: (1) intracellular or extra-cellular molecules with direct anti-pathogen effects, such as reactive oxygen species, antimicrobial peptides and proteins suppressing intracellular pathogen amplification and spreading; and (2) surface or extracellular molecules regulating the interactions between epithelial cells and inflammatory cells, including cytokines, chemokines, MHC-II genes, complement and adhesion proteins1-3. While interacting with receptors such as the Toll-like receptor family triggers activation of discrete parts of pulmonary host defense, the overall regulation of this diverse airway epithelial host defense system is not well understood.
In a genome-wide RNA sequencing analysis of human airway epithelium gene expression, we observed expression of POU domain class 2-associating factor 1 (POU2AF1; also named OCA-B, OBF-1 and BOB.1), a gene previously considered to be expressed specifically in lymphocytes, where it functions as a co-activator of octamer-binding OCT1 and OCT2 transcription factors to regulate immunoglobin expression and other host defense-related genes4-6. POU2AF1 has no intrinsic DNA binding activity, but by recognizing the POU domain of OCT1 and OCT2, it plays an essential role in B-cell responses to antigens and is required for the formation of germinal centers7-9.
Since POU2AF1 function in B cells relates to host defense, with the surprising observation that POU2AF1 is expressed in the airway epithelium, a cell population critical to pulmonary host defense, we asked: (1) is POU2AF1 really expressed in the human airway epithelium, not in lymphocytes contaminating the airway epithelial samples; and (2) if so, does POU2AF1 play a role in the airway epithelium by broadly regulating the expression of genes relevant to host defense as it does in B cells? Using in vivo and in vitro analyses, the data demonstrates that POU2AF1 is expressed in the human airway epithelium, it is normally up-regulated as airway basal stem/progenitor cells differentiate to a mucociliated epithelium and the expression of POU2AF1 in the airway epithelium correlates with the expression of a large number of genes relevant to airway epithelium host defense. Interestingly, cigarette smoking, a stress to the airway epithelium known to impair host defense, down-regulates airway epithelial expression of POU2AF1, and up-regulation of POU2AF1 in airway epithelium can attenuate smoking-related down-regulation of airway epithelial host-defense genes.
Methods
Human Airway Epithelial Cells
Airway epithelial cells were collected by brushing the epithelium using flexible bronchoscopy as previously described10. Subjects were recruited under a protocol approved by the Weill Cornell Medical College and Rockefeller University Institutional Review Boards Review Board, with written informed consent obtained from each volunteer before enrollment in the study.
The human airway basal cells (defined by expression of basal cells markers, lacking expression of ciliated and secretory cell markers and self-renewal and differentiation capacity) were obtained either from Lonza (Walkersville, MD) or isolated from brushed airway epithelial cells. The basal cell clones derived from a single cell were isolated with sterile cloning cylinders. The mucocilliary differentiation of airway basal cells was induced on ALI as described previously11. The BCi-NS1.1 immortalized human airway basal cell line has been reported previously12. The effect of cigarette smoke on gene expression was assessed on ALI model with CSE exposure, as described previously13,14.
Gene Expression Analysis of Clinical Samples
To assess normal POU2AF1 gene expression in vivo, RNA-Seq analysis was carried out on small airway epithelium [small airway epithelium (SAE), 10th to 12th order bronchi] from 5 healthy nonsmokers. The raw data has been deposited in the Short Read Archive (SRA) section of the NCBI SRA database (SRP005411). Data of POU2AF1 and all transcription factors with expression level >0.125 Reads per kilobase of exon per million mapped reads (RPKM) detection limit were extracted for comparison.
To assess the effect of smoking on POU2AF1 and its downstream gene expression in vivo, microarray data (Affymetrix U133 plus 2 microarray) from two SAE cohorts (cohort I, 60 healthy nonsmokers, 71 healthy smokers; cohort II, 16 healthy nonsmokers, 20 healthy smokers) were used. The raw data used for expression assessment has been deposited in the Gene Expression Omnibus site (GEO; GSE17905 and GSE43079; http://www.ncbi.nlm.nih.gov/geo/). POU2AF1 probeset 1569675_at was used. To confirm POU2AF1 expression change from the microarray study, SAE from randomly selected subjects (17 healthy nonsmokers, 21 healthy smokers) were used for TaqMan PCR validation.
To assess the expression of POU2AF1 downstream genes in undifferentiated human airway basal cells and differentiated airway epithelium in vivo, 12 large airway epithelial samples obtained by brushing and 5 purified large airway basal cells (all from healthy nonsmokers) were evaluated using Affymetrix U133 plus 2.0 microarrays. The raw data is public available in GEO (GSE24337; http://www.ncbi.nlm.nih.gov/geo/).
All microarray data were MAS5-processed and normalized by “per chip only” using GeneSpring version 7.3.1 (Agilent Technologies, Palo Alto, CA). Both hierarchical analysis and principal component analysis (PCA) were performed in Partek Genomics Suite (Partek, St. Louis, Missouri). Euclidean distance metric was used for hierarchical cluster analysis. Covariance matrix was used for PCA analysis.
Lentivirus Production and Infection
Lenti-POU2AF1, lenti-RFP (red fluorescent protein, as a control), lenti-KLF4 (as a control) expressing lentiviral plasmids were from Thermo Scientific Inc (Pittsburgh, PA). Lenti-OSGIN1 (as a control) was generated in our laboratory using standard methods. These vectors have some shared components including IRES, GFP, blasticidin S resistance gene and WPRE. The replication deficient lentiviruses were generated in 293A cells using compatible packaging vectors. The infectious titer of each virus was determined by flow cytometry via GFP positivity following infection of HT1080 cells (ATCC, Manassas, VA). For infection of the basal cells, recombinant lentiviruses at an equal multiplicity of infection (MOI) were added with 6 μg/ml of polybrene to aid viral infection. The media were changed every day. As part of quality control, to compare the transduction efficiency of different lentiviral vectors in basal cells, TaqMan PCR was used to assess the common lentiviral component WPRE at mRNA level in each sample after 3 days of infection.
RNA-Seq Data Analysis of Basal Cells with POU2AF1 Overexpression
Lonza airway basal cells (lot 314700, passage 1) were cultured in 24 well plate and then infected with lenti-POU2AF1 or lenti-RFP. Uninfected basal cells were used as control. After 72 hr infection (3 wells per group), the samples were collected and processed for RNA-seq. Samples were loaded onto an Illumina flowcell for paired-end sequencing reactions using the Illumina HiSeq 2000 as previously detailed15. The raw data has been deposited in the NCBI SRA database (; GSE60989; http://www.ncbi.nlm.nih.gov/geo/).
In order to obtain comparable quantification measurements between the lentivirus vector and POU2AF1-ORF, a custom Python script was written to process the alignments using Samtools to extract the read accounts corresponding to the POU2AF1-ORF and the lentivirus vector transcripts.
Only human protein encoding genes with average expression level >0.04 FPKM in at least one group were used for further analysis. The 2-tailed Student's t-test followed by multiple test correction (step up method, Partek Genome Suite) was used. A p value less than p< 0.05 was deemed significant. The top 50 genes induced by POU2AF1 overexpression in basal cells were determined by the fold-change of the average expression level of lenti-POU2AF1 vs lenti-RFP group. The hierarchical analysis and heatmap plotting were performed in Partek Genomics Suite using the Euclidean distance metric. The Gene ontology (GO) enrichment analysis (http://gather.genome.duke.edu/) was used to determine the dominant function of the top 50 genes induced by POU2AF1. Based on the enrichment analysis results, to get up to date information, the function of each gene was further assessed by a manual literature search.
Lentivirus-infected ALI Culture Followed by CSE Treatment
For infection of the basal cells on ALI cultures, recombinant lentiviruses at an equal multiplicity of infection (MOI) were added at the time of seeding the cells on transwell inserts (ALI-day −2). The following day, the infectious media was removed and the ALI culturing protocol continued as described above. In all experiments, the cells were infected with an MOI that allowed for ~90% infection of the cells as determined by GFP positivity. Cells were exposed to CSE (3%) between days 0 and 14 from the basolateral side of the transwell inserts. The medium was changed every 2 days, and each time a fresh CSE aliquot was thawed and diluted accordingly.
TaqMan Real-Time PCR, Staining and Western Analyses
Standard methods were used. Only TaqMan probes with best coverage were used (Life Technologies, Grand Island, NY). Antibodies are listed in Table S2.
Statistical Analysis
A 2-tailed Student’s t-test was used for statistical analyses of all in vitro experiments. In all analyses, a p value less than p<0.05 was deemed significant, unless specified. The analysis detail of microarray and RNAseq were described in the corresponding section above.
Results
Identification of POU2AF1 as a Human Airway Epithelium Gene
RNA-Seq has low noise and high specificity, and, importantly, provides quantitative information on mRNA transcript number16. Unexpectedly, in the process of seeking novel host defense regulators in the human airway epithelium, we detected expression in airway epithelium RNA-Seq data of POU2AF1, a transcription cofactor generally regarded as lymphocyte-restricted4,6 (Figure S1A). The expression of POU2AF1 in the human airway epithelium has an average of 6 RPKM, close to the average expression level of all the transcription factors in the airway epithelium (Figure S1B). Using a POU2AF1 antibody17, we documented POU2AF1 expression at the protein level is expressed in human tracheobronchial epithelium, with enriched expression in intermediate cells with elongated morphology and some ciliated cells (Figure S1C).
Ruling Out Lymphocyte Contamination as Source of POU2AF1 Expression
To rule out the possibility of lymphocyte contamination in the airway epithelium as a source of the POU2AF1 expression, we reasoned that gene expression of fully differentiated-airway epithelium derived from pure airway basal cells should be free of lymphocyte transcripts. Tracheal epithelium were brushed from 3 healthy nonsmokers and cultured in a flask to purify and expand airway basal cells. These cells were negative for the B cell maker CD20 and CD79B, while positive for the basal cell makers KRT5 and TP63 (not shown). The purified basal cells were then cultured on ALI to induce differentiation. By microarray analysis, the secretory cell marker gene SCGB1A1 (CC10) and ciliated cell marker gene DNAH5 were up-regulated during differentiation over time (Figure 1A), indicating successful mucociliary differentiation. The induction of differentiation was further supported by the cilia (beta tubulin) and secretory cell (SCGB1A1) staining (not shown). The microarray data demonstrated that POU2AF1 was up-regulated on ALI-day 7, reaching a peak on ALI-day21 (Figure 1B), slightly later than the peak of SCGB1A1 and DNAH5.
Figure 1.
Evidence that POU2AF1 expression in the airway epithelium is not secondary to lymphocyte contamination. A, B. Up-regulation of POU2AF1 during airway epithelium differentiation. Basal cells purified from brushed tracheal epithelial cells were cultured on an air liquid interface (ALI) to induce differentiation (n=3 subjects, 6 time points). Gene expression over time was assessed by Affymetrix U133 plus 2.0 microarrays. A. Control expression of differentiation-specific genes; SCGB1A1 (secretory cells), DNAH5 (ciliated cells). B. POU2AF1 expression. C, D. POU2AF1 expression in ALI differentiated cells derived from single clones of basal cells. Single-clone-derived basal cells were generated and cultured on ALI to induce differentiation. Gene expressions were assessed by TaqMan PCR. C. Single basal cell clone formation. One basal cell clone was tracked by phase microscopy from day 0 to 3. On day 8, the clone was stained with basal cell marker TP63 (green). D. Gene expression change of single clone-derived basal cells on ALI culture (n=8) compared to basal cells not plated on ALI (n=3). Shown are expression of various genes in basal cells alone vs. basal cells cultured on ALI. SCGB1A1 (secretory cells), DNAH5 (ciliated cells) and POU2AF1 are expressed in the differentiated cells derived from a single basal cell clone, but not basal cells. There is no expression of the B cell markers CD20 and CD79B or the T cell marker CD3E in the basal cells or the differentiated basal cells. (# = undetectable). E. Up-regulation of POU2AF1 in the BCi-NS1.1 immortalized human basal cell line during differentiation. ALI-day 0 and ALI-day 28 samples were assessed (5 paired samples). The secretory cell marker SCGB1A1 and ciliated cell marker DNAH5 was used as positive control for differentiation. All panels, data are mean ± standard deviation. **, p<0.01; ***, p<0.001.
As lymphocytes might survive in the culture medium of airway epithelial cells18, to further rule out minimum lymphocyte contamination, we induced airway epithelium differentiation from single-cell-derived basal cell clones (Figure 1C). Using TaqMan analysis, we confirmed that the ALI cultures demonstrated up-regulation of the secretory cell marker gene SCGB1A1, ciliated cell marker gene DNAH5 and POU2AF1 (Figure 1D). Importantly, there was no expression of B cell markers CD20, CD79B nor the T cell marker CD3E (Figure 1D). Finally, and consistent with this data, POU2AF1 was also up-regulated in an immortalized airway basal cell line12 during differentiation on ALI culture (Figure 1E). Together, these data provide compelling evidence that POU2AF1 is not only expressed in human airway epithelium but also up-regulated during airway epithelium differentiation.
Screening Downstream Genes of POU2AF1 in Human Airway Epithelial Cells
Given that POU2AF1 binds to OCT1/OCT2 to exert its biologic effect, we assessed OCT1/OCT2 expression in the human airway epithelium. The RNA-Seq data demonstrated that expression level of OCT1 and OCT2 were 28% and 17% respectively among all airway epithelial transcription factors, Western analysis confirmed that both OCT1 and OCT2 were expressed in human airway epithelial cells (not shown). Consistent with this data, immunofluorescence staining showed both OCT1 and OCT2 localized in the nucleus of airway epithelial cells (not shown).
As airway basal cells are undifferentiated and negative for POU2AF1 expression, we sought to use lentivirus-mediated POU2AF1 overexpression in basal cells as a sensitive way to screen for potential POU2AF1 downstream genes in airway epithelial cells. We achieved 90% gene transduction efficiency using lentiviral vectors (not shown). There was strong POU2AF1 staining (both cytoplasm and nucleus) in lenti-POU2AF1 infected basal cells (not shown). RNA-Seq was used to quantify the gene expression changes induced by POU2AF1. As a quality control to demonstrate that similar doses of gene delivery vehicles were used, we analyzed the expression levels of common components from the lentiviral vectors. The RNA-Seq data showed that while there were striking differences between POU2AF1 and RFP mRNA levels, expression of common components (IRES, GFP, blasticidin S resistance gene, WPRE) were similar between the lenti-POU2AF1 group and lenti-RFP group (Figure 2A). TaqMan PCR of WPRE further confirmed this finding (Figure 2B).
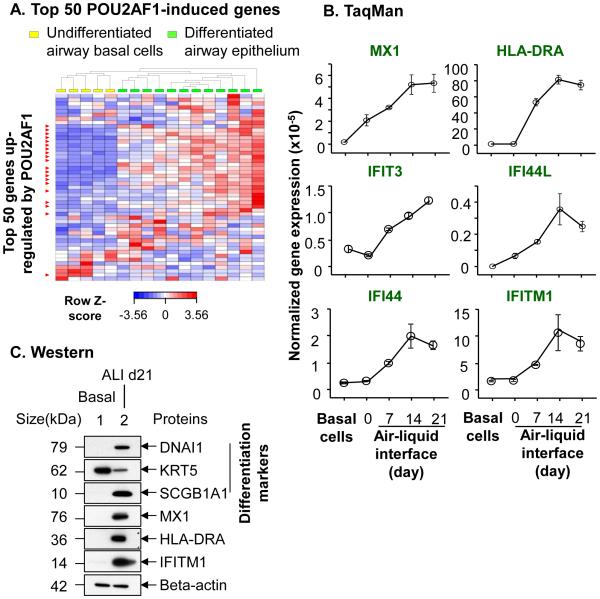
Figure 2.
POU2AF1-mediated up-regulation of host defense genes in human airway epithelial cells. Human primary airway basal cells were infected with a lenti-POU2AF1 or lenti-RFP (control) virus, using uninfected basal cells as a further control, with 3 samples per group. The basal cells were assessed after 3 days. A. mRNA expression of the lentivirus vector components. Top-schematic of the lentivirus vectors. The lenti-POU2AF1 and lenti-RFP virus had similar expression of the common components IRES, GFP, blasticidin S resistance gene and WPRE; the RFP vector expressed only RFP, not POU2AF1, while the POU2AF1 vector expressed POU2AF1, not RFP. B. TaqMan PCR assessment of WPRE expression. Both Lenti-POU2AF1 and Lenti-RFP groups had a similar expression level of WPRE. C. Unsupervised hierarchical clustering of the top 50 genes induced by POU2AF1. The top 50 genes were selected based upon the fold-changes between lenti-POU2AF1 and lenti-RFP viruses. The genes with a role in host defense are highlighted in red; 74% of the top 50 genes are host defense-related. D. Western analysis validation of POU2AF1 induced host defense genes. Examples include MX1, IFIT3 and IFITM1. GFP, a shared component of the lenti-RFP and lenti-POU2AF1vectors was used as control for the transduction efficiency and beta-actin as the loading control. E. Independent validation of POU2AF1 modulated expression of downstream genes. Airway basal cells from a different subject than panels C, D were used, with 3 samples per group. Six POU2AF1 induced genes (IFI44, HLA-DRA, IFIT3, IFITM1, IFI44L and MX1) were assessed by TaqMan PCR and the results assessed by unsupervised hierarchical clustering. Two additional vectors, lenti-KLF4 and lenti-OSGIN1 were used as controls to exclude unspecific effects.
In addition to modulation of the IgG gene in B cells, POU2AF1 has multiple known downstream genes with diverse functions in lymphocytes, such as cellular metabolism, cell survival, surface receptors, ion transport and cell adhesion4-6. Many of these genes have roles in host defense. We reviewed the literature and made an up-to-date POU2AF1 downstream gene list (not shown). Consistent with the literature4-6,19, POU2AF1 induced significant expression differences of known POU2AF1 downstream genes compared to lenti-RFP and uninfected controls (Figure S2), including ID219, ID319, HLA-DRA20, IL621, S100A1022 and KCNN422. Unsupervised hierarchical analysis of the top 50 genes up-regulated by POU2AF1 clearly segregated the lenti-POU2AF1 group from the control groups (lenti-RFP and uninfected; Figure 2C). Remarkably, gene ontology enrichment analysis revealed that “immune response,” “response to biotic stimulus” and “defense response” categories (all p<10−4) were the major functions of the top 50 genes. Assessment of gene functions demonstrated that 74% (37/50) of the genes (Table S1, also highlighted in Figure 2C) have host defense-related functions. These can be categorized as intracellular pathogen response (e.g., MX1, 144-fold; IFIT3, 79-fold; IFITM1, 32-fold); extracellular/surface host defense (e.g., CD53, 50-fold); cytokines and chemokines (CXCL10, 31-fold, IFNB1, 29-fold); complement (C1R, 23-fold); antigen presentation (HLA-DRA, 22-fold) and negative regulator of host defense response (USP18, 21-fold).
To validate the host defense genes induced by POU2AF1 at the protein level, Western analysis demonstrated host defense molecules MX1, IFIT3, and IFTM1, all were strikingly upregulated by POU2AF1 overexpression (Figure 2D). In contrast, there is no expression difference of GFP, a common component of the lenti-RFP and lenti-POU2AF1 vectors, suggesting similar efficiency of gene transduction was achieved. To further exclude nonspecific effects (e.g., genetic background or unsuitable controls), we repeated POU2AF1 overexpression study in basal cells from a different subject. Two more unrelated lentivirus vectors (lenti-KLF4 and lenti-OSGIN1) were used as additional controls. The results of TaqMan PCR confirmed that POU2AF1 induced marked up-regulation of host defense genes (Figure 2E). Similar results were achieved from two additional independent experiments.
To further exclude confounding factors related to the lentivirus preparations, independent preparations of lenti-POU2AF1 and lenti-GFP (same expression cassette as lenti-POU2AF1, expressing GFP only) viruses were prepared and used to assess host defense genes induction. TaqMan PCR confirmed our previous finding that lenti-POU2AF1 can up-regulate MX1, IFI44, IFI44L and IFITM1 (not shown). Collectively, these data suggest that POU2AF1 functions to broadly regulate host defense response genes in human airway epithelium.
Up-regulation of POU2AF1-regulated Host Defense Genes during Airway Epithelium Differentiation
As POU2AF1 is up-regulated during airway epithelium differentiation, we asked whether the host defense genes regulated by POU2AF1 have orchestrated changes in gene expression. By comparing the expression profile of 12 brushed large airway epithelium (fully differentiated) vs 5 cultured large airway basal cells (undifferentiated), we found that more than half of the POU2AF1-regulated host defense genes had significantly higher expression in the differentiated airway epithelium and clearly divided the differentiated and undifferentiated airway epithelium into two populations (Figure 3A). This observation was further assessed in the time course study of airway epithelium differentiation in vitro. In support of the microarray data, TaqMan PCR confirmed that MX1, HLA-DRA, IFIT3, IFI44, IFI44L and IFITM1 were up-regulated during airway epithelium differentiation over time (Figure 3B). Consistently, Western analysis proved that there was striking up-regulation of MX1, HLA-DRA and IFITM1 during airway epithelium differentiation (Figure 3C). Together, these data suggest that POU2AF1 is involved in maintaining a host defense “tone” in human airway epithelium.
Figure 3.
Assessment of in vitro differentiated human airway epithelium for the expression of the downstream genes induced by up-regulation of POU2AF1 in airway basal cells. A. Comparison of unsupervised hierarchical clustering of differentiated airway epithelium vs undifferentiated airway epithelium for the top 50 genes induced by lentivirus-mediated POU2AF1 up-regulation in basal cells. Affymetrix U133 plus 2 microarray data from 12 brushed large airway epithelium vs 5 cultured large airway basal cells were assessed. Shown in red are the significantly changed (p<0.05 after Benjamini and Hochberg correction) host defense genes, representing 40% of the top 50 POU2AF1 up-regulated genes. B. Validation of the microarray data using TaqMan analysis of POU2AF1-upregulated genes during airway epithelium differentiation over time. mRNAs from basal cells and matched ALI-day 0, day 7, day 14 and day 21 samples were used. Four POU2AF1 up-regulated host defense-related genes (MX1, HLA-DRA, IFIT3, IFI44, IFI44L and IFITM1) were assessed. 18s rRNA was used as an endogenous control. Shown is the result from one experiment, n=3 samples per time point. Data are mean ± standard deviation. Experiments were repeated 3 times with similar results. C. Western analysis of POU2AF1-upregulated genes during airway epithelium differentiation. Protein from basal cells and matched ALI-day 21 sample were assessed. Markers of ciliated (DNAI1), secretory (SCGB1A1) and basal (KRT5) cells were used as differentiation controls. Beta-actin was used as a loading control. Three POU2AF1 up-regulated host defense-related genes (MX1, HLA-DRA and IFITM1) were assessed. Experiments were repeated twice using cells from different subjects.
Down-regulation of POU2AF1 by Cigarette Smoking in Human Airway Epithelium
Given that smoking is one of the major environmental factors affecting disease-related airway epithelium differentiation and a known insult having a deleterious effect on the lung host defense response, we postulated that expression of POU2AF1 might be regulated by cigarette smoking. To determine whether cigarette smoke has a direct effect on POU2AF1 expression, we used cigarette smoke extract (CSE) to treat airway epithelium on ALI culture in vitro13,14. As a control, CSE induced up-regulation of the oxidative stress gene CYP1A1 (Figure 4, left panel). Interestingly, POU2AF1 expression was suppressed by CSE in a dose-dependent manner (Figure 4, right panel). Consistently, immunofluorescence staining demonstrated that CSE treatment suppressed POU2AF1 expression in airway epithelium on ALI, which was accompanied with loss of mucociliary differentiation as evidenced by the absence of β-tubulin IV (cilia marker) staining (Figure 4B).
Figure 4.
Down-regulation of the expression of POU2AF1 in the airway epithelium by cigarette smoking. A, B. Down-regulation of POU2AF1 by smoking in human airway epithelium in vitro. Airway epithelium derived from airway basal cells were cultured on ALI and treated with cigarette smoke extract (CSE) at different concentrations (0 to 6%). A. Gene expression changes assessed by TaqMan PCR. Left panel, CYP1A1, a smoking-related oxidative stress gene, was used as a control; Right panel, POU2AF1 expression. The data is a summary from 3 independent experiments. B. Protein level changes assessed by immunofluorescent staining. Red-POU2AF1; green-beta tublin IV (cilia marker); blue-nuclei. Same exposure time and image adjustment were used for microscopy. Bar = 20 μm. Top panel, control, no CSE treatment; Bottom panel, 6% CSE. Similar results were achieved from 3 independent experiments. C.D. In vivo down-regulation of POU2AF1 in the human airway epithelium by smoking. Human small airway epithelium was collected from healthy nonsmokers and healthy smokers by bronchoscopy. Gene expression was assessed by microarray. Each dot represents one subject. The p value was calculated after Benjamini and Hochberg correction of genome-wide comparison. C. POU2AF1 in small airway epithelium, microarray, initial cohort (60 healthy nonsmokers, 71 healthy smokers). D. POU2AF1 in small airway epithelium, microarray, validation cohort (16 healthy nonsmokers, 20 healthy smokers). E. TaqMan PCR validation of POU2AF1 expression of nonsmokers vs. small airway epithelium. Data is from 17 healthy nonsmokers, 21 healthy smokers. F. G. Principal component analysis of POU2AF1 downstream genes in the airway epithelium of nonsmokers and smokers. The list of the top 50 POU2AF1 downstream genes was from Figure 3C. The study population was the same as that in Figure 5C, D. F. PCA analysis, expression pattern of POU2AF1 and its downstream genes, initial cohort. G. PCA analysis, expression pattern of POU2AF1 and its downstream genes, validation cohort. Data represents mean ± standard deviation. Statistics was calculated by 2 tailed Student’s t-test. **, p<0.01; ***, p<0.001, ****, p<0.0001.
To assess the effect of smoking on POU2AF1 expression on the human airway in vivo, we used microarray to assess POU2AF1 expression in brushed airway epithelium on smokers compared to nonsmokers23. Consistent with the in vitro finding, smoking caused significantly down-regulated POU2AF1 expression in our initial cohort (−1.7 fold, p<10−4, Figure 4C). The down-regulation of POU2AF1 was unexpected as two known smoking-heightened pathways, NF-Kappa B and the endoplasmic reticulum stress (ER stress) are positive regulators of POU2AF1 expression in lymphocytes24,25. To verify these results, we assessed POU2AF1 expression in an independent microarray cohort of small airway epithelium26, observing a 1.9-fold down-regulation of POU2AF1 in smokers (Figure 4D). Furthermore, using a different technique (TaqMan PCR), down-regulation of POU2AF1 in the small airway epithelium by smoking was confirmed (3.4-fold down-regulated, p<10−4; Figure 4E).
We then asked whether POU2AF1-regulated host defense genes are globally deregulated by smoking. Interestingly, in both the initial and in a validation airway epithelium microarray cohort 23,26, expression of POU2AF1 and its downstream host defense genes segregated smokers and nonsmokers into two populations (Figure 4F, 4G), suggesting smoking disorders the POU2AF1-regulated host defense gene network in the human airway epithelium. Based on these data, we conclude that, in addition to previous known stimuli4,6, POU2AF1 gene expression is also regulated by cigarette smoking, and impaired expression of POU2AF1 likely contributes to the disordered airway host defense associated with smoking.
Attenuation of Smoking-induced Downregulation of Host Defense Genes by POU2AF1
Next, we assessed whether POU2AF1 can reverse the smoking-induced down-regulation of host defense genes using ALI cultures with CSE treatment, with lentivirus vectors used to mediate sustained expression of POU2AF1 in the ALI cultures. We focused on three POU2AF1-regulated host defense genes, MX1, HLA-DRA and IFITM1, all of which have good antibodies available for detection of expression. As expected, CSE exposure suppressed expression of MX1 (Figure 5A), HLA-DRA (Figure 5B) and IFITM1 (Figure 5C) at the mRNA level. Importantly, sustained POU2AF1 expression attenuated the smoking-induced down-regulation of MX1 and HLA-DRA (Figure 5A, B). Western analysis further confirmed these findings (Figure 5D). For IFIM1, the attenuation was more obvious at the protein level than the mRNA level (Figure 5C, D).
Figure 5.
Attenuation of CSE-induced down-regulation of host defense genes in airway epithelium by up-regulation of POU2AF1. Airway epithelium derived from basal cells on ALI with or without POU2AF1 overexpression were treated with 3% CSE. A-D. The expression of POU2AF1 regulated host defense genes (MX1, HLA-DRA and IFITM1) were assessed by TaqMan PCR and Western analysis. Experiments were repeated 3 times with similar results. Statistics were calculated by 2-tailed Student’s t-test. *, p<0.05. A-C. TaqMan PCR assessment of the host defense-related gene expression on CSE treated airway epithelium on ALI. A. MX1. B. HLA-DRA. C. IFITM1. D. Western analysis of the expression change of MX1, HLA-DRA and IFITM1 on CSE treated airway epithelium on ALI. Beta actin was used as loading control.
Discussion
It is well recognized that exposure to cigarette smoke is a substantial risk factor for lung infection27-29. Airway infection, in turn, can trigger exacerbations of smoking-induced lung diseases, such as COPD, the 3rd leading cause of death in the world (http://www.who.int/). Despite the accumulated epidemiologic observations, the mechanisms that mediate the smoking-induced dysfunctional host defense in the human airway epithelium have not been clearly defined. In the present human-based study, initiated from a genome-wide screening, POU2AF1 was identified as a novel host defense regulator in the human airway epithelium and that the impairment of host defense in the human airway epithelium induced by smoking is related, at least in part, to compromised expression of POU2AF1.
Expression and Regulation of POU2AF1 in Human Airway Epithelium
POU2AF1 has been previously considered to be a lymphocyte-specific gene4-9. It is constitutively expressed in B cells, and is inducible in T cells upon stimulation 4. The stimuli that can up-regulate POU2AF1 include IL4, LPS, CD40 and BCR signaling17 and phorbol myristate acetate plus ionomycin24. Sporadic literature, however, suggests POU2AF1 might have a role in cells other than lymphocytes. For example, in the murine intestinal follicle-associated epithelium, which includes M cells and is specialized for the uptake and transcytosis of macromolecules and microorganisms, there was higher POU2AF1 expression than in villous epithelium30,31. One common feature of these observations is that they are relevant to the interface between organs and the environment, which, together with the known functions, imply a defense role of POU2AF1.
Our genome wide screening identified that POU2AF1 has an average expression level among all transcription factors in human airway epithelium. Although lymphocyte infiltration is common in airway epithelium32, the up-regulation of POU2AF1 during differentiation of airway basal cells and an airway basal cell immortalized line, as well as cells derived from simple basal cells clones, provided conclusive evidence that the observed POU2AF1 expression in human airway epithelium could not be due to lymphocyte contamination. The function and expression pattern of POU2AF1 is reminiscent of another airway transcription factor, ELF3, which is also up-regulated during differentiation, not cell lineage specific and related to innate immunity33.
Airway epithelial cells and lymphocytes are usually localized in disparate milieus, which raise an intriguing question whether POU2AF1 has similar regulation mechanisms in both cell types. In the current study, we found that cigarette smoking is a novel repressor of POU2AF1 in human airway epithelium. Interestingly, microRNA miR-126, a negative regulator of POU2AF1 in the whole mouse airway (but in unknown cell types)34, is up-regulated by smoking in the human airway epithelium35. These data suggest that, in response to smoking in vivo, both genetic and epigenetic mechanisms are likely involved in the regulation of POU2AF1 in human airway epithelium. Finally, several COPD related cytokines and growth factors can differentially regulate POU2AF1, suggesting that the homeostasis of POU2AF1 is likely disordered in COPD.
Function of POU2AF1 in Human Airway Epithelium
POU2AF1 has been reported to regulate various genes, either in a direct or indirect manner. In lymphocytes, the gene expression regulated by POU2AF1 can be cell type or cell differentiation stage dependent4. Besides IgG, the downstream genes of POU2AF1 in lymphocytes belong to multiple categories, including cellular metabolism, cell survival and proliferation, ion channel, cell adhesion, cytokines and chemokines4,6.
Since POU2AF1 needs to bind to either OCT1 or OCT2 to take effect, we confirmed expression of OCT1 and OCT2 in human airway epithelium before assessing the functions of POU2AF1. Interestingly, we found many known POU2AF1 downstream genes expressed in B cells are also expressed in airway epithelium and regulated by POU2AF1. For example, ID2 is a transcription factor expressed in the distal lung tip epithelium36; S100A10 37 and KCNN438 are ion channel related genes, both of which have cystic fibrosis transmembrane conductance regulator-related functions in human airway epithelial cells; HLA-DRA is an MHC-II gene related to antigen presentation in human airway epithelium39; and BCL6 is an inflammation suppressor in mouse airway epithelium40. Together, this data suggests that the functions of POU2AF1 have some overlap between lymphocytes and airway epithelial cells.
Other than above known shared targets, the dominant function of the most highly induced genes by POU2AF1 in airway epithelial cells are related to host defense. Among the top 50 genes induced by POU2AF1, only HLA-DRA has been proved to be a downstream gene of POU2AF1 in lymphocytes. These genes function to defend against invasive pathogens through either directly suppressing pathogen survival/spreading (e.g., MX1, MX2, IFI44, IFI44L), or indirectly adjusting inflammatory cell responses (e.g., HLA-DRA, IL19, C1R). Many of these genes are involved in the interferon-mediated anti-virus/microbe response, including two type I interferon genes, IFNB and IFNL1 that are up-regulated by POU2AF1. However, IFNG (type II interferon), a direct target gene of POU2AF1 in B cells41 was not induced. As not all POU2AF1-modified genes are regulated by type I interferon, and OCT1/OCT2 binding sites are quite common in immunity-related genes20,21,42, it is likely that up-regulation of POU2AF1 enhanced an interactive network regulating host defense in human airway epithelium. Among all the lentiviral vectors (lenti-RFP, lenti-GFP, lenti-KLF4, lenti-OSGIN1) that we tested, only POU2AF1 induced such striking effects of regulation of host defense genes.
The role of POU2AF1 in regulating host defense genes is further supported by the finding that genes downstream of POU2AF1 have coordinated expression changes with POU2AF1 in human airway epithelium both in vitro and in vivo. Importantly, sustained expression of POU2AF1 attenuated the down-regulation of host defense genes by cigarette smoke. Together, these data suggest that suppressed expression of POU2AF1 may contribute to the dysfunction of host defense induced by cigarette smoking in human airway epithelium.
Besides mRNA, POU2AF1 can also regulate inflammation-related microRNAs in B cells43. MiR-146a is the only microRNA shown to be a direct target of POU2AF143. MiR-146a tunes inflammatory responses by targeting genes involved in the Toll-like receptor pathway43, an essential component of the host defense system3. Interestingly, consistent with the down-regulation of POU2AF1 by smoking, miRNA-146a is also expressed in human airway epithelium and down-regulated by smoking in vivo35. One limitation of current study is that, because of technical challenges, we were not able to test the host defense response of the human airway epithelium in the context of suppression of POU2AF1.
In summary, our data demonstrate that expression of POU2AF1 is not only expressed in lymphocytes but also in airway epithelium, where it is diminished by the stress of smoking. In the context that POU2AF1 is involved in the regulation of the host defense response of airway epithelium and that host defense dysfunction leads to vulnerability to respiratory infection, POU2AF1 and its related pathway might be therapeutic targets for smoking related airway diseases.
Supplementary Material
Acknowledgments
We thank N. Mohamed for help in preparing this manuscript.
Footnotes
Grant support: These studies were supported, in part, by R01 HL107882, P20 HL113443, UL1 TR000457 and UL1 RR0204143. GW, RW and AB were supported, in part, by T32 HL094284. For P20 HL113443, research reported in this publication was supported by NIH and the Family Smoking Prevention and Tobacco Control Act. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or the Food and Drug Administration.
References
- 1.Bals R, Hiemstra PS. Innate immunity in the lung: how epithelial cells fight against respiratory pathogens. Eur Respir J. 2004;23:327–33. doi: 10.1183/09031936.03.00098803. [DOI] [PubMed] [Google Scholar]
- 2.Parker D, Prince A. Innate immunity in the respiratory epithelium. Am J Respir Cell Mol Biol. 2011;45:189–201. doi: 10.1165/rcmb.2011-0011RT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vareille M, Kieninger E, Edwards MR, Regamey N. The airway epithelium: soldier in the fight against respiratory viruses. Clin Microbiol Rev. 2011;24:210–29. doi: 10.1128/CMR.00014-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brunner C, Wirth T. BOB.1/OBF.1 - A critical regulator of B cell function. Curr Immunol Rev. 2006;2:3–12. [Google Scholar]
- 5.Luo Y, Roeder RG. B-cell-specific coactivator OCA-B: biochemical aspects, role in B-cell development and beyond. Cold Spring Harb Symp Quant Biol. 1999;64:119–31. doi: 10.1101/sqb.1999.64.119. [DOI] [PubMed] [Google Scholar]
- 6.Teitell MA. OCA-B regulation of B-cell development and function. Trends Immunol. 2003;24:546–53. doi: 10.1016/j.it.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 7.Kim U, et al. The B-cell-specific transcription coactivator OCA-B/OBF-1/Bob-1 is essential for normal production of immunoglobulin isotypes. Nature. 1996;383:542–7. doi: 10.1038/383542a0. [DOI] [PubMed] [Google Scholar]
- 8.Nielsen PJ, Georgiev O, Lorenz B, Schaffner W. B lymphocytes are impaired in mice lacking the transcriptional co-activator Bob1/OCA-B/OBF1. Eur J Immunol. 1996;26:3214–8. doi: 10.1002/eji.1830261255. [DOI] [PubMed] [Google Scholar]
- 9.Schubart DB, Rolink A, Kosco-Vilbois MH, Botteri F, Matthias P. B-cell-specific coactivator OBF-1/OCA-B/Bob1 required for immune response and germinal centre formation. Nature. 1996;383:538–42. doi: 10.1038/383538a0. [DOI] [PubMed] [Google Scholar]
- 10.Harvey BG, Heguy A, Leopold PL, Carolan BJ, Ferris B, Crystal RG. Modification of gene expression of the small airway epithelium in response to cigarette smoking. J Mol Med (Berl) 2007;85:39–53. doi: 10.1007/s00109-006-0103-z. [DOI] [PubMed] [Google Scholar]
- 11.Hackett NR, et al. The human airway epithelial basal cell transcriptome. PLoS One. 2011;6:e18378. doi: 10.1371/journal.pone.0018378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walters MS, et al. Generation of a human airway epithelium derived basal cell line with multipotent differentiation capacity. Respir Res. 2013;14:135. doi: 10.1186/1465-9921-14-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brekman A, Walters MS, Tilley AE, Crystal RG. FOXJ1 Prevents Cilia Growth Inhibition by Cigarette Smoke in Human Airway Epithelium in Vitro. Am J Respir Cell Mol Biol. 2014 doi: 10.1165/rcmb.2013-0363OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang R, et al. Smoking-induced upregulation of AKR1B10 expression in the airway epithelium of healthy individuals. Chest. 2010;138:1402–10. doi: 10.1378/chest.09-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ryan DM, et al. Smoking dysregulates the human airway basal cell transcriptome at COPD risk locus 19q13.2. PLoS One. 2014;9:e88051. doi: 10.1371/journal.pone.0088051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hackett NR, et al. RNA-Seq quantification of the human small airway epithelium transcriptome. BMC Genomics. 2012;13:82. doi: 10.1186/1471-2164-13-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin XF, Reichlin A, Luo Y, Roeder RG, Nussenzweig MC. OCA-B integrates B cell antigen receptor-, CD40L- and IL 4-mediated signals for the germinal center pathway of B cell development. EMBO J. 1998;17:5066–75. doi: 10.1093/emboj/17.17.5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deppong CM, Xu J, Brody SL, Green JM. Airway epithelial cells suppress T cell proliferation by an IFNgamma/STAT1/TGFbeta-dependent mechanism. Am J Physiol Lung Cell Mol Physiol. 2012;302:L167–L173. doi: 10.1152/ajplung.00188.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bordon A, et al. Enforced expression of the transcriptional coactivator OBF1 impairs B cell differentiation at the earliest stage of development. PLoS One. 2008;3:e4007. doi: 10.1371/journal.pone.0004007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fontes JD, Jabrane-Ferrat N, Toth CR, Peterlin BM. Binding and cooperative interactions between two B cell-specific transcriptional coactivators. J Exp Med. 1996;183:2517–21. doi: 10.1084/jem.183.6.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karnowski A, et al. B and T cells collaborate in antiviral responses via IL-6, IL-21, and transcriptional activator and coactivator, Oct2 and OBF-1. J Exp Med. 2012;209:2049–64. doi: 10.1084/jem.20111504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim U, Siegel R, Ren X, Gunther CS, Gaasterland T, Roeder RG. Identification of transcription coactivator OCA-B-dependent genes involved in antigen-dependent B cell differentiation by cDNA array analyses. Proc Natl Acad Sci U S A. 2003;100:8868–73. doi: 10.1073/pnas.1033108100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang G, et al. Smoking-mediated up-regulation of GAD67 expression in the human airway epithelium. Respir Res. 2010;11:150. doi: 10.1186/1465-9921-11-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mueller K, et al. Octamer-dependent transcription in T cells is mediated by NFAT and NF-kappaB. Nucleic Acids Res. 2013;41:2138–54. doi: 10.1093/nar/gks1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen Y, Hendershot LM. Identification of ERdj3 and OBF-1/BOB-1/OCA-B as direct targets of XBP-1 during plasma cell differentiation. J Immunol. 2007;179:2969–78. doi: 10.4049/jimmunol.179.5.2969. [DOI] [PubMed] [Google Scholar]
- 26.Buro-Auriemma LJ, et al. Cigarette smoking induces small airway epithelial epigenetic changes with corresponding modulation of gene expression. Hum Mol Genet. 2013;22:4726–38. doi: 10.1093/hmg/ddt326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arcavi L, Benowitz NL. Cigarette smoking and infection. Arch Intern Med. 2004;164:2206–16. doi: 10.1001/archinte.164.20.2206. [DOI] [PubMed] [Google Scholar]
- 28.Bauer CM, Morissette MC, Stampfli MR. The influence of cigarette smoking on viral infections: translating bench science to impact COPD pathogenesis and acute exacerbations of COPD clinically. Chest. 2013;143:196–206. doi: 10.1378/chest.12-0930. [DOI] [PubMed] [Google Scholar]
- 29.Murin S, Bilello KS. Respiratory tract infections: another reason not to smoke. Cleve Clin J Med. 2005;72:916–20. doi: 10.3949/ccjm.72.10.916. [DOI] [PubMed] [Google Scholar]
- 30.Mach J, Hshieh T, Hsieh D, Grubbs N, Chervonsky A. Development of intestinal M cells. Immunol Rev. 2005;206:177–89. doi: 10.1111/j.0105-2896.2005.00281.x. [DOI] [PubMed] [Google Scholar]
- 31.Nakato G, Fukuda S, Hase K, Goitsuka R, Cooper MD, Ohno H. New approach for m-cell-specific molecules screening by comprehensive transcriptome analysis. DNA Res. 2009;16:227–35. doi: 10.1093/dnares/dsp013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeffery PK. Comparative morphology of the airways in asthma and chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 1994;150:S6–13. doi: 10.1164/ajrccm/150.5_Pt_2.S6. [DOI] [PubMed] [Google Scholar]
- 33.Wu J, et al. Regulation of epithelium-specific Ets-like factors ESE-1 and ESE-3 in airway epithelial cells: potential roles in airway inflammation. Cell Res. 2008;18:649–63. doi: 10.1038/cr.2008.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mattes J, Collison A, Plank M, Phipps S, Foster PS. Antagonism of microRNA-126 suppresses the effector function of TH2 cells and the development of allergic airways disease. Proc Natl Acad Sci U S A. 2009;106:18704–9. doi: 10.1073/pnas.0905063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang R, et al. Persistent changes in smoking-induced dysregulated small airway epithelium miRNA expression despite smoking cessation. Am J Respir Crit Care Med. 2013;187:A1206. [Google Scholar]
- 36.Rawlins EL, Clark CP, Xue Y, Hogan BL. The Id2+ distal tip lung epithelium contains individual multipotent embryonic progenitor cells. Development. 2009;136:3741–5. doi: 10.1242/dev.037317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borthwick LA, et al. The formation of the cAMP/protein kinase A-dependent annexin 2-S100A10 complex with cystic fibrosis conductance regulator protein (CFTR) regulates CFTR channel function. Mol Biol Cell. 2007;18:3388–97. doi: 10.1091/mbc.E07-02-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mall M, et al. Modulation of Ca2+-activated Cl-secretion by basolateral K+ channels in human normal and cystic fibrosis airway epithelia. Pediatr Res. 2003;53:608–18. doi: 10.1203/01.PDR.0000057204.51420.DC. [DOI] [PubMed] [Google Scholar]
- 39.Kalb TH, Chuang MT, Marom Z, Mayer L. Evidence for accessory cell function by class II MHC antigen-expressing airway epithelial cells. Am J Respir Cell Mol Biol. 1991;4:320–9. doi: 10.1165/ajrcmb/4.4.320. [DOI] [PubMed] [Google Scholar]
- 40.Seto T, et al. Bcl6 in pulmonary epithelium coordinately controls the expression of the CC-type chemokine genes and attenuates allergic airway inflammation. Clin Exp Allergy. 2011;41:1568–78. doi: 10.1111/j.1365-2222.2011.03836.x. [DOI] [PubMed] [Google Scholar]
- 41.Brunner C, Sindrilaru A, Girkontaite I, Fischer KD, Sunderkotter C, Wirth T. BOB.1/OBF.1 controls the balance of TH1 and TH2 immune responses. EMBO J. 2007;26:3191–202. doi: 10.1038/sj.emboj.7601742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lins K, et al. OBF1 enhances transcriptional potential of Oct1. EMBO J. 2003;22:2188–98. doi: 10.1093/emboj/cdg199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lindner JM, Kayo H, Hedlund S, Fukuda Y, Fukao T, Nielsen PJ. Cutting edge: The transcription factor Bob1 counteracts B cell activation and regulates miR-146a in B cells. J Immunol. 2014;192:4483–6. doi: 10.4049/jimmunol.1303022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.