Abstract
HIV-infected persons are at greater risk of developing tuberculosis (TB) even before profound CD4 loss occurs, suggesting that HIV alters CD4+T cell functions capable of containing bacterial replication. An effective immune response to Mycobacterium tuberculosis likely relies on the development of a balanced CD4 response, where distinct CD4+T helper subsets act in synergy to control the infection. To define the diversity of Mtb-specific CD4+Th subsets and determine whether HIV infection impacts such responses, the expression of lineage-defining transcription factors T-bet, Gata3, RORγt and Foxp3 was measured in Mtb-specific CD4+T cells in HIV-uninfected (n=20) and HIV-infected individuals (n=20) with latent TB infection. Our results show that upon 5 day restimulation in vitro, Mtb-specific CD4+T cells from healthy individuals have the ability to exhibit a broad spectrum of T helper subsets, defined by specific patterns of transcription factor co-expression. These transcription factor profiles were skewed in HIV-infected individuals where the proportion of T-bethighFoxp3+ Mtb-specific CD4+T cells was significantly decreased (p=0.002) compared to HIV-uninfected individuals, a change that correlated inversely with HIV viral load (p=0.0007) and plasma TNF-α (p=0.027). Our data demonstrate an important balance in T helper subset diversity defined by lineage-defining transcription factor co-expression profiles that is disrupted by HIV infection and suggest a role for HIV in impairing TB immunity by altering the equilibrium of Mtb-specific CD4+T helper subsets.
Introduction
CD4+ T helper (Th) subsets play a major role orchestrating immune responses to Mycobacterium tuberculosis (Mtb) (1-4). In mouse models, Th1 CD4+ T cell responses producing IFN-γ are necessary but not sufficient to control TB infection (5). Th17 CD4+ T cells can also confer partial protection against Mtb (6, 7), but an excess of Th17 cells may promote pathology, fueling inflammation and favoring the accumulation of pathogenic neutrophils (8, 9). Regulatory CD4+ T cells (Treg), endowed with suppressive functions, exert detrimental effects during active Mtb infection, by delaying the onset of adaptive responses (10, 11). However, during chronic infection, Tregs can contribute to the resolution of Mtb by preventing inflammation-mediated tissue damage (12). These findings suggest that the clinical outcome of Mtb infection relies on the host's ability to generate a diverse repertoire of Th responses with balanced effector and regulatory functions. In this model, pro-inflammatory responses enhance bacterial killing required to clear or control infection, while anti-inflammatory responses limit pathology and inflammation during initial infection and latency. However, the precise balance of CD4+ T cells needed to control Mtb growth and prevent TB disease remain unclear.
The lineage commitment of CD4+ T cells is regulated by the nature of the threat encountered, and the quality of the cytokine milieu at the time of T cell receptor engagement (13-15). The combination of these signals results in the expression of specific canonical lineage-defining transcription factors (TF), such as T-bet, Gata3, RORγt or Foxp3, leading to CD4 polarization into Th1, Th2, Th17 or Treg subsets, respectively. The development of distinct CD4+ Th subsets has long been thought to result in a fixed and stable commitment of CD4+ T cells controlled by a single lineage-defining regulator. However, over the past few years, murine model studies have revealed that this view is over-simplified. CD4+ T cell phenotypes are more diverse and flexible than previously appreciated (15). CD4+ T cells cytokine profiles can evolve upon changing environmental conditions and ‘mixed’ phenotypes characterized by co-expression of multiple transcription factors have been reported (reviewed in (16-18)). This suggests that transcription factors regulate lineage commitment as a network rather than as unique determinants (19-21). Few studies have described this phenomenon in human CD4+ T cells (22-24), and the spectrum of Th subsets of Mtb-specific CD4+ responses is largely unknown.
HIV is one of the major risk factors for TB reactivation. While HIV has been shown to impair both the innate and adaptive immune responses, the most obvious immune defect caused by HIV is a progressive reduction in absolute CD4+ T cell numbers that correlates with increasing risk of TB (25). However, shortly after HIV acquisition or when CD4+ T cell numbers improve upon HIV treatment, the risk of TB remains increased (26, 27). These observations suggest that, in addition to depleting Mtb-specific cells, HIV may also alter their function. Several potential mechanisms have been reported, including the preferential HIV infection of Mtb-specific CD4+ T cells (28, 29), a decrease in polyfunctional CD4+ responses (30, 31) and alteration in IL-10 regulatory pathways (32, 33). Moreover, HIV infection is characterized by generalized immune activation during which cytokine production is altered (34). Since the cytokine environment plays a major role in shaping CD4+ Th differentiation, it is conceivable that HIV infection could skew the lineage differentiation of Mtb-specific CD4+ T cells, favoring the development of TB. To explore this hypothesis, we defined the diversity of Mtb-specific CD4+ Th subsets in healthy individuals sensitized by Mtb, by measuring the expression of four canonical lineage-defining transcription factors, and tested how these profiles were altered in HIV-infected individuals.
Material and Methods
Study participants
Twenty HIV-uninfected and 20 HIV-infected individuals sensitized to Mtb were recruited from the Ubuntu clinic (Khayelitsha, Cape Town). Exclusion criteria included: antiretroviral treatment and previous history of chronic disease (including TB). Mtb sensitization was defined based on a positive interferon-γ release assay (Quantiferon, Cellestis), no symptoms of active TB disease, a negative TB culture and a normal chest X-ray. Absolute blood CD4+ cell counts were measured using a Flow-CARE PLG CD4 test (Beckman Coulter). For HIV-infected individuals, plasma HIV-1 RNA levels were quantified using Abbott m2000 RealTime HIV-1 assay. The clinical characteristics of each individual are presented in Table 1. The University of Cape Town institutional review board approved this study (158/2010) and all subjects provided informed consent for participation.
Table 1. Clinical characteristics of the study cohort.
|
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Ki67+ cells (% total CD4)B | ||||||||
|
| ||||||||
| PIDA | Gender | HIV status | Age | CD4 count (cells/mm3) | HIV Viral Load (RNA copies/ml) | ESAT/CFP response | PPD response | TT response |
| BAL-1010 | M | - | 23 | 580 | na | nd | 1.96 | nd |
| BAL-1066 | F | - | 39 | 621 | na | 0.55 | 4.05 | nd |
| BAL-1001 | M | - | 19 | 631 | na | 1.16 | 2.77 | 0.42 |
| BAL-1049 | F | - | 40 | 655 | na | 0.12 | 0.36 | 0.00 |
| BAL-1015 | M | - | 23 | 659 | na | 0.00 | 2.64 | 1.05 |
| BAL-1011 | M | - | 28 | 801 | na | nd | 0.90 | nd |
| BAL-1033 | F | - | 20 | 813 | na | 1.85 | 2.95 | nd |
| BAL-1028 | F | - | 44 | 814 | na | 1.43 | 1.74 | 0.94 |
| BAL-1094 | F | - | 29 | 827 | na | 0.00 | 1.09 | 0.00 |
| BAL-1054 | F | - | 21 | 832 | na | 0.09 | 0.43 | nd |
| BAL-1058 | F | - | 35 | 866 | na | 0.56 | 4.14 | 0.44 |
| BAL-1060 | F | - | 23 | 902 | na | 2.46 | 5.27 | 0.14 |
| BAL-1025 | F | - | 23 | 915 | na | 0.26 | 1.19 | 0.09 |
| BAL-1055 | M | - | 19 | 932 | na | 1.36 | 1.04 | 0.28 |
| BAL-1024 | M | - | 24 | 939 | na | 0.44 | 1.26 | nd |
| BAL-1070 | M | - | 25 | 1028 | na | nd | 3.10 | nd |
| BAL-1068 | F | - | 32 | 1130 | na | 0.12 | 0.25 | 0.17 |
| BAL-1031 | F | - | 19 | 1169 | na | 0.43 | 1.19 | nd |
| BAL-1029 | F | - | 22 | 1430 | na | 1.29 | 0.30 | 0.00 |
| BAL-1032 | F | - | 21 | nd | na | 0.42 | 1.32 | 0.06 |
|
| ||||||||
| Median | 23 | 832 | na | 0.44 | 1.29 | 0.15 | ||
| IQR | 21-31 | 659-939 | na | 0.19-1.79 | 0.46-2.33 | 0.01-0.43 | ||
|
| ||||||||
| BAL-1127 | F | + | 31 | 817 | 40 | 0.17 | 0.33 | nd |
| BAL-1151 | F | + | 27 | 749 | 40 | 0.79 | 1.42 | nd |
| BAL-1005 | F | + | 52 | 604 | 94 | 0.57 | 1.28 | nd |
| BAL-1150 | F | + | 26 | 894 | 311 | nd | 4.06 | nd |
| BAL-1143 | F | + | 26 | 656 | 618 | 0.32 | 0.58 | nd |
| BAL-1076 | F | + | 25 | 543 | 908 | 0.20 | 0.11 | nd |
| BAL-1154 | F | + | 34 | 988 | 1,848 | 0.23 | 0.21 | nd |
| BAL-1006 | F | + | 38 | 802 | 4,142 | 0.06 | 0.24 | nd |
| BAL-1075 | F | + | 31 | 965 | 5,923 | nd | 1.83 | nd |
| BAL-1134 | F | + | 37 | 599 | 6,383 | 0.12 | 0.20 | nd |
| BAL-1084 | F | + | 40 | 619 | 9,192 | 0.00 | 0.27 | nd |
| BAL-1152 | F | + | 30 | 571 | 9,697 | nd | 0.47 | nd |
| BAL-1073 | F | + | 26 | 632 | 12,275 | 0.23 | 0.78 | nd |
| BAL-1137 | F | + | 27 | 560 | 18,797 | 0.56 | 0.08 | nd |
| BAL-1020 | F | + | 41 | 406 | 31,146 | nd | 0.19 | nd |
| BAL-1079 | F | + | 34 | 591 | 32,485 | nd | 1.86 | nd |
| BAL-1126 | F | + | 30 | 433 | 32,485 | 0.00 | 0.12 | nd |
| BAL-1045 | F | + | 31 | 522 | 59,126 | 0.25 | 0.14 | nd |
| BAL-1142 | M | + | 53 | 441 | 544,849 | 0.71 | 1.53 | nd |
| BAL-1119 | M | + | 30 | 545 | 580,150 | nd | 1.84 | nd |
|
| ||||||||
| Median | 31 | 602 | 7,788 | 0.23 | 0.40 | nd | ||
| IQR | 27-38 | 543-788 | 690-32,150 | 0.12-0.71 | 0.19-1.34 | nd | ||
Patient Identification number
Five-day lymphoproliferative assay in response to ESAT-6/CFP-10 peptide pool, Mtb purified protein derivative (PPD) or tetanus toxoid (TT). Data corresponds to the proportion of Ki67 positive cells expressed as a percentage of total CD4+ T cells after background substraction.
nd: Not determined
na: Not applicable
Reagents
For the 5-day proliferation assay tuberculin purified protein derivative (PPD, Staten Serum Institute) was used at 5 μg/ml; Tetanus toxoid (TT, Sanofi Pasteur) at 1 IU/ml and Staphylococcal Enterotoxin B (SEB) at 0.2 μg/ml. Mtb peptide pool consisting of 15mers peptides overlapping by 10aa, spanning the entire ESAT-6 and CFP-10 proteins (Peptide Synthetics) was used at 2 μg/ml. The following conjugated monoclonal antibodies were used: CD14-APC-Alexa750 (Invitrogen), CD19-APC-Alexa750 (Invitrogen), CD3-BV650 (Biolegend), CD4-ECD (Beckman Coulter), CD8-V500 (BD), T-bet-PE-cy7 (e-Bioscience), RORγt-PE (e-Bioscience), Gata3-PerCPeFluor710 (e-Bioscience), Foxp3-PacBlue (Biolegend), Ki67-FITC (BD), IFN-γ-Alexa700 (BD), IL-2-BV605 (BD), TNF-α-APC (BD), IL-17-APC (BD). The Near Infra-Red amine reactive dye (Molecular Probes) was used as a marker of cell viability.
Cell activation and staining
Cryopreserved peripheral blood mononuclear cells, obtained by standard Ficoll-Hypaque density gradient centrifugation, were seeded at 2×106 cells/tube in complete RPMI media and stimulated with ESAT-6/CFP-10 peptide pool, PPD, TT or SEB. Cells were cultured for 5 days. When cytokine expression was assessed, Brefeldin-A (10 μg/ml, Sigma) was added 5 hours prior to the end of the assay. Afterwards, cells were first stained with a viability dye, then surface-stained with CD3, CD4, CD8, CD14 and CD19 antibodies. Cells were fixed and permeabilized using the Transcription Factor Buffer Set (BD) and stained with T-bet, RORγt, Gata3, Foxp3, Ki67 and cytokine antibodies in selected experiments. Approximately 1,000,000 events were acquired on a LSRII flow cytometer (BD) and analysis was performed using FlowJo (v9.7.6, Treestar). Antigen-specific CD4+ T cells were defined based on Ki67 expression. A positive response was defined as at least twice the background (no antigen stimulation), greater than 0.05% of total CD4+ T cells and at least 40 events (to minimize the possibility of error due to a low number of events when further subdividing these cells based on their TF profiles). The co-expression of TF was analyzed using a Boolean gating strategy using Pestle (v1.6.2) and Spice (v5.1) (35).
Plasma cytokine measurement
Ten cytokines/chemokines were measured in blood plasma of HIV-infected and HIV-uninfected individuals (n=18) by electrochemiluminescence (ECL) multiplex technology using a Meso Scale Discovery Multi-spot 10 analyte plate (IL-1β, IL-4, IL-6, IL-10, IL-13, IFN-γ, TNF-α, IL-2, IL-12p70 and IL-8) according to manufacturer's instructions (Meso Scale Diagnostics). The sensitivity of the kit ranged from 0.05 to 1.5 pg/ml depending on the analyte measured. Plasma samples were thawed and filtered by centrifugation using 0.2μm cellulose acetate filters prior to cytokine/chemokine measurements. ECL was performed by the MGH clinical lab core (Boston). Cytokine concentrations below the lower limits of detection were reported as the midpoint between the lowest concentration and zero for each cytokine measured.
Statistics
The Spearman Rank correlation was applied for assessing nonparametric associations. All tests were 2-tailed and p values ≤0.05 were considered significant. For plasma cytokine data, p values were adjusted using the Bonferroni step-down procedure (36) Regarding Spice analyses, comparison of distributions was performed using a Student's t-test and a partial permutation test (35).
Results
Mycobacterium tuberculosis (Mtb)-specific CD4+ T cells from healthy donors exhibit distinct co-expression profiles of lineage-defining transcription factors (TF) upon a 5-day restimulation
To assess the diversity of Th subsets in Mtb-specific memory CD4+ T cells, PBMC from Mtb-sensitized healthy individuals were activated for 5 days with an ESAT-6/CFP10 peptide pool. Mtb-responding CD4+ T cells were detected based on their proliferation capacity by measuring Ki67 up-regulation (37) (Figure 1A). Fifteen out of the 17 individuals tested had a detectable response to ESAT-6/CFP-10 (Table 1). The expression of Gata3, RORγt, Foxp3 and T-bet were assessed and the gating strategy is presented in Supplemental Figure S1. We focused on cells exhibiting a T-bet high phenotype, as T-bet high expression is a characteristic of a fully differentiated Th1 phenotype upon restimulation (38, 39) and is triggered by inflammatory signals (40). Figure 1B shows that all TF tested were upregulated in proliferating Mtb-specific CD4+ T cells, compared to non-proliferating cells. A median of 94% of Mtb-specific CD4+ T cells expressed a high level of T-bet (IQR: 69-95.1), 58.4% were Foxp3 positive (IQR: 55.2-69.8), 39% were RORγthigh (IQR: 30.4-51.7) and 12% (IQR: 8.9-17.6) expressed high level of Gata3 (Figure 1C). Using a Boolean gating strategy, we defined TF co-expression in Mtb-specific CD4+ T cells. Almost 70% of Mtb-specific CD4+ T cells co-expressed two, three or four TF simultaneously (Figure 1D). The most prevalent subsets were Foxp3+RORγthighT-bethigh, Foxp3+T-bethigh and T-bethigh, each representing approximately a median of 20% of Mtb-responding CD4+ T cells. Moreover, a median of 6% (IQR: 2-9) of Mtb-specific CD4+ T cells expressed all four TF tested. Cells expressing high levels of RORγt or Gata3 alone were barely detectable. Of note, following a short-term stimulation (15 hours), most (∼70%) Mtb-specific CD4+ T cells (defined by cytokine expression, Figure 2A), did not express transcription factors apart from T-bet at a low level. Moreover, co-expression of transcription factors was marginal, with ∼1% of Mtb-specific cells co-expressing T-bet and RORγt. Similar results were obtained in response to PPD and SEB stimulation (Figure 2 B and C).
Figure 1. Lineage-defining transcription factor (TF) expression profile in Mtb-specific CD4+ T cells upon a 5 day in vitro stimulation in healthy Mtb-sensitized individuals.
PBMC from HIV-uninfected individuals were cultured for 5 days in the presence of ESAT-6/CFP-10 peptide pool. (A) Detection of Mtb-specific CD4+ T cells based on Ki67 expression. NS: non-stimulated. (B) Representative dot plots showing the level of expression of Foxp3, RORγt, Gata3 and T-bet in Mtb-specific CD4+ T cells (top panel). Representative histograms of TF expression levels in Ki67+ cells (red) and Ki67- cells (black). (C) Proportion of TF positive cells within Mtb-specific CD4+ T cells expressed as a percentage of Ki67+CD4+ T cells. Each dot represents an individual. (D) Proportion of Mtb-specific CD4+ T cells expressing 4, 3, 2, 1 or none of the TF measured. The pie and bar chart show the proportion of each possible combination of TF expression in Mtb-specific CD4+ T cells. Each slice of the pie corresponds to a distinct combination of TF. The arc illustrates the proportion of Mtb-specific CD4+ T cells expressing multiple (≥ 2) transcription factors.
Figure 2. Lineage-defining transcription factor expression in Mtb- and SEB-specific CD4+ T cells upon short-term (15 hours) stimulation.
(A) Dot plots of IFN-γ, IL-2 and IL-17 production in response to a 15-hour stimulation with Mtb peptides (ESAT-6 and CFP-10) or SEB. NS: No stimulation. (B) Expression of TF in cytokine producing CD4+ T cells in response to Mtb peptides (left panel) or SEB (right panel). Numbers correspond to the proportion of transcription factor positive cells within cytokine producing cells (IFN-γ, IL-2 or IL-17). (C) Proportion of ESAT/CFP- (black, n=4), PPD- (dark grey, n=5) and SEB- (light gray, n=7) specific CD4+ T cells expressing 4, 3, 2, 1 or none of the TF measured. The pie and bar chart show the proportion of each possible combination of TF expression in antigen-specific CD4+ T cells. Bars represent median and interquartile ranges. Statistical comparisons were performed using a Student's t-test. *<0.05, **<0.01, ***<0.001.
To define whether such diversity was observed in resting CD4+ T cells, we measured TF expression in CD4+ T cells ex vivo (Figures 3). In the absence of any stimulation, only a marginal fraction of CD4+ T cells expressed Gata3, RORγt, Foxp3 (<0.5%) or T-bet (median 2.5%, range: 1-7.7%; Figures 3B-C). However, actively proliferating ex vivo CD4+ T cells (as measured by Ki67 expression) exhibited significantly elevated expression of TF compared to non-proliferating cells (p=0.0007, Figure 3C). Importantly, while no co-expression of TF was observed in Ki67-CD4+ T cells, low frequencies of proliferating Ki67+CD4+ T cells expressed multiple TF namely Foxp3+T-bethigh, Foxp3+Gata3high or T-bethighRORγthigh. These data show that CD4+ T cells co-expressing TF can be found in vivo and suggest that such flexible CD4+ T cells are confined to cycling cells.
Figure 3. Co-expression of lineage-defining transcription factor in proliferating CD4+ T cells ex vivo.
(A) Representative dot plot of Ki67 expression in ex vivo CD4+ T cells. The median frequency and range of Ki67+CD4+ T cells are indicated in the gate (n=7). (B) Representative histograms of TF expression levels in Ki67+ cells (red) and Ki67- cells (black). (C) Proportion of Ki67- (Top panel) and Ki67+ (bottom panel) CD4+ T cells expressing 4, 3, 2, 1 or none of the TF measured. The pie and bar chart show the proportion of each possible combination of TF expression. Each slice of the pie corresponds to a distinct combination of TF.
Overall, these data suggest that the proportion of circulating ex vivo CD4+ T cells maintained as “committed” (i.e. expressing detectable levels of TF) is limited. Upon 5-day restimulation, memory Mtb-specific CD4+ T cells exhibit a broad and diverse profile of lineage-defining transcription factors, revealing flexibility in the Mtb-specific CD4 immune response.
The diversity of lineage-defining transcription factor expression profiles in CD4+ T cells varies according to pathogen specificity
To define whether CD4+ Th diversity was dependent on the nature of the antigen recognized, we compared the TF expression profiles of CD4+ T cells in response to Mtb peptide pool (ESAT-6/CFP-10), Mtb-PPD, tetanus toxoid (TT) and SEB. Figure 4A shows antigen-responding Ki67+CD4+ T cells in one healthy donor. Mtb PPD-specific responses were detected in all 20 healthy individuals tested, ESAT-6/CFP-10-specific responses in 15/17 individuals tested and TT-specific responses in 8/11 subjects tested (Figure 4B). ESAT-6/CFP-10- and PPD-specific CD4+ T cells exhibited similar expression of T-bet, Foxp3, RORγt and Gata3 (Figure 4C). In contrast, TT-specific CD4+ T cells expressed significantly lower T-bet compared to ESAT-6/CFP-10- and PPD-specific CD4+ T cells (p=0.009 and p=0.004, respectively, Figure 4C and data not shown). We next compared the co-expression of TF in CD4+ T cells of different specificities. PPD-specific CD4+ T cells exhibited a similar TF expression pattern as ESAT-6/CFP-10-specific cells but both were significantly different from TT-specific (p<0.0001 and p=0.005, respectively) and SEB-reactive CD4+ T cells (p<0.0001) (Figure 4D). Detailed analysis of TF combinations expressed by antigen-specific CD4+ T cells revealed that the most prevalent subsets within PPD-specific CD4+ T cells were Foxp3+RORγthighT-bethigh, Foxp3+T-bethigh, and T-bethigh (each subset representing approximately 20% of antigen-responding cells). In response to TT stimulation, the spectrum of TF expression was broader, with ∼20% of cells expressing Foxp3 and T-bet, Foxp3 alone or none of the TF tested, and ∼10% of cells being Foxp3+Gata3high or T-bethigh. SEB-responding cells consisted predominantly of T-bethigh cells (∼40%), Foxp3+T-bethigh (∼20%), or cells negative for all TF tested (∼20%) (Figure 4E).
Figure 4. Diversity of T helper CD4 subsets in response to different pathogens upon a 5-day in vitro stimulation in healthy Mtb-sensitized individuals.
(A) Representative dot plots of Ki67 expression in CD4+ T cells in response to a 5 day-stimulation in the presence of Mtb peptide pool (ESAT-6/CFP-10), Mtb purified protein derivative (PPD) or tetanus toxoid (TT) within the same individual. The frequency of antigen-specific CD4+ T cells is indicated in the gate. (B) Frequencies of proliferating antigen-specific CD4+ T cells after background subtraction. Twenty individuals were tested for PPD, 17 for ESAT6/CFP10 and 11 for TT responses. Bars represent the median. The number of responders is indicated at the bottom of the graph. (C) Representative overlay histograms of Foxp3, RORγt, Gata3 and T-bet expression levels in Ki67+CD4+ T cells in response to ESAT-6/CFP-10 peptide pool (blue), PPD (red) and TT (green) stimulation within the same individual. The black line corresponds to Ki67- CD4+ T cells. (D) Pie charts showing the median proportion of each possible TF combination in ESAT-6/CFP-10-, PPD-, TT- and SEB-specific Ki67+CD4+ T cells. The number of individuals analyzed for each stimulus is indicated. Statistical comparisons were performed using the pie statistic tool integrated in the Spice software. (E) Proportions of cells expressing 1, 2, 3, 4 or none of the lineage-defining transcription factors tested in ESAT-6/CFP-10- (blue), PPD- (red), TT- (green) and SEB- (grey) specific CD4+ T cells using a Boolean gating strategy. Bars represent median and interquartile ranges. Statistical comparisons were performed using a Student's t-test. *<0.05, **<0.01, ***<0.001.
These data demonstrate that the spectrum of TF expression in CD4+ T cells varies according to pathogen specificity, indicating that CD4 polarization into distinct Th subsets depends on the nature of the antigen recognized.
PPD-specific CD4+ T cells with different lineage-defining transcription factor profiles exhibit distinct functional capacities
To determine whether cytokine secretion capacity varied according to TF expression pattern, we measured the ability of PPD-specific CD4+ T cells to produce IFN-γ and TNF-α. Since PPD responses were of greater magnitude and exhibited a similar TF profile to ESAT-6/CFP-10 peptide responses, we focused on PPD responses to assess the functional capacity of Mtb-specific CD4+ T cells expressing different TF combinations. PBMC from healthy individuals (n=6) were stimulated with PPD for 5 days and cytokine secretion was blocked using Brefeldin during the 5 last hours of the stimulation. Since only a low frequency of PPD-specific CD4+ T cells expressed Gata3, we concentrated on three TF for these experiments. The co-expression profile of T-bet, RORγt and Foxp3 in PPD-specific CD4+ T cells is presented in Figure 5A. A median of 35% (range: 35% to 41%) of proliferating PPD-specific CD4+ T cells produced TNF-α (Figure 5B), while 19% (range: 17% to 25%) were positive for IFN-γ (Figure 5C). We next assessed TNF-α and IFN-γ secretion in PPD-specific CD4+ T cells exhibiting distinct TF expression profiles (Figure 5D). Given the low frequency (<2.5%) of PPD-specific CD4+ T cells with a T-betlowRORγthighFoxp3+ or T-betlowRORγthighFoxp3- phenotype (Figure 5A), these two subsets were excluded from analysis. The highest TNF-α production (∼60%) was observed in T-bethighRORγthighFoxp3+ PPD-specific CD4+ T cells, or co-expressing T-bet and RORγt. Foxp3+T-bethigh and T-bethigh cells moderately expressed TNF-α, with a median of 27% and 35% of TNF-α positive cells, respectively. The lowest TNF-α production (∼10%) was observed in cells expressing only Foxp3, or none of the TF tested. Whilst the overall proportion of PPD-specific CD4+ T cells producing IFN-γ was significantly lower than cells producing TNF-α (p=0.028), a similar secretion profile was observed among the different subsets (Figures 5D-E). Further analyses showed that PPD-specific CD4+ T cells co-expressing T-bet, RORγt and Foxp3, or T-bet and RORγt, were significantly more bi-functional than Foxp3+T-bethigh cells or T-bethigh single positive cells (Figure 5F). Of note, it is worth noting that by assessing the cytokine secretion profile of Mtb-responsive CD4+ T cells (Ki67+cells) after a 5-day proliferation assay, this may not recapitulate their full functional potential since these cells will more likely have a broader cytokine secretion profile at the onset of activation, prior to TF up-regulation.
Figure 5. Functional capacity of PPD-specific CD4+ T cells varies according to their transcription factor profile.
PBMC from HIV-uninfected Mtb-sensitized individuals (n=6) were stimulated with PPD for 5 days. Brefeldin A was added for the last 5 hours and the expression level of T-bet, RORγt, Foxp3, IFN-γ and TNF-α were assessed by flow cytometry. (A) Proportion of cells expressing 3, 2, 1 or no transcription factors in PPD-specific CD4+ T cells using a Boolean gating strategy. Expression of each of the 8 possible combinations of transcription factors is shown on the x-axis. Bars represent median and interquartile ranges. Above the dotted horizontal line are the subsets that represent >2.5% of the total PPD-specific CD4+ T cells. The pie chart summarizes the data showing the median proportion of each possible TF combination in PPD-specific CD4+ T cells. The arc illustrates the proportion of PPD-specific CD4+ T cells co-expressing two or three transcription factors. (B and C) Representative dot plots of TNF-α (B) and IFN-γ (C) expression in PPD-specific Ki67+CD4+ T cells in response to a 5 day-stimulation in the presence of PPD within the same individual. The median and range of the proportion of TNF-α and IFN-γ positive cells within PPD-specific Ki67+CD4+ T cells, for the six individuals analyzed, is indicated. NS: No Stimulation. (D) Proportion of TNF-α+ cells (top panel) or IFN-γ+ cells (bottom panel) within the Ki67+CD4+ T cells expressing different combinations of TF. Bars represent the median and interquartile range. Only subsets representing > 2.5% of total Ki67+ CD4+ T cells are depicted on the graph. (E) Representative overlay dot plots of TNF-α (top) and IFN-γ (bottom) expression in Ki67+CD4+ T cells (grey) and PPD-specific CD4+ T cells exhibiting distinct TF profiles, namely T-bethighRORγthighFoxp3+ (red); T-bethighRORγthighFoxp3- (orange); T-bethighRORγtlowFoxp3+ (green); T-bethighRORγtlowFoxp3- (blue) and T-betlowRORγtlowFoxp3- (purple). The percentage of cytokine positive cells within the PPD-specific Ki67+ population is shown on each dot plot. Ki67-CD4+ T cells are shown in grey. (F) Proportion of IFN-γ+TNF-α+ cells (black), IFN-γ-TNF-α+ (dark grey), IFN-γ+TNF-α- (light grey) and IFN-γ-TNF-α- (white) cells within PPD-specific Ki67+CD4+ T cells exhibiting distinct TF profiles.
We also assessed the expression of IL-2 and IL-17, these cytokines were not detectable in response to PPD. Thus, to define whether PPD-specific CD4+ T cells retained the capacity to produce these cytokines, cells were re-stimulated with PMA/Ionomycin as described in Figure 6A. Whilst a small fraction of PPD-specific Ki67+CD4+ T cells were able to secrete IL-2 (∼10%), no IL-17 was detected in response to mitogen re-stimulation (Figure 6B). Moreover, in response to PMA/Ionomycin restimulation, cycling PPD-specific Ki67+CD4+ T cells exhibited a broad spectrum of TF expression profile while the majority of PMA/Ionomycin-responding CD4+ T cells did not express any of the measured transcription factors, regardless of their cytokine expression profile (Figure 6C). It is worth mentioning that similar data where obtained when cells were restimulated using PPD (added 3 hours prior to Brefeldin addition), showing that sufficient Mtb antigens were still present at saturating levels over the culture period in our experimental condition. These data could be explained by distinct kinetics of production of these cytokines (IL-2 and IL-17) as compared to IFN-γ or TNF-α in our experimental setting.
Figure 6. Cytokine secretion potential of PPD-responsive CD4+ T cells upon PMA/ionomycin re-stimulation.
(A) Experimental design. PBMC were stimulated with PPD or unstimulated (Stim 1) for 5 days. Five hours before the end of the stimulation, brefeldin was added alone or in the presence PMA and Ionomycin (Stim 2). After 5 hours, cells were stained and analyzed by flow cytometry. (B) Representative examples of cytokine expression (IFN-γ, IL-2 and IL-17) in PPD-responding CD4+ T cells after a 5-day stimulation and in response to PMA and Ionomycin re-stimulation. (C) Transcription factor expression profile of PMA/ionomycin- or PPD-responding CD4+ T cells. The pies show the median proportion of each possible combination of TF. Each slice of the pie corresponds to a distinct combination of TF. Data are representative of 3 independent experiments.
Additionally, a sizeable proportion of TNF-α and IFN-γ-producing cells were detected within the Ki67-negative CD4+ T cell population in response to PPD [for TNF-α: a median of 43.3% of total TNF-α-producing cells (ranging from 20% to 57%), Figure 5B; and for IFN-γ, a median of 23% of total IFN-γ-producing cells (ranging from 8% to 39%), Figure 5C]. This indicates that not all PPD-responding cells actively proliferated upon a 5-day stimulation. We characterized the TF expression profile of PPD-responding non-proliferating CD4+ T cells. In these cells, the degree of TF co-expression was significantly lower compared to Ki67+IFN-γ+ cells (∼25% vs ∼80%) (Supplemental Figure S2). Since no up-regulation of TF was observed after short-term activation, these data suggest that the up-regulation of TF could require prolonged engagement of the TCR in combination with an appropriate “polarizing” cytokine milieu.
Overall, these data show that depending on the combination of TF expressed, PPD-specific CD4+ T cells have distinct functional capacities, suggesting that these cells are endowed with different effector functions.
HIV infection alters the transcription factor profile of PPD-specific CD4+ Th subsets
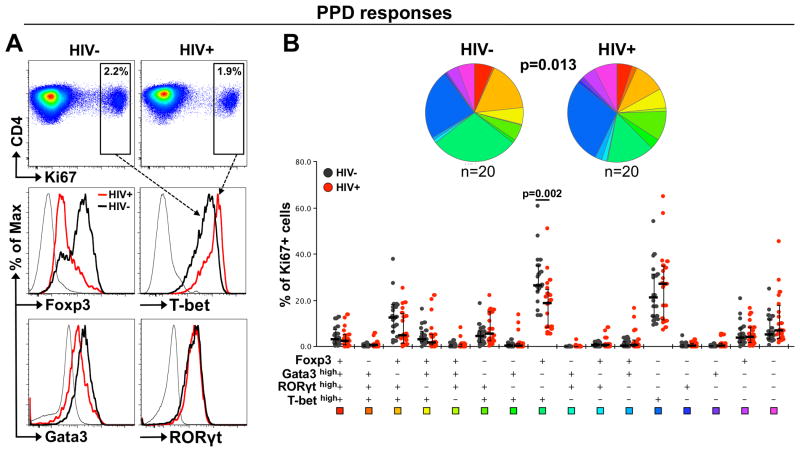
To determine whether HIV infection affects the lineage commitment of the CD4 response to TB, we compared the TF expression profiles of PPD-specific CD4+ T cells in HIV-uninfected and HIV-infected individuals. The HIV-infected individuals included in this study had well-maintained CD4 counts (median: 602 cells/mm3), variable HIV plasma viral loads (median: 7,788 RNA copies/ml) and were ART naïve (Table 1). A representative example of Foxp3, T-bet, Gata3 and RORγt expression levels in PPD-specific CD4+ T cells in an HIV-uninfected and HIV-infected subject is presented in Figure 7A. The overall TF expression profiles in PPD-specific CD4+ T cells were significantly different between HIV-uninfected and HIV-infected individuals (p=0.013). One specific subset of PPD-responding CD4+ T cells was altered in HIV-infected individuals where a significantly lower proportion of cells co-expressing Foxp3 and T-bethigh was observed compared to HIV-uninfected subjects (p=0.002) (Figure 7B). Of note, similar results were obtained for ESAT-6/CFP-10 responding CD4+ T cells, while no difference in the TF profile of SEB-responding CD4+ T cells was observed between HIV-uninfected and HIV-infected individuals (Supplemental Figure S3). Since PBMC were cultured for 5 days, these differences could have arisen as a result of in vitro HIV replication. However, stimulation in the presence of antiretroviral drugs did not affect the TF profile observed in PPD-specific CD4+ T cells in HIV-infected individuals (data not shown). This suggests that the skewing of TF profiles in PPD-specific CD4+ T cells in HIV-infected individuals was not an artifact of cell culture and reflects in vivo changes to the CD4+ Th response to TB.
Figure 7. Comparison of TF expression profiles in PPD-specific CD4+ T cells in HIV-infected and HIV-uninfected individuals.
(A) Top panel: Representative dot plots of Ki67 expression in response to a 5 day PPD stimulation in an HIV-infected and an HIV-uninfected individual. Bottom panel: Representative overlay histograms of Foxp3, RORγt, Gata3 and T-bet expression levels in PPD-specific Ki67+ CD4+ T cell responses in an HIV-infected (red line) and an HIV-uninfected (black line) individual. (B) Top panel: Pie charts showing the median proportion of each possible TF combination within PPD-specific Ki67+CD4+ T cells in 20 HIV-infected and 20 HIV-uninfected individuals. Statistical comparisons were performed using the pie statistic tool integrated in the Spice software. Bottom panel: Proportions of cells expressing each possible TF combination in PPD-specific CD4+ T cells using a Boolean gating strategy. HIV-infected individuals are depicted with red dots and HIV-uninfected individuals with black dots. Bars represent medians and interquartile ranges. Statistical comparisons were performed using the Student's t-test.
We next examined the relationship between TF expression profiles in PPD-specific CD4+ T cells and HIV viral load. Figure 8A shows T-bet and Foxp3 expression levels in PPD-specific CD4+ T cells in individuals with differing HIV viral loads. We found a striking significant inverse correlation between the proportion of Foxp3+T-bethigh PPD-specific CD4+ T cells and HIV viral load (r=-0.69, p=0.0007, Figure 8B), suggesting that the reduction in this subset may be related to in vivo HIV viral replication. Of note, the proportion of Foxp3+T-bethigh PPD-specific CD4+ T cells also positively associate with absolute CD4 count in HIV-infected individuals (p=0.01, r=0.56, data not shown). To define potential factors that impact the lineage differentiation of Mtb-specific CD4+ T cells during HIV infection, we investigated the relationship between markers of chronic inflammation and the TF profile of PPD-specific CD4+ T cells. Concentrations of 10 cytokines/chemokines were measured in the plasma of HIV-infected (n=18) and HIV-uninfected subjects (n=18). The proportion of Foxp3+T-bethigh PPD-specific CD4+ T cells inversely correlated with plasma concentrations of TNF-α and IFN-γ (Figure 8C). After adjusting for multiple comparisons, the association between plasma TNF-α concentration and PPD-specific Foxp3+T-bethigh CD4+ T cells remained significant (p=0.019, r=-0.5).
Figure 8. Association of the TF profile of PPD-specific CD4+ T cells with HIV viral load and IFN-γ and TNF-α plasma concentrations.
(A) Representative dot pots of the expression profile of Foxp3 and T-bet in PPD-specific Ki67+CD4+ T cells (red) overlaid on total Ki67-CD4+ T cells in four HIV-infected individuals with varying HIV viral loads (VL). The percentage of Ki67+CD4+ T cells is indicated in each quadrant. (B) Relationship between the proportion of PPD-specific Foxp3+T-bethighKi67+CD4+ T cells and HIV viral load. (C) Relationship between the proportion of PPD-specific Foxp3+T-bethigh Ki67+CD4+ T cells and the plasma concentration of IFN-γ and TNF-α in HIV-infected (red dots) and HIV-uninfected (black dots) individuals. Cytokine measurements were available for 18 of the HIV-infected and 18 uninfected individuals. Correlations were tested by a two-tailed non-parametric Spearman Rank test. For plasma cytokines, p-values were adjusted for multiple comparisons using the Bonferroni step-down (Holm) procedure. Crude and adjusted (Adj) p-values are reported.
Overall, these data suggest that HIV infection impairs the lineage commitment of Mtb-specific CD4+ T cells, leading to a significant decrease of one particular subset, cells co-expressing Foxp3 and T-bet. The contraction of this CD4+ T cell subset correlated with HIV viral load and plasma TNF-α, suggesting that the host inflammatory status may play an important role in controlling the fate of Mtb-specific CD4+ T cells.
Discussion
To better understand the CD4 immune response to Mtb, we evaluated the diversity of Mtb-specific-CD4+ Th subsets. The expression of four canonical transcription factors, T-bet, Gata3, RORγt and Foxp3 was measured simultaneously within Mtb-specific CD4+ T cells. Our data indicate that Mtb-specific CD4+ T cells exhibit a hitherto unappreciated wide spectrum of Th subsets, defined by the co-expression of lineage-defining transcription factors. Since HIV infection is a major risk factor for TB, we also examined the extent to which HIV impacts the diversity of Mtb-specific CD4+ Th subsets. Our results show that HIV infection significantly alters the transcriptional profiles of these cells, most likely by promoting an inflammatory environment; these changes in the equilibrium of Mtb-specific CD4+ Th subsets during HIV infection may have implications for the development and/or maintenance of protective TB immunity.
Based on the “the master regulator” model where the expression of a single transcription factor unilaterally controls the fate of CD4 lineage commitment, the co-expression of multiple transcription factors that we observed in Mtb-specific CD4+ T cells may seem surprising. However, over the past few years a mounting body of evidence has revealed that lineage-defining transcription factors act as a network more often than they do in isolation, and co-expression of TF in CD4+ T cells allows for the diversity and flexibility of CD4+ Th subsets (reviewed in (17)). The co-expression of RORγt and T-bet has been reported in both mice (41-43) and humans (23, 44, 45). These cells, referred as Th1/Th17, have the ability to secrete IFN-γ and IL-17, and exhibit pathogenic potential in several disease models. Additionally, the stable co-expression of Foxp3 with other lineage-defining transcription factors, such as T-bet, Gata3, RORγt or Bcl-6, has been described in specialized Treg subpopulations; these cells co-express the transcription factor of the very cells they are able to regulate (46-48). These observations illustrate the complexity of the transcriptional regulation of CD4+ Th differentiation and suggest that the diversity of Th subsets may be more varied than described to date. While the majority of circulating Mtb-specific CD4+ T cells does not co-express transcription factors, they retain the potential to up-regulate several transcription factors after in vitro expansion. These data suggest that the majority of memory CD4+ T cells are not maintained as committed to a specific lineage in the absence of antigen stimulation and that several rounds of cell division are necessary to induce TF upregulation. In this context, the quality and quantity of cytokines produced at the onset of T cell activation, which are dependent on the nature of the threat encountered, most likely dictates the array of Th subsets generated upon antigen re-exposure. Depending on the combination of TF expressed, Mtb-specific CD4+ T cells exhibited a different potential to secrete IFN-γ or TNF-α, suggesting that these distinct subsets have different effector functions. Further analyses will be needed to define in more detail the cytokine secretion potential of these different subsets.
What is the relevance of such CD4 diversity in the protection against TB? Th1, Th17 and Treg cells have been involved in TB immunity (5-12), suggesting that the control of Mtb replication depends on the generation of a balanced CD4 response, reaching an equilibrium between effector and regulatory cells. Our data support this hypothesis where, upon Mtb encounter, memory CD4+ T cells differentiate into a spectrum of Th subsets with distinct functions. Moreover, we showed that in HIV-infected individuals with well-preserved CD4 counts, the TF profile of Mtb-specific CD4+ T cells is significantly altered compared to HIV-uninfected individuals, with a reduction in one specific subset co-expressing Foxp3 and T-bet. If we consider this particular subset to exhibit regulatory functions, its contraction could impair bacterial control by promoting premature cell exhaustion (49), enhancing pro-inflammatory signals and possible tissue damage (12, 50) and favoring an unbalanced Teff/Treg ratio. Alternatively, it is also possible that the up-regulation of Foxp3 is a transient phenomenon induced in response to TCR triggering. In this context, Foxp3 up-regulation has been associated with hypo-responsiveness of activated T cells, but was not directly correlated with their suppressive capabilities (51). Thus, an impaired Th1 response may also compromise bacterial control and this would also suggest that the CD4 response towards Mtb in HIV-infected individuals is of a more inflammatory nature compared to HIV-uninfected individuals.
Several mechanisms could be responsible for the alteration of Mtb-specific CD4+ Th subsets in the context of HIV infection. Firstly, the composition of the cytokine milieu, at the time of TCR engagement, plays a major role in regulating the lineage commitment of CD4+ T cells. It has been demonstrated that the polyfunctional capacities of Mtb-specific CD4+ T cells are altered during HIV infection, where the proportion of IL-2 secreting Mtb-specific CD4+ T cells inversely correlate with HIV viral load (30). Moreover, HIV infection is characterized by generalized dysregulated cytokine production, partially driven by viral replication (34). Thus, HIV infection could create a biased cytokine environment leading to aberrant differentiation of Mtb-specific CD4+ T cells. Our results are in accordance with this hypothesis, showing that the proportion of Mtb-specific CD4+ T cells co-expressing Foxp3 and T-bet was inversely correlated with HIV viral load and the plasma concentration TNF-α. This suggests that HIV-induced chronic inflammation could dramatically impact the differentiation profile and functionality of TB-specific CD4+ T cells responses (and, indeed, responses to other pathogens). Since inflammatory signals are known to modulate the epigenetic state of cells (19, 52, 53), we could hypothesize that a sustained inflammatory environment may “imprint“ the potential of CD4+ T cells for plasticity by modulating their epigenetic signature. The effect of bystander inflammation on the establishment of memory T cells has been previously reported in a LCMV mouse model (54), where prolonged inflammation disrupts the development of memory CD8+T cells; a phenomenon regulated by T-bet and Blimp-1. Secondly, there is some evidence that HIV preferentially infects Mtb-specific CD4+ T cells (25, 55). Therefore, the contraction of Foxp3+T-bethigh Mtb-specific CD4+ T cells could also result from the increased permissiveness to HIV infection of this particular subset.
It is worth noting that, by assessing the transcription factor profile of Mtb-responsive CD4+ T cells (Ki67+cells) after a 5-day proliferation assay, we may have excluded other relevant subsets of effector cells that contribute to the immune response to Mtb but do not proliferate and it remains to be determined whether such flexibility will be observed in vivo. The TF profile described in this experimental setting may only partially recapitulate the global ex vivo Mtb-specific CD4+ T cell response. Nevertheless, our results show that in Mtb-sensitized individuals, a portion of Mtb-specific CD4+ T cells in the periphery may be present in a quiescent state and long-term reactivation allowed us to reveal their flexible potential. Moreover, Further experiments will be needed to define whether the HIV-induced alteration of Mtb-specific CD4+ T cells is restricted to this particular pathogen or also observed in CD4+ T cells of other specificities.
In conclusion, these studies have implications not only for the understanding of the specific attributes of Mtb-specific responses required for an efficient immune response to Mtb but could shed further light on the mechanism by which HIV increases TB susceptibility. We hypothesize that HIV-induced sustained inflammation may promote the generation of a TB-specific CD4 response of the right specificity but the wrong type, where CD4 functional capacity shifts towards a more pathogenic/inflammatory profile, which impair efficient control of bacterial growth.
Supplementary Material
Acknowledgments
We thank Rubina Bunjun and Tracey Müller for assistance with sample processing, and Kathryn Norman for administrative assistance and Dr Mario Roederer for scientific input.
This work was funded by the National Research Foundation of South Africa (NRF, 92558 to CR), Poliomyelitis Research Foundation (PRF, 14/20 to CR), the National Institutes of Health, the Office of the Director (OD) (NIH, R21AI115977 to CR and EJW); and European and Developing Countries Clinical Trials Partnership (EDCTP; TA_08_40200_020 to WAB). RJW is supported by the Wellcome Trust (104803), the Medical Research Council (MRC, U1175.02.002.00014.01), the European Union (FP7-Health-F3-2012-305578) and FP7-PEOPLE-2011-IRSES. WAB is a Wellcome Trust Intermediate Fellow in Public Health and Tropical Medicine (089832/Z/09/Z).
Footnotes
Contributions: CR and WAB designed the study; CR, NS and APS performed the experiments; BC and DSK generated the plasma cytokine data; CR analyzed the data and wrote the paper. RJW, EJW and WAB contributed to the data discussion and participated in the preparation of the manuscript.
Disclosure: The authors have no conflict of interest.
References
- 1.Ernst JD. The immunological life cycle of tuberculosis. Nature reviews Immunology. 2012;12:581–591. doi: 10.1038/nri3259. [DOI] [PubMed] [Google Scholar]
- 2.Lin PL, Flynn JL. Understanding latent tuberculosis: a moving target. Journal of immunology. 2010;185:15–22. doi: 10.4049/jimmunol.0903856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O'Garra A, Redford PS, McNab FW, Bloom CI, Wilkinson RJ, Berry MP. The immune response in tuberculosis. Annual review of immunology. 2013;31:475–527. doi: 10.1146/annurev-immunol-032712-095939. [DOI] [PubMed] [Google Scholar]
- 4.Prezzemolo T, Guggino G, La Manna MP, Di Liberto D, Dieli F, Caccamo N. Functional Signatures of Human CD4 and CD8 T Cell Responses to Mycobacterium tuberculosis. Frontiers in immunology. 2014;5:180. doi: 10.3389/fimmu.2014.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desvignes L, Wolf AJ, Ernst JD. Dynamic roles of type I and type II IFNs in early infection with Mycobacterium tuberculosis. Journal of immunology. 2012;188:6205–6215. doi: 10.4049/jimmunol.1200255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wozniak TM, Saunders BM, Ryan AA, Britton WJ. Mycobacterium bovis BCG-specific Th17 cells confer partial protection against Mycobacterium tuberculosis infection in the absence of gamma interferon. Infection and immunity. 2010;78:4187–4194. doi: 10.1128/IAI.01392-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chatterjee S, Dwivedi VP, Singh Y, Siddiqui I, Sharma P, Van Kaer L, Chattopadhyay D, Das G. Early secreted antigen ESAT-6 of Mycobacterium tuberculosis promotes protective T helper 17 cell responses in a toll-like receptor-2-dependent manner. PLoS pathogens. 2011;7:e1002378. doi: 10.1371/journal.ppat.1002378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cruz A, Fraga AG, Fountain JJ, Rangel-Moreno J, Torrado E, Saraiva M, Pereira DR, Randall TD, Pedrosa J, Cooper AM, Castro AG. Pathological role of interleukin 17 in mice subjected to repeated BCG vaccination after infection with Mycobacterium tuberculosis. The Journal of experimental medicine. 2010;207:1609–1616. doi: 10.1084/jem.20100265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nandi B, Behar SM. Regulation of neutrophils by interferon-gamma limits lung inflammation during tuberculosis infection. The Journal of experimental medicine. 2011;208:2251–2262. doi: 10.1084/jem.20110919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scott-Browne JP, Shafiani S, Tucker-Heard G, Ishida-Tsubota K, Fontenot JD, Rudensky AY, Bevan MJ, Urdahl KB. Expansion and function of Foxp3-expressing T regulatory cells during tuberculosis. The Journal of experimental medicine. 2007;204:2159–2169. doi: 10.1084/jem.20062105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shafiani S, Tucker-Heard G, Kariyone A, Takatsu K, Urdahl KB. Pathogen-specific regulatory T cells delay the arrival of effector T cells in the lung during early tuberculosis. The Journal of experimental medicine. 2010;207:1409–1420. doi: 10.1084/jem.20091885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McBride A, Konowich J, Salgame P. Host defense and recruitment of Foxp3(+) T regulatory cells to the lungs in chronic Mycobacterium tuberculosis infection requires toll-like receptor 2. PLoS pathogens. 2013;9:e1003397. doi: 10.1371/journal.ppat.1003397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 2010;327:1098–1102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu J, Paul WE. Peripheral CD4+ T-cell differentiation regulated by networks of cytokines and transcription factors. Immunological reviews. 2010;238:247–262. doi: 10.1111/j.1600-065X.2010.00951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakayamada S, Takahashi H, Kanno Y, O'Shea JJ. Helper T cell diversity and plasticity. Current opinion in immunology. 2012;24:297–302. doi: 10.1016/j.coi.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murphy KM, Stockinger B. Effector T cell plasticity: flexibility in the face of changing circumstances. Nature immunology. 2010;11:674–680. doi: 10.1038/ni.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oestreich KJ, Weinmann AS. Master regulators or lineage-specifying? Changing views on CD4+ T cell transcription factors. Nature reviews Immunology. 2012;12:799–804. doi: 10.1038/nri3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evans CM, Jenner RG. Transcription factor interplay in T helper cell differentiation. Briefings in functional genomics. 2013;12:499–511. doi: 10.1093/bfgp/elt025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vahedi G, Takahashi H, Nakayamada S, Sun HW, Sartorelli V, Kanno Y, O'Shea JJ. STATs shape the active enhancer landscape of T cell populations. Cell. 2012;151:981–993. doi: 10.1016/j.cell.2012.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanno Y, Vahedi G, Hirahara K, Singleton K, O'Shea JJ. Transcriptional and epigenetic control of T helper cell specification: molecular mechanisms underlying commitment and plasticity. Annual review of immunology. 2012;30:707–731. doi: 10.1146/annurev-immunol-020711-075058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tripathi SK, Lahesmaa R. Transcriptional and epigenetic regulation of T-helper lineage specification. Immunological reviews. 2014;261:62–83. doi: 10.1111/imr.12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang YH, Voo KS, Liu B, Chen CY, Uygungil B, Spoede W, Bernstein JA, Huston DP, Liu YJ. A novel subset of CD4(+) T(H)2 memory/effector cells that produce inflammatory IL-17 cytokine and promote the exacerbation of chronic allergic asthma. The Journal of experimental medicine. 2010;207:2479–2491. doi: 10.1084/jem.20101376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zielinski CE, Mele F, Aschenbrenner D, Jarrossay D, Ronchi F, Gattorno M, Monticelli S, Lanzavecchia A, Sallusto F. Pathogen-induced human TH17 cells produce IFN-gamma or IL-10 and are regulated by IL-1beta. Nature. 2012;484:514–518. doi: 10.1038/nature10957. [DOI] [PubMed] [Google Scholar]
- 24.Ueno A, Jijon H, Chan R, Ford K, Hirota C, Kaplan GG, Beck PL, Iacucci M, Fort Gasia M, Barkema HW, Panaccione R, Ghosh S. Increased prevalence of circulating novel IL-17 secreting Foxp3 expressing CD4+ T cells and defective suppressive function of circulating Foxp3+ regulatory cells support plasticity between Th17 and regulatory T cells in inflammatory bowel disease patients. Inflammatory bowel diseases. 2013;19:2522–2534. doi: 10.1097/MIB.0b013e3182a85709. [DOI] [PubMed] [Google Scholar]
- 25.Geldmacher C, Zumla A, Hoelscher M. Interaction between HIV and Mycobacterium tuberculosis: HIV-1-induced CD4 T-cell depletion and the development of active tuberculosis. Current opinion in HIV and AIDS. 2012;7:268–275. doi: 10.1097/COH.0b013e3283524e32. [DOI] [PubMed] [Google Scholar]
- 26.Badri M, Wilson D, Wood R. Effect of highly active antiretroviral therapy on incidence of tuberculosis in South Africa: a cohort study. Lancet. 2002;359:2059–2064. doi: 10.1016/S0140-6736(02)08904-3. [DOI] [PubMed] [Google Scholar]
- 27.Sonnenberg P, Glynn JR, Fielding K, Murray J, Godfrey-Faussett P, Shearer S. How soon after infection with HIV does the risk of tuberculosis start to increase? A retrospective cohort study in South African gold miners. The Journal of infectious diseases. 2005;191:150–158. doi: 10.1086/426827. [DOI] [PubMed] [Google Scholar]
- 28.Geldmacher C, Ngwenyama N, Schuetz A, Petrovas C, Reither K, Heeregrave EJ, Casazza JP, Ambrozak DR, Louder M, Ampofo W, Pollakis G, Hill B, Sanga E, Saathoff E, Maboko L, Roederer M, Paxton WA, Hoelscher M, Koup RA. Preferential infection and depletion of Mycobacterium tuberculosis-specific CD4 T cells after HIV-1 infection. The Journal of experimental medicine. 2010;207:2869–2881. doi: 10.1084/jem.20100090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diedrich CR, Flynn JL. HIV-1/mycobacterium tuberculosis coinfection immunology: how does HIV-1 exacerbate tuberculosis? Infection and immunity. 2011;79:1407–1417. doi: 10.1128/IAI.01126-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Day CL, Mkhwanazi N, Reddy S, Mncube Z, van der Stok M, Klenerman P, Walker BD. Detection of polyfunctional Mycobacterium tuberculosis-specific T cells and association with viral load in HIV-1-infected persons. The Journal of infectious diseases. 2008;197:990–999. doi: 10.1086/529048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jambo KC, Sepako E, Fullerton DG, Mzinza D, Glennie S, Wright AK, Heyderman RS, Gordon SB. Bronchoalveolar CD4+ T cell responses to respiratory antigens are impaired in HIV-infected adults. Thorax. 2011;66:375–382. doi: 10.1136/thx.2010.153825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mattila JT, Diedrich CR, Lin PL, Phuah J, Flynn JL. Simian immunodeficiency virus-induced changes in T cell cytokine responses in cynomolgus macaques with latent Mycobacterium tuberculosis infection are associated with timing of reactivation. Journal of immunology. 2011;186:3527–3537. doi: 10.4049/jimmunol.1003773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomlinson GS, Bell LC, Walker NF, Tsang J, Brown JS, Breen R, Lipman M, Katz DR, Miller RF, Chain BM, Elkington PT, Noursadeghi M. HIV-1 infection of macrophages dysregulates innate immune responses to Mycobacterium tuberculosis by inhibition of interleukin-10. The Journal of infectious diseases. 2014;209:1055–1065. doi: 10.1093/infdis/jit621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reuter MA, Pombo C, Betts MR. Cytokine production and dysregulation in HIV pathogenesis: lessons for development of therapeutics and vaccines. Cytokine & growth factor reviews. 2012;23:181–191. doi: 10.1016/j.cytogfr.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roederer M, Nozzi JL, Nason MC. SPICE: exploration and analysis of post-cytometric complex multivariate datasets. Cytometry Part A : the journal of the International Society for Analytical Cytology. 2011;79:167–174. doi: 10.1002/cyto.a.21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Statistics in medicine. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 37.Soares A, Govender L, Hughes J, Mavakla W, de Kock M, Barnard C, Pienaar B, Janse van Rensburg E, Jacobs G, Khomba G, Stone L, Abel B, Scriba TJ, Hanekom WA. Novel application of Ki67 to quantify antigen-specific in vitro lymphoproliferation. Journal of immunological methods. 2010;362:43–50. doi: 10.1016/j.jim.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hale JS, Youngblood B, Latner DR, Mohammed AU, Ye L, Akondy RS, Wu T, Iyer SS, Ahmed R. Distinct memory CD4+ T cells with commitment to T follicular helper- and T helper 1-cell lineages are generated after acute viral infection. Immunity. 2013;38:805–817. doi: 10.1016/j.immuni.2013.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lazarevic V, Glimcher LH, Lord GM. T-bet: a bridge between innate and adaptive immunity. Nature reviews Immunology. 2013;13:777–789. doi: 10.1038/nri3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaech SM, Cui W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nature reviews Immunology. 2012;12:749–761. doi: 10.1038/nri3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou L, Lopes JE, Chong MM, Ivanov, Min R, Victora GD, Shen Y, Du J, Rubtsov YP, Rudensky AY, Ziegler SF, Littman DR. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature. 2008;453:236–240. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ghoreschi K, Laurence A, Yang XP, Tato CM, McGeachy MJ, Konkel JE, Ramos HL, Wei L, Davidson TS, Bouladoux N, Grainger JR, Chen Q, Kanno Y, Watford WT, Sun HW, Eberl G, Shevach EM, Belkaid Y, Cua DJ, Chen W, O'Shea JJ. Generation of pathogenic T(H)17 cells in the absence of TGF-beta signalling. Nature. 2010;467:967–971. doi: 10.1038/nature09447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee Y, Awasthi A, Yosef N, Quintana FJ, Xiao S, Peters A, Wu C, Kleinewietfeld M, Kunder S, Hafler DA, Sobel RA, Regev A, Kuchroo VK. Induction and molecular signature of pathogenic TH17 cells. Nature immunology. 2012;13:991–999. doi: 10.1038/ni.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Annunziato F, Cosmi L, Santarlasci V, Maggi L, Liotta F, Mazzinghi B, Parente E, Fili L, Ferri S, Frosali F, Giudici F, Romagnani P, Parronchi P, Tonelli F, Maggi E, Romagnani S. Phenotypic and functional features of human Th17 cells. The Journal of experimental medicine. 2007;204:1849–1861. doi: 10.1084/jem.20070663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kastirr I, Maglie S, Paroni M, Alfen JS, Nizzoli G, Sugliano E, Crosti MC, Moro M, Steckel B, Steinfelder S, Stolzel K, Romagnani C, Botti F, Caprioli F, Pagani M, Abrignani S, Geginat J. IL-21 Is a Central Memory T Cell-Associated Cytokine That Inhibits the Generation of Pathogenic Th1/17 Effector Cells. Journal of immunology. 2014;193:3322–3331. doi: 10.4049/jimmunol.1400775. [DOI] [PubMed] [Google Scholar]
- 46.Feuerer M, Hill JA, Mathis D, Benoist C. Foxp3+ regulatory T cells: differentiation, specification, subphenotypes. Nature immunology. 2009;10:689–695. doi: 10.1038/ni.1760. [DOI] [PubMed] [Google Scholar]
- 47.Josefowicz SZ, Rudensky A. Control of regulatory T cell lineage commitment and maintenance. Immunity. 2009;30:616–625. doi: 10.1016/j.immuni.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Campbell DJ, Koch MA. Phenotypical and functional specialization of FOXP3+ regulatory T cells. Nature reviews Immunology. 2011;11:119–130. doi: 10.1038/nri2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barber DL, Mayer-Barber KD, Feng CG, Sharpe AH, Sher A. CD4 T cells promote rather than control tuberculosis in the absence of PD-1-mediated inhibition. Journal of immunology. 2011;186:1598–1607. doi: 10.4049/jimmunol.1003304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sakai S, Mayer-Barber KD, Barber DL. Defining features of protective CD4 T cell responses to Mycobacterium tuberculosis. Current opinion in immunology. 2014;29:137–142. doi: 10.1016/j.coi.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang J, Ioan-Facsinay A, van der Voort EI, Huizinga TW, Toes RE. Transient expression of FOXP3 in human activated nonregulatory CD4+ T cells. Eur J Immunol. 2007;37:129–138. doi: 10.1002/eji.200636435. [DOI] [PubMed] [Google Scholar]
- 52.Backdahl L, Bushell A, Beck S. Inflammatory signalling as mediator of epigenetic modulation in tissue-specific chronic inflammation. The international journal of biochemistry & cell biology. 2009;41:176–184. doi: 10.1016/j.biocel.2008.08.023. [DOI] [PubMed] [Google Scholar]
- 53.Natoli G. Maintaining cell identity through global control of genomic organization. Immunity. 2010;33:12–24. doi: 10.1016/j.immuni.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 54.Stelekati E, Shin H, Doering TA, Dolfi DV, Ziegler CG, Beiting DP, Dawson L, Liboon J, Wolski D, Ali MA, Katsikis PD, Shen H, Roos DS, Haining WN, Lauer GM, Wherry EJ. Bystander chronic infection negatively impacts development of CD8(+) T cell memory. Immunity. 2014;40:801–813. doi: 10.1016/j.immuni.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu H, Nau M, Ehrenberg P, Chenine AL, Macedo C, Zhou Y, Daye ZJ, Wei Z, Vahey M, Michael NL, Kim JH, Marovich M, Ratto-Kim S. Distinct gene-expression profiles associated with the susceptibility of pathogen-specific CD4 T cells to HIV-1 infection. Blood. 2013;121:1136–1144. doi: 10.1182/blood-2012-07-446278. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.