Abstract
Background
L‐Leu and its metabolite β‐hydroxy‐β‐methylbutyrate (HMB) stimulate muscle protein synthesis enhancing the phosphorylation of proteins that regulate anabolic signalling pathways. Alterations in these pathways are observed in many catabolic diseases, and HMB and L‐Leu have proven their anabolic effects in in vivo and in vitro models. The aim of this study was to compare the anabolic effects of L‐Leu and HMB in myotubes grown in the absence of any catabolic stimuli.
Methods
Studies were conducted in vitro using rat L6 myotubes under normal growth conditions (non‐involving L‐Leu‐deprived conditions). Protein synthesis and mechanistic target of rapamycin signalling pathway were determined.
Results
Only HMB was able to increase protein synthesis through a mechanism that involves the phosphorylation of the mechanistic target of rapamycin as well as its downstream elements, pS6 kinase, 4E binding protein‐1, and eIF4E. HMB was significantly more effective than L‐Leu in promoting these effects through an activation of protein kinase B/Akt. Because the conversion of L‐Leu to HMB is limited in muscle, L6 cells were transfected with a plasmid that codes for α‐keto isocaproate dioxygenase, the key enzyme involved in the catabolic conversion of α‐keto isocaproate into HMB. In these transfected cells, L‐Leu was able to promote protein synthesis and mechanistic target of rapamycin regulated pathway activation equally to HMB. Additionally, these effects of leucine were reverted to a normal state by mesotrione, a specific inhibitor of α‐keto isocaproate dioxygenase.
Conclusion
Our results suggest that HMB is an active L‐Leu metabolite able to maximize protein synthesis in skeletal muscle under conditions, in which no amino acid deprivation occurred. It may be proposed that supplementation with HMB may be very useful to stimulate protein synthesis in wasting conditions associated with chronic diseases, such as cancer or chronic heart failure.
Keywords: β‐Hydroxy‐β‐methylbutyrate; Protein synthesis; mTOR, leucine; Skeletal muscle
Introduction
Protein synthesis in mammalian cells is regulated by hormones, growth factors, amino acids, and carbohydrates through well‐known mechanisms.1 Phosphorylation of the kinase mechanistic target of rapamycin (mTOR) plays a central role in the regulation of protein synthesis.2 Mechanistic target of rapamycin is activated through protein kinase B (PKB/Akt)1 or extracellular signal‐regulated kinase (ERK1/2) pathways.3 4E binding protein‐1 (4E‐BP1) and p70 S6 kinase (S6K1) are key regulatory proteins modulated by mTOR that are involved in the initiation of mRNA translation.4, 5 Once activated, mTOR phosphorylates 4E‐BP1, which accelerates the release of eukaryotic initiation factor 4E (eIF4E) from 4E‐BP1 or directly activates S6K1.1, 2
Amino acids, particularly branched chain amino acids, are capable of increasing protein synthesis in skeletal muscle.6, 7, 8, 9 Their mechanism of action is dual; they provide substrates required for polypeptide synthesis and modulate signalling pathways responsible for translation initiation.10, 11 Among them, leucine is the most effective amino acid in stimulating protein synthesis through a direct activation of mTOR. Phosphorylation of mTOR drives to a 4E‐BP1 activation and an induction of S6K1 signalling.6, 10, 11, 12 Most studies on the beneficial effects of leucine enhancing protein synthesis have been carried out on leucine deficient media6, 7, 13 in in vitro studies or in models of muscle atrophy such as immobilization14 or experimental cachexia15, 16 involving a limited food intake period.
β‐Hydroxy‐β‐methylbutyrate (HMB), a metabolite of leucine, has been proposed as partially responsible for the effects exerted by leucine on protein synthesis.17, 18 Studies suggest that HMB administration attenuates the loss of skeletal muscle mass by reducing protein degradation and increasing protein synthesis in models of muscle atrophy induced by cachexia,17, 19, 20 ageing,21 or glucocorticoid treatment.22 Recently, it has been suggested that HMB could be more effective than leucine in attenuating body weight loss in an experimental model of cachexia.16
The aim of the present study was to compare the effects of L‐Leu and HMB on protein synthesis using an in vitro model of L6 rat myotubes under normal growth conditions (non‐involving L‐Leu‐deprived conditions). Our results point out that HMB is more potent than L‐Leu, increasing protein synthesis, and its signalling. In addition, because the muscle has a limited capacity to produce HMB from L‐Leu,23 we over‐expressed the enzyme α‐keto isocaproate dioxygenase (KICD) necessary for the conversion of HMB from L‐Leu in L6 cells. Effectively, in these transfected cells, L‐Leu potentiated its effects on protein synthesis. In conclusion, our results appear to indicate that L‐Leu has to be converted into HMB to achieve full effects on protein synthesis in muscle under our experimental setting.
Materials and methods
Materials
HMB free acid, D‐Leu and L‐Leu, LY294002, and PD98059 were obtained from Sigma (St. Louis, MO, USA). Mesotrione (2‐(4‐Mesyl‐2‐nitrobenzoyl)‐1,3‐cyclohexanedione)‐Pestanal©, catalogue no. 33855 was from Fluka (St. Louis, MO, USA). Tissue culture media and supplements were from Sigma and Invitrogen (Carlsbad, CA, USA). Foetal calf serum (FCS) was from Cultek (Madrid, Spain). Antibodies against PKB/Akt, phospho‐PKB/Akt (Ser473), mTOR, phospho‐mTOR (Ser2448), 4E‐BP1, S6K1, and phospho‐S6K1 (Thr389) were from Cell Signaling (Beverly, MA, USA). eIF4E and phospho‐eIF4E (Ser209) antibodies were from SAB Signalway Antibody (Pearland, TX, USA). Horseradish peroxidase‐conjugated secondary antibodies were from Sigma. 4‐Hydroxyphenylpyruvate dioxygenase antibody (H‐300) was from Santa Cruz Biotechnology (Santa Cruz, CA, USA). This antibody has been used given that KICD enzymatic activity is associated with this enzyme.24
Cell culture
The L6.C11 rat skeletal muscle myoblast line (European Collection of Cell Cultures no. 92102119) was grown in Dulbecco's Modified Eagle's Medium supplemented with 10% (v/v) FCS, 2 mM glutamine, plus 100 units/mL penicillin, and 0.1 mg/mL streptomycin and was maintained at sub‐confluent densities. This medium contains 0.8 mM L‐Leu. When the myoblasts reached about 80% confluency, they were differentiated into myotubes by exchanging the growth medium with a differentiation medium Dulbecco's Modified Eagle's Medium supplemented with 2% (v/v) FCS for 5–6 days. After differentiation, myotubes were starved in FCS‐free medium for 18 h prior to each treatment and the experiment was carried out in this starvation medium, except in the measurement of protein synthesis. Ten millimolars of L‐Leu and 50 μM HMB were selected as appropriate concentrations for these experiments because their use has been considered suitable to achieve the greatest stimulatory effects.16, 25
Determination of protein synthesis
Protein synthesis was measured as previously described26 with the following modifications. L6 cells were plated on 48‐well tissue culture plates, differentiated for 5 days, and then starved 18–24 h in FCS free medium. Cells were treated with 10 mM L‐Leu, 50 μM HMB, or 50 nM insulin for 2 h in media with 10% FCS and 0.8 mM tyrosine. Ten micromolars PD98059, an inhibitor of mitogen‐activated protein kinase kinase activation, or 20 μM LY294002, a specific inhibitor of PI3K, were added 30 min prior to incubation with effectors. Cells were then spiked with 1 μCi/mL of L‐[ring‐3,5‐3H]‐tyrosine (Perkin Elmer, Waltham MA) and incubated for 1 h. Reaction was stopped by placing the plates on ice. Wells were thoroughly washed two times with ice cold phosphate‐buffered saline media containing 2 mM non‐radioactive tyrosine, and cells were then lysed in 0.1 mM NaOH/0.1% sodium deoxycholate. Proteins were precipitated adding cold 20% tricarboxylic acid cycle (Sigma). This mixture was then incubated at 4°C for 15 min. Following centrifugation (16 000 × g for 10 min), the pellet was thoroughly washed with cold 10% tricarboxylic acid cycle and then precipitated proteins were dissolved in 0.1 mL of 1 M NaOH. An aliquot (5 μL) of the NaOH solubilized material was used for total protein quantification, and the remaining dissolved proteins were neutralized with 1 M HCl and mixed with ReadySafe scintillation fluid (Beckman Coulter, Brea CA). The radiolabel was determined with a scintillation counter (Beckman Coulter). Data were computed as disintegrations per minute per microgram of proteins.
Analysis of protein phosphorylation
L6 myotubes were treated with 10 mM L‐Leu or 50 μM HMB in Dulbecco's Modified Eagle's Medium without FCS for 30 min. Following this treatment, plates were processed as described.27, 28 The protein concentration of the supernatants was measured using a bicinchoninic acid method (Bio‐Rad, Madrid, Spain). Proteins (40 µg) were separated by sodium dodecyl sulphate–polyacrylamide gel electrophoresis, transferred onto nitrocellulose membranes, and immunoblotted with specific antibodies; the immunoblots were developed by an enhanced chemiluminescence detection method. Antibodies against non‐phosphorylated kinases were used to determine loading levels.
Cloning and eukaryotic expression of rat liver α‐keto isocaproate dioxygenase
The cDNA corresponding to the coding sequence of the rat KICD gene (Genebank adhesion no. AF082834) was amplified by reverse transcription polymerase chain reaction (RT‐PCR) from 5 µg of rat liver total RNA using a oligo‐NotI (dT) primer (First Strand Synthesis kit, GE Healthcare, Uppsala, Sweden). Polymerase chain reaction amplification was accomplished using the primers KICD‐F and KICD‐R with introduced XhoI and BamHI restriction endonuclease sites (shown in italics), respectively: KICD‐F ‐ CTCGAG CT ATG ACA ACC TAC AGC AAC AAA GG and KICD‐R ‐ GGATCC TTA CAT TCC GA CCT CAC ACC G (sequences corresponding to the start and stop codons are shown in bold). The DNA fragments were sub‐cloned in pSTblue‐1 (Novagen, Darmstadt, Germany), and the sequence was verified by automated DNA sequencing. To over‐express the rat KICD in eukaryotic cells, the KICD coding sequence was cloned in a modified pEGFP‐N3 plasmid (Clontech, Palo Alto, CA), where the coding sequence for the enhanced green fluorescent protein gene was removed in order to generate an mRNA coding for only the native KICD under the control of the cytomegalovirus promoter.
For transfection experiments, L6 myoblasts were used at 60–80% confluence. The transfection was performed using LipofectAMINE 2000 (Invitrogen) according to the manufacturer's instructions. Following removal of the transfection reagent, fresh medium was added to the cultures. Stably transfected cells were selected by addition of 700 µg/mL geneticin (Invitrogen) to the normal growth medium, 48 h after transfection. To differentiate transfected cultures, differentiation medium was applied 24 h later and during the next 5–6 days.
Reverse transcription polymerase chain reaction
Specific mRNA presence was assayed by RT‐PCR. Total RNA was isolated from the cells or tissues using a guanidinium thiocyanate method.29 Five micrograms of total RNA was used as a template to generate the cDNA by reverse transcription with the First‐strand cDNA synthesis kit (GE Helthcare Life Sciences, Uppsala, Sweden) using an oligodT‐NotI primer, according to the manufacturer's instructions. The reverse transcription products were amplified by polymerase chain reaction using the primers described in the preceding texts for the cloning of KICD. For the β‐actin, primers forward 5′‐GGCCAACCGTGAAAAGATG‐3′ and reverse 5′‐GGATCTTCATGAGGTAGTCTGTC‐3′ were used.
Statistical analysis
Results are expressed as mean ± SEM. Statistical analysis was performed by one‐way analysis of variance followed by the Tukey test as appropriate. P < 0.05 was considered statistically significant.
Results
L‐Leu and HMB effects on protein synthesis and on mTOR mediated signalling
L6 cells were grown, differentiated, and incubated in a medium with a basal concentration of 0.8 mM L‐Leu. At no time did the cells experience a restriction on the content of L‐Leu before or during the incubation in the absence (Control: C) or presence of effectors: 10 mM L‐Leu, 50 μM HMB, or 50 nM insulin. HMB significantly increased protein synthesis, while L‐Leu failed to increase it (Figure 1A). Furthermore, the effect of HMB on protein synthesis was similar to that obtained using insulin.
Figure 1.
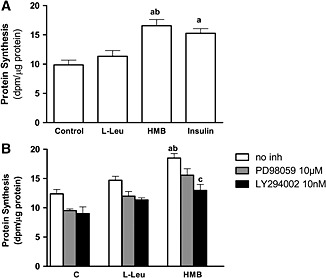
Effects of L‐Leu and HMB on protein synthesis and mTOR activation in L6 myotubes. Protein synthesis was measured following 2 h of 10 mM L‐Leu, 50 μM HMB, or 50 nM insulin supplementation in growth medium (A) and in the absence or presence of PD98059 and LY294002 (B). Inhibitors were added 30 min prior to the L‐Leu or HMB addition. Obtained data were expressed as dpm/µg of proteins (n = 5). Results are expressed as mean ± SEM. a P < 0.05, when compared with untreated control cells; b P < 0.05 compared with leucine treated cells; c P < 0.05 compared with HMB treated cells.
It has been suggested that HMB effects on muscle could be mediated via the PI3K/Akt and mitogen‐activated protein kinase kinase (MAPK)/ERK pathways.19, 30 At the same time, L‐Leu effects on protein synthesis have been associated with Akt independent routes.31, 32 To test the involvement of either PI3K/Akt or MAPK/ERK on the L‐Leu and HMB effects, L6 myotubes were pre‐incubated with inhibitors of mitogen‐activated protein kinase kinase phosphorylation (10 μM PD98059) or PI3K (20 μM LY294002) prior to the treatment with L‐Leu or HMB (Figure 1B). While the inhibition of the ERK1/2 signalling pathway did not change either L‐Leu or HMB effects on protein synthesis, the blockade of PI3K signalling significantly diminished the effects of HMB.
Next, we assayed the effects of L‐Leu and HMB on the phosphorylation status of PKB/Akt. Analysis of the phosphorylation of PKB/Akt, showed that incubation with HMB increased the phosphorylation of PKB/Akt, while L‐Leu failed to exert any effects (Figure 2A).
Figure 2.
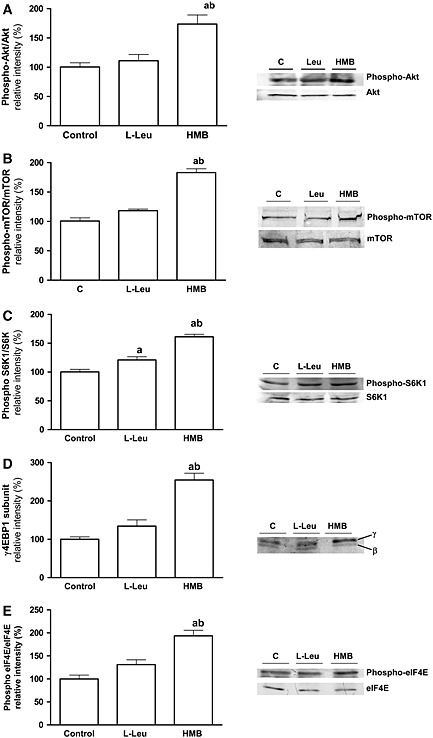
Regulation of mTOR signalling by 10 mM L‐Leu and 50 μM HMB in L6 myotubes. Phosphorylation of PKB/Akt (A), mTOR (B) S6K1 (C), 4E‐BP1 (D), and eIF4E (E) was assayed by western blot. Results are expressed as mean ± SEM (n = 4). a P < 0.05, when compared with untreated control cells; b P < 0.05 compared with leucine treated cells.
Finally, the effects of L‐Leu and HMB on the activation of mTOR (Figure 2B) showed that only HMB was able to significantly increase the phosphorylation of mTOR, results that match those described for the protein synthesis.
Thereafter, the phosphorylation status of key elements downstream of mTOR was assayed. As shown in Figure 2C, addition of HMB to L6 myotubes increased S6K1 phosphorylation at Thr389. HMB augmented the slower migrating γ band of 4E‐BP1, indicating an increase in 4E‐BP1 phosphorylation (Figure 2D).17, 18 HMB supplementation also increased the phosphorylation of eIF4E at Ser209 (Figure 2E). On the contrary, the addition of L‐Leu only produced an increase in the activation of S6K1 without affecting the phosphorylation status of 4E‐BP1 and eIF4E. However, the increase of S6K1 in response to L‐Leu was significantly lower compared with HMB treated cells.
Over‐expression of rat α‐keto isocaproate dioxygenase in L6 myotubes
HMB is a metabolite of L‐Leu. It has been described that in both humans and rats, blood circulating levels of HMB are mainly because of the hepatic metabolism of leucine by the KICD.33 On the contrary, in muscle, the main catabolic pathway is regulated by the branched chain α‐keto acid dehydrogenase complex34 to obtain isovaleryl‐CoA, limiting the conversion of L‐Leu to HMB.
Considering the described organ distribution of L‐Leu metabolism to HMB, a suggestive hypothesis is that the moderate effects of L‐Leu on protein synthesis and mTOR phosphorylation under our in vitro experimental conditions would be because of the limited rate of conversion of L‐Leu to HMB in muscle, pointing to HMB as an active L‐Leu metabolite.
First, we assayed the expression of KICD on differentiated L6 myotubes (Figure 3A). The expression profile of the rat KICD was probed with an anti‐4‐hydroxyphenylpyruvate dioxygenase antibody23 as well as by RT‐PCR. The results indicated that KICD is absent in L6 differentiated muscle cells. Furthermore, we have confirmed the distribution of the KICD enzyme in rat tissues by western blot and RT‐PCR (Figure 3B). KICD was mainly expressed in the liver and kidney, while it was below our detection limit in the brain, skeletal muscle (soleus), and the diaphragm.
Figure 3.
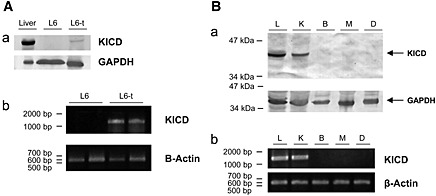
Expression profile of the rat KICD. (A) Expression of KICD in L6 myotubes (a) western blot corresponding to 20 µg of proteins from liver, L6 cells (L6) and L6 cells transfected with pKICD plasmid (L6‐t). Protein loading amounts were normalized using a GAPDH antibody. (b) RT‐PCR of KICD from total RNA of non‐transfected and transfected L6 myotubes is shown using β‐actin as reference gene. (B) Expression of KICD in rat tissues. (a) Western blot corresponding to 20 µg of proteins from the liver (L), kidney (K), brain (B), soleus muscle (M), or diaphragm (D) probed with an anti‐4HPPD antibody. Arrows in the figure mark the expected size band for the rat KICD and rat GADPH. (b) RT‐PCR of KICD from total RNA of the same tissues is shown using β‐actin as a reference gene.
To analyse the effects of a higher rate of conversion of L‐Leu to HMB, an expression plasmid, termed pKICD coding for the rat enzyme, was transfected in L6 muscle cells and the expression of KICD was confirmed by western blot and RT‐PCR (Figure 3A). pKICD L6 transfected cells showed higher levels of KICD than untransfected cells.
Protein synthesis and mTOR activation in L6 muscle cells over‐expressing KICD
The effects of L‐Leu or HMB supplementation on protein synthesis were assayed on L6 cells over‐expressing KICD under non‐amino‐acid deprivation. In these transfected cells, HMB retains the capability to increase protein synthesis and mTOR phosphorylation. Interestingly, incubations with 10 mM L‐Leu, but not 10 mM D‐Leu, produced a significant increase in protein synthesis (Figure 4A). This effect is remarkable considering that L‐Leu addition to L6 non‐transfected myotubes promoted a significantly lower effect compared with HMB in protein synthesis (Figure 1A).
Figure 4.
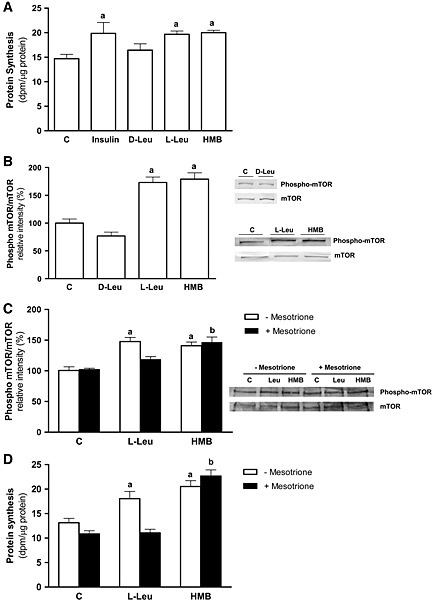
Effects of L‐Leu and HMB on protein synthesis and mTOR phosphorylation in KICD L6 transfected myotubes. (A) Protein synthesis rates in the presence of 10 mM D‐Leu, 10 mM L‐Leu, 50 μM HMB, or 50 nM insulin. Data were computed as dpm/µg of proteins (n = 8). (B) Phosphorylation of mTOR by 10 mM D‐Leu, 10 mM L‐Leu, or 50 μM HMB. (C) Phosphorylation of mTOR and protein synthesis by 10 mM L‐Leu and 50 μM HMB in (D) the absence and presence of 1 μM mesotrione. Results are expressed as mean ± SEM (n = 4). a P < 0.05, when compared with untreated control transfected cells; b P < 0.05, when compared with mesotrione treated control transfected cells.
Following this, we assayed the effects of L‐Leu or HMB supplementation on mTOR phosphorylation (Figure 4B) in the pKICD transfected cells. In these cells, while incubation with D‐Leu did not change the phosphorylation status of the kinase, incubation of the cells with L‐Leu produced an mTOR phosphorylation similar to those obtained with HMB.
In addition, while pre‐incubation with mesotrione, a specific KICD inhibitor,34 did not have any significant effect on the HMB‐induced phosphorylation of the kinase, it significantly decreased the effects of L‐Leu on mTOR phosphorylation to the levels obtained in the untransfected cells (Figure 4C). To confirm the importance of KICD on leucine‐mediated effects, protein synthesis was measured in the absence and presence of 1 μM mesotrione in the L6‐transfected cells (Figure 4D). Pre‐incubation with mesotrione blocked the increase in protein synthesis because of L‐Leu, while it has no effect on HMB‐treated cells.
Once the effects of L‐Leu and HMB on the pKICD transfected cells on mTOR phosphorylation were established, the effects of both effectors on the related signalling pathway were analysed in the KICD over‐expressing cells. At this point, supplementation with L‐Leu or HMB produced a significant and similar increase in the phosphorylation of PKB/Akt (Figure 5A). Finally, in the transfected cells, L‐Leu increased the phosphorylation of S6K1 (Figure 5B), 4E‐BP1 (Figure 5C), and eIF4E (Figure 5D) in a similar extent to HMB.
Figure 5.
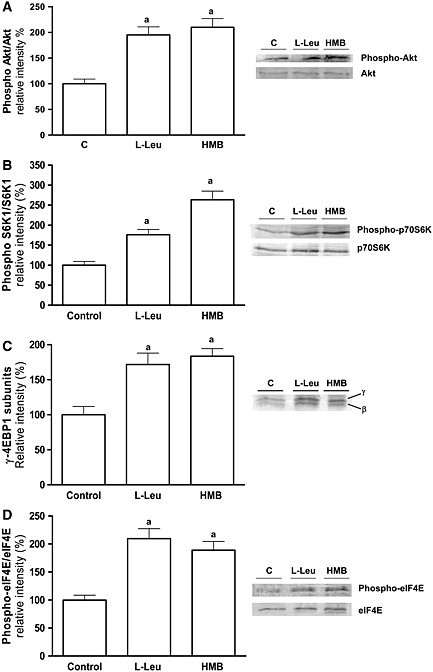
Upstream and downstream signalling by L‐Leu or HMB in KICD transfected L6 myotubes. Phosphorylation of PKB/Akt (A), S6K1 (B), 4E‐BP1 (C), and eIF4E (D) by 10 mM L‐Leu and 50 μM HMB was assayed by western‐Blot. Results are expressed as mean ± SEM (n = 4). a P < 0.05, when compared with untreated control transfected cells.
Discussion
L‐Leu stimulates protein synthesis in muscle cells through at least two distinct mechanisms: by stimulation of mRNA translation initiation machinery13, 35 and by activation of intracellular signalling pathways linked to mTOR.6, 36, 37 The positive effects of L‐Leu supplementation on protein synthesis have been widely studied in situations where the subjects were either in deficient or fasting conditions,8, 11, 12, 13, 14, 15 and therefore under a catabolic stimulus.
However, in catabolic diseases where circulating L‐Leu levels are restored by nutritional support, information on whether additional supplementation of L‐Leu could further increase protein synthesis still remains elusive. Crozier et al.12 showed that while administration of low doses of L‐Leu‐stimulated protein synthesis in muscle in food‐deprived mature rats, further supplementation with L‐Leu failed to elicit an additional response. Verhoeven et al.37 also reported that long‐term supplementation of L‐Leu in well‐nourished healthy elderly men did not improve muscular mass and strength. Additionally, Dirk et al.38 concluded that dietary protein supplementation did not reduce muscle loss during a short‐term period of disuse in healthy older men. Finally, in a recent report25 using a C2C12 muscle cell line, L‐Leu was not shown to produce a dose–response curve in protein synthesis nor in mTOR phosphorylation from 1.5 to 16.1 mM range of concentration. These results raise additional questions on the effectiveness of L‐Leu supplementation in the absence of catabolic stimuli such as amino acid deprivation.
Several studies have proposed that L‐Leu metabolite HMB may be responsible for the effects of L‐Leu on protein synthesis.23, 39 HMB has positive effects promoting strength and lean body mass in sport performance.40 Also, as a therapeutic supplement (especially in atrophic conditions), HMB stimulates muscle protein synthesis and reduces protein degradation.9, 17, 18, 21, 41 The increase in protein synthesis by HMB has been correlated with the activation of mTOR and two of its substrates 4E‐BP1 and S6K1, proteins involved in the translation of mRNA and protein synthesis.14, 40
In this work, to compare the effects of L‐Leu and HMB supplementation on protein synthesis in the absence of any catabolic stimuli, we have used a rat differentiated L6 muscle cell in culture. This well characterized in vitro muscle cell line42 has allowed us to determine the muscle specific effects of L‐Leu and HMB supplementation avoiding any systemic interference inherent with the use of animal models, for example the need to rule out any effects because of increased insulin secretion as a consequence of the administration of L‐Leu in animal models.43 Cells were maintained in a culture media containing L‐Leu concentration (0.8 mM) as described previously.44 As effectors, we chose a concentration of 10 mM L‐Leu based on a recent report,25 in which a leucine threshold has been demonstrated. A concentration of 50 μM HMB was selected because it has been considered appropriate in different studies on induction of protein synthesis16, 17, 18 or prevention of protein degradation.21, 22
Under these conditions, the effects of L‐Leu and HMB have been compared with protein synthesis and mTOR signalling in the L6 rat myotubes. L‐Leu did not significantly stimulate protein synthesis and mTOR phosphorylation. These results are in agreement with those that demonstrate that supplementation with L‐Leu failed to elicit an additional response in rats12 or in well‐nourished healthy elderly men.37 Interestingly, our results showed that HMB was a significantly more active effector than L‐Leu under these conditions. HMB increased protein synthesis 1.5‐fold and mTOR phosphorylation 1.6‐fold. These results are in agreement with other authors who have demonstrated a similar increase in protein synthesis and mTOR phosphorylation in response to HMB in murine myotubes.15, 21
The activation of mTOR, as a key element in protein synthesis, could be modulated by PI3K/Akt and MAPK/ERK1, 2, 3 signalling pathways. Therefore, we measured the effects of L‐Leu and HMB on protein synthesis in the presence of specific inhibitors of these pathways. Our results showed that the blockade of PI3K/Akt signalling blunted the effects of HMB to control levels; on the contrary, the inhibition of the MAPK/ERK pathway had no significant effects on protein synthesis in response to L‐Leu or HMB.
A cornerstone of the PI3K‐mediated signalling is PKB/Akt, and therefore, we analysed the L‐Leu and HMB effects on PKB/Akt phosphorylation. Incubation with L‐Leu did not modify the phosphorylation status of PKB/Akt, a result in concordance with studies that propose a PKB/Akt independent activation of the anabolic actions of L‐Leu.2, 31, 32 In contrast, HMB significantly enhanced PKB/Akt phosphorylation, in agreement with previous studies.19, 21 Therefore, under our experimental conditions, our results would support the idea that HMB acts mainly via the class I PI3K/Akt pathway,19 whereas it is described that L‐Leu exerts its action via the class III PI3K2, 31 that could mediate the activation of mTOR. In fact, it has been described that branched amino acids stimulate directly mTOR through a mechanism involving Rag GTPases.45 This alternate pathway has been described under amino acid deprivation,45 but during our experimental conditions without leucine starvation, there was no increase in protein synthesis nor in mTOR phosphorylation. Up to this point, under non‐amino‐acid deprivation, our results indicated HMB as a more active inductor of protein synthesis. The higher HMB activity might be because of its capacity to activate an mTOR pathway in a dissimilar manner compared with L‐Leu.
To further confirm the higher HMB effects on mTOR signalling, kinases downstream of mTOR and directly involved in the initiation of protein translation such as S6K1 and 4E‐BP14, 5 were analysed. HMB increased the phosphorylation of both kinases while L‐Leu only activated S6K1. S6K1 was activated by L‐Leu 1.2‐fold compared with untreated cells. This result is lower than that reported by Gran and Cameron11 in primary human myotubes and observed in rodent muscle cells.6 This discrepancy could be attributed to their experimental design, in which muscle cells were amino acid starved prior the addition of L‐Leu. A recent study in C2C12 cells has demonstrated that S6K1 is the only kinase in the mTOR signalling pathway with capacity to be activated in a dose–response manner.25 HMB activated S6K1 1.6‐fold, a value significantly higher than the effect obtained with L‐Leu. These results were similar to those obtained by Pimentel et al. 46 and are in agreement with the main mechanism of action reported for HMB that involved the activation of mTOR and S6K1.
The translational repressor 4E‐BP1 is bound to eIF4E forming an inactive complex. The 4E‐BP1 hyperphosphorylation by mTOR results in the release of eIF4E allowing the initiation of translation.17, 18 During sodium dodecyl sulphate–polyacrylamide gel electrophoresis, 4E‐BP1 was resolved into multiple electrophoretic bands, with the phosphorylated γ‐form having the slowest electrophoretic mobility. Also, in agreement with Wilkinson et al.,41 there was a significant difference in the γ‐4E‐BP1 band expression in response to HMB compared with L‐Leu or control cells.
The catabolism of leucine is very well described.40 Leucine is transaminated in muscle by branched‐chain aminotransferase to α‐ketoisocaproate. Then, α‐ketoisocaproate is decarboxylated to isovaleryl‐CoA by the mitochondrial enzyme branched‐chain α‐ketoacid dehydrogenase and finally acyl‐CoA derivatives are formed and entered into the citric acid cycle.36 The anticatabolic actions of L‐Leu metabolites such as α‐ketoisocaproate have been known for 35 years.47
An alternative metabolic pathway for leucine has been shown in the liver. The cytosolic enzyme α‐ketoisocaproate dioxygenase (or 4‐hydroxyphenylpyruvate dioxygenase)48 produces HMB from L‐Leu. KICD is mainly expressed in the liver, kidney, and neurons, but it is absent in muscle tissues,49 although in a recent study,41 this enzyme has been shown to be expressed in skeletal muscle at low levels. This enzyme provides a direct pathway for the synthesis of HMB, and this pathway accounts for up to 5% of the L‐Leu catabolism in humans.23
Therefore, the use of this muscle cell line has allowed us to differentiate between systemic and muscle specific L‐Leu and HMB effects. In addition, transfected L6 rat myotubes expressing the enzyme KICD, needed to produce HMB from Leu, have been used to analyse the function of L‐Leu and HMB in the regulation of protein synthesis. Because our results indicate that HMB is a more efficient effector to promote protein synthesis in non‐deprived amino acid conditions, we have transfected L6 myotubes with a plasmid pKICD to over‐express this key enzyme for the conversion of L‐Leu to HMB. These transfected cells would allow us to confirm the hypothesis that L‐Leu conversion into HMB in muscle cells is the limiting step in order to obtain a complete response on protein synthesis and mTOR signalling.
The over‐expression of KICD in L6 myotubes played an important role in the potentiation of L‐Leu effects because in these transfected cells, L‐Leu and HMB promoted protein synthesis and the signalling pathways involved in the initiation of translation in a similar manner. These results could be specifically associated with the conversion of L‐Leu to HMB; since first, D‐Leu failed to elicit any response, and second, the effects on protein synthesis and mTOR phosphorylation obtained with L‐Leu were blocked by the pre‐incubation with mesotrione, a specific inhibitor of KICD.50 Furthermore, in the cells over‐expressing KICD, L‐Leu exerts similar effects compared with HMB regarding protein synthesis and all its signalling pathways, PKB/Akt, mTOR, S6K1, 4E‐BP1, and eIF4E. These results clearly support HMB as the active leucine metabolite that regulates protein synthesis and highlights the key role of KICD as an essential enzyme mediating L‐Leu action. Our findings could suggest that under pathological conditions, in which KICD activity may be impaired, L‐Leu action could not be effective enough to promote mTOR activation and protein synthesis to counteract muscle loss.
In summary, our study points out that HMB is more active than L‐Leu increasing protein synthesis and mTOR signalling in non‐deprived amino acid conditions. The over‐expression of KICD in L6 cells promotes L‐Leu response and highlights the relevance of L‐Leu metabolism to HMB in the increase of protein synthesis in muscle. These results indicate that under our experimental conditions, efficient conversion of L‐Leu into HMB is required for complete mTOR phosphorylation through a straightforward PKB/Akt activation. Bearing in mind the described effects of HMB on protein synthesis, it may be suggested that HMB supplementation in patients suffering from wasting diseases—such as cancer, chronic obstructive pulmonary disease, and chronic heart failure—may be a good therapeutic strategy to stimulate protein synthesis in order to counteract muscle loss because of increased protein degradation, characteristic of these pathological conditions.
Conflict of interest
This work was funded by Abbott Nutrition R&D. María D. Girón, José D. Vílchez, Rafael Salto, Natalia Sevillano, and Josep M. Argilés declare that they have no conflict of interest.
Manuel Manzano, Ricardo Rueda, Nefertiti Campos, and Jose M. López‐Pedrosa are employees of the sponsor company Abbott Nutrition R&D, Granada, Spain.
Acknowledgements
The authors of this manuscript certify that they comply with the ethical guidelines for authorship and publishing in the Journal of Cachexia, Sarcopenia, and Muscle 2010; 1:7–8 (von Haehling S, Morley JE, Coats AJ, Anker SD).
The author contributions are as follows: M.D.G., R.S., M.M., R.R., and J.M.L.P. conceived and designed the experiments. M.D.G., J.D.V., R.S., and N.S. conducted the research. M.D.G., R.S., M.M., R.R., and J.M.L.P analysed the data. M.D.G., R.S., M.M., N.C., and J.M.L.P. wrote the paper.
We thank Luciana Whizar Toscano for checking the article.
Girón, M. D. , Vílchez, J. D. , Salto, R. , Manzano, M. , Sevillano, N. , Campos, N. , Argilés, J. M. , Rueda, R. , and López‐Pedrosa, J. M. (2016) Conversion of leucine to β‐hydroxy‐β‐methylbutyrate by α‐keto isocaproate dioxygenase is required for a potent stimulation of protein synthesis in L6 rat myotubes. Journal of Cachexia, Sarcopenia and Muscle, 7: 68–78. doi: 10.1002/jcsm.12032.
References
- 1. Weigl LG. Lost in translation: regulation of skeletal muscle protein synthesis. Curr Opin Pharmacol. 2012;12:377–382. [DOI] [PubMed] [Google Scholar]
- 2. Wang X, Proud CG. The mTOR pathway in the control of protein synthesis. Physiology. 2006;21:362–369. [DOI] [PubMed] [Google Scholar]
- 3. Ma L, Chen Z, Erdjument‐Bromage H, Tempst P, Pandolfi PP. Phosphorylation and functional inactivation of TSC2 by ERK implications for tuberous sclerosis and cancer pathogenesis. Cell. 2005;121:179–193. [DOI] [PubMed] [Google Scholar]
- 4. Howell JJ, Manning BD. mTOR couples cellular nutrient sensing to organismal metabolic homeostasis. Trends Endocrinol Metab. 2011;22:94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yoshizawa F, Mochizuki S, Sugahara K. Differential dose response of mTOR signaling to oral administration of leucine in skeletal muscle and liver of rats. Biosci Biotechnol Biochem. 2013;77:839–842. [DOI] [PubMed] [Google Scholar]
- 6. Atherton PJ, Smith K, Etheridge T, Rankin D, Rennie MJ. Distinct anabolic signalling responses to amino acids in C2C12 skeletal muscle cells. Amino Acids. 2010;38:1533–1539. [DOI] [PubMed] [Google Scholar]
- 7. Maki T, Yamamoto D, Nakanishi S, Iida K, Iguchi G, Takahashi Y, et al. Branched‐chain amino acids reduce hindlimb suspension‐induced muscle atrophy and protein levels of atrogin‐1 and MuRF1 in rats. Nutr Res. 2012;32:676–683. [DOI] [PubMed] [Google Scholar]
- 8. Miura Y, Nakazawa T, Yagasaki K. Possible involvement of calcium signaling pathways in L‐leucine‐stimulated protein synthesis in L6 myotubes. Biosci Biotechnol Biochem. 2006;70:1533–1536. [DOI] [PubMed] [Google Scholar]
- 9. Wang X, Campbell LE, Miller CM. Proud CG Amino acid availability regulates p70 S6 kinase and multiple translation factors. Biochem J. 1998;334:261–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Escobar J, Frank JW, Suryawan A, Nguyen HV, Van Horn CG, Hutson SM, et al. Leucine and alpha‐ketoisocaproic acid, but not norleucine, stimulate skeletal muscle protein synthesis in neonatal pigs. J Nutr. 2010;140:1418–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gran P, Cameron‐Smith D. The actions of exogenous leucine on mTOR signalling and amino acid transporters in human myotubes. BMC Physiol. 2011;11:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Crozier SJ, Kimball SR, Emmert SW, Anthony JC, Jefferson LS. Oral leucine administration stimulates protein synthesis in rat skeletal muscle. J Nutr. 2005;135:376–382. [DOI] [PubMed] [Google Scholar]
- 13. Boutry C, El‐Kadi SW, Suryanan A, Wheatley SM, Orellana RA, Kimball SR, et al. Leucine pulses enhance skeletal muscle protein synthesis during continuous feeding in neonatal pigs. Am J Physiol Endocrinol Metab. 2013;305:E620–E631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Savary‐Auzeloux I, Magne H, Migné C, Oberli M, Breuillé D, Faure M, et al. A dietary supplementation with leucine and antioxidants is capable to accelerate muscle mass recovery after immobilization in adult rats. PLoS One. 2012;8:e81495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Elley HL, Russell ST, Tisdale MJ. Effect of branched‐chain amino acids on muscle atrophy in cancer cachexia. Biochem J. 2007;407:113–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mirza KA, Pereira SL, Voss AC, Tisdale MJ Comparison of the anticatabolic effects of leucine and Ca‐β‐hydroxy‐β‐methylbutyrate in experimental models of cancer cachexia. Nutrition. 2014;30:807–813. [DOI] [PubMed] [Google Scholar]
- 17. Eley HL, Russell ST, Baxter JH, Mukerji P, Tisdale MJ. Signaling pathways initiated by beta‐hydroxy‐beta‐methylbutyrate to attenuate the depression of protein synthesis in skeletal muscle in response to cachectic stimuli. Am J Physiol Endocrinol Metab. 2007;293:E923–931. [DOI] [PubMed] [Google Scholar]
- 18. Eley HL, Russell ST, Tisdale MJ. Attenuation of depression of muscle protein synthesis induced by lipopolysaccharide, tumor necrosis factor, and angiotensin II by beta‐hydroxy‐beta‐methylbutyrate. Am J Physiol Endocrinol Metab. 2008;295:E1409–1416. [DOI] [PubMed] [Google Scholar]
- 19. Kornasio R, Riederer I, Butler‐Browne G, Mouly V, Uni Z, Halevy O. Beta‐hydroxy‐beta‐methylbutyrate (HMB) stimulates myogenic cell proliferation, differentiation and survival via the MAPK/ERK and PI3K/Akt pathways. Biochim Biophys Acta. 2009;1793:755–763. [DOI] [PubMed] [Google Scholar]
- 20. Smith HJ, Mukerji P, Tisdale MJ. Attenuation of proteasome‐induced proteolysis in skeletal muscle by {beta}‐hydroxy‐{beta}‐methylbutyrate in cancer‐induced muscle loss. Cancer Res. 2005;65:277–283. [PubMed] [Google Scholar]
- 21. Kimura K, Cheng XW, Inoue A, Hu L, Koike T, Kuzuya M. β‐Hydroxy‐β‐methylbutyrate facilitates PI3K/Akt‐dependent mammalian target of rapamycin and FoxO1/3a phosphorylations and alleviates tumor necrosis factor α/interferon γ‐induced MuRF‐1 expression in C2C12 cells. Nutr Res. 2014; 34: 368–374. [DOI] [PubMed]
- 22. Aversa Z, Alamdari N, Castillero E, Muscaritoli M, Rossi Fanelli F, Hasselgren PO. β‐Hydroxy‐beta‐methylbutyrate (HMB) prevents dexamethasone‐induced myotube atrophy. Biochem Biophys Res Commun. 2012;423:739–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nissen S, Sharp R, Ray M, Rathmacher JA, Rice D, Fuller JC Jr, et al Effect of leucine metabolite beta‐hydroxy‐beta‐methylbutyrate on muscle metabolism during resistance‐exercise training. J Appl Physiol. 1996;81:2095–2104. [DOI] [PubMed] [Google Scholar]
- 24. Crouch NP, Lee MH, Iturriagagoitia‐Bueno T, MacKinnon CH. Cloning, expression, and purification of mammalian 4‐hydroxyphenylpyruvate dioxygenase/alpha‐ketoisocaproate dioxygenase. Methods Enzymol. 2000;324:342–355. [DOI] [PubMed] [Google Scholar]
- 25. Areta JL, Hawley JA, Ji‐Ming Y, Stanley‐Chan MH, Vernon GC. Increasing leucine concentration stimulates mTOR signaling and cell growth in C2C12 skeletal muscle cells. Nutr Res. 2014;34:1000–1007. [DOI] [PubMed] [Google Scholar]
- 26. Gulve EA, Dice JF. Regulation of protein synthesis and degradation in L8 myotubes. Effects of serum, insulin and insulin‐like growth factors. Biochem J 1989;260:377–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Giron MD, Sevillano N, Salto R, Haidour A, Manzano M, Jiménez ML, et al. Salacia oblonga extract increases glucose transporter 4‐mediated glucose uptake in L6 rat myotubes: role of mangiferin. Clin Nutr. 2009;28:565–574. [DOI] [PubMed] [Google Scholar]
- 28. Giron MD, Sevillano N, Vargas AM, Domínguez J, Guinovart JJ, Salto R. The glucose‐lowering agent sodium tungstate increases the levels and translocation of GLUT4 in L6 myotubes through a mechanism associated with ERK1/2 and MEF2D. Diabetologia. 2008;51:1285–1295. [DOI] [PubMed] [Google Scholar]
- 29. Chomczynski P, Sacchi N. Single‐step method of RNA isolation by acid guanidinium thiocyanate‐phenol‐chloroform extraction. Anal Biochem. 1987;162:156–159. [DOI] [PubMed] [Google Scholar]
- 30. Alway SE, Pereira SL, Edens NK, Hao Y, Bennett BT. β‐Hydroxy‐β‐methylbutyrate (HMB) enhances the proliferation of satellite cells in fast muscles of aged rats during recovery from disuse atrophy. Exp Gerontol. 2013;48:973–984. [DOI] [PubMed] [Google Scholar]
- 31. Nobukini T, Joaquin M, Roccio M, Dann SG, Kim SY, Gulati P, et al. Amino acids mediate mTOR/raptor signalling through activation of class 3 phosphatidylinositol 3OH‐kinase. Proc Natl Acad Sci USA 2005;102:14238–14243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Drummond MJ, Rasmussen BB. Leucine‐enriched nutrients and the regulation of mammalian target of rapamycin signalling and human skeletal muscle protein synthesis. Curr Opin Clin Nutr Metab Care. 2008;11:222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xu M, Nakai N, Ishigure K, Nonami T, Nagasaki M, Obayashi M, et al. The alpha‐ketoisocaproate catabolism in human and rat livers. Biochem Biophys Res Commun. 2013;276:1080–1084. [DOI] [PubMed] [Google Scholar]
- 34. Shimomura Y, Fujii H, Suzuki M, Murakami T, Fujitsuka N, Nakai N. Branched‐chain alpha‐keto acid dehydrogenase complex in rat skeletal muscle: regulation of the activity and gene expression by nutrition and physical exercise. J Nutr. 1995;125:1762S–1765S. [DOI] [PubMed] [Google Scholar]
- 35. Anthony JC, Yoshizawa F, Anthony TG, Vary TC, Jefferson LS, Kimball SR. Leucine stimulates translation initiation in skeletal muscle of postabsorptive rats via a rapamycin‐sensitive pathway. J Nutr. 2000;130:2413–2419. [DOI] [PubMed] [Google Scholar]
- 36. Lynch CJ, Halle B, Fujii H, Vary TC, Wallin R, Damuni Z, et al. Potential role of leucine metabolism in the leucine‐signaling pathway involving mTOR. Am J Physiol Endocrinol Metab. 2003;285:E854–E863. [DOI] [PubMed] [Google Scholar]
- 37. Verhoeven S, Vanschoonbeek K, Verdijk LB, Koopman R, Wodzig WK, Dendale P, et al. Long‐term leucine supplementation does not increase muscle mass or strength in healthy elderly men. Am J Clin Nutr. 2009;89:1468–1475. [DOI] [PubMed] [Google Scholar]
- 38. Dirks ML, Wall BT, Nilwik R, Weerts DH, Verdijk LB, van Loon LJ. Skeletal muscle disuse atrophy is not attenuated by dietary protein supplementation in healthy older men. J Nutr. 2014;144:1196–1203. [DOI] [PubMed] [Google Scholar]
- 39. Slater GJ, Jenkins D. Beta‐hydroxy‐beta‐methylbutyrate (HMB) supplementation and the promotion of muscle growth and strength. Sport Med. 2000;30:105–116. [DOI] [PubMed] [Google Scholar]
- 40. Zanchi NE, Gerlinger‐Romero F, Guimaraes‐Ferreira L, de Siqueira Filho MA, Felitti V, Lira FS, et al. HMB supplementation: clinical and athletic performance‐related effects and mechanisms of action. Amino Acids. 2011;40:1015–1025. [DOI] [PubMed] [Google Scholar]
- 41. Wilkinson DJ, Hossain T, Hill DS, Phillips BE, Crossland H, Williams J, et al. Effects of leucine and its metabolite β‐hydroxy‐β‐methylbutyrate on human skeletal muscle protein metabolism. J Physiol. 2013;591:2911–2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Richler C, Yaffe D. The in vitro cultivation and differentiation capacities of myogenic cell lines. Dev Biol. 1970;23:1–22. [DOI] [PubMed] [Google Scholar]
- 43. Yang J, Chi Y, Burkhardt BR, Guan Y, Wolf BA. Leucine metabolism in regulation of insulin secretion from pancreatic beta cells. Nutr Rev. 2010;68:270–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tischler ME, Desautels M, Goldberg AL. Does leucine, leucyl‐tRNA, or some metabolite of leucine regulate protein synthesis and degradation in skeletal and cardiac muscle? J Biol Chem. 1982;257:1613–1621. [PubMed] [Google Scholar]
- 45. Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar‐Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTOR. Science. 2008;320:1496–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pimentel GD, Rosa JC, Lira FS, Zanchi NE, Ropelle ER, Oyama LM, et al. β‐Hydroxy‐β‐methylbutyrate (HMβ) supplementation stimulates skeletal muscle hypertrophy in rats via the mTOR pathway. Nutr Metab. 2011;8:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hider RC, Fern EB, London DR. Relationship between intracellular amino acids and protein synthesis in the extensor digitorum longus muscle of rats. Biochem J. 1960;114:171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Van Koevering M, Nissen S. Oxidation of leucine and alpha‐ketoisocaproate to beta‐hydroxy‐beta‐methylbutyrate in vivo. Am J Physiol. 1992;262:E27–31. [DOI] [PubMed] [Google Scholar]
- 49. Neve S, Aarenstrup L, Tornehave D, Rahbek‐Nielsen H, Corydon TJ, Roepstorff P, et al. Tissue distribution, intracellular localization and proteolytic processing of rat 4‐hydroxyphenylpyruvate dioxygenase. Cell Biol Int. 2003;27:611–624. [DOI] [PubMed] [Google Scholar]
- 50. Hall MG, Wilks MF, Provan WM, Eksborg S, Lumholtz B. Pharmacokinetics and pharmacodynamics of NTBC [2‐(2‐nitro‐4‐fluoromethylbenzoyl)‐1,3‐cyclohexanedione] and mesotrione, inhibitors of 4‐hydroxyphenyl pyruvate dioxygenase (HPPD) following a single dose to healthy male volunteers. Br J Clin Pharmacol. 2001;52:169–177. [DOI] [PMC free article] [PubMed] [Google Scholar]


