Summary
Human/simian immunodeficiency virus (HIV/SIV) infection can cause severe depletion of CD4+ T cells in both plasma and mucosa; it also results in damage to the gut mucosa barrier, which makes the condition more conducive to microbial translocation. In this study, we used SIV‐infected Chinese rhesus macaques to quantify the extent of microbial translocation and the function of immune cells in the entire gastrointestinal tract and to compare their differences between rapid and slow progressors. The results showed that in the slow progressors, microbial products translocated considerably and deeply into the lamina propria of the gut; the tissue macrophages had no significant differences compared with the rapid progressors, but there was a slightly higher percentage of mucosal CD8+ T cells and a large amount of extracellular microbial products in the lamina propria of the intestinal mucosa of the slow progressors. The data suggested that although microbial translocation increased markedly, the mucosal macrophages and CD8+ T cells were insufficient to clear the infiltrated microbes in the slow progressors. Also, therapies aimed at suppressing the translocation of microbial products in the mucosa could help to delay the progression of SIV disease.
Keywords: lamina propria, microbial translocation, mucosa, simian immunodeficiency virus
Abbreviations
- GI
gastrointestinal
- LP
lamina propria
- LPS
lipopolysaccharides
- p.i.
post infection
- pVL
viral load in plasma
- SIV
simian immunodeficiency virus
- TLR
Toll‐like receptor
Introduction
The gastrointestinal (GI) tract is the predominant structural and immunological barrier against the invasion of toxic substances, antigens and bacteria. In addition, the mucosal epithelium, which is a single‐cell layer formed by enterocytes, plays an important role in nutrient absorption and immune defence. More than 40% of lymphocytes in the body are found in the GI tract,1 and most of these are CCR5 and activated memory CD4+ T lymphocytes.2, 3 CCR5 is one of the co‐receptors that human/ simian immunodeficiency viruses (HIV/SIV) use to invade cells, and activated memory CD4+ T lymphocytes are the first candidates of attack by the virus; hence, the GI tract is the preferred target for HIV/SIV replication.4 Infection with HIV/SIV can cause a series of GI conditions, including diarrhoea, weight loss, nutrient malabsorption, inflammatory infiltrates, mucosal damage and opportunistic infections. It also causes severe depletion of lamina propria (LP) CD4+ T cells in both productively infected and bystander cells,5, 6, 7 coinciding with increases in intestinal CD8+ T cells.8, 9
In a normal state of health, the GI tract already contains an inconceivable number of normal microbiota, which live within humans, adopting them as hosts; most of these are commensal and beneficial bacteria that play an important role in shaping the immune system.10 However, the disruption of the homeostatic balance between the host and the intestinal microflora is part of the pathogenesis of HIV/SIV infection, and the composition of the GI microbiota generally changes after infection; for example, the genera Bacteroides, Odoribacter and Parabacteroides distasonis significantly decrease, whereas the genus Prevotella, of the family Paraprevotellaceae, notably increases after infection.11 Microbial products could invade the LP through a break in the GI mucosa, even translocating from the intestinal lumen into the periphery in the late acute phase of infection; the extent of translocation is correlated with the integrity of the GI epithelium.12
Previous studies have suggested that the GI tract is a major site of viral replication and that HIV/SIV infection can cause several histopathological changes in gut mucosa. Most of the studies have focused on the mechanism of microbial translocation or the activation of the immune system induced by microbial translocation. However, little is known about whether and how changes in the levels of microbial translocation and the amount of immune cells in the entire GI tissue take place and how these differ with the disease duration after HIV/SIV infection. In this study, we used SIVmac239‐infected Chinese rhesus macaques as models and divided them into the following two groups as previously described:13 rapid progressors, which died within 25 weeks after infection; and slow progressors, which died over 106 weeks after infection. In our research, animals 07099 and 04039 were classified as rapid progressors, and 04039 and 00067 as slow progressors. We provide for the first time direct immunohistochemical evidence from the study of all sections of the GI tracts from SIVmac239‐infected Chinese rhesus macaques, including stomach, duodenum to ileum, and caecum to rectum. Quantitative image analysis was used to confirm the distinct histological changes in rapid and slow progressors. In the slow progressors, we found that the extent of microbial products traversing the damaged epithelium of the intestinal mucosa was greater and that the functions of macrophages and CD8+ T cells in the GI tract deteriorated after infection.
Materials and methods
Animals and sample collection
The four adults, 5‐ to 7‐year‐old, colony‐bred male Chinese rhesus macaques used in this study were from the Kunming Primate Research Centre, Chinese Academy of Sciences. They were housed and cared for in accordance with the guidelines of the Committee on Animals of the Kunming Institute of Zoology, Chinese Academy of Sciences, and the Animal Welfare Act. The GI tract segments were sampled at necropsy and placed into a fixative. The GI biopsy samples were then subjected to haematoxylin & eosin and immunohistochemical staining as described below.
Plasma viral loads
The levels of viral RNA in plasma were measured by real‐time RT‐PCR (TaqMan) assay for SIV gag as previously described.14 Briefly, virus RNA was extracted by using the High Pure Viral RNA kit (Roche, Indianapolis, Indiana, USA) according to the manufacturer's instructions. Quantitative RT‐PCR assay was performed with an RNA‐direct Real‐time PCR master mix (Toyobo, Osaka, Honshu, Japan). The primers used were 5′‐TCGGTCTTAGCTCCATTAGTGCC‐3′ and 5′‐GCTTCCTCAGTGTGTTTCACTTTC‐3′. The TagMan probe sequence was 5′‐FAM‐CTTCTGCGTGAATGCACCAGATGACGC‐TAMRA‐3′. PCR were carried out on an ABI Vii7 Sequence Detection System under the following conditions: one cycle at 90° for 30 seconds and at 60° for 20 min, followed by 45 cycles at 90° for 15 seconds and at 60° for 1 min. The quantification limit was determined to be 50 copies/ml by a standard curve constructed from serial dilutions of an appropriate RNA transcript.
Histological examination
The GI tissues were fixed in 4% paraformaldehyde, embedded in paraffin, and stained according to the conventional haematoxylin & eosin technique.
Immunofluorescence and immunohistochemistry
Tissues were fixed in paraformaldehyde, embedded in paraffin, consecutively cut into 5‐µm sections, and mounted on glass slides for immunohistochemical analysis. The sections were deparaffinized in xylene and rehydrated through a graded ethanol series; for antigen retrieval, sections immersed in saline sodium citrate buffer were microwave heated for 6 min thrice. This was followed by cooling to room temperature, washing with 1 × PBS containing 0·05% Tween‐20 (1 × PBST), and incubation with 0·1% Triton X‐100 for 10 min at room temperature. The sections were then washed, treated with 3% hydrogen peroxide for 10 min at room temperature, washed again, and blocked with 10% BSA at 37° for 60 min. Primary antibodies were diluted in BSA and incubated overnight at 4° or 37° for 60 min. Then, the sections were washed and incubated with secondary antibodies in BSA for 60 min at room temperature. For immunofluorescence, the secondary antibodies were conjugated with different fluoresceins, and sections were directly counterstained with 5 µg/ml DAPI for 5 min at room temperature, washed, and then mounted. For immunohistochemistry, the secondary antibodies were conjugated with horseradish peroxidase; 3,3′‐diaminobenzidine tetrahydrochloride was used as a substrate to yield a deep brown colour. This was followed by counterstaining with haematoxylin and then mounting. The primary antibodies used were: rabbit anti‐P27 polyclonal antibodies generated in our laboratory, mouse anti‐lipopolysaccharide (LPS) core (Hycult Biotech, Plymouth Meeting, PA), mouse anti‐macrophage (Dako, Carpinteria, CA), polyclonal rabbit anti‐Escherichia coli (Dako), purified mouse anti‐human CD4 (BD, Franklin Lakes, NJ), purified mouse anti‐human CD8 (BD), and polyclonal rabbit anti‐human CD3 (Dako).
Quantitative image analysis
High‐powered images (× 200 or × 400 magnification) were obtained with a Leica fluorescence inverted microscope. To quantify the level of microbial products (E. coli) that invaded the LP, we randomly selected five image fields of equal area in each tissue section and used photoshop and image J software for post‐processing and quantitative image analysis.
Statistical analysis
All data were analysed using graphpad prism version 5.0 (GraphPad, San Diego, CA, USA). Paired Student's t‐test was used to compare the amount of microbes and immune cells in each GI tissue between rapid and slow progressors. Statistical significance was set at P < 0·05.
Results
Viral load and blood T‐cell counts in Chinese rhesus macaques after infection with SIVmac239
The dynamics of the viral load and T‐cell counts in blood were investigated for each animal; the viral load and T‐cell counts in the early phase of infection in slow progressors were also studied in our previous work.14 Specimens were collected and examined at different time‐points after infection. The viral load in plasma (pVL) peaked almost at day 14 post infection (p.i.) and then gradually and persistently declined; however, it was continuously detected at each time‐point in all animals. The pVL rebounded and sustained its increase to a high level before death, but it remained below the peak level, as shown in Fig. 1(a). The count of blood CD4+ and CD8+ T cells significantly decreased in the two groups on days 7–14 p.i. and then increased, fluctuating under the normal level. A different picture was observed for macaque 04039; its CD4+ and CD8+ T‐cell counts exceeded the normal pre‐infection level at some time‐points, which may be explained by the extremely low viral load at those times. The T‐cell counts dropped sharply before death in all the animals, especially in the slow progressors (Fig. 1b, c).
Figure 1.
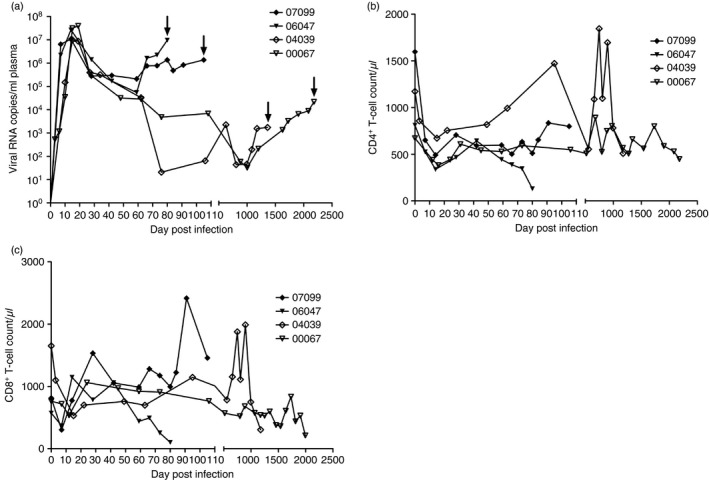
Dynamics of the viral load and CD4+ T‐cell and CD8+ T‐cell counts in Chinese rhesus macaques after SIVmac239 infection. (a) Plasma viral load in SIVmac239‐infected Chinese rhesus macaques; the arrows indicate the date of natural death. (b) Blood CD4+ T‐cell counts during infection. (c) Blood CD8+ T‐cell counts during infection.
All four macaques died a natural death at different time‐points after SIVmac239 infection; their tissues were then collected. Animal 07099 died at day 105 p.i. (pVL = 1·36 × 106, blood CD4+ T cells = 799·18 cells/µL, blood CD8+ T cells = 1456·35 cells/μL), 06047 died at day 80 p.i. (pVL = 0·96 × 107, blood CD4+ T cells = 131·24 cells/μL, blood CD8+ T cells = 100·41 cells/μL), 04039 died at day 1360 p.i. (pVL = 1·73 × 103, blood CD4+ T cells = 509·03 cells/μL, blood CD8+ T cells = 304·22 cells/μL), and 00067 died at day 2180 p.i. (pVL = 2·18 × 104, blood CD4+ T cells = 448·91 cells/μL, blood CD8+ T cells = 211·71 cells/μL).
p27 is a highly conserved protein of the SIV capsid and shares antigenicity with most SIV strains; as a result, it is commonly used as a detection marker for SIV. To determine whether the viral particles were also present in the mucosal tissues from the two groups after infection, we sought evidence by p27 staining of paraffin‐embedded GI tissue sections from rapid and slow progressors. A large number of SIV‐positive cells were detected in the LP of all intestinal tissues from the slow progressors. In contrast, SIV‐infected cells were rarely observed in the intestinal sections from the rapid progressors (Fig. 2a). A further evaluation of the staining results based on the quantitative analysis of p27 was performed (Fig. 2b). The results indicated that the p27 staining in most GI tract sections from the slow progressors was recorded as SIV‐positive, whereas it was defined as SIV‐negative in the rapid progressors. These findings suggested that the virus, which had settled in the GI mucosa in the early phase after infection, could accumulate in the late phase, implying that the intestinal mucosa might be a major reservoir for HIV/SIV after infection.
Figure 2.
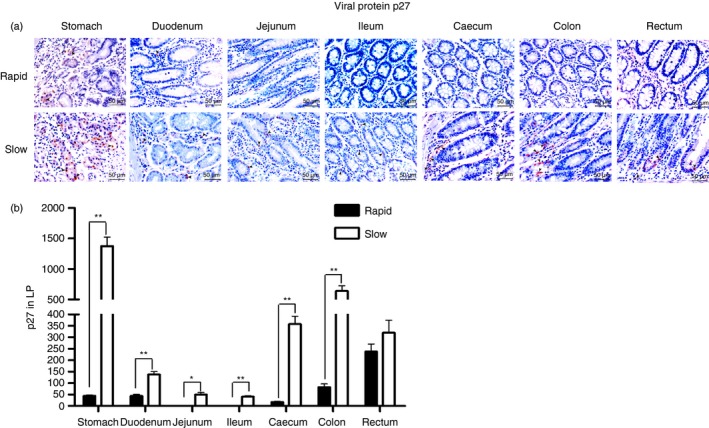
Detection of the viral protein p27 (brown) in the gastrointestinal (GI) tracts of rapid and slow progressors. (a) Representative images (400 ×) of the GI tract stained for p27 antigen (light red) are shown. (b) Quantitative analysis of p27 (* : P ≤ 0.05, ** : 0.05 < P ≤ 0.01).
Pathological manifestations of GI mucosa after SIV infection
Gastrointestinal complications are commonly seen after SIV infection and may occur during the early phase of the disease. Histopathological changes due to SIV infection in the intestinal mucosa include epithelium damage, inflammatory infiltrates, and lymphatic follicle aggregation, among others.15 These are intimately associated with the translocation of microbial products and even have a considerable effect on the progression of disease. Herein, we described and illustrated lesions in the GI tract that might be the result of a direct effect of SIV infection; the major characteristics of mucosal disease observed in the GI tracts of the two groups were mucosal erosion, inflammation accompanied by haemorrhage and oedema. The mucosal erosion was more severe in the GI tracts of the slow progressors, especially in the caecum of animal 00067, in which a focal ulcer was observed. The amount of inflammatory cells that infiltrated the LP of the stomach of 00067 and the caecum of 04039 was much higher than that in the rapid progressors; these cells congregated and formed a large follicle. However, the duodenums of 04039 and 00067 were unaffected by the viral infection, which might protect them from the attack of microbes. Hence, pathological changes of GI mucosa in slow progressors might be more severe than those in the rapid progressors.
Microbial translocation in GI tissues
Previous studies have reported that microbial translocation occurs during progressive HIV or SIV infection and that it correlates with the integrity of the epithelial barrier.16, 17 To investigate whether different levels of microbial translocation exist in the entire GI tract during varying stages of infection, we compared the level of LPS that translocated into the LP of GI tissues between rapid and slow progressors. Numerous LPS+ cells were observed within all GI tissues in the slow progressors, but only a few LPS+ cells were found within the LP of the ileum and colon in the rapid progressors (Fig. 3a, b).
Figure 3.
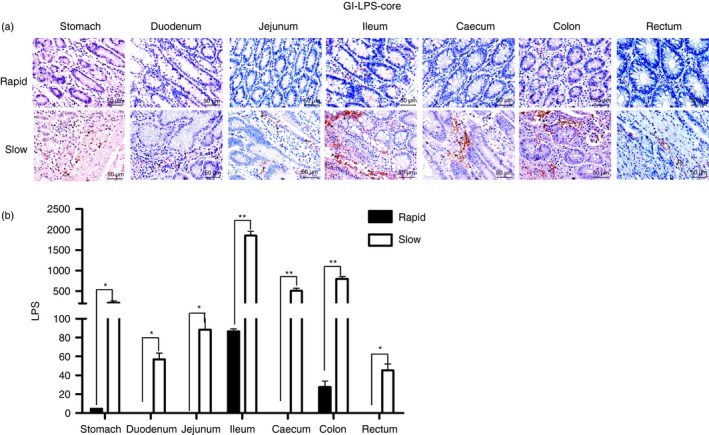
Detection of microbial translocation in the gastrointestinal (GI) tract of SIVmac239‐infected Chinese rhesus macaques. (a) Representative images (400 ×) of the GI tract stained for lipopolysaccharide (LPS)‐core antigen (light red) are shown. (b) Quantitative analysis of LPS (* : P ≤ 0.05, ** : 0.05 < P ≤ 0.01).
To find support for the hypothesis that microbial translocation accumulates in the GI tissues of the slow progressors, we used a rabbit polyclonal antibody against E. coli, which reacts with the majority of proteins present in E. coli lysates. We obtained similar results to those described above; massive numbers of E. coli invaded the LP of the stomach of 04039, but a small amount of non‐cell‐associated E. coli was detected within the LP of the stomach of 06047 (Fig. 4a). Moreover, quantitative analysis showed a statistically significant difference between the rapid and slow groups when we compared the level of increased microbial translocation in almost the entire GI tract, except the duodenum. This was probably because the integrity of the duodenum in the slow progressors was not destroyed, and the integrity of mucosal epithelium could protect tissues from the invasion of exotic microorganisms (Fig. 4b). Collectively, these data indicated that microbial translocation occurred in both rapid and slow progressors, but it was more serious in the latter group, which might contribute to accelerating the development of disease.
Figure 4.
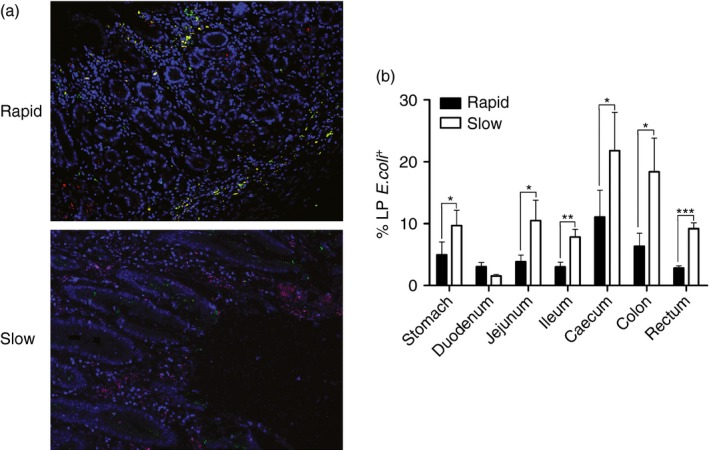
Detection of microbial translocation in the gastrointestinal (GI) tract of SIVmac239‐infected Chinese rhesus macaques. (a) Representative images (200 ×) of the GI tract stained for Escherichia coli constituents (red), macrophages (green), and DAPI (nuclei; blue). (b) Quantitative analysis of the proportion of E. coli that stained positively for the markers (* : P ≤ 0.05, ** : 0.05 < P ≤ 0.01, *** : 0.01 < P ≤ 0.001).
Phagocytosis by intestinal macrophages
The GI mucosa is the largest reservoir of macrophages in the body, and the macrophages in the intestinal LP are generally the first phagocytic cells to interact with microbial products that translocate through the breached mucosal epithelium.18, 19 Abundant extracelluar microbial products were observed in the slow progressors; hence, we doubted whether the phagocytic activity of these macrophages in slow progressors was undermined. To evaluate this finding, we performed spatial co‐localization of E. coli with macrophages in the GI tract of the two groups. The frequencies of macrophages had no significant decrease in the slow progressors compared with the rapid progressors (Fig. 5a); however, very few of the microbial products that translocated into the mucosa were found within HAM56+ macrophages in the slow progressors (Fig. 4a). By using the same quantitative analysis, we found that the percentage of macrophages that associated with microbial products in the GI tract was markedly lower in the slow progressors than in the rapid progressors, except in the duodenum section (Fig. 5b, c). Taken together, these data implied that the phagocytosis of intestinal macrophages was significantly weakened in the slow progressors.
Figure 5.
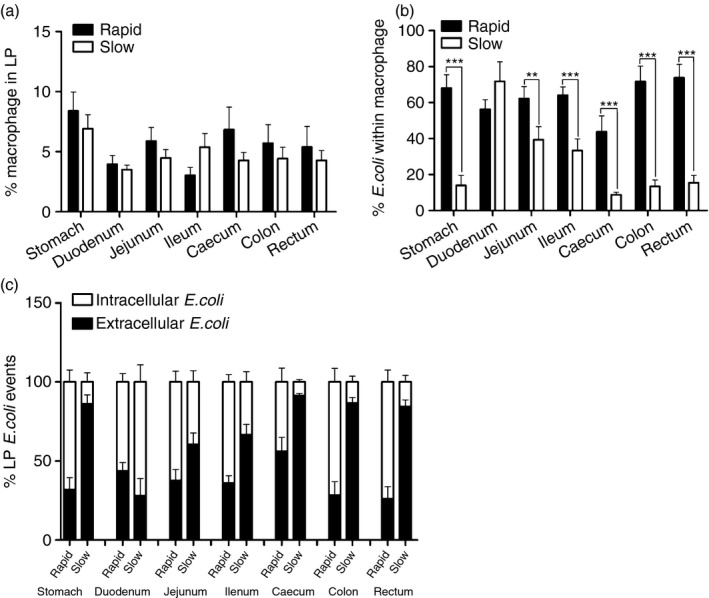
Changes in the number and function of macrophages during simian immunodeficiency virus (SIV) infection. (a) Proportion of macrophages from the lamina propria (LP) of rapid (black bar) and slow progressors (white bar). (b) The capacity of macrophages for phagocytosis of Escherichia coli in rapid (black bar) and slow progressors (white bar) (** : 0.05 ≤ P < 0.01, *** : 0.01 < P ≤ 0.001). (c) The frequency of intracellular (white bar) and extracellular (black bar) E. coli events in gastrointestinal (GI) tissues from rapid and slow progressors.
Quantification of CD4+ and CD8+ T lymphocytosis in the GI tract after SIV infection
Mucosal CD4+ T cells seem to be the first productively infected cells and lead to viral spread after infection. Monitoring of the quantity of CD4+ T cells in tissues is frequently used to evaluate the pathogenic changes after HIV/SIV infection.20 Hence, we quantified the CD3+ CD4+ T cells in the intestinal tissues by immunofluorescence staining to determine whether their prevalence changes during different stages of disease progression. Figure 6(a) shows the representative results (the stomach of 06047 was represented for rapid progressors, the stomach of 04039 for slow progressors). The histopathological analysis indicated that the slow progressors had a significantly smaller number of mucosal CD4+ T cells compared with the rapid progressors (Fig. 6b).
Figure 6.
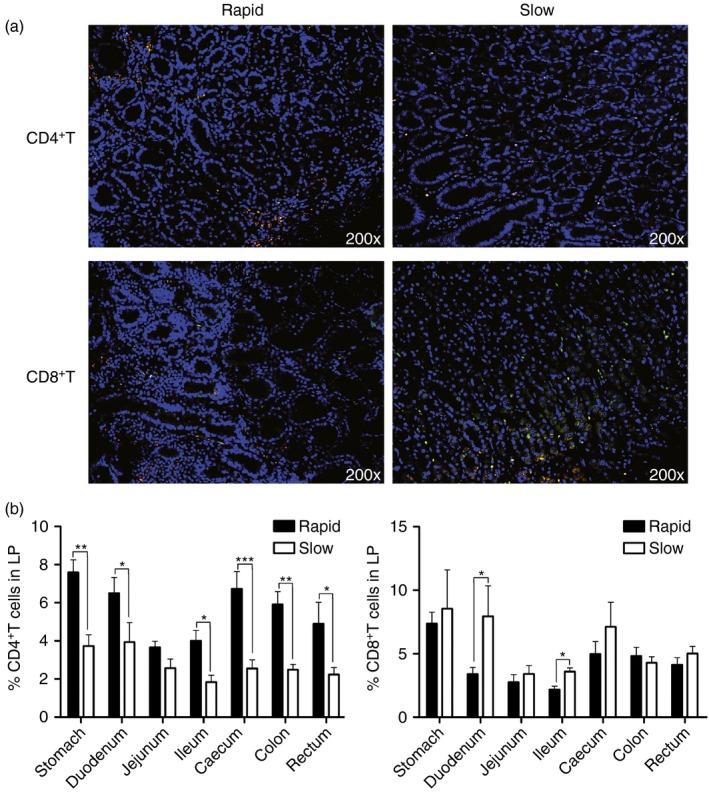
Dynamics of CD4+ and CD8+ T cells in the gastrointestinal (GI) tract after simian immunodeficiency virus (SIV) infection. (a) Representative images (200 ×) of the GI tract stained for CD4 or CD8 (green), CD3 (red), and DAPI (nuclei; blue). (b) Quantitative analysis showing the proportion of CD4+ and CD8+ T cells in the lamina propria (LP) of the GI tract of rapid and slow progressors (* : P ≤ 0.05, ** : 0.05 < P ≤ 0.01, *** : 0.01 < P ≤ 0.001).
CD8+ T cells have been shown to play an important role in the control of viral replication, and they also act as a critical factor in the effective clearance of bacterial pathogens; hence, depletion of mucosal CD8+ T cells should impair such a protective effect. However, the number of CD8+ T cells that were detected in the mucosal LP of the slow progressors was slightly higher than that in the rapid progressors, although only the duodenum and ileum sections showed a statistically significant difference between the two groups (Fig. 6c, d). In general, the number of mucosal CD4+ T cells in the slow progressors decreased markedly after HIV/SIV infection; the increasing microbial translocation may be one of the causes of this depletion. In contrast, the number of CD8+ T cells in the GI tract increased slightly; however, its antimicrobial activities might be weakened during the progress of infection.
Discussion
Previous studies have implied that microbial translocation in the intestinal mucosa is a major cause of pathological immune activation during HIV/SIV infection. The bacterial products could stimulate innate immune cells to produce pro‐inflammatory mediators through pattern recognition receptors; however, excessive production of pro‐inflammatory mediators might lead to a series of pathological changes, such as capillary leakage, tissue injury and multiple organ failure.16, 21 There is a lack of direct evidence on how the levels of microbial translocation and the functions of immune cells in the entire GI tract differ with the duration of disease. Here, we used immunohistological analysis to directly compare distinct pathological changes between rapid and slow progressors. Our analysis showed that: (i) the viral load in plasma could be detected at all time‐points in all animals, but the virus could only be found in some sections of the GI tract in the slow progressors, (ii) the level of translocated microbial products increased considerably in almost all GI sections in the slow progressors, (iii) the macrophage clearing of microbial products that invaded the LP of GI mucosa weakened in the slow progressors, and (iv) the amount of CD4+ T cells in mucosal tissues decreased markedly, whereas the number of mucosal CD8+ T cells increased in the slow progressors, but only the duodenum and ileum sections showed a statistically significant difference.
In our study, we used SIVmac239‐infected Chinese rhesus macaques to compare the different pathological changes between rapid and slow progressors. First, we determined the viral load and the number CD4+ and CD8+ T cells in blood from the two groups. We found that the viral load before death was much higher in the rapid progressors than in the slow progressors; this coincided with a notable decrease in CD4+ and CD8+ T cells in blood from the two groups, especially in animal 06047. These might be the major factors that caused the quick death of the rapid progressors. Because CD4+ T cells are the ‘first responders’ to SIV infection, they can produce antiviral cytokines, such as interferon‐γ, interleukin‐2 and tumour necrosis factor‐α, or help other immune effector cells to indirectly restrict the virus. The depletion of blood CD4+ T cells resulted in the absence of a post‐peak viral decline and accelerated the disease progression; this indicated that the antiviral response of blood CD4+ T cells might play an important role in limiting SIV replication and extending the lifespan of virus‐infected animals.22, 23 CD8+ cytotoxic T lymphocytes have been shown to protect against viral infection in animals by the direct cytolysis of infected cells or the delivery of active granzyme B to SIV‐infected targets, which then eliminates the virus‐infected cells; CD8+ cytotoxic T lymphocytes are also correlated with protection from SIV infection.24, 25
Second, the microbial translocation in HIV‐infected individuals and SIV‐infected rhesus macaques has been described in several studies,26, 27, 28 which showed that in the presence of increasing microbial translocation in plasma and intestinal mucosa, the integrity of the intestinal epithelium is destroyed after HIV/SIV infection. In addition, it has been reported that microbial translocation through the damaged mucosal epithelium is a major driver of immune activation and that excessive immune activation is one of the strongest predictors of disease progression; this means that the microbial translocation measured in the mucosa may be a useful parameter for evaluating disease progression. However, most of the current works only focus on one section of the GI tract. In the present study, we examined for the first time the translocation of microbial products in the entire GI tract of SIV‐infected Chinese rhesus macaques during different disease durations. We used two distinct methods to measure the translocation of microbes in the intestinal mucosa, which showed that LPS+ cells could be found only in the slow progressors, whereas E. coli + cells could be detected in both the slow and the rapid progressors; this might be a result of the broad spectrum of polyclonal antibodies against E. coli. Taken together, the findings indicated that microbial translocation started in the early phase of infection and then increased considerably in the intestinal mucosa as the illness progressed.
Third, we took much interest in whether the host responses were affected by the increasing microbial translocation. The GI mucosa contained the largest reservoir of macrophages, and these intestinal macrophages could recognize and eliminate invading microorganisms and noxious macromolecules through phagocytosis and killing; hence, they were considered as the ‘gatekeeper’ of the GI tract, restricting its infiltration by microbes.18, 29 Several groups have reported that intestinal macrophages not only restricted HIV/SIV replication30 but also equipped the host defence against microorganisms by scavenging for foreign bacteria in the LP; however, their capability for phagocytosis is apparently affected, especially in the late stages of AIDS.12 In our study, we used direct immunofluorescence staining to detect bacterial products and macrophages. The results confirmed that there was no significant difference in the percentage of tissue macrophages between rapid and slow progressors; however, the extracellular bacterial constituents in the LP of intestinal mucosa markedly increased in the slow progressors. Our data strongly suggested that the function of intestinal macrophages had been gradually impaired during the progression of infection, resulting in weakened phagocytosis or clearing of translocated microorganisms.
Fourth, in this research, we also detected the CD4+ and CD8+ T cells in plasma from all the animals at different time‐points. Taken as a whole, the data showed similar trends in changes during infection. However, the intestinal mucosa was the preferred target for HIV/SIV infection, and mucosal CD4+ and CD8+ T cells played an important role in the innate immune system. We assumed that the mucosal CD4+ and CD8+ T cells had distinct differences between different courses of the disease, which might be partially affected by the translocation of microbial products. We compared the proportions of these cells between rapid and slow progressors by directly staining the CD3+ CD4+ and CD3+ CD8+ T cells in the mucosal tissues. The results supported the hypothesis that the amount of mucosal CD4+ T cells was significantly higher in rapid progressors than in slow progressors; however, the opposite was true for the amount of mucosal CD8+ T cells. These results were not unexpected because mucosal CD4+ T cells are the primary attack and kill targets of the infected virus in tissues. Besides, mucosal CD4+ T cells expressed Toll‐like‐receptor 4 (TLR4), which is the receptor for LPS; triggering TLR4 activates various signalling pathways, including the extrinsic and intrinsic pathways of apoptosis, which then leads to cell death.31 This might imply that greater microbial translocation would result in more mucosal CD4+ T‐cell deaths. TLRs are an important component of the innate immune system. They can sense a microbial infection and trigger antimicrobial responses, as well as induce the migration of immune cells to the infection sites to kill and halt the spread of invading pathogens.32 Translocated bacteria contain a variety of TLR ligands, including bacterial DNA, LPS and peptidoglycan.33 Hence, the infiltrated microbes in the intestinal mucosa could indirectly activate and recruit specific CD8+ T cells, which might accumulate in the GI mucosa and mediate cytotoxic T lymphocytes to protect against viral infection and foreign invaders; this could make them the main contributor to the increasing number of CD8+ T cells in the mucosa after infection. However, in the slow progressors, although the mucosal CD8+ T cells increased, the percentage of infiltrated microbial products still increased. We speculated that the intestinal CD8+ T cells in the slow progressors had been deficient in clearing the translocated microbes, resulting in the marked increase in extracellular microbial products. Unfortunately, we could not conclusively verify the injurious effect of mucosal CD8+ T cells on the translocated microbes because of the lack of LP mononuclear cells isolated from fresh GI tissues.
In summary, our study found that microbial translocation occurred in the early stage of infection and then increased considerably as the disease progressed. In the slow progressors, mucosal CD8+ T cells accumulated in the GI tract, but the tissue macrophages showed no significant change. However, the activities of the CD8+ T cells and macrophages in tissues were disrupted, weakening their ability to clear the translocated microbial products. Our report could provide a useful strategy for slowing the disease progression by reducing or inhibiting the microbial translocation in mucosa after HIV/SIV infection.
Disclosure
The authors declare no financial or commercial conflict of interest.
Acknowledgements
This work was supported in part by grants from the National Basic Research Programme of China (2012CBA01305), the National Natural Science Foundation of China (81172876, 81471620, 81273251, U1202228, 81571606), the Key Scientific and Technological Programme of China (2012ZX10001‐007, 2013ZX10001‐002, 2014ZX10005‐002‐006), Natural Science Foundation of Yunnan Province (2014FB181) and the Knowledge Innovation Programme of the Chinese Academy of Sciences (KJZD‐EW‐L10‐02).
References
- 1. Schieferdecker HL, Ullrich R, Hirseland H, Zeitz M. T cell differentiation antigens on lymphocytes in the human intestinal lamina propria. J Immunol 1992; 149:2816–22. [PubMed] [Google Scholar]
- 2. Cavarelli M, Scarlatti G. HIV‐1 infection: the role of the gastrointestinal tract. Am J Reprod Immunol 2014; 71:537–42. [DOI] [PubMed] [Google Scholar]
- 3. Lackner AA, Mohan M, Veazey RS. The gastrointestinal tract and AIDS pathogenesis. Gastroenterology 2009; 136:1966–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Veazey RS, DeMaria M, Chalifoux LV, Shvetz DE, Pauley DR, Knight HL et al Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science 1998; 280:427–31. [DOI] [PubMed] [Google Scholar]
- 5. Li Q, Duan L, Estes JD et al Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature 2005; 434:1148–52. [DOI] [PubMed] [Google Scholar]
- 6. Steele AK, Lee EJ, Manuzak JA, Dillon SM, Beckham JD, McCarter MD et al Microbial exposure alters HIV‐1‐induced mucosal CD4+ T cell death pathways ex vivo . Retrovirology 2014; 11:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guadalupe M, Reay E, Sankaran S, Prindiville T, Flamm J, McNeil A et al Severe CD4+ T‐cell depletion in gut lymphoid tissue during primary human immunodeficiency virus type 1 infection and substantial delay in restoration following highly active antiretroviral therapy. J Virol 2003; 77:11708–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. George MD, Reay E, Sankaran S, Dandekar S. Early antiretroviral therapy for simian immunodeficiency virus infection leads to mucosal CD4+ T‐cell restoration and enhanced gene expression regulating mucosal repair and regeneration. J Virol 2005; 79:2709–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Veazey RS, Gauduin MC, Mansfield KG, Tham IC, Altman JD, Lifson JD et al Emergence and kinetics of simian immunodeficiency virus‐specific CD8+ T cells in the intestines of macaques during primary infection. J Virol 2001; 75:10515–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shu Z, Ma J, Tuerhong D, Yang C, Upur H. How intestinal bacteria can promote HIV replication. AIDS Rev 2013; 15:32–7. [PubMed] [Google Scholar]
- 11. Lozupone CA, Rhodes ME, Neff CP, Fontenot AP, Campbell TB, Palmer BE. HIV‐induced alteration in gut microbiota: driving factors, consequences, and effects of antiretroviral therapy. Gut Microbes 2014; 5:562–70. [DOI] [PubMed] [Google Scholar]
- 12. Estes JD, Harris LD, Klatt NR et al Damaged intestinal epithelial integrity linked to microbial translocation in pathogenic simian immunodeficiency virus infections. PLoS Pathog 2010; 6:e1001052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Durudas A, Milush JM, Chen HL, Engram JC, Silvestri G, Sodora DL. Elevated levels of innate immune modulators in lymph nodes and blood are associated with more‐rapid disease progression in simian immunodeficiency virus‐infected monkeys. J Virol 2009; 83:12229–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xia HJ, Zhang GH, Ma JP, Dai ZX, Li SY, Han JB et al Dendritic cell subsets dynamics and cytokine production in SIVmac239‐infected Chinese rhesus macaques. Retrovirology 2010; 7:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dandekar S. Pathogenesis of HIV in the gastrointestinal tract. Curr HIV/AIDS Rep 2007; 4:10–5. [DOI] [PubMed] [Google Scholar]
- 16. Klatt NR, Funderburg NT, Brenchley JM. Microbial translocation, immune activation, and HIV disease. Trends Microbiol 2013; 21:6–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nowroozalizadeh S, Mansson F, da Silva Z et al Microbial translocation correlates with the severity of both HIV‐1 and HIV‐2 infections. J Infect Dis 2010; 201:1150–4. [DOI] [PubMed] [Google Scholar]
- 18. Schenk M, Mueller C. Adaptations of intestinal macrophages to an antigen‐rich environment. Semin Immunol 2007; 19:84–93. [DOI] [PubMed] [Google Scholar]
- 19. Smythies LE, Sellers M, Clements RH, Mosteller‐Barnum M, Meng G, Benjamin WH et al Human intestinal macrophages display profound inflammatory anergy despite avid phagocytic and bactericidal activity. J Clin Invest 2005; 115:66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Malzahn J, Shen C, Caruso L, Ghosh P, Sankapal SR, Barratt‐Boyes S et al Effect of early anti‐retroviral therapy on the pathogenic changes in mucosal tissues of SIV infected rhesus macaques. Virol J 2012; 9:269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brenchley JM, Douek DC. Microbial translocation across the GI tract. Annu Rev Immunol 2012; 30:149–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ortiz AM, Klatt NR, Li B et al Depletion of CD4+ T cells abrogates post‐peak decline of viremia in SIV‐infected rhesus macaques. J Clin Invest 2011; 121:4433–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang X, Xu H, Pahar B, Lackner AA, Veazey RS. Divergent kinetics of proliferating T cell subsets in simian immunodeficiency virus (SIV) infection: SIV eliminates the “first responder” CD4+ T cells in primary infection. J Virol 2013; 87:7032–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Freel SA, Saunders KO, Tomaras GD. CD8+ T‐cell‐mediated control of HIV‐1 and SIV infection. Immunol Res 2011; 49:135–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mendoza D, Migueles SA, Rood JE et al Cytotoxic capacity of SIV‐specific CD8+ T cells against primary autologous targets correlates with immune control in SIV‐infected rhesus macaques. PLoS Pathog 2013; 9:e1003195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Marchetti G, Bellistrì GMM, Borghi E et al Microbial translocation is associated with sustained failure in CD4+ T‐cell reconstitution in HIV‐infected patients on long‐term highly active antiretroviral therapy. AIDS 2008; 22:2035–8. [DOI] [PubMed] [Google Scholar]
- 27. Leinert C, Stahl‐Hennig C, Ecker A, Schneider T, Fuchs D, Sauermann U et al Microbial translocation in simian immunodeficiency virus (SIV)‐infected rhesus monkeys (Macaca mulatta). J Med Primatol 2010; 39:243–51. [DOI] [PubMed] [Google Scholar]
- 28. Dyavar Shetty R, Velu V, Titanji K, Bosinger SE, Freeman GJ, Silvestri G et al PD‐1 blockade during chronic SIV infection reduces hyperimmune activation and microbial translocation in rhesus macaques. J Clin Invest 2012; 122:1712–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Smith PD, Ochsenbauer‐Jambor C, Smythies LE. Intestinal macrophages: unique effector cells of the innate immune system. Immunol Rev 2005; 206:149–59. [DOI] [PubMed] [Google Scholar]
- 30. Shen R, Meng G, Ochsenbauer C et al Stromal down‐regulation of macrophage CD4/CCR5 expression and NF‐κB activation mediates HIV‐1 non‐permissiveness in intestinal macrophages. PLoS Pathog 2011; 7:e1002060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mburu S, Marnewick JL, Abayomi A, Ipp H. Modulation of LPS‐induced CD4+ T‐cell activation and apoptosis by antioxidants in untreated asymptomatic HIV infected participants: an in vitro study. Clin Dev Immunol 2013; 2013:631063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Iwasaki A, Medzhitov R. Toll‐like receptor control of the adaptive immune responses. Nat Immunol 2004; 5:987–95. [DOI] [PubMed] [Google Scholar]
- 33. Paulos CM, Wrzesinski C, Kaiser A et al Microbial translocation augments the function of adoptively transferred self/tumor‐specific CD8+ T cells via TLR4 signaling. J Clin Invest 2007; 117:2197–20. [DOI] [PMC free article] [PubMed] [Google Scholar]


