Abstract
Hypoxia is a major cause of radiation resistance, which may predispose to local recurrence after radiation therapy (RT). While hypoxia increases tumor cell survival after RT because there is less oxygen to oxidize damaged DNA, whether signaling pathways triggered by hypoxia contribute to radiation resistance is poorly understood. For example, intratumoral hypoxia can increase hypoxia inducible factor 1 alpha (HIF-1α), which may regulate pathways that contribute to radiation sensitization or radiation resistance. To clarify the role of HIF-1α in regulating tumor response to radiation therapy, we generated a novel genetically engineered mouse model of soft tissue sarcoma with an intact or deleted HIF-1α. Deletion of HIF-1α sensitized primary sarcomas to RT in vivo. Moreover, cell lines derived from primary sarcomas lacking HIF-1α, or in which HIF-1α was knocked down, had decreased clonogenic survival in vitro, demonstrating that HIF-1α can promote radiation resistance in a cell autonomous manner. In HIF-1α intact and deleted sarcoma cells, radiation-induced reactive oxygen species (ROS), DNA damage repair, and activation of autophagy were similar. However, sarcoma cells lacking HIF-1α had impaired mitochondrial biogenesis and metabolic response after radiation which might contribute to radiation resistance. These results show that HIF-1α promotes radiation resistance in a cell autonomous manner.
INTRODUCTION
Tissue hypoxia, defined by physiologically low oxygen availability, is one of the most important cellular stresses due to the vital role of oxygen as the final electron acceptor in the electron transport chain. Importantly, tissue hypoxia is observed in solid tumors [1, 2], and has long been recognized to negatively affect solid tumor response to radiation therapy. For example, animal experiments in the mid-20th century showed that increasing oxygen concentration in the tumor by providing subjects with high pressure oxygen to inspire during radiation therapy increased both tumor and normal tissue sensitivity to radiation [3–5]. Moreover, a number of clinical trials have been conducted in which cancer patients were treated with concurrent radiation therapy and either hyperbaric oxygen, other modifiers of hypoxia such as carbogen-nicotinamide, or radiobiologic mimics of oxygen such as metronidazole. A 2007 meta-analysis of all 61 published clinical trials to date revealed that 87% of the studies demonstrated that modifiers of tumor hypoxia during radiation therapy resulted in either improvement in loco-regional control, overall survival, or both [6]. Interestingly, in a phase III trial of advanced head and neck cancer patients randomized to chemo-radiation with or without the hypoxic cytotoxin tirapazamine, in the subset of patients that had hypoxia imaging by PET prior to treatment, only the patients with hypoxic tumors benefited from tirapazamine [7]. One mechanism that contributes to the oxygen enhancement ratio is that molecular oxygen can interact with radiation-induced radicals in the cell to form adducts in carbohydrates, proteins, or lipids. Most importantly, radiation induces oxygen-DNA adducts that are difficult or impossible to repair, which can lead to cell death.
In addition to acting as an acceptor of electrons from ionized molecules, oxygen can also regulate radiation response by altering cellular gene expression via hypoxia inducible factors (HIFs) [8]. HIFs are heterodimeric basic Helix-Loop-Helix PER-ARNT-SIM (bHLH-PAS) family transcription factors composed of an alpha and a beta subunit. When dimerized, HIFs bind to hypoxia responsive elements (HRE) in the promoters or enhancers of target genes. The alpha subunit is regulated by oxygen tension. At high oxygen levels (>8–10%), HIF alpha subunits become less abundant at the protein level due to prolyl hydroxylase-mediated degradation, which for HIF-1α decreases the half-life to 5–10 minutes despite continued transcription. Under hypoxia, the alpha subunits are stabilized, so that HIF proteins are able to rapidly alter pathways such as cellular metabolism [9], angiogenesis [10], and survival [11]. Although hypoxia in tumors activates HIF-1α, it is not clear whether activation of HIF-1α signaling contributes to radiation resistance in primary cancers.
Previous studies investigating the role of HIF-1α in tumor response to radiation therapy using transplanted tumor model systems suggest that HIF-1α may both promote radiation resistance and radiation sensitivity. For example, HIF-1α can sensitize tumor cells to radiation by promoting ATP metabolism, proliferation, and p53 activation, but HIF-1α can also cause resistance to radiation therapy by promoting endothelial cell survival [12]. Other studies have suggested that HIF-1α promotes resistance to radiation by increasing the transcription of genes that inhibit apoptosis [13]. However, to our knowledge the role of a HIF-1α in mediating tumor response to radiation therapy in a primary mouse tumor model has not been performed. Here, we generated a novel primary mouse model of soft tissue sarcoma to dissect the role of HIF-1α function within tumor cells in the response of primary cancers to radiation therapy. We show that deletion of HIF-1α in this sarcoma model sensitizes tumor cells to radiation therapy in a cell-autonomous manner.
RESULTS
Human soft tissue sarcomas upregulate HIF-1α after chemo-radiation therapy
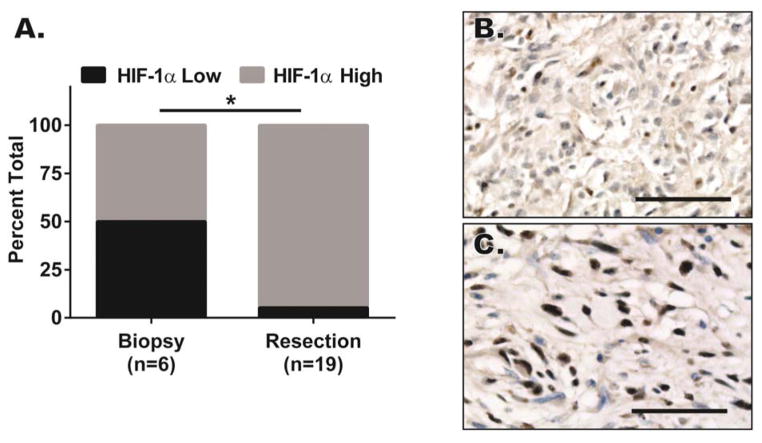
To examine whether HIF-1α levels change in human soft tissue sarcomas in response to treatment, we obtained a tissue microarray (TMA) of soft tissue sarcoma samples that included biopsies before treatment or at the time of surgical resection after interdigitated neoadjuvant MAID chemotherapy (mesna, doxorubicin, ifosfamide, and dacarbazine) and 44 Gy radiation therapy [14]. HIF-1α immunostaining was performed on the TMA and the staining intensity was scored for HIF-1α by a sarcoma pathologist blinded to the timing of chemo-radiation therapy relative to the biopsy. An increase in HIF-1α nuclear intensity and quantity of HIF-1α-positive nuclei in tumor cells were observed in samples following chemo-radiation therapy (Figure 1A–C).
Figure 1. Human soft tissue sarcomas accumulated nuclear HIF-1α after interdigitated chemo-radiation therapy.
(A–C) Formalin-fixed paraffin embedded (FFPE) tissue microarray from human soft tissue sarcoma biopsies prior to chemo-radiation therapy and after chemo-radiation therapy was stained with antibody against HIF-1α. The HIF-1α stained tissue microarray was subsequently read by a pathologist blinded to the samples and scored based on HIF-1α staining intensity. A) HIF-1α nuclear accumulation score increased significantly with interdigitated chemo-radiation therapy using the Fisher’s exact test. B–C) Paired pre- and post-therapy human soft tissue sarcoma biopsies stained with an antibody to HIF-1α. Scalebar = 50μm. * p<0.05.
Generation and characterization of a novel mouse model of soft tissue sarcoma
We previously showed that some tumors generated by injection of tamoxifen into Pax7Cre-ER-T2/+; KrasLSL-G12D/+; p53FL/FL mice resembled human rhabdomyosarcoma (RMS) [15, 16]. Because NRAS is more commonly mutated in human RMS than KRAS [17–19], we generated Pax7Cre-ER-T2/+; NrasLSL-G12D/+; p53FL/FL; ROSA26mTmG/mTmG (P7NP) mice to model mutations that more frequently occur in human RMS. The ROSA26mTmG allele is a fluorescent reporter allele that in the absence of Cre recombinase expresses membrane-tagged red fluorescent protein (tdTomato) from the ubiquitous ROSA26 promoter, but in the presence of Cre recombinase deletes tdTomato to express membrane-tagged green fluorescent protein (eGFP). In this model, the expression of the fusion protein CreER-T2 is driven by the endogenous Pax7 promoter, which in the adult mouse is expressed in skeletal muscle satellite cells [20]. Exposure of CreER-T2 to 4-hydroxytamoxifen (4-OHT), which is the active metabolite of tamoxifen, leads to the accumulation of CreER-T2 in the nucleus and recombination of loxP flanked alleles to activate expression of the NrasG12D oncogene, delete both alleles of p53, delete tdTomato, and express eGFP. Therefore, the fluorescent proteins will mark the tumor cells that have undergone Cre-mediated recombination green, but the normal cells of the stroma that have not undergone Cre-mediated recombination will remain red. Intramuscular (IM) hind limb injection of 4-OHT in the Pax7CreER-T2/+; NrasLSL-G12D/+; p53Fl/Fl; ROSA26mTmG/mTmG mice caused sarcomas to form at the site of injection after 1–3 months with 100% penetrance(Figure 2A).
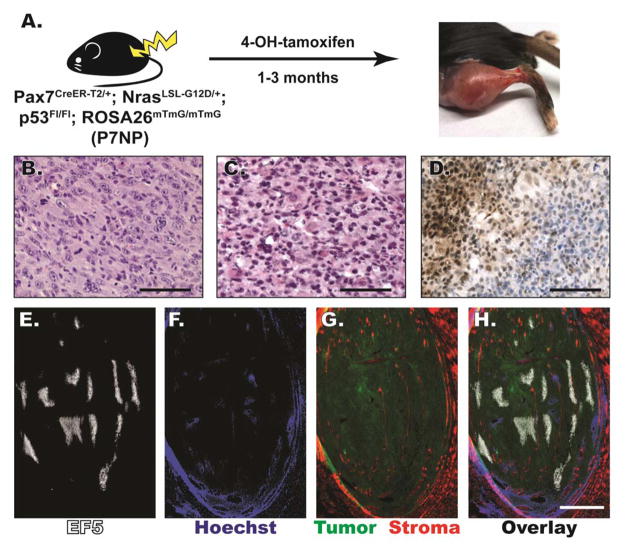
Figure 2. P7NP sarcomas were either UPS or RMS by histology, and had regions of HIF-1α accumulation and tumor hypoxia.
A) Schematic of novel Pax7Cre-ER-T2/+; NrasLSL-G12D/+; p53FL/FL; ROSA26mTmG/mTmG (P7NP) mouse model of soft tissue sarcoma generated by intramuscular 4-hydroxytamoxifen injection. (B, C) FFPE sections of primary soft tissue sarcomas from P7NP mice were stained with hematoxylin and eosin. B) P7NP sarcoma with histology resembling undifferentiated pleomorphic sarcoma (UPS). Scalebar = 100μm. C) P7NP sarcoma with histology resembling rhabdomysarcoma (RMS). Scalebar = 100μm. D) Representative FFPE sarcoma section stained with antibody against HIF-1α and signal-amplified with DAB. HIF-1α staining of sarcomas from P7NP mice revealed regional heavy nuclear accumulation of HIF-1α. Scalebar = 200μm. (E–H) Representative frozen sarcoma whole tumor cross-section in a P7NP mouse that received intraperitoneal injection of EF5 and intravenous injection of Hoechst 33342 prior to tumor harvest. E) EF5 distribution in a whole cross-section of P7NP sarcoma showed areas of tumor hypoxia. F) Hoechst 33342 perfusion showed well-perfused tumor periphery and surrounding normal tissue, and poorly perfused areas in the tumor core. G) eGFP and tdTomato visualization showed eGFP positive tumor with surrounding tdTomato positive normal tissue as well as tdTomato positive stromal infiltration within the tumor. H) Overlay of E-G shows distribution of tissue hypoxia and perfusion relative to tumor and surrounding normal tissue. Scalebar = 2mm.
Consistent with previous findings from our group, sarcomas derived from adult skeletal muscle satellite cells with activation of oncogenic RAS and deletion of p53 fall in a spectrum of histological subtypes ranging from undifferentiated pleomorphic sarcoma (UPS, Figure 2B) to RMS (Figure 2C). Whereas UPS does not show any specific line of differentiation and is a diagnosis of exclusion, RMS contains rhabdomyoblasts, which are cells with eosinophilic cytoplasm and eccentric nuclei [21]. Regardless of histological subtype, these sarcomas showed regions of localized HIF-1α staining by immunohistochemistry (Figure 2D) that suggested the presence of tumor hypoxia. The presence of hypoxic regions and their relationship to vascular perfusion within the tumor were further examined with EF5 (Figure 2E) and Hoechst 33342 perfusion (Figure 2F) staining. Overall, these results show that soft tissue sarcomas arising in Pax7CreER-T2/+; NrasLSL-G12D/+; p53Fl/Fl; ROSA26mTmG/mTmG mice have regions of tumor hypoxia as demonstrated by positive EF5 staining and nuclear localization of HIF-1α.
Deletion of HIF-1α in primary soft tissue sarcomas in P7NP mice does not affect sarcoma subtype or degree of tumor hypoxia
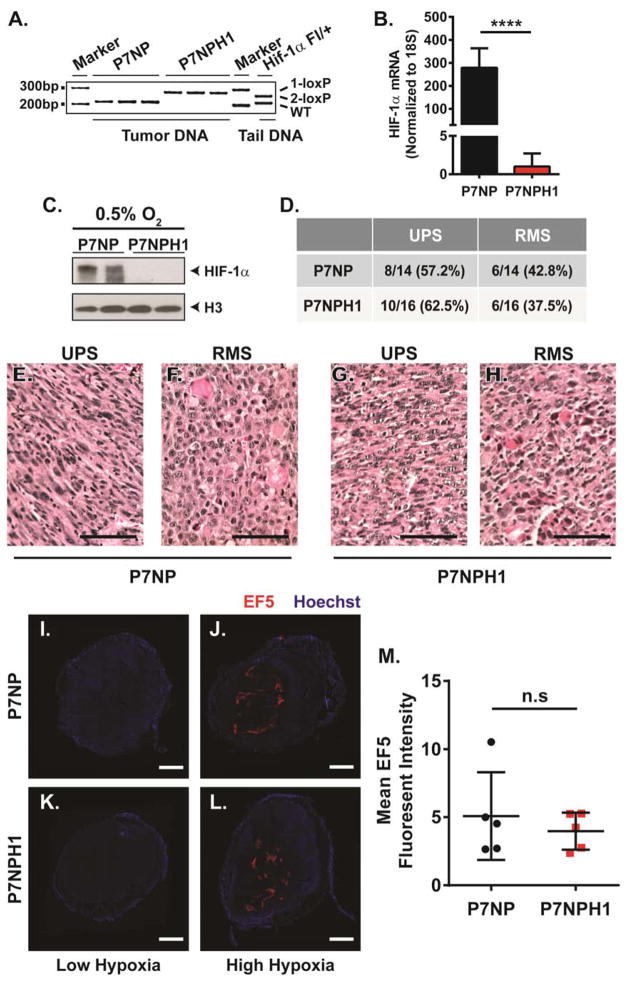
To determine the role of HIF-1α in primary sarcomas treated with radiation therapy, Pax7Cre-ER-T2/+; NrasLSL-G12D/+; p53FL/FL; Hif-1α+/+; ROSA26mTmG/mTmG (P7NP) mice were crossed to Hif-1αFL/FL mice to generate Pax7Cre-ER-T2/+; NrasLSL-G12D/+; p53FL/FL; Hif-1αFL/FL; ROSA26mTmG/mTmG (P7NPH1) mice. Hif-1αFL/FL mice delete HIF-1α function in the presence of Cre. P7NP and P7NPH1 mice were injected with IM 4-OHT into the hind limb. Sarcoma cells from tumors from P7NP and P7NPH1 mice were dissociated and cultured to deplete stromal cells. When DNA was extracted from tumor cells after in vitro culture, 50 cycles of PCR showed that the Hif-1αFL alleles were efficiently recombined (1-loxP) at the genomic level (Figure 3A). qRT-PCR was performed using exon 2-specific primers for Hif-1α on cDNA synthesized from RNA extracted from P7NP and P7NPH1 tumor cells, which confirmed the lack of Hif-1α transcript in P7NPH1 tumor cells (Figure 3B). Finally, P7NP and P7NPH1 tumor cells were cultured under hypoxic conditions (0.5% oxygen) for 16 hours and nuclear lysates were extracted. Western Blot of those lysates confirmed lack of HIF-1α protein accumulation in P7NPH1 cells following hypoxic stimulus (Figure 3C). As HIF-1α was ablated at the start of tumorigenesis, this could potentially modify tumor subtype, such as tumor differentiation status. Therefore, tumors were collected from P7NP and P7NPH1 mice and hematoxylin and eosin (H&E) stained sections were evaluated by a sarcoma pathologist blinded to genotype. Regardless of HIF-1α status, ~60% of the tumors generated resembled UPS by H&E staining, while ~40% of the tumors generated resembled RMS by H&E (Figure 3D–H).
Figure 3. HIF-1α deletion in sarcomas from P7NPH1 mice was efficient and did not change tumor subtype or degree of tumor hypoxia.
(A–C) Primary cells from P7NP and P7NPH1 tumors were dissociated into single cell suspensions with tissue proteases, and cultured deplete stromal cells. After 2–4 serial passages, primary tumor cells were collected to verify HIF-1α status by genomic DNA, RNA expression, and protein expression. A) Using primers specific for the HIF-1α conditional allele, the loxP flanked exon 2 of the Hif1a conditional allele was efficiently recombined (1-loxP) at the genomic level in sarcomas from P7NPH1 mice. B) qRT-PCR specific for exon 2-containing Hif1a mRNA transcript showed significant loss of expression in tumor cells from P7NPH1 mice when normalized to 18S by a two-tailed student t-test. C) Western Blot using antibody against HIF-1α in denatured cell lysates from tumors in P7NPH1 mice showed no detectable HIF-1α protein in the nuclear protein extracts after culture under 0.5% oxygen for 16 hours. Antibody probing against histone H3 served as nuclear protein loading control. (D–H) FFPE sections of primary P7NP and P7NPH1 tumors were stained with hematoxylin and eosin and scored as UPS or RMS by a pathologist blinded to the genotypes of the tumors. D) Histologic examination of P7NP and P7NPH1 tumors indicated deletion of HIF-1α did not alter the distribution of tumor histologic subtype. E–H) Representative hematoxylin and eosin stained FFPE sections of UPS and RMS from P7NP and P7NPH1 tumors. Scalebar = 50μm. (I–M) After primary tumors reached 600mm3, P7NP and P7NPH1 mice received intraperitoneal injection of EF5 and intravenous injection of Hoechst 33342 prior to tumor harvest. Tumors were evaluated for the presence of hypoxia by EF5 staining and for vascular perfusion by Hoechst 33342. (I–L) Representative frozen sarcoma whole tumor cross-sections with low levels of hypoxia and high levels of hypoxia in P7NP and P7NPH1 mice. Scalebar = 2mm. M) EF5 staining intensity of tumors using whole tumor cross-sections, normalized to tumor area, showed no significant variations between degree of tumor hypoxia from P7NP and P7NPH1 mice by a two-tailed student t-test. n.s. = not significant. **** p<0.0001.
To determine how HIF-1α may affect tumor microenvironmental hypoxia in primary sarcomas generated in P7NP and P7NPH1 mice, whole tumor cross-sections were obtained from nonirradiated P7NP and P7NPH1 mice. Tumor hypoxia was evaluated with EF5 and tumor perfusion was evaluated with Hoechst. Both tumors from P7NP and P7NPH1 mice ranged from containing low hypoxia to high hypoxia content (Figure 3I–L). Furthermore, there was no difference between the range of hypoxia content between tumors from P7NP and P7NPH1 mice (Figure 3M). This suggests that the tumor hypoxic content varies, and the loss of HIF-1α does not alter the presence of tumor hypoxia in this mouse model of primary soft tissue sarcoma.
Deletion of HIF-1α sensitizes tumor cells to radiation therapy in a cell autonomous manner
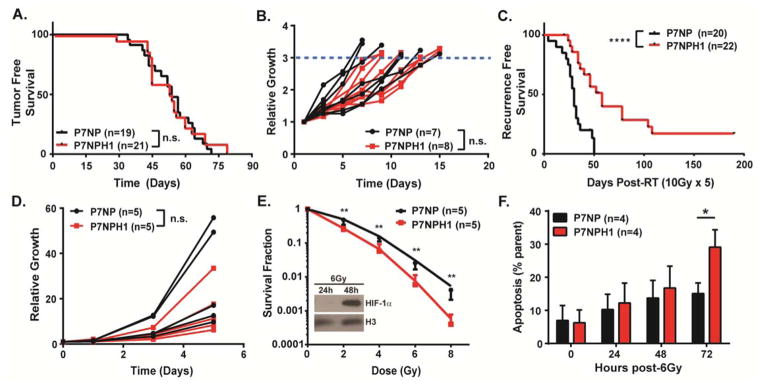
Similar to our previous results in which deleting HIF-1α did not affect tumor initiation or growth in primary soft tissue sarcomas initiated by expression of KrasG12D and p53 deletion [22], we did not observe a significant difference in time to tumor initiation after 4-OHT injection (Figure 4A) or tumor growth (Figure 4B) between sarcomas in P7NP and P7NPH1 mice. To investigate the impact of deleting HIF-1α in tumor cells on radiation response of primary sarcomas, we generated a separate cohort of sarcomas in P7NP and P7NPH1 littermate mice. When these tumors reached approximately 200mm3, we used image-guided radiation therapy to deliver 5 daily fractions of 10Gy. After irradiation, these tumors were followed for 180 days or until tumors tripled from their initial volume at the time of the first dose of irradiation. P7NP sarcomas (n=20) recurred between 2–50 days after the last dose of irradiation; in contrast, P7NPH1 sarcomas (n=22) did not start to recur until 40 days after irradiation with 13.6% surviving to 180 days without any evidence of tumor recurrence (Figure 4C). Taken together, the data indicate that although HIF-1α has diverse targets that may play opposing roles in response to radiation therapy [23], the overall effect of deleting HIF-1α in vivo in this mouse model of soft tissue sarcoma is radiosensitization.
Figure 4. Deletion of HIF-1α in the P7NP mouse model of soft tissue sarcoma did not affect in vivo tumor initiation but did sensitize primary tumors to radiation therapy both in vivo and in vitro.
(A–C) P7NP and P7NPH1 mice were injected with intramuscular 4-hydroxytamoxifen in the hindlimb and followed for tumor formation and tumor growth kinetics. After the tumors reached 200mm3, a cohort of nonirradiated tumors was measured for tumor growth. A separate cohort underwent daily irradiation to 10Gy for 5 days. These tumors were followed for local control. A) Deletion of HIF-1α (P7NPH1) did not affect primary tumor onset as compared to tumors from P7NP mice by a log-rank test. B) Deletion of HIF-1α (P7NPH1) did not affect nonirradiated tumor growth kinetics compared to tumors from P7NP mice as measured by time to tumor tripling by a two-tailed student t-test. C) Deletion of HIF-1α (P7NPH1) sensitized tumors to fractionated radiation therapy as compared to tumors from P7NP mice by a log-rank test. (D–F) Primary cells from P7NP and P7NPH1 tumors were isolated as described in Figure 3. The growth rate and radiosensitivity as measured by clonogenic survival and apoptosis after radiation for primary tumor cells were compared in vitro under 21% oxygen. D) The growth rates were measured by cell counting at indicated times. Deletion of HIF-1α (P7NPH1) did not affect sarcoma cell proliferation as compared to sarcoma cells from P7NP mice in vitro by a two-tailed student t-test. E) Deletion of HIF-1α (P7NPH1) increased sarcoma cell radiosensitivity as compared to sarcoma cells from P7NP mice in vitro by a two-tailed student t-test. Inset: P7NP cells irradiated with 6Gy accumulate nuclear HIF-1α at 24 and 48 hours after irradiation. Histone H3 served as a control for the nuclear extract. F) The percentages of apoptotic cells after 6Gy were measured by incubating P7NP and P7NPH1 cells at indicated times with Annexin V antibody and evaluated with flow cytometry. Deletion of HIF-1α (P7NPH1) sarcoma cells resulted increased apoptosis 72 hours following 6Gy irradiation as compared to sarcoma cells from P7NP mice by a two-tailed student t-test. n.s. = not significant. * p<0.05. ** p<0.01. **** p<0.0001.
The radiosensitization phenotype in P7NPH1 sarcomas in vivo may be dependent on tumor cell HIF-1α signaling within the tumor cells themselves (cell-autonomous) and/or through extracellular remodeling of tumor stroma (non-cell-autonomous). In order to determine whether a cell-autonomous mechanism contributed to the radiosensitization phenotype of P7NPH1 sarcomas in vivo, we isolated tumor cells from unirradiated P7NP and P7NPH1 mice and cultured them in vitro. After serial passaging the sarcoma cells to deplete stromal cells, we confirmed deletion of HIF-1α in the sarcoma cells by PCR as in Figure 3A. Sarcoma cell proliferation rates were measured in vitro between the cell lines from P7NP and P7NPH1 mice by cell counting. There was no difference in sarcoma cell proliferation in vitro between the cell lines from P7NP and P7NPH1 mice (Figure 4D). Next, we assessed the impact of HIF-1α deletion on clonogenic survival of these cell lines following irradiation in vitro. We first assessed HIF-1α nuclear accumulation under 21% oxygen culture after irradiation in P7NP cells by collecting nuclear protein lysates at 24 and 48 hours after 6 Gy irradiation (Figure 4E, inset). By Western Blot, we showed that there was already weak HIF-1α accumulation at 24 hours after irradiation, which became prominent at 48 hours after irradiation. This suggests that HIF-1α can accumulate in sarcoma cells under normoxia with radiation treatment in a time-dependent manner. When these sarcoma cells were exposed to increasing doses of radiation under normoxia, at all doses of radiation tested, P7NP sarcoma cells were more capable of forming colonies than P7NPH1 sarcoma cells (Figure 4E). The dose-modifying factor (DMF) was calculated from these results to be 1.31 using methods as previously described [24]. To complement the clonogenic assay, we also measured apoptosis by staining for Annexin V and 7AAD. Because the difference in clonogenic survival between P7NP and P7NPH1 cells lines after 6 Gy was approximately 5 fold (Figure 4E), this dose was selected for the apoptosis experiment. The percentage of Annexin V-positive P7NPH1 cells was higher than P7NP cells 72 hours following irradiation (Figure 4F), suggesting that the increase in apoptotic rate contributes to the increase in P7NPH1 sarcoma cell radiosensitivity.. These results support a tumor cell-autonomous role for HIF-1α in promoting tumor cell survival after radiation therapy.
Deletion of HIF-1α does not affect DNA damage repair, cellular ROS response or autophagy following irradiation
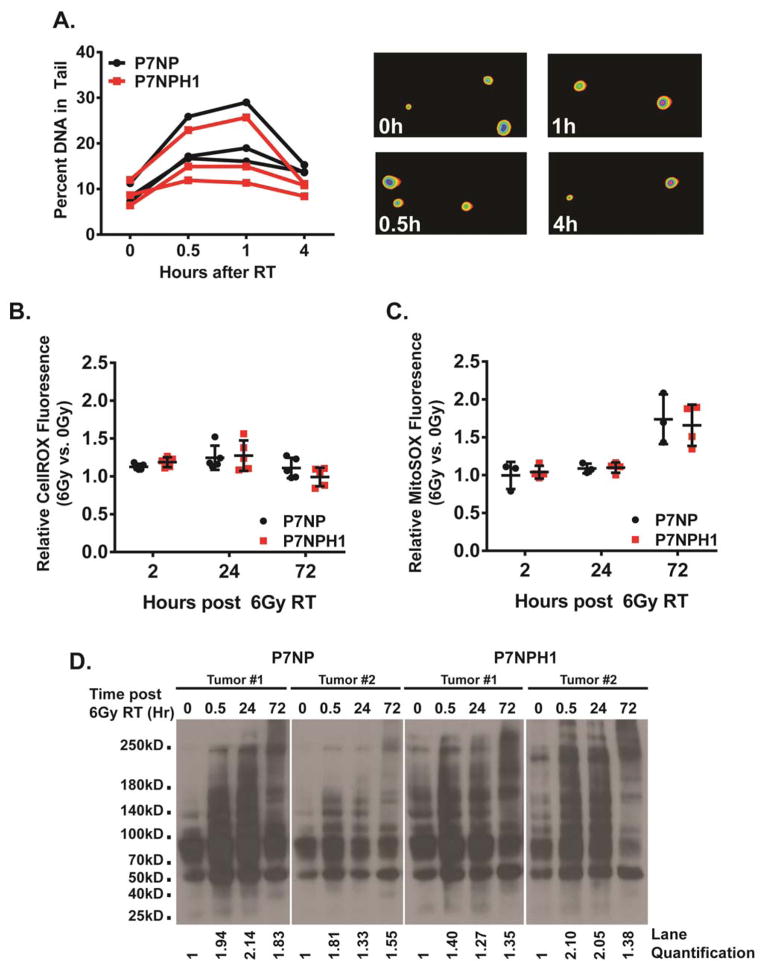
Insufficient repair of radiation-induced DNA double strand breaks can lead to increased cell death after radiation. To determine if decreased DNA repair of P7NPH1 cells contributed to enhanced radiation sensitivity, we performed the comet assay [25]. To reduce the basal presence of the tail due to incompletely synthesized DNA strands, cells were cultured in normal media with 0.1% fetal bovine serum to induce G1 arrest. G1 arrested cells were irradiated with 10Gy under 21% oxygen conditions and collected at 0, 0.5, 1, and 4 hours after irradiation to measure the length of the comet tail. Both P7NP and P7NPH1 tumor cells had similar basal tail lengths without irradiation, and irradiation induced an increase in tail length (Figure 5A). After irradiation, the tail resolved with similar kinetics in P7NP and P7NPH1 cells suggesting that cells with or without HIF-1α were able to repair DNA through non-homologous end joining with similar efficiency.
Figure 5. Deletion of HIF-1α did not affect DNA damage repair or cellular ROS clearance after irradiation.
Primary cells from P7NP and P7NPH1 tumors were isolated as described in Figure 3. All experiments were performed with in vitro cultures under 21% oxygen. A) DNA damage repair after 10Gy irradiation was measured with the Comet assay. Cells were irradiated and collected at indicated times, and embedded in agarose. DNA damage was measured by the percentage of visible tail after electrophoresis. Deletion of HIF-1α (P7NPH1) did not affect non-homologous end-joining repair as compared to sarcoma cells from P7NP mice by a two-tailed student t-test. Representative images of the DNA tail were shown in the right panels. B) Cells were irradiated with 6Gy. Accumulation of reactive oxygen species (ROS) was measured first by incubating nonirradiated and irradiated cells with CellRox DeepRed reagent at 2, 24, and 72 hours, then by performing flow cytometry. Relative ratios of ROS were calculated by normalizing the irradiated samples to nonirradiated controls at each time point. Deletion of HIF-1α (P7NPH1) did not affect ROS accumulation as compared to sarcoma cells from P7NP mice by a two-tailed student t-test. C) Cells were irradiated with 6Gy. Accumulation of mitochondrial superoxide was measured first by incubating nonirradiated and irradiated cells with MitoSOX reagent at 2, 24, and 72 hours, then by performing flow cytometry. Relative ratios of mitochondrial superoxide were calculated by normalizing the irradiated samples to nonirradiated controls at each time point. Deletion of HIF-1α (P7NPH1) did not affect mitochondrial superoxide accumulation as compared to sarcoma cells from P7NP mice by a two-tailed student t-test. D) Two P7NP and P7NPH1 primary sarcoma cell lines were irradiated with 6Gy. Damage to cellular structures by ROS was measured by protein carbonylation at indicated times after irradiation using OxyBlot per the manufacturer’s instructions. Deletion of HIF-1α (P7NPH1) did not affect the rate of accumulation or resolution of ROS-induced protein carbonylation. ImageJ quantification of cabonylated proteins for each time point was calculated as a ratio to the 0h time point, and listed below each lane.
Radiation-induced reactive oxygen species (ROS) and subsequent ROS-related cellular damage can cause cell death following radiation exposure. Because the rate of repair for radiation-induced DNA damage was similar between P7NP and P7NPH1 cells (Figure 5A), radiation induced ROS and subsequent clearance of ROS-induced adducts to cellular macromolecules were examined to determine if they may contribute to the increased radiation sensitivity observed in P7NPH1 cells. To determine how the level of ROS accumulation changes in response to radiation, P7NP and P7NPH1 cells were first irradiated with 6 Gy. At 2, 24, and 72 hours after irradiation, intracellular ROS was measured in both irradiated cells and non-irradiated controls by CellROX Deep Red fluorescence by FACS. Intracellular accumulation of ROS was similar in P7NP and P7NPH1 cells at both 2 and 24 hours after irradiation as compared to baseline (Figure 5B), and this accumulation largely returned to baseline at 72 hours after irradiation. Mitochondria-specific accumulation of superoxide was also examined using MitoSOX fluorescence by FACS to further assess the intracellular ROS content after irradiation. Similar to CellROX measurements of total intracellular ROS after irradiation, P7NP and P7NPH1 cells have similar levels of mitochondrial superoxide generation at 2, 24, and 72 hours after irradiation (Figure 5C). Because there was no difference in ROS accumulation either by measurement of total intracellular ROS or mitochondrial superoxide between P7NP and P7NPH1 cells after irradiation, accumulation of ROS does not appear to account for the increased radiosensitivity of P7NPH1 cells.
Although P7NP and P7NPH1 cells accumulate similar levels of ROS after irradiation, P7NPH1 cells may be more sensitive to ROS-induced cellular damage due to defects in repairing and clearing of damaged cellular macromolecules. Oxygen radicals not only cause DNA damage, but they also cause damage to proteins and lipids [26, 27]. Oxidation-mediated changes in proteins can be detected by protein-carbonyl adducts. To examine the extent of ROS-induced cellular damage in P7NP and P7NPH1 cells after irradiation, we assayed protein carbonylation as an indicator for cellular repair of ROS-mediated macromolecule damage. To do so, two of each of P7NP and P7NPH1 primary sarcoma cell lines were irradiated with 6 Gy and protein lysates were collected at baseline and at 0.5, 24, and 72 hours after irradiation. Western Blotting for ROS-induced protein carbonylation and the quantifications of the bands in the lanes showed an immediate accumulation of protein carbonylation at 0.5 hours after irradiation in both P7NP and P7NPH1 cells, and this persisted at 24 hours after irradiation (Figure 5D). However, protein carbonylation was resolving for some of the cell lines by 72 hours after irradiation in both P7NP and P7NPH1 cells as indicated by a decrease in carbonyl group staining intensity in the first (P7NP tumor #1) and fourth (P7NPH1 tumor #2) panels at 72 hours. Together, these results indicate that P7NP and P7NPH1 sarcoma cells accumulate similar levels of ROS and also similar levels of ROS-induced protein damage after irradiation. Moreover, both P7NP and P7NPH1 cells resolve the ROS accumulation and ROS-induced damage after irradiation with similar kinetics. Thus, direct ROS-mediated effects do not appear to be the primary mechanism causing radiosensitivity in P7NPH1 cells.
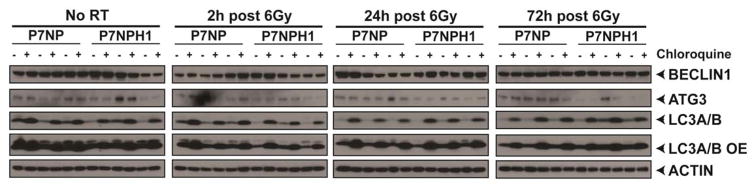
Clearance of damaged cellular macromolecules depends on a functional autophagy pathway, but autophagy also has other functions including catabolism of non-damaged cellular components that may promote survival under stress [28, 29]. Thus, we examined possible defects in autophagy flux as a mechanism for radiation sensitization in P7NPH1 cells. During autophagy, LC3-I is modified to LC3-II on autophagosomes. Therefore, we assayed LC3A/B-I and LC3A/B-II accumulation by Western Blot to measure autophagosome formation after irradiation. Whole cell lysates from P7NP and P7NPH1 cells were collected at baseline, 24, and 48 hours after 6 Gy. The LC3A/B-I and LC3A/B-II accumulation were normalized against actin. LC3A/B-II accumulated after irradiation in a time-dependent manner after irradiation in both P7NP and P7NPH1 cells (Figure 6). To determine if LC3A/B-II accumulation was due to an increase in autophagic flux or an inability to complete the autophagy pathway leading to the accumulation of autophagosomes, chloroquine was used to inhibit the progression of autophagic flux. Increased accumulation of LC3A/B-II after chloroquine treatment (Figure 6) indicated that both P7NP and P7NPH1 cells up-regulated autophagic flux in response to radiation and there was no discernable difference in functional autophagic flux between P7NP and P7NPH1 cells. We also examined the accumulation of BECLIN1 and ATG3, two other important components of the autophagy pathway. Similar to LC3A/B-II accumulation after irradiation, BECLIN1 and ATG3 demonstrated tumor cell line-specific changes that were not dependent of HIF-1α status. These results suggest that differences in autophagic flux do not contribute significantly to the radiation sensitivity phenotype of P7NPH1 cells.
Figure 6. Deletion of HIF-1α did not affect changes in autophagy after irradiation.
Primary cells from P7NP and P7NPH1 tumors were isolated as described in Figure 3. All experiments were performed with in vitro cultures under 21% oxygen. Cells were irradiated with 6Gy with and without adding 25μM chloroquine 2 hours prior to protein collection at the indicated times. Lysates were probed with an antibody against LC3A/B-I (top band) and LC3A/B-II (bottom band) to measure changes in autophagic flux, and BECLIN1 and ATG3 to measure other components of the autophagy pathway. Deletion of HIF-1α (P7NPH1) did not affect the changes in autophagic flux after irradiation.
Deletion of HIF-1α alters metabolic response following irradiation
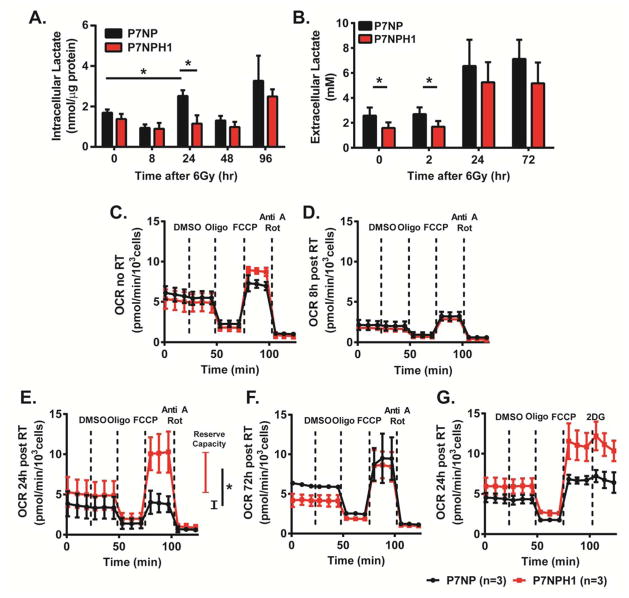
Because HIF-1α is known to regulate metabolic pathways and because changes in cellular metabolism may contribute to radiation sensitivity, we examined how deletion of HIF-1α affects the metabolic response to radiation. We first assessed how intracellular lactate changed in response to radiation in P7NP and P7NPH1 cells using mass spectrometry (Figure 7A). This showed that while P7NP cells had increased lactate accumulation 24 hours after 6 Gy irradiation, this accumulation was not evident in P7NPH1 cells. We also assessed the extracellular accumulation of lactate in the culture medium after irradiation using a commercial lactate kit (Figure 7B). This showed that while extracellular lactate concentration was higher for P7NP cells when compared to P7NPH1 cells at baseline and 2 hours after irradiation, both P7NP and P7NPH1 cells have increased extracellular lactate by 24 hours.
Figure 7. Loss of HIF-1α was correlated with metabolic changes in sarcoma cells in response to irradiation.
Primary cells from P7NP and P7NPH1 tumors were isolated as described in Figure 3. All experiments were performed with in vitro cultures under 21% oxygen. A) Cells were irradiated with 6Gy and lysates were collected at the indicated times after irradiation, and intracellular lactate was measured with mass spectrometry. The results showed P7NP cells had significantly increased intracellular lactate content at 24 hours after irradiation, both compared to nonirradiated P7NP cells and compared to P7NPH1 cells by a two-tailed student t-test. B) Cells were irradiated with 6Gy and lysates were collected at the indicated times after irradiation, and extracellular lactate was measured with the lactate colorimetric kit. The results showed P7NP cells had significantly increased extracellular lactate content at 0 and 2 hours after irradiation as compared to P7NPH1 cells by a two-tailed student t-test. (C–G) Oxygen consumption rates (OCR) were measured in cells that were irradiated at 6Gy at indicated times after irradiation. DMSO, oligomycin (Oligo), FCCP, antimycin A (Anti A), rotenone (Rot), and 2-deoxyglucose (2-DG) were added at indicated times. C) P7NP and P7NPH1 cells had similar OCR at baseline. D) P7NP and P7NPH1 cells both had reduced OCR at 8 hours following irradiation. E) P7NPH1 cells restored maximal electron transport chain (ETC) activity at 24 hours following irradiation and has more reserve capacity as compared to P7NP cells by two-tailed student t-test. F) P7NP cells restored maximal ETC activity at 72 hours following irradiation. G) Respiration in P7NP or P7NPH1 cells at 24 hours after irradiation was not inhibited by 2-DG. * p<0.05.
We then assessed oxygen consumption rates (OCR) of P7NP and P7NPH1 cells at baseline, 8, 24, and 72 hours after irradiation (Figure 7C–G). Drugs were added at different times during the OCR measurement to define the contribution from different components of oxidative phosphorylation (OXPHOS). DMSO was added first as a control because it was used as the vehicle for the other drugs. After generating baseline OCR measurements with and without the DMSO control, we added oligomycin (Oligo) to inhibit ATP synthase activity to measure the contribution of ATP synthesis to the OCR. After the OCR stabilized following the addition of oligomycin, carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) was added to cause dissociation of the proton gradient and uncouple the electron transport chain (ETC) from the proton gradient to measure the maximal capacity of the ETC. Finally, antimycin A (Anti A) and rotenone (Rot) were added to inhibit the ETC to quantify residual OCR from non-mitochondrial sources. These experiments showed that at baseline, OCR measurements were similar between P7NP and P7NPH1 sarcoma cells (Figure 7C). Eight hours after 6 Gy, OCR decreased to a similar extent in both P7NP and P7NPH1 cells indicating radiation-induced inhibition of mitochondrial OXPHOS (Figure 7D). At 24 hours after 6 Gy, P7NPH1 cells regained baseline ETC activities while those of P7NP cells were still defective as shown by differences in OCR after FCCP administration (Figure 7E). Furthermore, P7NPH1 cells have increased reserve capacity as compared to P7NP cells at 24 hours after irradiation. The ETC defect of P7NP cells was restored by 72 hours after irradiation (Figure 7F). The OCR results suggest that at 24 hours after irradiation, although the baseline OCR was similar between P7NP and P7NPH1 cells, P7NPH1 cells may have increased flux or accumulation of ETC substrates such as NADH or succinate that result in increased maximal oxygen consumption and reserve capacity after FCCP-mediated ETC and ATP synthase dissociation. Taken together, these results further support that P7NP and P7NPH1 cells elicit different metabolic changes 24 hours after irradiation. To determine the metabolic source of ETC activity, P7NP and P7NPH1 cells were irradiated and OCR was measured 24 hours later, but instead of using Anti A and Rot to inhibit the ETC at the final step, 2-deoxyglucose (2-DG) was added to inhibit the glycolytic pathway at the level of hexokinase (Figure 7G). Addition of 2-DG did not significantly inhibit OCR generated by the ETC following FCCP administration in either P7NPH1 or P7NP cells, suggesting that glucose was not the source for increased ETC substrate in P7NPH1 cells at 24 hours after irradiation.
Deletion of HIF-1α alters mitochondrial density following irradiation
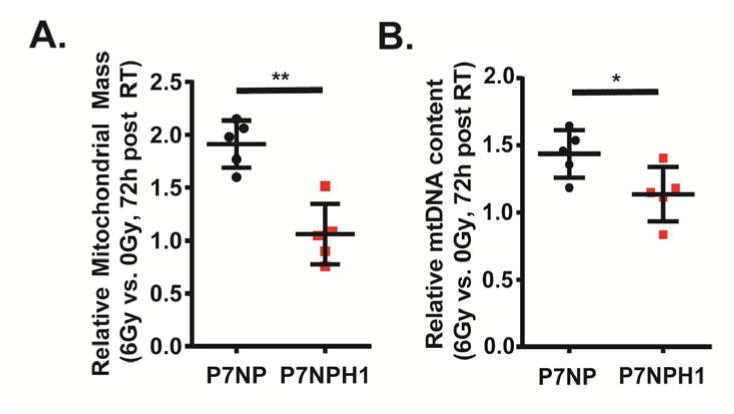
Because we observed metabolic changes in HIF-1α-deficient sarcoma cells following irradiation, we assessed mitochondrial density in P7NP and P7NPH1 cells by measuring mitochondrial mass before and after irradiation using flow cytometry with MitoTracker Deep Red and mtDNA qRT-PCR. Using flow cytometry, we found that at 72 hours after irradiation, P7NP cells had a significantly higher mitochondrial mass than P7NPH1 cells (Figure 8A). Similarly, P7NP cells with intact HIF-1α had increased mtDNA content as measured by qPCR compared to P7NPH1 cells 72 hours after irradiation (Figure 8B). These results show that HIF-1α-wild type and -deficient sarcoma cells regulate mitochondrial biogenesis differentially after irradiation and this may play a role in the observed radiation sensitivity in HIF-1α-deficient cells.
Figure 8. Loss of HIF-1α was correlated with decreased mitochondrial biogenesis in sarcoma cells in response to radiation.
Primary cells from P7NP and P7NPH1 tumors were isolated as described in Figure 3. All experiments were performed with in vitro cultures under 21% oxygen. A) The mitochondrial density in P7NP and P7NPH1 cells in response to 6Gy irradiation was measured first by incubating nonirradiated and irradiated cells with the MitoTracker DeepRed reagent after 72 hours, then by performing flow cytometry. Signal from irradiated cells were normalized to nonirradiated controls. The P7NPH1 cells had significantly decreased mitochondrial density at 72 hours following irradiation when compared to P7NP cells by a two-tailed student t-test. B) The mitochondrial DNA (mtDNA) content was measured via real-time PCR using DNA extracted from purified P7NP and P7NPH1 sarcoma cells at 72 hours after 6Gy irradiation, and normalized to 18S and nonirradiated controls. P7NP cells had more mtDNA content at 72 hours following irradiation as compared to P7NPH1 cells by a two-tailed student t-test. * p<0.05. ** p<0.01.
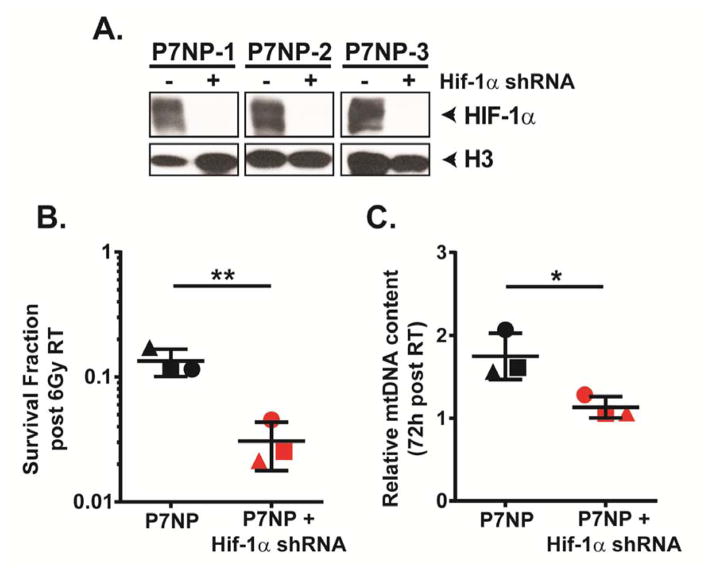
Knockdown of Hif-1α in P7NP sarcoma cells causes radio-sensitization and a mitochondrial biogenesis defect following irradiation
We next tested whether the radio-sensitization and mitochondrial biogenesis defect after irradiation in P7NPH1 sarcoma cells could be recapitulated in P7NP sarcoma cells stably expressing a shRNA to Hif-1α. First, we assessed the degree of Hif-1α knockdown in sarcoma cells isolated from 3 individual P7NP primary tumors after culturing in 0.5% oxygen for 24 hours. We showed that after Hif-1α knockdown, the level of residual HIF-1α nuclear protein was reduced to below the level of detection by Western Blot (Figure 9A). After confirming successful depletion of HIF-1α protein in P7NP sarcoma cells, we irradiated parental P7NP sarcoma cells and corresponding Hif-1α knockdown daughter cells with 6Gy and assessed clonogenic survival. Similar to the radiation response of P7NPH1 cells, P7NP cells with Hif-1α knockdown also demonstrated increased sensitization to radiation as compared to P7NP controls (Figure 9B). Additionally, P7NP cells with knockdown of Hif-1α also have significantly decreased mtDNA content as measured by qPCR than the parental P7NP controls at 72 hours after 6Gy (Figure 9C). These results further support a role for HIF-1α in mediating resistance of sarcoma cells to radiation in a cell-autonomous manner, which is associated with an increase in mitochondrial biogenesis after irradiation.
Figure 9. Knockdown of HIF-1α sensitizes P7NP cells to radiation and leads to a radiation-induced mitochondrial biogenesis defect.
Primary cells from P7NP tumors were isolated as described in Figure 3. All experiments were performed with in vitro cultures under 21% oxygen. A) Efficiency of Hif-1α knockdown by shRNA was measured by measuring HIF-1α protein level in nuclear extracts of parental P7NP sarcoma cells and P7NP cells stably expressing Hif-1α shRNA cultured under 0.5% oxygen for 24 hours. This showed that the efficiency of Hif-1α knockdown in P7NP sarcoma cells obtained from 3 different tumors was beyond the level of detection by Western Blot. B) Knockdown of Hif-1α by shRNA resulted in significantly increased radiation sensitivity compared to the parental controls after exposure to 6Gy under 21% oxygen as measured by clonogenic survival, by a two-tailed student t-test. Each symbol shape represents a parental control P7NP cells and its corresponding knockdown daughter sarcoma cells. C) The mitochondrial DNA (mtDNA) content was measured via real-time PCR using DNA extracted from parental P7NP and P7NP cells with Hif-1α knockdown at 72 hours after 6Gy irradiation, and normalized to 18S. P7NP cells had more mtDNA content at 72 hours following irradiation as compared to P7NP cells with Hif-1α shRNA, by a two-tailed student t-test. Each symbol shape represents a parental control P7NP and its corresponding knockdown daughter sarcoma cells. * p<0.05. ** p<0.01.
DISCUSSION
Intratumoral hypoxia causes radiation resistance in a number of cancers including soft tissue sarcomas [30–33]. Molecular oxygen promotes radiation sensitivity by reacting with DNA radicals to create adducts that are difficult for the cell to repair, which leads to increased cell death. Thus, under hypoxic conditions, ionized DNA is more easily repaired and cells are more likely to survive. In addition, hypoxia activates signaling pathways, such as HIFs, which may also contribute to radiation resistance. In this scenario, these signaling pathways could be targeted to sensitize hypoxic tumors. In addition, HIF-1α is activated by radiation [34] and chemotherapy [35] even under normoxic conditions. Indeed, we observed increased expression of HIF-1α in human soft tissue sarcomas after interdigitated chemotherapy and radiation therapy. Therefore, such survival pathways may also regulate tumor response in normoxic tumors. Previous studies using transplanted model systems have implicated HIF-1α in radiation resistance via non-cell autonomous mechanisms [11, 12, 36]. For example, activation of HIF-1α in tumor cells increase secretion of cytokines such as VEGF, which promotes survival of tumor endothelial cells [11]. Moreover, following radiation therapy HIF-1α can also increase the expression of SDF1, which recruits bone marrow derived macrophages to tumors to promote vasculogenesis and tumor regrowth [36]. However, the role of HIF-1α in mediating the response of primary cancers to radiation therapy had not previously been investigated.
In this study, we generated a novel primary mouse model of soft tissue sarcoma expressing oncogenic NRAS with p53 deletion that resembles human RMS and UPS. Other studies including those from our group [15, 37] have shown that by introducing driver mutations in PAX7-expressing adult skeletal muscle stem cells, primary soft tissue sarcomas develop in a continuum of RMS and UPS, suggesting that these entities may be more similar than previously anticipated.
We then deleted HIF-1α in the tumor cells of these sarcomas at the time of sarcoma initiation. Complete deletion of HIF-1α was confirmed by PCR for recombination of the genomic Hif-1αFL DNA, qRT-PCR for the exon 2-containing Hif-1α transcript, and by Western Blot for HIF-1α protein in stroma-depleted tumor cells. Deletion of HIF-1α in sarcomas in P7NP mice, did not affect the timing of sarcomagenesis or sarcoma growth kinetics. However, sarcomas lacking HIF-1α in the tumor cells were more sensitive to 50 Gy of fractionated radiation therapy (10Gy x 5) in vivo. Although five fractions of radiation therapy may not recapitulate the most frequently used radiation therapy schedule for soft tissue sarcomas, it is still clinically relevant as emerging evidence suggests that hypofractionation with five fractions may have similar rates of local control and toxicity profiles [38]. Moreover, sarcoma cells lacking HIF-1α by either genomic ablation or shRNA knockdown were also sensitized to radiation in vitro by clonogenic assays. Taken together, these results indicate that in addition to non-cell autonomous mechanisms, HIF-1α can promote radiation resistance in primary cancers through cell autonomous mechanisms.
A number of cell autonomous pathways can regulate the response of tumor cells to radiation including ROS scavenging, autophagy, and DNA damage repair [39–41]. However, when we examined these pathways in sarcoma cells with an intact or deleted HIF-1α, no significant differences in these pathways were observed after irradiation in 21% oxygen in vitro. These results suggest that under these conditions, HIF-1α does not promote radiation resistance through these cell autonomous pathways. However, these results do not exclude the possibility that under hypoxic conditions or in vivo that these pathways could contribute to HIF-1α mediated resistance to radiation therapy.
In contrast, when we measured mitochondrial density and changes in oxidative phosphorylation after irradiation, we found that consistent with previous studies [42, 43] following 6 Gy sarcoma cells with an intact HIF-1α increased their mitochondrial density and decreased their mitochondrial OXPHOS. However, sarcoma cells without HIF-1α did not increase their mitochondrial density and recovered mitochondrial respiration much more quickly after irradiation. Furthermore, this result was recapitulated by irradiating sarcoma cells with successful Hif-1α knockdown by shRNA. These results suggest that HIF-1α regulates mitochondrial respiration and density after irradiation, which is one potential cell autonomous mechanism that may contribute to radiation resistance. Future studies will seek to identify HIF-1α transcriptional targets that regulate mitochondrial respiration and density after irradiation. If these pathways do contribute to radiation resistance, then they may provide therapeutic targets to sensitize tumors to radiation therapy.
In summary, we have established a novel mouse model of soft tissue sarcoma in which deletion of HIF-1α increases the sensitivity of tumors to fractionated radiation therapy in vivo. This is the first study to show that deletion of HIF-1α increased the sensitivity of cancer to radiation therapy in an autochthonous tumor model. Moreover, we find that sarcoma cells lacking HIF-1α are more sensitive to radiation in vitro with a DMF of 1.31. Therefore, HIF-1α promotes radiation resistance via cell autonomous mechanisms, which may include altering mitochondrial metabolism. Identifying HIF-1α targets that regulate the response of mitochondria to radiation may identify novel pathways that regulate radiation resistance that could be targeted to improve radiation sensitivity.
METHODS
Generation of the Mouse Model of Soft Tissue Sarcoma
All animal experiments were performed according to protocols approved by the Duke University Institutional Animal Care and Use Committee. Mouse models of soft tissue sarcoma were generated in a mixed 129/SVJae and C57BL/6 background using a combination of previously described alleles: Pax7Cre-ER-T2 (provided by Chen-Ming Fan, Carnegie Institution for Science) [20], p53FL (provided by Anton Berns, The Netherlands Cancer Institute) [44], Hif1aFL (provided by Randall Johnson, University of Cambridge) [45], and NrasLSL-G12D [46] and ROSA26mTmG [47] which were obtained from The Jackson Laboratory. (Z)-4-hydroxytamoxifen (Sigma Aldrich), which is referred to as 4-OHT, was first dissolved in 100% ethanol at a concentration of 250mg/ml, and then diluted with corn oil (Sigma Aldrich) to a final working solution concentration of 25mg/ml. Primary mouse soft tissue sarcomas were generated in the mouse hind limb by intramuscular (IM) injection of 20μl 4-OHT working solution via a 27.5 gauge insulin syringe (Terumo).
Image-Guided Fractionated Radiation Therapy
Hind limb irradiation was performed on mice with a tumor-bearing hind limb to model radiation therapy when the tumor reached 200mm3 as measured by calipers using the formula V = (pi*x*y*z)/6, where V defines volume, and x, y, and z define length, width, and height of the tumor. At the time of radiation delivery, the animal was placed on the irradiation stage in the X-RAD 225Cx micro-CT/micro-irradiator (Precision X-ray). Continuous 2% isoflurane anesthesia was delivered by nosecone at a rate of 2L of oxygen per minute. A 40mm × 40mm square collimator was used to deliver radiation therapy to the hind limb of the mouse with fluoroscopy-guided radiation therapy using 225kVp and 13mA using two opposed fields, as previously described [48]. Subsequent fractions were delivered with 24 hour intervals between fractions. Mice were followed for tumor recurrence as defined by tumor growth past tripling the initial volume on the day of the first treatment.
Injection of EF5 and Hoechst 33342
Tumor-bearing mice were anesthetized by intraperitoneal (IP) injection of pentobarbital at 85mg/kg. The hypoxia marker EF5 (Cameron Koch, University of Pennsylvania) was delivered by IP injection at 26.5ml/kg from a 10mM stock solution in saline. The injection of 100μl of 20mg/ml Hoechst 33342 (Sigma Aldrich) diluted in saline occurred via tail vein injection in the anesthetized animals, 1 min before surgical removal of the tumors. The tumor was surgically removed from the deeply anesthetized animal and was snap frozen over liquid nitrogen.
Tumor Cell Isolation and Culture
Tumor tissue from untreated sarcomas was removed from mice and digested with 5mg/ml collagenase IV (Worthington), 2.5U/ml dispase (BD), and 0.05% trypsin (Life Technologies) at 37°C for 40 minutes. The resulting mixture was strained through a 70μm filter (BD), and filtered cells were re-suspended in culture medium consisting of MEM (Life Technologies) with 10% fetal bovine serum (Sigma), 5.6mM glucose, and 0.6mM glutamax (Life Technologies). Cells were serially passaged to deplete stromal (non-tumor) cells. Tumor cell purity was verified with 50 cycles of polymerase chain reaction (PCR) of bulk cell DNA for the presence of wild type and conditionally deleted alleles. Cell counting was performed using the Cellometer auto 2000 (Nexcelom) counter. For hypoxic tissue culture, cells were placed in the INVIVO2 500 (Ruskinn) hypoxia chamber with 0.5% O2 and 5% CO2 for 24 hours. For in vitro irradiation, cells were irradiated in culture plates either with an X-RAD 320 (Precision X-ray) irradiator at 320kVp and 10mA, except for the DNA end-joining repair assay which utilized an X-RAD 160 (Precision X-ray) at 160kVp and 18mA, for the indicated doses. Irradiated cells were returned to the culture chamber for the indicated times. For shRNA knockdown, lentiviral particles were generated using Hif-1α shRNA (Thermo Scientific, TRCN000054450) and Mission Lentiviral Packaging Mix (Sigma Aldrich, SHP001) in 293T cells per manufacturer’s specifications. P7NP sarcoma cells were transduced with polybrene (4μg/ml) and supernatant medium containing lentiviral particles, and clones were positively selected in puromycin (3μg/ml) for 10 days prior to assessment of HIF-1α knockdown efficiency by hypoxic culture and Western Blot.
PCR, qRT-PCR
DNA was isolated using the DNeasy Blood and Tissue Kit (Qiagen) and PCR experiments were performed with a C1000 Touch Thermal Cycler (Bio-Rad). RNA was isolated using the RNeasy Mini Kit (Qiagen), cDNA was synthesized using the iScript cDNA Synthesis Kit (Bio-Rad), and qPCR experiments were performed with an iCycler (Bio-Rad). Nucleic acid concentrations were measured using NanoDrop 1000. Primers that were used are as follows: 5′-TTGGGGATGAAAACATCTGC -3′ (Hif1a Genomic DNA FWD1), 5′-GGAGCTATCTCTCTAGACC-3′ (Hif1a Genomic DNA FWD2), 5′-GCAGTTAAGAGCACTAGTTG-3′ (Hif1a Genomic DNA REV), 5′-TCTCGGCGAAGCAAAGAGTCTGAA-3′ (Hif1a cDNA FWD), 5′-TAGACCACCGGCATCCAGAAGTTT-3′ (Hif1a cDNA REV), 5′-CCTATCACCCTTGCCATCAT-3′ (mtDNA FWD), 5′-GAGGCTGTTGCTTGTGTGAC-3′ (mtDNA REV), 5′-GCCCGAAGCGTTTACTTTGA-3′ (18S FWD), 5′-TCCATTATTCCTAGCTGCGGTATC-3′ (18S REV).
Western Blot, Immunofluorescence, Immunohistochemistry and Fluorescence-Activated Cell Sorting
For Western Blots, whole cell lysates were generated by direct application of 2x Laemmli buffer (Bio-Rad) with β-mercaptoethanol (Sigma) onto the cell monolayers. Nuclear extracts were generated using the NE-PER kit (Pierce) per the manufacturer’s instructions. Protein lysates from cells treated by hypoxia were collected directly in the hypoxia chamber with 800μM deferoxamine (Sigma Aldrich) added to all lysis buffers. Protein concentrations were measured using either the BCA protein assay kit (Pierce) or the 660nm protein assay kit (Pierce). Protein lysates were electrophoresed on mini-PROTEAN pre-cast gels (Bio-Rad) in the mini-PROTEAN tetra cell system (Bio-Rad), and transferred onto PVDF membranes. For protein carbonylation blots, experiments were performed using the OxyBlot protein oxidation detection kit (Millipore) per the manufacturer’s instructions. For frozen sectioning, 10μm sections were generated via a cryostat (Leica) and stored unfixed at −80°C. For paraffin sectioning, tumor specimens were fixed in 10% formalin and 70% EtOH. After paraffin embedding, 4μm sections were generated using a manual rotary microtome (Leica) Immunohistochemistry on paraffin embedded tissues was performed using ready-to-use target retrieval solution (DAKO), antibody diluent (DAKO), the Vectastain Elite ABC Kit (Vector Labs) and 3,3′-Diaminobenzidine tetrahydrochloride (Sigma Aldrich). For frozen sections, unfixed slides were first fixed in 4% paraformaldehyde, and immunohistochemistry proceeded as described above except without the usage of target retrieval solution. For immunofluorescence experiments, frozen sections were obtained as described above and stained as previously described [48]. Images were captured either with a Leica DM5500B microscope using Leica Application Suite software, an Axioskop 2 plus microscope (Zeiss) with computerized scanning stage (Marzhauser), or a custom Axioskop microscope (Zeiss) in black box with a 16 bit CCD camera with photon counting capabilities (Andor). Image analyses were performed using ImageJ software. For fluorescence-activated cell sorting, cells were re-suspended in PBS with 2% fetal bovine serum in a single cell suspension in 5ml polystyrene round-bottom tubes (BD). Cells were sorted or analyzed with a FACSVantage cell sorter (BD). Final analyses on the raw data were performed using FlowJo software. Antibodies and other reagents used are listed in Supplementary Table 1.
Tissue Microarrays
Use of human sarcoma tissue microarray was approved by the Institutional Review Board at Duke University Medical Center. Tissue microarrays were generated by the Radiation Therapy Oncology Group (RTOG) using pre- and post- treatment biopsies of soft tissue sarcomas from Massachusetts General Hospital. In total, there were 23 evaluable cases with 8 samples of paired pre- and post-treatment biopies, 1 sample with pre-treatment biopsy only, and 14 samples of post-treatment only. TMAs were stained for HIF-1α as above and scored semi-quantitatively on a scale of 0–3+ by a sarcoma pathologist (DMC) blinded to sample identification and treatment. A median score was used for each tumor as each sample was present in triplicate on the TMA. Scores were correlated with pre- and post-treatment status using the Fisher’s exact test.
Targeted Metabolite Analysis
Adherent cells were scraped and pelleted in ice-cold PBS. Cell pellets were re-suspended in ice-cold deionized water containing 0.6% formic acid. Acetonitrile was added to the re-suspended cell solution in a 1:1 ratio. Samples were stored in −80°C and were analyzed as previously described [49].
For the extracellular lactate assay, culture medium at indicated time points were filtered through 10kD polyethersulfone spin columns (BioVision), snap frozen in liquid nitrogen, and stored in in −80°C. Analysis was performed using a commercially available lactate colorimetric assay kit (BioVision) per manufacturer’s instructions.
Seahorse Analysis
Adherent cells were grown and treated in Seahorse 24-well plates (Seahorse Bioscience). 24 hours prior to experimentation, 1ml calibrant solution was placed in each well of the sensor cartridge plate, and the sensor cartridge immersed in calibrant solution was incubated at 37°C overnight. Prior to experimentation, cell media was changed to 600μl XF assay media (Seahorse Bioscience) and incubated in 37°C for at least 60 minutes. 30μl of 20X pre-warmed compounds were loaded into each of the 4 ports per well in the sensor cartridge, resulting in final concentrations of drugs as indicated as follows: 2-deoxyglucose (2-DG) 20mM, oligomycin (Oligo) 1μM, carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) 0.5μM, antimicin A (Anti A) 1.5μM, and rotenone (Rot) 0.75μM. The sensor cartridge and cells were placed in the Seahorse XF24 analyzer for oxygen consumption and extracellular acidification readings.
DNA End-Joining Repair Assay
The comet assay was performed using the Trevigen comet assay kit. Prior to irradiation, cells were cultured for 24 hours in complete medium substituted with 0.1% fetal bovine serum to stimulate G1 arrest. At the indicated time after irradiation with 10Gy, adherent cells were scraped and pelleted in ice-cold PBS. Cell pellets were re-suspended in ice-cold PBS at a concentration of 1×105cells/ml. 25μl of cell suspension was added to 250μl low-melting point agarose, and 50μl of cells in agarose was added and solidified on each well of the slide. Cells in solidified agarose on comet assay slides were lysed at 4°C in 1x lysis buffer for 1 hour, followed by neutral buffer for 30 minutes. Slides were electrophoresed for 45min at 21V, and DNA was precipitated in DNA precipitation buffer for 30 minutes at room temperature following electrophoresis. Slides were then fixed in 70% ethanol for 30 minutes and dried overnight prior to DNA staining with SYBR Gold. Images were taken by epifluorescence microscopy and DNA tails were measured using the Trevigen software package.
Supplementary Material
Acknowledgments
We thank Loretta Woodlief, Laura Jeffords, Michael Reinsvold, and Hanna Norris for assistance with mouse care and irradiation. We thank other members of the Kirsch lab for advice and support. For reagents and advice regarding hypoxia and HIF-1α, we thank Thies Schroeder, Ken Young, Karin Eisinger-Mathason and M. Celeste Simon. This work was supported by NIH grants F30 CA180680 (M.Z.), R01 CA169220 (D.G.K.), and partially by the Cancer Center Support Grant P30 CA014236.
Footnotes
CONFLICT OF INTEREST
The authors declare that there are no conflicts of interest.
References
- 1.Mohyeldin A, Garzon-Muvdi T, Quinones-Hinojosa A. Oxygen in stem cell biology: a critical component of the stem cell niche. Cell Stem Cell. 2010;7(2):150–61. doi: 10.1016/j.stem.2010.07.007. Epub 2010/08/05 S1934-5909(10)00341-3 [pii] [DOI] [PubMed] [Google Scholar]
- 2.Thomlinson RH, Gray LH. The histological structure of some human lung cancers and the possible implications for radiotherapy. Br J Cancer. 1955;9(4):539–49. doi: 10.1038/bjc.1955.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Den Brenk HA. Effect of high pressure oxygen on radiosensitivity of Ehrlich’s tumour in mice after “immunological approximation”. Br J Cancer. 1961;15:61–84. doi: 10.1038/bjc.1961.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vandenbrenk HA, Elliott K, Hutchings H. Further Observations on Radiocurability of a Solid Ehrlich and Tissue Reactions in the Mouse with Fractionated Radiation Doses and the Effects of Oxygen. British journal of cancer. 1963;17:281–6. doi: 10.1038/bjc.1963.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gray LH, Conger AD, Ebert M, Hornsey S, Scott OC. The concentration of oxygen dissolved in tissues at the time of irradiation as a factor in radiotherapy. Br J Radiol. 1953;26(312):638–48. doi: 10.1259/0007-1285-26-312-638. [DOI] [PubMed] [Google Scholar]
- 6.Overgaard J. Hypoxic radiosensitization: adored and ignored. J Clin Oncol. 2007;25(26):4066–74. doi: 10.1200/JCO.2007.12.7878. [DOI] [PubMed] [Google Scholar]
- 7.Rischin D, Hicks RJ, Fisher R, Binns D, Corry J, Porceddu S, Peters LJ Trans-Tasman Radiation Oncology Group S. Prognostic significance of [18F]-misonidazole positron emission tomography-detected tumor hypoxia in patients with advanced head and neck cancer randomly assigned to chemoradiation with or without tirapazamine: a substudy of Trans-Tasman Radiation Oncology Group Study 98.02. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2006;24(13):2098–104. doi: 10.1200/JCO.2005.05.2878. [DOI] [PubMed] [Google Scholar]
- 8.Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell. 2010;40(2):294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Semenza GL, Roth PH, Fang HM, Wang GL. Transcriptional regulation of genes encoding glycolytic enzymes by hypoxia-inducible factor 1. J Biol Chem. 1994;269(38):23757–63. [PubMed] [Google Scholar]
- 10.Rey S, Lee K, Wang CJ, Gupta K, Chen S, McMillan A, Bhise N, Levchenko A, Semenza GL. Synergistic effect of HIF-1alpha gene therapy and HIF-1-activated bone marrow-derived angiogenic cells in a mouse model of limb ischemia. Proc Natl Acad Sci U S A. 2009;106(48):20399–404. doi: 10.1073/pnas.0911921106. Epub 2009/12/02 0911921106 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moeller BJ, Cao Y, Li CY, Dewhirst MW. Radiation activates HIF-1 to regulate vascular radiosensitivity in tumors: role of reoxygenation, free radicals, and stress granules. Cancer Cell. 2004;5(5):429–41. doi: 10.1016/s1535-6108(04)00115-1. Epub 2004/05/18 S1535610804001151 [pii] [DOI] [PubMed] [Google Scholar]
- 12.Moeller BJ, Dreher MR, Rabbani ZN, Schroeder T, Cao Y, Li CY, Dewhirst MW. Pleiotropic effects of HIF-1 blockade on tumor radiosensitivity. Cancer Cell. 2005;8(2):99–110. doi: 10.1016/j.ccr.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 13.Liu J, Zhang J, Wang X, Li Y, Chen Y, Li K, Zhang J, Yao L, Guo G. HIF-1 and NDRG2 contribute to hypoxia-induced radioresistance of cervical cancer Hela cells. Exp Cell Res. 2010;316(12):1985–93. doi: 10.1016/j.yexcr.2010.02.028. [DOI] [PubMed] [Google Scholar]
- 14.Mullen JT, Kobayashi W, Wang JJ, Harmon DC, Choy E, Hornicek FJ, Rosenberg AE, Chen YL, Spiro IJ, DeLaney TF. Long-term follow-up of patients treated with neoadjuvant chemotherapy and radiotherapy for large, extremity soft tissue sarcomas. Cancer. 2012;118(15):3758–65. doi: 10.1002/cncr.26696. [DOI] [PubMed] [Google Scholar]
- 15.Blum JM, Ano L, Li Z, Van Mater D, Bennett BD, Sachdeva M, Lagutina I, Zhang M, Mito JK, Dodd LG, Cardona DM, Dodd RD, Williams N, Ma Y, Lepper C, Linardic CM, Mukherjee S, Grosveld GC, Fan C-M, Kirsch DG. Distinct and Overlapping Sarcoma Subtypes Initiated from Muscle Stem and Progenitor Cells. Cell Reports. 2013 doi: 10.1016/j.celrep.2013.10.020. http://dx.doi.org/10.1016/j.celrep.2013.10.020. [DOI] [PMC free article] [PubMed]
- 16.Van Mater D, Ano L, Blum JM, Webster MT, Huang W, Williams N, Ma Y, Cardona DM, Fan CM, Kirsch DG. Acute tissue injury activates satellite cells and promotes sarcoma formation via the HGF/c-MET signaling pathway. Cancer Res. 2014 doi: 10.1158/0008-5472.CAN-14-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinelli S, McDowell HP, Vigne SD, Kokai G, Uccini S, Tartaglia M, Dominici C. RAS signaling dysregulation in human embryonal Rhabdomyosarcoma. Genes Chromosomes Cancer. 2009;48(11):975–82. doi: 10.1002/gcc.20702. Epub 2009/08/15. [DOI] [PubMed] [Google Scholar]
- 18.Chen X, Stewart E, Shelat AA, Qu C, Bahrami A, Hatley M, Wu G, Bradley C, McEvoy J, Pappo A, Spunt S, Valentine MB, Valentine V, Krafcik F, Lang WH, Wierdl M, Tsurkan L, Tolleman V, Federico SM, Morton C, Lu C, Ding L, Easton J, Rusch M, Nagahawatte P, Wang J, Parker M, Wei L, Hedlund E, Finkelstein D, Edmonson M, Shurtleff S, Boggs K, Mulder H, Yergeau D, Skapek S, Hawkins DS, Ramirez N, Potter PM, Sandoval JA, Davidoff AM, Mardis ER, Wilson RK, Zhang J, Downing JR, Dyer MA. St Jude Children’s Research Hospital-Washington University Pediatric Cancer Genome P. Targeting oxidative stress in embryonal rhabdomyosarcoma. Cancer Cell. 2013;24(6):710–24. doi: 10.1016/j.ccr.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shern JF, Chen L, Chmielecki J, Wei JS, Patidar R, Rosenberg M, Ambrogio L, Auclair D, Wang J, Song YK, Tolman C, Hurd L, Liao H, Zhang S, Bogen D, Brohl AS, Sindiri S, Catchpoole D, Badgett T, Getz G, Mora J, Anderson JR, Skapek SX, Barr FG, Meyerson M, Hawkins DS, Khan J. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer discovery. 2014;4(2):216–31. doi: 10.1158/2159-8290.CD-13-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lepper C, Conway SJ, Fan CM. Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature. 2009;460(7255):627–31. doi: 10.1038/nature08209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guillou L, Aurias A. Soft tissue sarcomas with complex genomic profiles. Virchows Arch. 2010;456(2):201–17. doi: 10.1007/s00428-009-0853-4. Epub 2010/03/11. [DOI] [PubMed] [Google Scholar]
- 22.Eisinger-Mathason TS, Zhang M, Qiu Q, Skuli N, Nakazawa MS, Karakasheva T, Mucaj V, Shay JE, Stangenberg L, Sadri N, Pure E, Yoon SS, Kirsch DG, Simon MC. Hypoxia-dependent modification of collagen networks promotes sarcoma metastasis. Cancer Discov. 2013;3(10):1190–205. doi: 10.1158/2159-8290.CD-13-0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moeller BJ, Dewhirst MW. HIF-1 and tumour radiosensitivity. Br J Cancer. 2006;95(1):1–5. doi: 10.1038/sj.bjc.6603201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koch CJ, Howell RL. Combined radiation-protective and radiation-sensitizing agents: II. Radiosensitivity of hypoxic or aerobic Chinese hamster fibroblasts in the presence of cysteamine and misonidazole: implications for the “oxygen effect” (with appendix on calculation of dose-modifying factors) Radiation research. 1981;87(2):265–83. [PubMed] [Google Scholar]
- 25.Olive PL, Banath JP. The comet assay: a method to measure DNA damage in individual cells. Nature protocols. 2006;1(1):23–9. doi: 10.1038/nprot.2006.5. [DOI] [PubMed] [Google Scholar]
- 26.Oliver CN. Inactivation of enzymes and oxidative modification of proteins by stimulated neutrophils. Archives of biochemistry and biophysics. 1987;253(1):62–72. doi: 10.1016/0003-9861(87)90637-0. [DOI] [PubMed] [Google Scholar]
- 27.Slater TF. Mechanisms of protection against the damage produced in biological systems by oxygen-derived radicals. Ciba Foundation symposium. 1978;(65):143–76. doi: 10.1002/9780470715413.ch9. [DOI] [PubMed] [Google Scholar]
- 28.Yang A, Rajeshkumar NV, Wang X, Yabuuchi S, Alexander BM, Chu GC, Von Hoff DD, Maitra A, Kimmelman AC. Autophagy Is Critical for Pancreatic Tumor Growth and Progression in Tumors with p53 Alterations. Cancer discovery. 2014;4(8):905–13. doi: 10.1158/2159-8290.CD-14-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mathew R, Khor S, Hackett SR, Rabinowitz JD, Perlman DH, White E. Functional Role of Autophagy-Mediated Proteome Remodeling in Cell Survival Signaling and Innate Immunity. Molecular cell. 2014 doi: 10.1016/j.molcel.2014.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brizel DM, Sibley GS, Prosnitz LR, Scher RL, Dewhirst MW. Tumor hypoxia adversely affects the prognosis of carcinoma of the head and neck. Int J Radiat Oncol Biol Phys. 1997;38(2):285–9. doi: 10.1016/s0360-3016(97)00101-6. [DOI] [PubMed] [Google Scholar]
- 31.Hockel M, Knoop C, Schlenger K, Vorndran B, Baussmann E, Mitze M, Knapstein PG, Vaupel P. Intratumoral pO2 predicts survival in advanced cancer of the uterine cervix. Radiotherapy and oncology : journal of the European Society for Therapeutic Radiology and Oncology. 1993;26(1):45–50. doi: 10.1016/0167-8140(93)90025-4. [DOI] [PubMed] [Google Scholar]
- 32.Gatenby RA, Kessler HB, Rosenblum JS, Coia LR, Moldofsky PJ, Hartz WH, Broder GJ. Oxygen distribution in squamous cell carcinoma metastases and its relationship to outcome of radiation therapy. Int J Radiat Oncol Biol Phys. 1988;14(5):831–8. doi: 10.1016/0360-3016(88)90002-8. [DOI] [PubMed] [Google Scholar]
- 33.Brizel DM, Scully SP, Harrelson JM, Layfield LJ, Bean JM, Prosnitz LR, Dewhirst MW. Tumor oxygenation predicts for the likelihood of distant metastases in human soft tissue sarcoma. Cancer research. 1996;56(5):941–3. [PubMed] [Google Scholar]
- 34.Kim YH, Yoo KC, Cui YH, Uddin N, Lim EJ, Kim MJ, Nam SY, Kim IG, Suh Y, Lee SJ. Radiation promotes malignant progression of glioma cells through HIF-1alpha stabilization. Cancer letters. 2014 doi: 10.1016/j.canlet.2014.07.048. [DOI] [PubMed] [Google Scholar]
- 35.Cao Y, Eble JM, Moon E, Yuan H, Weitzel DH, Landon CD, Nien CY, Hanna G, Rich JN, Provenzale JM, Dewhirst MW. Tumor cells upregulate normoxic HIF-1alpha in response to doxorubicin. Cancer Res. 2013;73(20):6230–42. doi: 10.1158/0008-5472.CAN-12-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kioi M, Vogel H, Schultz G, Hoffman RM, Harsh GR, Brown JM. Inhibition of vasculogenesis, but not angiogenesis, prevents the recurrence of glioblastoma after irradiation in mice. J Clin Invest. 2010;120(3):694–705. doi: 10.1172/JCI40283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rubin BP, Nishijo K, Chen HI, Yi X, Schuetze DP, Pal R, Prajapati SI, Abraham J, Arenkiel BR, Chen QR, Davis S, McCleish AT, Capecchi MR, Michalek JE, Zarzabal LA, Khan J, Yu Z, Parham DM, Barr FG, Meltzer PS, Chen Y, Keller C. Evidence for an unanticipated relationship between undifferentiated pleomorphic sarcoma and embryonal rhabdomyosarcoma. Cancer Cell. 2011;19(2):177–91. doi: 10.1016/j.ccr.2010.12.023. Epub 2011/02/15 S1535-6108(10)00532-5 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kosela-Paterczyk H, Szacht M, Morysinski T, Lugowska I, Dziewirski W, Falkowski S, Zdzienicki M, Pienkowski A, Szamotulska K, Switaj T, Rutkowski P. Preoperative hypofractionated radiotherapy in the treatment of localized soft tissue sarcomas. European journal of surgical oncology : the journal of the European Society of Surgical Oncology and the British Association of Surgical Oncology. 2014;40(12):1641–7. doi: 10.1016/j.ejso.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 39.Moding EJ, Kastan MB, Kirsch DG. Strategies for optimizing the response of cancer and normal tissues to radiation. Nature reviews Drug discovery. 2013;12(7):526–42. doi: 10.1038/nrd4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Apel A, Herr I, Schwarz H, Rodemann HP, Mayer A. Blocked autophagy sensitizes resistant carcinoma cells to radiation therapy. Cancer Res. 2008;68(5):1485–94. doi: 10.1158/0008-5472.CAN-07-0562. [DOI] [PubMed] [Google Scholar]
- 41.Paglin S, Hollister T, Delohery T, Hackett N, McMahill M, Sphicas E, Domingo D, Yahalom J. A novel response of cancer cells to radiation involves autophagy and formation of acidic vesicles. Cancer Res. 2001;61(2):439–44. [PubMed] [Google Scholar]
- 42.Nugent SM, Mothersill CE, Seymour C, McClean B, Lyng FM, Murphy JE. Increased mitochondrial mass in cells with functionally compromised mitochondria after exposure to both direct gamma radiation and bystander factors. Radiation research. 2007;168(1):134–42. doi: 10.1667/RR0769.1. [DOI] [PubMed] [Google Scholar]
- 43.Zhou X, Li N, Wang Y, Wang Y, Zhang X, Zhang H. Effects of X-irradiation on mitochondrial DNA damage and its supercoiling formation change. Mitochondrion. 2011;11(6):886–92. doi: 10.1016/j.mito.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 44.Marino S, Vooijs M, van Der Gulden H, Jonkers J, Berns A. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes & development. 2000;14(8):994–1004. [PMC free article] [PubMed] [Google Scholar]
- 45.Ryan HE, Poloni M, McNulty W, Elson D, Gassmann M, Arbeit JM, Johnson RS. Hypoxia-inducible factor-1alpha is a positive factor in solid tumor growth. Cancer research. 2000;60(15):4010–5. [PubMed] [Google Scholar]
- 46.Haigis KM, Kendall KR, Wang Y, Cheung A, Haigis MC, Glickman JN, Niwa-Kawakita M, Sweet-Cordero A, Sebolt-Leopold J, Shannon KM, Settleman J, Giovannini M, Jacks T. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat Genet. 2008;40(5):600–8. doi: 10.1038/ng.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45(9):593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 48.Moding EJ, Lee CL, Castle KD, Oh P, Mao L, Zha S, Min HD, Ma Y, Das S, Kirsch DG. Atm deletion with dual recombinase technology preferentially radiosensitizes tumor endothelium. J Clin Invest. 2014;124(8):3325–38. doi: 10.1172/JCI73932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jensen MV, Joseph JW, Ilkayeva O, Burgess S, Lu D, Ronnebaum SM, Odegaard M, Becker TC, Sherry AD, Newgard CB. Compensatory responses to pyruvate carboxylase suppression in islet beta-cells. Preservation of glucose-stimulated insulin secretion. J Biol Chem. 2006;281(31):22342–51. doi: 10.1074/jbc.M604350200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.