Abstract
Gram-negative bacteria, such as Escherichia coli, frequently utilize tripartite efflux complexes in the resistance-nodulation-division (RND) family to expel diverse toxic compounds from the cell. These complexes span both the inner and outer membranes of the bacterium via an α-helical, inner membrane transporter; a periplasmic membrane fusion protein; and a β-barrel, outer membrane channel. One such efflux system CusCBA is responsible for extruding biocidal Cu(I) and Ag(I) ions. To remove these toxic ions, the CusC outer membrane channel must form a β-barrel structural domain, which creates a pore and spans the entire outer membrane. We here report the crystal structures of wild-type CusC as well as two CusC mutants, suggesting that the first N-terminal cysteine residue plays an important role in protein-membrane interactions and is critical for the insertion of this channel protein into the outer membrane. These structures provide insight into the mechanisms on CusC folding and transmembrane channel formation. It is also found that the interactions between CusC and membrane may be crucial for controlling the opening and closing of this β-barrel, outer membrane channel.
INTRODUCTION
Integral membrane proteins are very hydrophobic and cannot be dissolved in aqueous solution. These proteins can be divided into two distinct structural classes: α-helical bundles and β-barrels. The β-barrel membrane proteins are often found in the outer membranes of Gram-negative bacteria and mitochondria, whereas the α-helical membrane proteins are commonly located in the inner membranes of bacteria and in plasma membranes. Approximately a third of all proteomes accounted for are embedded in biological membranes.1
Efflux pumps of the resistance-nodulation-cell division (RND) superfamily are ubiquitous in bacteria, archaea and eukaryotes. In Gram-negative bacteria, these RND pumps play major roles in the intrinsic and acquired tolerance of antibiotics and toxic compounds.2,3 They are the key components of Gram-negative pathogens to utilize in overcoming toxic environments that are otherwise unfavorable for their survival. An RND-type efflux pump4–11 is an α-helical, inner membrane protein. It typically works with a periplasmic membrane fusion protein12–18 as well as a β-barrel, outer membrane channel to form a functional tripartite protein complex.19,21 The resulting efflux complex spans both the inner and outer membranes of Gram-negative bacterium to export substrates directly from the cell.2,3 This process is driven by proton import, which is catalyzed by the inner membrane RND efflux pump.
E. coli CusA is a large α-helical, inner membrane RND-type heavy-metal efflux pump that is responsible for extruding the biocidal Cu(I) and Ag(I) ions.22,23 CusA operates with a periplasmic membrane fusion protein CusB, and a β-barrel, outer membrane channel CusC to form a functional protein complex. The resulting CusCBA three-part efflux system spans the entire cell envelope and confers resistance to Cu(I) and Ag(I) by exporting these metal ions directly out of the cell.22,23
The crystal structures of each individual component of this tripartite complex system have been determined. The structure of CusA suggests that this RND pump exists as a homotrimer.11 Each subunit of CusA consists of 12 transmembrane α-helices (TM1–TM12) and a large periplasmic domain formed by two periplasmic loops between TM1 and TM2, and TM7 and TM8, respectively. The periplasmic domain of CusA can be divided into a pore domain (comprising sub-domains PN1, PN2, PC1 and PC2) and a CusC docking domain (containing sub-domains DN and DC). The structures indicate that this transporter utilizes methionine pairs and clusters to bind and export Cu(I) and Ag(I) ions.11
Overall, the structure of CusB demonstrates that this adaptor protein is folded into a four-domain elongated structure, ~120 Å long and ~40 Å wide.16 The first three domains (domains 1–3) of the protein are mostly β-strands. However, the fourth domain (domain 4) is all α-helices and is folded into a three-helix bundle structure.
Interestingly, the co-crystal structure of the CusBA adaptor-transporter reveals that the trimeric CusA pump associates with six CusB molecules to form the CusB6-CusA3 complex.24 Thus, the entire tripartite efflux assembly is expected to be in the form of CusC3-CusB6-CusA3, which span both the inner and outer membranes of E. coli to export Cu(I) and Ag(I) ions. This assemblage is indeed in good agreement with the predicted 3:6:3 polypeptide ratios of these tripartite complexes.25,26
Recently, the crystal structure of the CusC channel has also been resolved,21 suggesting that the architecture of this protein resembles those of TolC19 and OprM.20 The trimeric CusC channel consists of a membrane-anchoring β-barrel domain and an elongated periplasmic α-helical tunnel.21 The periplasmic tunnel is ~100 Å long with an outermost diameter of ~35 Å at the tip of the tunnel.
It is interesting to note that the N-terminal end of CusC forms an elongated loop. This loop extends from the membrane surface and leads down to the middle section (equatorial domain) of the α-helical periplasmic domain. The first N-terminal residue of CusC is a cysteine (Cys1). It has been observed that this residue is covalently linked to the lipid elements at the inner leaflet of the outer membrane. We reasoned that this Cys1 residue may play an important role in protein-membrane interaction and could be critical for the insertion of this channel protein into the outer membrane. We thus removed the Cys1 residue of CusC to form the ΔC1 mutant. We also replaced this residue by a serine to create the C1S mutant channel. Here we report the crystal structures of the wild-type CusC outer membrane channel as well as the ΔC1 and C1S mutant channels. In comparison with these three structures, it is suggested that the Cys1 residue indeed plays a crucial role in anchoring the transmembrane β-barrel onto the outer membrane. These structures also indicate that the ΔC1 and C1S mutants should represent the unstructured intermediate state of these β-barrel channel proteins.
RESULTS
Crystal structure of the wild-type CusC channel protein
We cloned, expressed and purified the wild-type, ΔC1 and C1S CusC proteins. Each of these proteins contains a 6xHis at the C-terminus. We obtained crystals of all these three channels using vapor diffusion. Data collection and refinement statistics of these CusC crystals are summarized in Table 1.
Table 1.
Data collection, phasing and structural refinement statistics of the CusC, ΔC1 and C1S proteins.
| CusC (WT) | ΔC1 | ΔC1 (SeMet) | C1S | |
|---|---|---|---|---|
| Data Collection | ||||
| Space group | R32 | P21 | P21 | P21 |
| Cell dimensions | ||||
| a, b, c (Å) | 88.49, 88.49, 474.72.60 | 62.02, 104.47, 72.03 | 61.93, 104.41, 71.95 | 61.88, 105.03, 72.36 |
| α, β, γ (°) | 90, 90, 120 | 90, 101.03, 90 | 90, 101.03, 90 | 90, 101.06, 90 |
| Wavelength (Å) | 0.979 | 0.979 | 0.979 | 0.979 |
| Resolution (Å) | 50-2.09 (2.17–2.09) | 50-2.53 (2.63–2.53) | 50-2.8 (2.90–2.80) | 50-2.69 (2.84–2.69) |
| Rsym (%) | 8.3 (40.4) | 6.9 (41.7) | 11.6 (45.9) | 10.2 (30.4) |
| Average I/σI | 12.7 (1.8) | 12.1 (1.7) | 10.5 (2.16) | 9.3 (3.4) |
| Completeness (%) | 97.3 (97.2) | 94.0 (91.6) | 99.9 (99.8) | 98.5 (95.0) |
| Redundancy | 2.4 (2.4) | 2.1 (2.0) | 2.8 (2.8) | 3.8 (3.7) |
| Total reflections | 640,300 | 426,465 | 654,832 | 94,904 |
| Unique reflections | 43,397 | 30,236 | 22,466 | 24,874 |
| Phasing | ||||
| Number of sites | 10 | |||
| Figure of merit (acentric/centric) | 0.821/0.797 | |||
| Refinement | ||||
| Resolution (Å) | 50-2.09 | 50-2.53 | 50-2.69 | |
| No. reflections | 54,129 | 31,197 | 24,838 | |
| Rwork/Rfree (%) | 20.60/23.50 | 20.77/26.71 | 19.08/24.61 | |
| B-factors (Å2) | ||||
| Protein chain A | 30.6 | 43.7 | 46.3 | |
| Protein chain B | 50.7 | 51.8 | ||
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.007 | 0.008 | 0.009 | |
| Bond angles (°) | 0.924 | 1.105 | 1.143 | |
| Ramachandran | ||||
| most favored | 96.4 | 93.4 | 92.8 | |
| additional allowed | 3.6 | 6.6 | 6.2 | |
| generously allowed | 0 | 0 | 0.7 | |
| disallowed | 0 | 0 | 0.3 | |
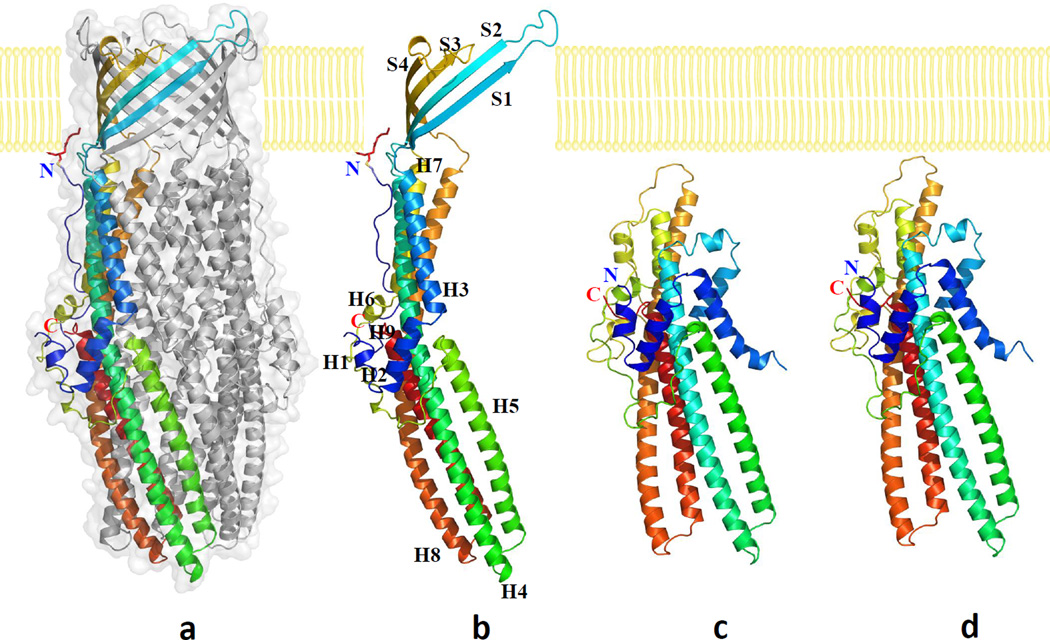
The crystal structure of the wild-type CusC channel was resolved to a resolution of 2.09 Å (Fig. 1a). The final structure is nearly identical to the structure of CusC (pdb code: 3PIK)21 determined by Kulathila et al. Superimposition of these two structures results in an RMSD of 0.28 Å for 429 Cα atoms. CusC exists as a homotrimer that forms a ~130 Å long α/β barrel. Each subunit of CusC contains four β-strands (contributing to the 12-stranded outer membrane β-barrel) and nine α-helices (forming the elongated periplasmic α-barrel) (Figs. 1b, S1 and S2). The trimeric CusC channel creates a large cylindrical internal cavity of ~28,000 Å3. Like the previous crystal structure of CusC,21 our x-ray structure suggests that the N-terminal Cys1 residue is covalently linked to the outer membrane via a thioester bond. Thus, the trimeric CusC channel is most likely triacylated through the Cys1 residue to secure the anchoring of this protein onto the outer membrane.
Fig. 1.
Structures of the CusC channel proteins. (a) Side view of the trimeric CusC channel. One of the protomers of CusC is in rainbow colors. The other two molecules of CusC are colored gray. (b) Ribbon diagram of the structure of a CusC protomer. The CusC protomer is acylated (red sticks) through the Cys1 residue to anchor onto the outer membrane. (c) Ribbon diagram of the structure of a protomer of the ΔC1 mutant. (d) Ribbon diagram of the structure of a protomer of the C1S mutant. The molecules are colored using a rainbow gradient from the N-terminus (blue) to the C-terminus (red).
Crystal structure of the ΔC1 CusC mutant
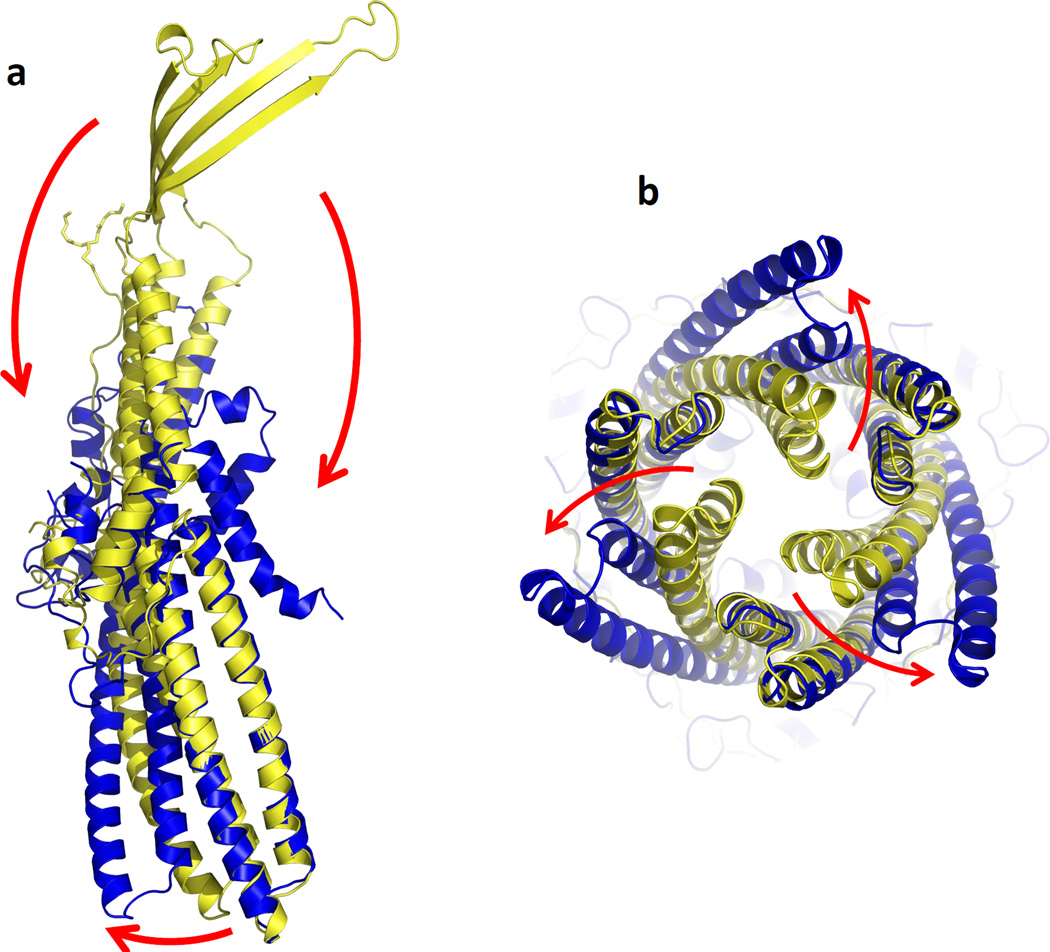
The crystal structure of the ΔC1 CusC channel was determined to a resolution of 2.53 Å (Figs. 1c and S3). The final ΔC1 mutant model consists of the complete mature protein sequence, with the exception of the disordered residues 21–31 and 99–129. Intriguingly, the overall structure of the ΔC1 mutant is very distinct from that of wild-type CusC. Superimposition of a protomer of these two structures gives a high RMSD of 22.49 Å (for 378 Cα atoms), suggesting highly significant differences between these two channels (Fig. 2). In ΔC1 CusC, the amino acids which form the four transmembrane β-strands in the wild-type structure adopt a dramatically different conformation. The majority of these residues appear to be unstructured and form two large random loops. Residues 290–326, which form the transmembrane β-strands S3 and S4 in the wild-type structure, are found to flip down to the outermost surface of the periplasmic α-helical tunnel. In addition, the top portion of the vertical periplasmic α-helices, H7 and H8, make substantial changes and are found to bend downward to accommodate for the change in conformation. Residues N276 of H7 and N334 of H8 appear to form hinges for the bending.
Fig. 2.
Comparison of the conformations of wild-type CusC and ΔC1. (a) Superimposition of a monomer of wild-type CusC onto that of the ΔC1 mutant. The structures of the wild-type CusC and ΔC1 protomers are colored yellow and blue, respectively. The arrows indicate the drastic changes in positions and secondary structures when comparing the conformations of the wild-type and ΔC1 CusC. (b) Superimposition of each protomer of the wild-type CusC trimer with the ΔC1 mutant. Each arrow indicates the shift in position of H8 and H9 in each protomer when comparing the wild-type and ΔC1 structures.
Interestingly, the drastic changes in conformation described above are accompanied by the structural changes of residues 84–116, which form the transmembrane β-strands S1 and S2 of the wild-type CusC channel. In the ΔC1 mutant, the electron density map in the region between residues 82 and 111 is very unclear, suggesting that the majority of the secondary structure of this area is disordered. Thus, this region, located at the innermost surface of the periplasmic α-helical tunnel, also creates a large random loop in the ΔC1 mutant. Similar to the case of helices H7 and H8, the top portion of the vertical periplasmic α-helices, H3 and H4, also bend downward and the hinges are found in nearby residues N76 and T137.
Excluding these areas, the remaining tertiary fold of the ΔC1 CusC monomer is very similar to that of a promoter of the wild-type CusC. However, ΔC1 crystallized as a monomer with two molecules in the asymmetric unit (Figs. S3 and S4). The structure suggests that the ΔC1 mutant is monomeric. This monomeric ΔC1 CusC mutant is mostly α-helical, forming a ~95 Å long secondary structure.
Crystal structure of the C1S CusC mutant
The crystal structure of the C1S mutant was determined to a resolution of 2.69 Å (Figs. 1d and S5). Overall, the structural model of the C1S mutant is nearly identical to that of the ΔC1 CusC channel. Like ΔC1, gel filtration suggests that the C1S mutant is also monomeric in nature (Fig. S6). Superimposition of these two structures gives an overall RMSD of 0.37 Å (for 381 Cα atoms). Again, the monomer’s four-stranded antiparallel β-sheet, which packs against one another to form the 12-stranded transmembrane β-barrel of the wild-type trimeric CusC channel, does not seem to exist in this mutant structure. Similar to the ΔC1 mutant, the residues that are supposed to form the four-stranded β-sheet contribute to two independent random loops. One of these loops is found at the position that is supposed to form the upper portion of the innermost core of the periplasmic α-helical tunnel. Another loop is located at the outermost core of the upper portion of this periplasmic tunnel.
The structural difference between wild-type CusC and ΔC1 or C1S suggests that the α-helical tunnels of the wild-type and these mutant channels are in different conformational states. Superimposition of the periplasmic α-helical domains of wild-type CusC and ΔC1 indicates that the conformation of helices H8 and H9 of these two structures are significantly different. It is found that helices H8 and H9 shift their positions in the ΔC1 mutant in comparison to those of the wild-type. When each protomer of the wild-type CusC trimer is superimposed with the ΔC1 mutant, the change in conformation between the wild-type and ΔC1 proteins can be interpreted as an outward swing of H8 and H9 at the periplasmic tip of the α-helical tunnel from the structure of the wild-type CusC trimer to adopt the conformational state of ΔC1 (Fig. 2). This conformational shift results in the opening of the channel of the periplasmic tunnel. As the structure of the wild-type CusC trimer represents a closed form of the channel,21 the conformation of the α-helical tunnel of the ΔC1 and C1S mutants may correspond to the open conformational state of the channel. If this is the case, then the interaction between the β-barrel residues and outer membrane may be critical for controlling the opening and closing of the CusC channel.
DISCUSSION
In our protein expression system, a CusC signaling peptide was included at the N-terminus of each target protein. Thus, each protein, including the wild-type CusC and mutants ΔC1 and C1S, should secret to the periplasm. To ensure that the harvested CusC proteins were attached to or anchored to the E. coli outer membrane, and not the inner membrane, we performed a pre-extraction procedure using 0.5% sodium lauroyl sarcosinate27 to selectively dissolve and remove proteins of the inner membrane (see Methods). The undissolved outer membrane constituents, containing the corresponding CusC outer membrane proteins, were then collected by ultracentrifugation before protein extraction. As all of our CusC proteins were extracted from the outer membrane pellets, with the use of detergents, these proteins should be associated with the outer membrane, viz., peripheral outer membrane proteins.
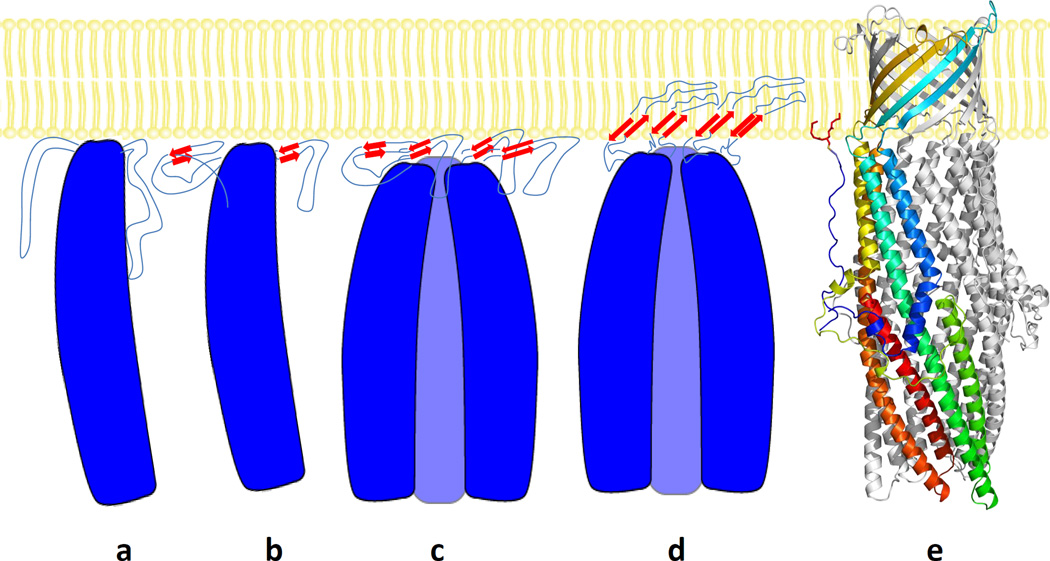
In comparison with the structural differences between the monomeric ΔC1 and C1S CusC mutants as well as trimeric wild-type CusC channel, these structural data provide molecular detail into the mechanisms of assembly and folding of this outer membrane channel (Fig. 3). In the case of the OmpA outer membrane protein,28 in vitro study has been demonstrated that the insertion and folding of this protein into the lipid bilayer occur spontaneously without the need of accessory proteins, including the β-barrel assembly machinery (BAM) complex.29,30 In the aqueous phase, OmpA is unstructured and does not have any β-signature. It encounters the inner leaflet of the outer membrane from the periplasm to form an intermediate IMI.31 At this state, the protein is still disordered and does not yet appear to have any β-structure. Upon membrane association, two more sequential intermediates, including molten disk (IM2) and molten global (IM3), are formed prior to the completion of the mature folded β-barrel channel.31 Some fractions of the β-structure have been found to develop in the IM2 intermediate state. However, these structures appear to localize on the membrane surface, and no correct tertiary β-contact or penetration into the membrane has yet been made. In the IM3 molten globule state, the β-hairpin loops have partially translocated into the lipid bilayer. This intermediate is more globular, but the correct tertiary fold is needed to complete to form the native structure. Finally, the native structure is achieved through an extensive rearrangement of side chain contacts and formation of backbone hydrogen bonds between strands to form a transmembrane β-channel30, spanning the outer membrane.
Fig. 3.
Model of folding and membrane insertion of the CusC channel protein. This includes the (a) unstructured IM1 intermediate, (b) monomeric molten disk (mono-IM2) intermediate, (c) trimeric molten disk (tri-IM2) intermediate, (d) trimeric IM3 molten globule intermediate, and (e) mature trimeric CusC protein.
In this respect, the monomeric ΔC1 and C1S structures should correspond to the unstructured IM1 intermediate of the β-channel membrane proteins. As we isolated these two mutant proteins from the membrane fractions, they should interact with the membrane even though they failed to form a fully inserted β-barrel. This may support the idea that the unstructured IM1 intermediate of CusC is associated with inner leaflet of the outer membrane. Interestingly, the architecture of the α-helical secondary structures of the periplasmic domain of this channel has already been achieved at this state. It has been proposed that the monomeric LukF structure,32 containing a partial β-strand signature, represents the molten disk form of the α-hemolysin heptamer.33 Thus, the next step for CusC folding may be the formation of a monomeric molten disk (mono-IM2) intermediate, where a partial β-strand conformation has been established at this state (Fig. 3). Upon oligomerization at the interface of the inner leaflet of the outer membrane, the subsequent step may be the transition to the trimeric molten disk (tri-IM2) intermediate, where the β-hairpins of each monomer are still localized at the membrane interface and have not yet penetrated into the membrane. Then, the next step should well be the formation of a trimeric IM3 molten globule intermediate, in which the β-hairpins have been inserted into the membrane and the partial β-barrel has been achieved. Finally, the native state of trimeric CusC should be formed and the mature β-barrel channel should be completed, allowing the β-structural elements to span the entire outer membrane.
Although the phenomenon of outer membrane protein folding has been studied extensively using fluorescence method, the three-dimensional structures of these unfolded intermediates are difficult to obtain by x-ray crystallography and NMR because of their intrinsically unstructured and disordered nature. This study provides snapshot of the conformation of these outer membrane channel proteins in their unstructured form. The structures of CusC have allowed us to unmask the sequential transition of conformations leading to the folding and membrane insertion of this channel. During the course of channel formation, it is believed that specific protein-protein and protein-lipid interactions play a critical role in the assembly of this trimeric transmembrane β-barrel channel.
Methods
Cloning, expression and purification of the CusC, C1S and ΔC1 channel proteins
Briefly, the full-length CusC membrane protein containing a 6xHis tag at the C-terminus was overproduced in E. coli C43(DE3)/pBAD22bΩcusC cells. Cells were grown in 12 L of LB medium with 100 µg/ml ampicillin at 37°C. When the OD600 reached 0.5, the culture was cooled down to 20°C and then treated with 0.2% arabinose to induce cusC expression. Cells were harvested after shaking for 16 h at 20°C. The collected bacteria were resuspended in buffer containing 20 mM Na-HEPES (pH 7.5), 300 mM NaCl and 1 mM phenylmethanesulfonyl fluoride (PMSF), and then disrupted with a French pressure cell. The membrane fraction was collected by ultracentrifugation, followed by a pre-extraction procedure by incubating in buffer containing 0.5% sodium lauroyl sarcosinate, 20 mM Na-HEPES (pH 7.5) and 50 mM NaCl for 0.5 h at room temperature. The membrane was collected and washed twice with buffer containing 20 mM Na-HEPES (pH 7.5) and 50 mM NaCl. The membrane protein was then solubilized in 2 % (w/v) n-dodecyl-β-D-maltoside (DDM). Insoluble material was removed by ultracentrifugation at 100,000 × g. The extracted protein was purified with a Ni2+-affinity column. The purity of the CusC protein (>95%) was judged using 10% SDS-PAGE stained with Coomassie Brilliant Blue. The purified protein was then dialyzed and concentrated to 15 mg/ml in buffer containing 20 mM Na-HEPES (pH 7.5), 200 mM NaCl and 0.05% DDM.
The C1S CusC protein that contains a 6xHis tag at the C-terminus was overproduced in E. coli C43(DE3) cells using the pBAD22bΩcusC(C1S) expression vector. The procedures for expressing and purifying the C1S mutant channel were identical to those of 6xHis CusC. The purified C1S protein was concentrated to 15 mg/ml in buffer containing 20 mM Na-HEPES (pH 7.5), 200 mM NaCl and 0.05% DDM for crystallization.
The ΔC1 CusC protein containing a 6xHis tag at the C-terminus was overproduced in E. coli C43(DE3) cells/pBAD22bΩcusC(ΔC1). The procedures for cell growth and protein expression were identical to those of 6xHis CusC. For protein purification, the procedures were nearly identical to those of the 6xHis CusC channel, except that extracted outer membrane was solubilized in 2% (w/v) 6-cyclohexyl-1-hexyl-β-D-maltoside (Cymal-6). Insoluble material was removed by ultracentrifugation at 100,000 × g. The ΔC1 protein was purified with a Ni2+-affinity column. The purity of the ΔC1 protein (>95%) was judged using 10% SDS-PAGE stained with Coomassie Brilliant Blue. The purified ΔC1 protein was then dialyzed and concentrated to 15 mg/ml in buffer containing 20 mM Na-HEPES (pH 7.5), 200 mM NaCl and 0.05% Cymal-6.
For 6xHis SeMet-ΔC1, the protein was expressed in E. coli BL21Star(DE3) cells possessing pET15bΩcusC(ΔC1). In brief, a 10 ml LB broth overnight culture containing E. coli BL21star(DE3)/pET15bΩcusC(ΔC1) cells was transferred into 120 ml of LB broth containing 100 µg/ml ampicillin and grown at 37°C. When the OD600 value reached 1.2, cells were harvested by centrifugation at 6000 rev/min for 10 min, and then washed two times with 20 ml of M9 minimal salts solution. The cells were re-suspended in 120 ml of M9 media and then transferred into a 12 L pre-warmed M9 solution containing 100 µg/ml ampicillin. The cell culture was incubated at 37°C with shaking. When the OD600 reached 0.4, 100 mg/l of lysine, phenylalanine and threonine, 50 mg/l isoleucine, leucine and valine, and 60 mg/l of L-selenomethionine were added. The culture was induced with 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) after 15 min. Cells were then harvested within 3 h after induction. The collected bacteria were resuspended in low salt buffer containing 100 mM sodium phosphate (pH 7.2), 10 % glycerol and 1 mM PMSF, and then disrupted with a French pressure cell. The membrane fraction was collected and washed twice with high salt buffer containing 20 mM sodium phosphate (pH 7.2), 2 M KCl, 10 % glycerol, 1 mM EDTA and 1 mM PMSF, and once with 20 mM Na-HEPES buffer (pH 7.5) containing 1 mM PMSF. The membrane protein was then solubilized in 2 % (w/v) Cymal-6. Insoluble material was removed by ultracentrifugation at 100,000 × g. The extracted protein was purified with a Ni2+-affinity column. The purity of the SeMet-ΔC1 protein (>95%) was judged using 10% SDS-PAGE stained with Coomassie Brilliant Blue. The purified protein was then dialyzed and concentrated to 20 mg/ml in a buffer containing 20 mM Na-HEPES (pH 7.5), 200 mM NaCl and 0.05% Cymal-6.
Crystallization of the CusC, ΔC1 and C1S proteins
Crystals of the 6xHis CusC were obtained using sitting-drop vapor diffusion. The CusC crystals were grown at room temperature in 24-well plates with the following procedures. A 2 µl protein solution containing 15 mg/ml CusC protein in 20 mM Na-HEPES (pH 7.5), 200 mM NaCl and 0.05% (w/v) DDM was mixed with a 2 µl of reservoir solution containing 8% PEG 3350, 0.05 M sodium acetate (pH 4.0), 0.2 M (NH4)2SO4, 1% JM 600 and 2% OG. The resultant mixture was equilibrated against 500 µl of the reservoir solution. Crystals of CusC grew to a full size in the drops within two weeks. Typically, the dimensions of the crystals were 0.2 mm × 0.2 mm × 0.2 mm. Cryoprotection was achieved by raising the glycerol concentration stepwise to 30% with a 5% increment in each step.
Crystals of the 6xHis ΔC1 mutant were obtained using sitting-drop vapor diffusion. Briefly, a 2 µl protein solution containing 15 mg/ml ΔC1 protein in 20 mM Na-HEPES (pH 7.5), 200 mM NaCl and 0.05% (w/v) Cymal-6 was mixed with a 2 µl of reservoir solution containing 10% PEG 2000, 0.1 M Na-HEPES (pH 7.5) and 0.1 M KSCN. The resultant mixture was equilibrated against 500 µl of the reservoir solution. Crystals of ΔC1 grew to a full size in the drops within three days with the dimensions 0.2 mm × 0.2 mm × 0.2 mm. Cryoprotection was achieved by raising the glycerol concentration stepwise to 30% with a 5% increment in each step.
The crystallization conditions for SeMet-ΔC1 were the same as those for the native ΔC1 protein.
The procedures for crystallizing the 6xHis C1S mutant were nearly identical to those of 6xHis CusC. The reservoir solution used to crystallize C1S contained 12% PEG 3350, 0.1 M sodium citrate (pH 6.5), 0.1 M sodium acetate and 15% butane-2,3-diol. Crystals of C1S also grew to a full size in the drops within two weeks. Cryoprotection was achieved by raising the glycerol concentration stepwise to 30% with a 5% increment in each step.
Data collection, structural determination and refinement
All diffraction data were collected at 100K at beamline 24ID-C located at the Advanced Photon Source, using an ADSC Quantum 315 CCD-based detector. Diffraction data were processed using DENZO and scaled using SCALEPACK.34 Crystals of 6xHis CusC belong to space group R32 (Table S1). Based on the molecular weight of CusC (49.3 kDa), a single molecule per asymmetric unit with a solvent content of 67.8%. The structure of the CusC channel protein was phased using molecular replacement (MR), utilizing the published CusC structure (pdb code: 3PIK) as the search model. Structural refinement was then performed using PHENIX35 and CNS36 by refining the model against our 2.09 Å-resolution diffraction data (Table 1).
Crystals of 6xHis ΔC1 took a space group P21 (Table 1). In the asymmetric unit, two molecules of ΔC1 were found with a solvent content of 47.1%. Single anomalous dispersion (SAD) phasing using the program PHASER37 was employed to obtain experimental phases. Phases were then subjected to density modification and phase extension to 2.53 Å-resolution using the program RESOLVE.38 The ΔC1 CusC protein contains six methionine residues, five selenium sites per ΔC1 molecule (10 selenium sites per asymmetric unit) were identified. The SeMet data not only augmented the experimental phases, but also helped in tracing the molecules by anomalous difference Fourier maps where we could ascertain the proper registry of SeMet residues. After tracing the initial model manually using the program Coot,39 the model was refined against the native data at 2.53 Å-resolution using PHENIX35 and CNS.36 The conformation of the two ΔC1 molecules in the asymmetric unit are very similar. Superimposition of these two molecules gives an overall RMSD of 1.0 Å (for 372 Cα atoms).
Crystals of 6xHis C1S CusC belong to space group P21 (Table 1). Two molecules per asymmetric unit were found in the crystal with a solvent content of 47.3%. The structure of the C1S protein was phased using MR, utilizing the ΔC1 structure as the search model. The model was then refined against the C1S data at 2.69 Å-resolution using the same procedures for the full-length CusC and ΔC1 structures. The structures of the two C1S molecules in the asymmetric unit are nearly identical to each other. Superimposition of these two molecules gives an overall RMSD of 0.9 Å (for 350 Cα atoms).
Gel filtration
A protein liquid chromatography Superdex 200 16/60 column (Amersham Pharmacia Biotech) with a mobile phase containing 20 mM Na-HEPES (pH 7.5), 200 mM NaCl and 0.05% DDM was used in the gel filtration experiments. Blue dextran (Sigma-Aldrich) was used to determine the column void volume, and proteins for use as gel filtration molecular weight standards were cytochrome C (Mr 12,400), carbonic anhydrase (Mr 29,000), albumin bovine serum (Mr 66,000), alcohol dehydrogenase (Mr 150,000), and β-Amylase (Mr 200,000). All these standards were purchased from Sigma-Aldrich. The molecular weights of the experimental samples were determined following the protocols supplied by the manufacturers.
Data Deposition
Atomic coordinates and structure factors have been deposited with the Protein Data Bank under codes 4K7R (wild-type CusC), 4K7K (ΔC1) and 4K34 (C1S).
Supplementary Material
Acknowledgments
This work is supported by an NIH Grant R01GM086431 (E.W.Y.). This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines of the Advanced Photon Source, supported by award RR-15301 from the National Center for Research Resources at the National Institutes of Health. Use of the Advanced Photon Source is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357.
References
- 1.Wallin E, von Heijine G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Prot. Sci. 1998;7:1029–1038. doi: 10.1002/pro.5560070420. (1998) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tseng TT, Gratwick KS, Kollman J, Park D, Nies DH, Goffeau A, Saier MH., Jr The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development protein. J. Mol. Microbiol. Biotechnol. 1999;1:107–125. [PubMed] [Google Scholar]
- 3.Nies DH. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 2003;27:313–339. doi: 10.1016/S0168-6445(03)00048-2. [DOI] [PubMed] [Google Scholar]
- 4.Murakami S, Nakashima R, Yamashita E, Yamaguchi A. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature. 2002;419:587–593. doi: 10.1038/nature01050. [DOI] [PubMed] [Google Scholar]
- 5.Yu EW, McDermott G, Zgruskaya HI, Nikaido H, Koshland DE., Jr Structural basis of multiple drug binding capacity of the AcrB multidrug efflux pump. Science. 2003;300:976–980. doi: 10.1126/science.1083137. [DOI] [PubMed] [Google Scholar]
- 6.Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature. 2006;443:173–179. doi: 10.1038/nature05076. [DOI] [PubMed] [Google Scholar]
- 7.Seeger MA, Schiefner A, Eicher T, Verrey F, Dietrichs K, Pos KM. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science. 2006;313:1295–1298. doi: 10.1126/science.1131542. [DOI] [PubMed] [Google Scholar]
- 8.Sennhauser G, Amstutz P, Briand C, Storchengegger O, Grütter MG. Drug export pathway of multidrug exporter AcrB revealed by DARPin inhibitors. PLoS Biol. 2007;5:e7. doi: 10.1371/journal.pbio.0050007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu EW, Aires JR, McDermott G, Nikaido H. A periplasmic drug-binding site of the AcrB multidrug efflux pump: a crystallographic and site-directed mutagenesis study. J. Bacteriol. 2005;187:6804–6815. doi: 10.1128/JB.187.19.6804-6815.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sennhauser G, Bukowska MA, Briand C, Grütter MG. Crystal structure of the multidrug exporter MexB from Pseudomonas aeruginosa. J. Mol. Biol. 2009;389:134–145. doi: 10.1016/j.jmb.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 11.Long F, Su C-C, Zimmermann MT, Boyken SE, Rajashankar KR, Jernigan RL, Yu EW. Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport. Nature. 2010;467:484–488. doi: 10.1038/nature09395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Higgins MK, Bokma E, Koronakis E, Hughes C, Koronakis V. Structure of the periplasmic component of a bacterial drug efflux pump. Proc. Natl. Acad. Sci. USA. 2004;101:9994–9999. doi: 10.1073/pnas.0400375101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akama H, Matsuura T, Kashiwag S, Yoneyama H, Narita S, Tsukihara T, Nakagawa A, Nakae T. Crystal structure of the membrane fusion protein, MexA, of the multidrug transporter in Pseudomonas aeruginosa. J. Biol. Chem. 2004;279:25939–25942. doi: 10.1074/jbc.C400164200. [DOI] [PubMed] [Google Scholar]
- 14.Mikolosko J, Bobyk K, Zgurskaya HI, Ghosh P. Conformational flexibility in the multidrug efflux system protein AcrA. Structure. 2006;14:577–587. doi: 10.1016/j.str.2005.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Symmons M, Bokma E, Koronakis E, Hughes C, Koronakis V. The assembled structure of a complete tripartite bacterial multidrug efflux pump. Proc. Natl. Acad. Sci. USA. 2009;106:7173–7178. doi: 10.1073/pnas.0900693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Su C-C, Yang F, Long F, Reyon D, Routh MD, Kuo DW, Mokhtari AK, Van Ornam JD, Rabe KL, Hoy JA, Lee YJ, Rajashankar KR, Yu EW. Crystal structure of the membrane fusion protein CusB from Escherichia coli. J. Mol. Biol. 2009;393:342–355. doi: 10.1016/j.jmb.2009.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yum S, Xu Y, Piao S, Sim S-H, Kim H-M, Jo W-S, Kim K-J, Kweon H-S, Jeong M-H, Lee K, Ha N-C. Crystal structure of the periplasmic component of a tripartite macrolide-specific efflux pump. J. Mol. Biol. 2009;387:1286–1297. doi: 10.1016/j.jmb.2009.02.048. [DOI] [PubMed] [Google Scholar]
- 18.De Angelis F, Lee JK, O’Connell JD, III, Miercke LJW, Verschueren KH, Srinivasan V, Bauvois C, Govaerts C, Robbins RA, Ruysschaert J-M, Stroud RM, Vandenbussche G. Metal-induced conformational changes in ZneB suggest an active role of membrane fusion proteins in efflux resistance systems. Proc. Natl. Acad. Sci. USA. 2010;107:11038–11043. doi: 10.1073/pnas.1003908107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koronakis V, Sharff A, Koronakis E, Luisi B, Hughes C. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature. 2000;405:914–919. doi: 10.1038/35016007. [DOI] [PubMed] [Google Scholar]
- 20.Akama H, Kanemaki M, Yoshimura M, Tsukihara T, Kashiwag T, Yoneyama H, Narita S, Nakagawa A, Nakae T. Crystal structure of the drug discharge outer membrane protein, OprM, of Pseudomonas aeruginosa: dual modes of membrane anchoring and occluded cavity end. J. Biol. Chem. 2004;279:52816–52819. doi: 10.1074/jbc.C400445200. [DOI] [PubMed] [Google Scholar]
- 21.Kulathila R, Kulathila R, Indic M, van den Berg B. Crystal structure of Escherichia coli CusC, the outer membrane component of a heavy-metal efflux pump. PLoS One. 2011;6:e15610. doi: 10.1371/journal.pone.0015610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franke S, Grass G, Nies DH. The product of the ybdE gene of the Escherichia coli chromosome is involved in detoxification of silver ions. Microbiol. 2001;147:965–972. doi: 10.1099/00221287-147-4-965. [DOI] [PubMed] [Google Scholar]
- 23.Franke S, Grass G, Rensing C, Nies DH. Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J. Bacteriol. 2003;185:3804–3812. doi: 10.1128/JB.185.13.3804-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su C-C, Long F, Zimmermann MT, Rajashankar KR, Jernigan RL, Yu EW. Crystal structure of the CusBA heavy-metal efflux pump of Escherichia coli. Nature. 2011;467:484–488. doi: 10.1038/nature09743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rensing C, Pribyl T, Nies DH. New functions for the three subunits of the CzcCBA cation-proton antiporter. J. Bacteriol. 1997;179:6871–6879. doi: 10.1128/jb.179.22.6871-6879.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stegmeier JF, Polleichtner G, Brandes N, Hotz C, Andersen C. Importance of the adaptor (membrane fusion) protein hairpin domain for the functionality of multidrug efflux pumps. Biochemistry. 2006;45:10303–10312. doi: 10.1021/bi060320g. [DOI] [PubMed] [Google Scholar]
- 27.Filip C, Fletcher G, Wulff JL, Earhart CF. Solubilization of the cytoplasmic membrane of Escherichia coli by the ionic detergent sodium-lauryl sarcosinate. J. Bacteriol. 1973;115:717–722. doi: 10.1128/jb.115.3.717-722.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pautsch A, Schulz GE. Structure of the outer membrane protein A transmembrane domain. Nature Struct. Biol. 1998;5:1013–1017. doi: 10.1038/2983. [DOI] [PubMed] [Google Scholar]
- 29.Kleinschmidt JH, Tamm LK. Folding intermediates of a beta-barrel membrane protein. Kinetic evidence for a multi-step membrane insertion mechanism. Biochemistry. 1996;35:12993–13000. doi: 10.1021/bi961478b. (1996) [DOI] [PubMed] [Google Scholar]
- 30.Tamm LK, Arora A, Kleinschmidt JH. Structure and Assembly of β-Barrel Membrane Proteins. J. Biol. Chem. 2001;276:32399–32402. doi: 10.1074/jbc.R100021200. [DOI] [PubMed] [Google Scholar]
- 31.Tamm LK, Hong H, Liang B. Folding and assembly of beta-barrel membrane proteins. Biochem. Biophys. Acta. 2004;1666:250–263. doi: 10.1016/j.bbamem.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 32.Olson R, Nariya H, Yokota K, Kamio Y, Gouaux E. Crystal structure of staphylococcal LukF delineates conformational changes accompanying formation of a transmembrane channel. Nature Struct. Biol. 1999;6:134–140. doi: 10.1038/5821. [DOI] [PubMed] [Google Scholar]
- 33.Song L, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE. Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science. 1996;274:1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 34.Otwinowski Z, Minor M. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 35.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCroy AJ, Moriarty NW, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 36.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 37.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Terwilliger TC. Maximum-likelihood density modification using pattern recognition of structural motifs. Acta Cryst. 2001;D57:1755–1762. doi: 10.1107/S0907444901013737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.