Abstract
Collagenases are the principal enzymes responsible for the degradation of collagens during embryonic development, wound healing, and cancer metastasis. However, the mechanism by which these enzymes disrupt the highly chemically and structurally stable collagen triple helix remains incompletely understood. We used a single-molecule magnetic tweezers assay to characterize the cleavage of heterotrimeric collagen I by both the human collagenase matrix metalloproteinase-1 (MMP-1) and collagenase from Clostridium histolyticum. We observe that the application of 16 pN of force causes an 8-fold increase in collagen proteolysis rates by MMP-1, but does not affect cleavage rates by Clostridium collagenase. Quantitative analysis of these data allows us to infer the structural changes in collagen associated with proteolytic cleavage by both enzymes. Our data support a model in which MMP-1 cuts a transient, unwound conformation of its recognition site. In contrast, our findings suggest that Clostridium collagenase is able to cleave the fully wound collagen triple helix, accounting for its lack of force sensitivity and low sequence specificity. We observe that the cleavage of heterotrimeric collagen is less force sensitive than the proteolysis of a homotrimeric collagen model peptide, consistent with studies suggesting that the MMP-1 recognition site in heterotrimeric collagen I is partially unwound at equilibrium.
Introduction
Extracellular matrix (ECM) remodeling by MMPs is a critical process during embryonic development,1–4 cancer metastasis,5 aneurysm formation,6 and atherosclerosis.7 Collagen I is an abundant extracellular matrix (ECM) protein whose primary purpose is to give structure to tissues and organs.8 Collagen I commonly assembles into homotrimeric α1(I)3 or heterotrimeric α1(I)2α2(I) triple helices.9 While the heterotrimer is the common assembly found in healthy adult tissues,9 the homotrimer is observed in fetal9,10 and fibrotic tissues,9,11–13 as well as cancer cell cultures.9,14–18 At present the physiological significance of the differences in homo- and heterotrimeric collagen distribution is unclear.
Collagen I is cleaved by multiple MMPs, including MMP-1, MMP-8, MMP-13 and MMP-14.9,19 Structural and biochemical data indicate that the MMP-1 active site is too small to accommodate the collagen triple helix, which must be disrupted prior to proteolysis.9,20 Two previously proposed models are that collagen may unwind prior to proteolysis, potentially presenting a flat ribbon to MMP that can fit edge-wise into the active site,20 or that spontaneously formed collagen loops are captured and cut by MMPs.21 Both models account for the correlation between thermodynamic stability and resistance to MMP-mediated proteolysis observed in previous studies.22 However, the models predict different geometries for the catalytically competent MMP-collagen complex.
Previous studies have examined the effect of mechanical load on the proteolytic degradation of collagen gels and collagen-containing tissue explants.23–33 However, a mechanistic interpretation of these studies is complicated due to the structural complexity of collagen I, which assembles into fibrils containing many thousands of trimeric units. Recent studies have also investigated the effect of mechanical load on the proteolysis of collagen trimers at the single molecule level.34,35 In a previous publication, we used magnetic tweezers to examine the mechanism by which MMP-1 cleaves an engineered homotrimeric collagen model peptide.34 Engineered collagen peptide trimers have been extensively used to study both collagen conformational dynamics and MMP proteolytic mechanisms.22,36,37 Consistent with inference from bulk measurements,9 we found that the collagen triple helix unwinds prior to proteolysis.34 Moreover, unwinding is exquisitely sensitive to mechanical stretch: 13 pN of extensional force (similar to forces likely experienced by individual trimers in vivo) exerted on a homotrimeric collagen I model protein induced an 80-fold increase in proteolysis rates.34
The relevance of the unwinding model to the proteolysis of full-length, post-translationally modified heterotrimeric collagen I, and indeed to the many other collagen-cleaving enzymes in addition to MMP-1, has not been directly tested. Here we characterize the effect of applied load on the proteolysis of full-length, post-translationally modified heterotrimeric α1(I)2α2(I) collagen I by MMP-1 and by collagenase isolated from Clostridium histolyticum.38,39 Human MMP-1 cleaves collagen I at a specific recognition site. In contrast, bacteria from the genus Clostridium secrete collagenases with much lower sequence specificities in order to dissolve tissue in the context of gas gangrene and other necrotic diseases.40,41 The comparison of these two enzymes thus offers a potential means to probe the structural and biophysical origins of collagenase activity and sequence specificity.
Materials and Methods
Materials
Recombinant, post-translationally modified human collagen I was purchased from Fibrogen (San Francisco, CA). Pyridoxal 5’-phosphate (PLP), Biotin hydrazide, collagenase from Clostridium histolyticum (C1639), collagenase substrate (N-[3-(2-Furyl)acryloyl]-Leu-Gly-Pro-Ala) and bovine serum albumin (BSA, part number A2153) were purchased from Sigma Aldrich (St. Louis, MO). Dynabeads MyOne T1 and M280 superparamagnetic beads were purchased from Invitrogen (Carlsbad, CA). An antibody targeting a sequence near the collagen C-terminus42 was purchased from Millipore (clone 5D8-G9; Billerica, MA). The MMP-1 gene was purchased from the Harvard Plasmid Database. A Zeiss Axiovert 100TV with a 10× objective and equipped with a CMOS camera (Thorlabs) was used for the single-molecule proteolysis experiments. MMP-1 was expressed, purified and activated, and the magnetic tweezers apparatus was calibrated as described previously in Adhikari et al.34 The N–terminus of the collagen trimer was biotinylated using PLP and biotin hydrazide as described earlier.43–45
Single-molecule assay
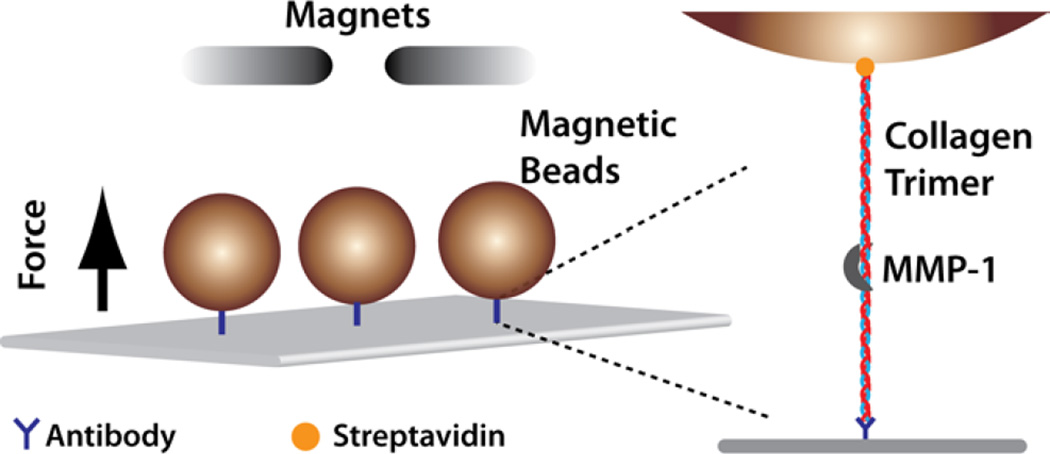
Microfluidic flow chambers were assembled as described in the Supporting Information. Collagen molecules were attached to the glass coverslip by an anti-C-terminus collagen antibody that binds specifically to intact trimeric collagen.42 The biotinylated N-terminus was attached to streptavidin-coated superparamagnetic beads (Figure 1).
Figure 1.
Force-dependent collagen cleavage assay. Collagen trimers are attached to the glass surface via an antibody targeting a sequence close to the C-terminus, and to a streptavidin-coated magnetic bead via N-terminal biotinylation. Force is applied to the collagen by modulating the height of the permanent magnets above the flow cell (Supporting Information).
Control experiments were performed to ensure specific attachment of the beads to the coverslip surface via collagen trimers. We systematically removed the attachment moieties one at a time and quantified the number of attached beads. A significant number of beads were observed to be attached to the coverslip surface only when both antibody and biotinylated collagen were present (Supporting Information). We further probed the specificity of attachment by quantifying the number of attached beads as a function of collagen concentration. The number of surface-bound beads per field of view increased approximately linearly with increasing collagen concentration, demonstrating that the availability of collagen trimers limited the recruitment of beads, and implying a preponderance of attachments through single trimers (Supporting Information). We observe that a single, rate-limiting step precedes MMP-1 mediated bead detachment (see Results). This observation provides independent confirmation that the beads are tethered by single trimers.
To confirm that bead detachment was the result of collagen proteolysis, we characterized the effect of both MMP-1 and Clostridium collagenase on both the collagen antibody and streptavidin using SDS PAGE (Supporting Information), and found that protease cuts neither the antibody nor the streptavidin (Supporting Information). We also monitored bead detachment as a function of applied force in the absence of any protease (Supporting Information). The slow detachment rate observed at higher forces (15 pN) in the absence of proteinase was comparable to the rate extrapolated for force-mediated biotin-streptavidin dissociation,46 and is small compared to the proteinase-mediated detachment rates observed at these forces.
Prior to proteolysis, the flow cell was introduced into the magnetic trap under low force (~1–2 pN) to remove loose beads. Any beads that detached were washed out with 1× PBS. To initiate the proteolysis experiments, either MMP-1 (3 µM) or Clostridium collagenase (0.9 units ml−1) was introduced into the flow cell. Several fields of view were sampled at each time point in order to observe an adequate number of tethered beads, yielding 100–1000 beads observed per experiment. Measurements were taken until no further bead detachment was observed.
Results
In our assay collagen cleavage results in bead detachment from the coverslip, as monitored using brightfield microscopy. A minority of beads remain attached to the coverslip even at long times. These beads were considered to be permanently and non-specifically attached to the coverslip (Supporting Information). Excluding non-specifically attached beads, the fraction of beads remaining at the coverslip surface is well-described by a single exponential:
| eqn 1 |
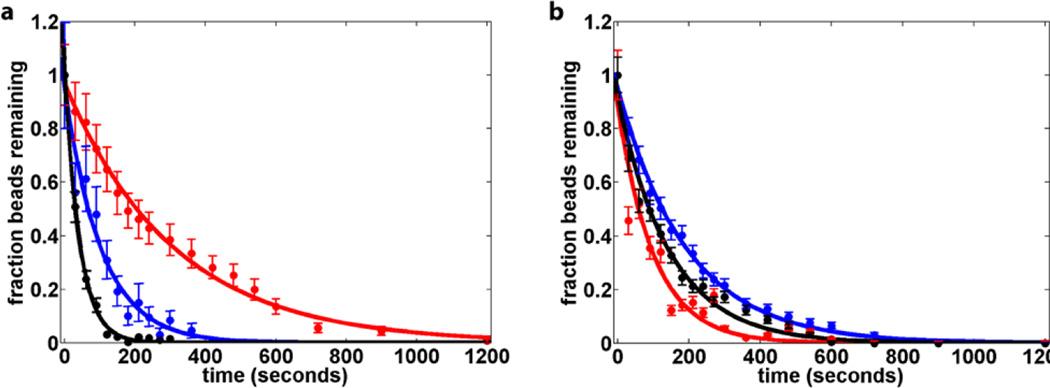
Where ft is the fraction of beads at time t, f0 is the fraction of beads at time t0, and k is the bead detachment rate in s−1 (Figure 2). The experiment was repeated for forces between 0.2 pN and 16.5 pN with 3 µM MMP-1. 16.5 pN of applied force leads to an 8-fold increase in the observed proteolysis rate. The observed rates, kobs, are well described by the Bell equation47 (Figure 3):
| eqn 2 |
Figure 2.
Single-molecule collagen proteolysis kinetics. (a) The cleavage rate of heterotrimeric collagen by MMP-1 increases upon application of force (red 0.25 pN, blue 10.7 pN, black 16.7 pN). (b) The rate of proteolysis by Clostridium collagenase remains essentially unaltered upon the application of force (red 0.25 pN, blue 5.2 pN, black 15pN).
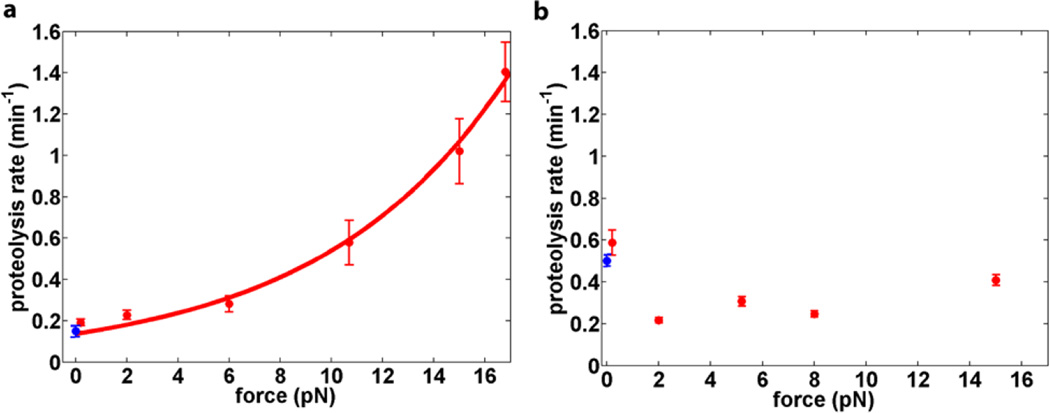
Figure 3.
Effect of force on collagen heterotrimer proteolysis. (a) MMP-1; (b) Clostridium collagenase. Collagen proteolysis by MMP-1 (a) is fit to the Bell equation, yielding a characteristic distance D = 0.57±0.01 nm. The proteolysis rate measured in bulk solution (blue; see Supporting Information), and closely matches the extrapolated single-molecule rate at zero force. Proteolysis by Clostridium collagenase (b) does not show obvious force dependence, with D ≈ 0 n m. The bulk proteolysis rate (blue) is consistent with rates measured in the single-molecule assay. Clostridium collagenase and MMP-1 concentrations were used such that the proteolysis rates at zero force are roughly comparable.
Equivalently, eqn. 2 can be recast in terms of D:
| eqn 3 |
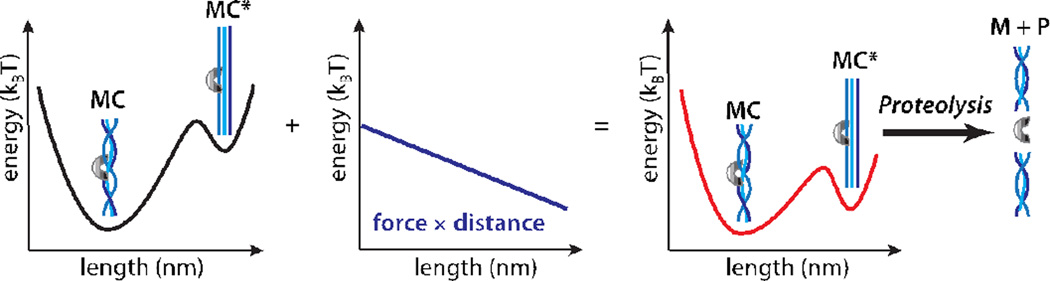
Here F is the applied load, D is a distance parameter that describes the degree to which applied load alters the observed rate constant, and kBT is the thermal energy. Although it represents a simplification of the complex dynamics that characterize biological macromolecules, eqn. 2 has proven successful in describing the effect of mechanical load on nucleic acid hairpin unfolding,48–50 protein-protein51 and protein-ligand interactions,46,51–53 protein unfolding,54,55 and motor protein kinetics.56–60 In general, the magnitude of D corresponds to the size of a change in conformation that accompanies the rate-limiting step. In the case of collagen proteolysis, a reasonable interpretation of D is that it reflects the change in collagen length that accompanies proteolysis. Eqn. 2 thus relates observed kinetic rates to the effect of mechanical load on the underlying energetic landscape (Figure 4).
Figure 4.
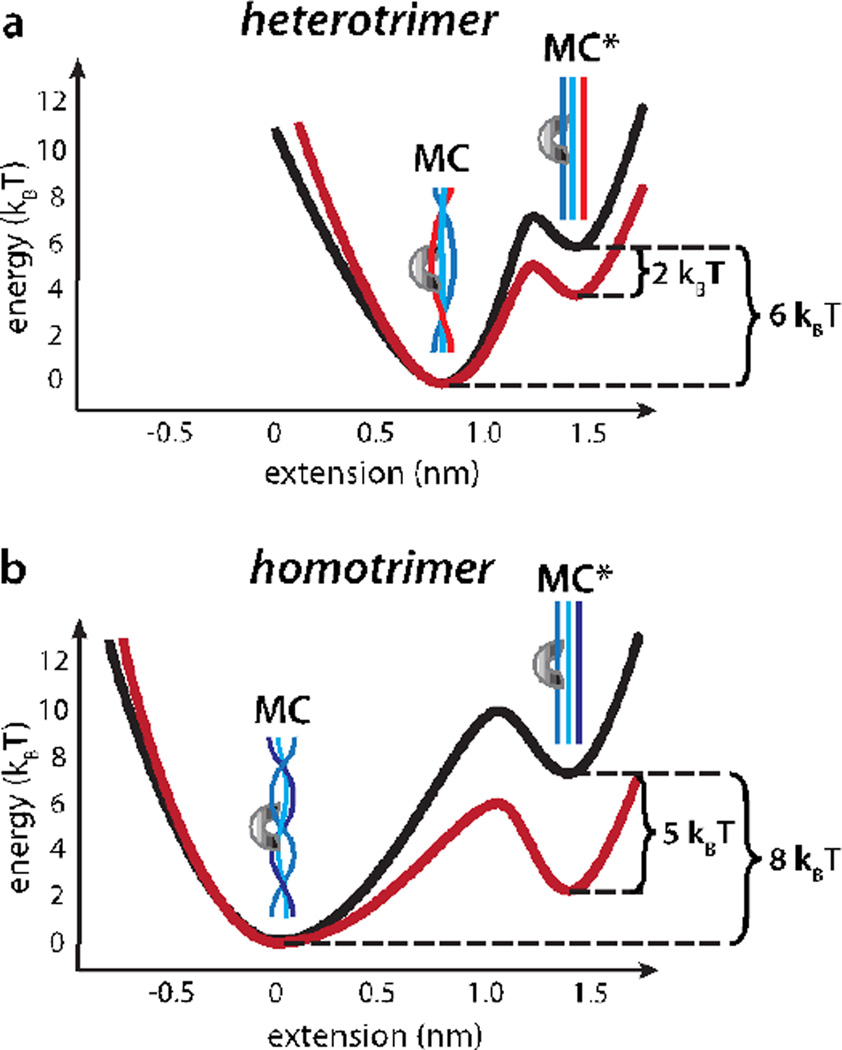
Energy landscapes illustrating the effect of force on collagen proteolysis by MMP-1. The MMP-1/collagen complex exists in an equilibrium between the MC and stretched MC* states (left). The affinity of both states for MMP-1 is similar, but MMP-1 preferentially cleaves MC* due to the increased accessibility of the individual collagen strands. Because the overall length increases in MC*, application of an external load adds a biasing energetic potential (middle), that stabilizes MC*(right), thus facilitating proteolysis.
The data were fit to eqn 2 to yield D = 0.57 ± 0.01 nm and kF=0= 0.14 ± 0.01 min−1. The value of kF=0 is identical to the bead detachment rate measured at 0.25 pN within the error of the measurement. Moreover, the fit value for kF=0 is also in good agreement (within experimental error) with the rate extrapolated from the bulk proteolysis measurements in solution (0.15 ± 0.01 min−1; Supporting Information). In the case of MMP-1, the single-exponential kinetics that we observe indicate that a single, rate-determining step governs the overall rate of proteolysis, consistent with selective cleavage at the MMP-1 recognition site.37
We performed analogous experiments with crude collagenase from Clostridium histolyticum. The application of force does not appreciably alter proteolysis rates for this enzyme (Figure 2; Figure S7). Measurement of collagen cleavage in a bulk solution experiment yields an extrapolated cutting rate of 0.50 ± 0.03 min−1, which is in good agreement with rates observed in our single molecule assay at 0.25 pN force (0.58 ± 0.07 min−1).
Discussion
Previous studies have shown that collagen trimer proteolysis by MMP-1 follows Michaelis-Menten-type kinetics.9 The single exponential kinetics observed in our data suggest the presence of a single rate-limiting step. While force dramatically increases cutting rates, it does not appreciably alter the apparent affinity of MMP-1 for the collagen trimer.34 We therefore infer that the force-dependent step follows MMP-1 binding, but precedes strand cleavage. Although more comprehensive kinetic models are possible,34 a kinetic framework with one force-sensitive step is adequate to describe the collagen proteolysis rates we observe in this and our previous study:
Here M is MMP-1, C is the collagen trimer, MC is the initial collagen-MMP complex, MC* is the cleavage competent complex, and P is the proteolyzed product. The discrete helical and stretched conformations assumed here are consistent with the generally cooperative nature of collagen unfolding.61 The kinetic model above implies that a rapid and force-dependent conformational equilibrium (K2(F), defined as k2(F)/k−2(F)), likely corresponding to the unwinding of the MMP-1 binding site, precedes a slower cutting step (kcut). Assuming a quasi-steady-state equilibrium (d[MC]/dt and d[MC*]/dt ≈ 0) and sub-saturating MMP concentrations, the rate of collagen disappearance is:
| eqn 4 |
Here KD is a dissociation constant defined as k−1/k1. Eqn. 4 yields the apparent first-order rate constant kobs:
| eqn 5 |
The force-dependent equilibrium K2(F) can be expressed as:
| eqn 6 |
where K2(F=0) is the equilibrium constant in the absence of force and D is the increase in length accompanying the transition from MC to MC*. Substitution into eqn 5 yields
| eqn 7 |
Eqn 7 is identical to eqn 2 with kF=0 = kcutK2(F=0)[M]/KD. In summary, mechanical force shifts the equilibrium towards the proteolytically vulnerable conformation by increasing the fraction of collagen trimers in the stretched, C* state. This results because mechanical load tilts the energy landscape by subtracting an energetic term of F×D (Figure 4).
In our earlier report, we observed that proteolysis of a homotrimeric collagen model peptide is accompanied by a force-sensitive increase in length (D) of 1.4 ± 0.25 nm, in excellent agreement with that expected for the complete unwinding of the MMP-1 recognition site in the collagen triple helix.34 In contrast, cleavage of heterotrimeric collagen is accompanied by a length increase of D = 0.57 ± 0.01 nm. Previous results show that the MMP-1 binding site in heterotrimeric collagen is both thermodynamically less stable and more susceptible to proteolysis than that of homotrimeric collagen.9 Computational modeling and NMR measurements of conformational dynamics likewise indicate that the heterotrimeric MMP recognition site is destabilized relative to the canonical collagen triple helix.21,62 The smaller D, relative structural instability, and relative proteolytic susceptibility of the heterotrimeric MMP-1 recognition site support the conclusion that it is approximately half unwound even in the absence of applied load (Figure 5).
Figure 5.
Energetic landscape for collagen proteolysis by MMP-1. (a) The heterotrimeric MMP-1 binding site is partially unwound even in the absence of applied load. Approximately 6 times the thermal energy (6 kBT) separates the MC and MC* states (black; see text). A 0.57 nm increase in length accompanies the transition from MC to MC*. 15 pN external load stabilizes MC* by 8.6 pN·nm, or ~2 kBT (red). (b) Approximately 8 kBT separate MC and MC* for the homotrimeric MMP-1 recognition site (black; see text). A 1.4 nm length increase accompanies the transition to the stretched state, resulting in ~5 kBT stabilization of MC* at 15 pN of load (red).
NMR,63 bulk kinetics,64 and computational modeling21,62 all suggest that the energy difference between the partially wound and fully disrupted heterotrimeric MMP-1 binding site is approximately 10–15 kJ mol−1, or ~6 kBT (Figure 5a, black trace). Since the apparent affinity of MMP-1 for trimeric collagen is not noticeably load-dependent, we assume that ~6 kBT also separates MC and MC* for the heterotrimeric MMP-1 recognition site. Bulk kinetics show that the cleavage of homotrimeric collagen I is approximately 10-fold slower than the heterotrimer.9,22 Assuming that this difference in rates reflects an increase in unwinding energy (i.e. kcut is the same for homo- and heterotrimers but K2 is not) the additional energy required to unwind the homo- vs. heterotrimer recognition site is ΔΔG = kBTln(K2,hetero/K2,homo), where K2,homo and K2,hetero are the unwinding equilibrium constants for the homo- and heterotrimers. ΔΔG thus calculated is ~2 kBT, implying that approximately 8 kBT separates the MC and MC* states for the homotrimer (Figure 5b, black trace). This calculation is consistent with the greater thermodynamic stability of the homotrimeric MMP-1 recognition site.65
The present model explains why the homotrimeric MMP-1 recognition site is less susceptible to proteolysis in the absence of force, but much more sensitive to the effect of mechanical load. The homotrimeric recognition site is fully wound at equilibrium, thus making it more difficult for MMP-1 to isolate and cleave one strand at a time. In contrast, our data suggest that the heterotrimer is already partially unwound, thus making local disruption of the triple helix by MMP-1 more thermodynamically feasible. However, a larger increase in length accompanies complete unwinding of the homotrimer versus the heterotrimer (Figure 5). Thus, a force of 15 pN stabilizes the unwound state of the homotrimer by 15 pN× 1.4 nm, or 5 kBT (Figure 5b; red trace). In contrast, a comparable load stabilizes the unwound heterotrimeric MMP-1 recognition site by only 15 pN×0.57nm, or 2 kBT (Figure 5a, red trace). In summary, although homotrimeric collagen is more stable than heterotrimeric collagen under the absence of load, it is much more sensitive to the effect of mechanical stretch.
While applied strain may influence proteolysis by Clostridium collagenase in other circumstances, in our assay the rate of collagen proteolysis by Clostridium collagenase has no noticeable dependence on applied force, thus suggesting that negligible changes in collagen length accompany proteolysis. In contrast to MMP-1, Clostridium collagenase has low sequence specificity (Supporting Information).39 In addition, a recent crystal structure for Clostridium collagenase G suggests that this enzyme may catalyze conformational transitions in the context of the full collagen fibril that are not probed in our assay, for example, a fibril “squeezing” mechanism where in collagenase G rearranges the collagen microfibril to capture a single trimer prior to proteolysis.66 These observations suggest that Clostridium collagenase cuts collagen independent of the unwinding transition that is apparently a prerequisite for proteolysis by MMP-1. This scenario is consistent with the function of Clostridium collagenases, which is to dissolve tissue in the context of bacterial infection.
Camp et al. report that force decreases the rate of collagen trimer proteolysis by Clostridium collagenase approximately 10 fold.35 Although the two assays are ostensibly similar, it is conceivable that subtle differences in experimental conditions may account for the difference in outcomes. For example, the Clostridium collagenase used in both our study and by Camp et al. consists of a cocktail of several enzymes with distinct activities. Further investigation may prove interesting in understanding both collagen and Clostridium collagenase structural dynamics.
A large body of previous work, including single-molecule studies,34 NMR data,36 x-ray spectroscopy67 bulk enzymology9,20,37,64,68 and computational modeling21,62 indicate that disruption of the collagen trimer likely precedes proteolysis by MMP-1. However, the majority of previous studies on bulk collagenous materials indicate that mechanical load results in a modest, ~2-fold reduction in proteolytic degradation rates,24,26,30 whereas our single-molecule measurements observe an opposite effect. Two possible explanations may account for this apparent discrepancy. At the single molecule level, effects stemming from changes in kinetic rate constants dominate. However, at the bulk scale, diffusive transport can play a decisive role in modulating the rate of enzymatic degradation.69 For example, mechanical load leads to a ~10-fold reduction in the rate of fibrin gel degradation by plasmin.70 This decrease in proteolytic susceptibility likely reflects changes in the diffusive transport of plasmin rather than changes in the proteolytic susceptibility of the fibrin molecules as a function of applied strain.71 It is possible that a similar mechanism might hold for collagen-based materials. Indeed, previous investigators have observed marked changes in diffusive transport in intervertebral disk72,73 and bovine coccygeal annulus fibrosus74 in response to externally applied strains. We note, however, that indirect measurements did not demonstrate changes in diffusive transport for Clostridium collagenase in response to external load.24,26
Alternatively, the collagen conformational dynamics present in fully assembled collagen I fibrils may fundamentally differ from those of the isolated trimer. In support of this model, mechanical tension appears to protect isolated collagen I fibrils from degradation by Clostridium collagenase.75 Clostridium collagenase G has been proposed to actively pull individual collagen trimers away from microfibrils.66 Such a mechanism is distinct from the force-dependent conformational changes that we observe, and could thus offer an elegant explanation for the differences between our single-molecule measurements and previous work in bulk samples. Understanding how mechanical strain influences ECM deposition and degradation remains a fascinating challenge for the future.
Conclusions
The information afforded by our single-molecule assay has allowed us to construct a biophysical model for the cleavage of collagen by MMP-1 and Clostridium collagenase. Both enzymes cleave collagen with a single rate-limiting step under the conditions assayed. This observation implies that the cleavage of one strand, likely the first to be cut, is rate limiting, and that both enzymes likely cut the trimer in a single encounter. Force markedly increases the rate of MMP-1 catalyzed cleavage for both homo- and heterotrimeric collagen, indicating that a stretching structural transition precedes proteolysis by this enzyme. This stretching transition is consistent with the proposal that collagen unwinds prior to proteolysis by MMPs.20
Our results suggest that MMP-1 and collagen I have mutually evolved to regulate proteolysis based on the narrow geometry of the MMP-1 active site and the differing accessibilities of the homo- and heterotrimeric recognition sites (Figure 5). This may account for the differing effects of mechanical load on the proteolysis of isolated collagen trimers vs. intact collagen I fibrils. At the fibrillar level, applied load may hinder a conformational transition that precedes proteolysis, thus protecting load-bearing collagen from degradation (see Discussion).
It is possible that the sensitivity to proteolytic degradation that we observe for isolated trimers may have physiological significance. Many collagens, for example collagen IV, are trimers in their physiological state.76 Fibrillar collagens, for instance collagen I, fray to produce partially exposed trimers during breakdown.77,78 Membrane-bound MMP colocalizes with integrins,79 and colocalization is required for fibrillar collagen80 and localized gelatin, i.e. partially denatured collagen, degradation.81 Single integrins can exert >20 pN of force on the ECM.82 A combination of cell-generated traction forces and MMP activity may thus help to degrade isolated collagen trimers (and possibly other ECM molecules) in the context of ECM remodeling. The involvement of MMPs in cancer and heart disease, both of which exhibit marked ECM remodeling during their progression, is consistent with this hypothesis. The involvement of MMPs in cancer metastasis in particular has led to a concerted effort to develop MMP inhibitors as potential pharmaceuticals.83,84 Small molecules which influence collagen structure, rather than MMP activity, may offer an alternate means of modulating ECM remodeling in a wide variety of disease states.
Supplementary Material
Acknowledgments
The authors thank Jack Chai for help with magnetic tweezers calibration and James Spudich for the loan of microscopy equipment. This research was funded by the Stanford Graduate Fellowship (ASA), the NIH New Innovator Award 1DP2OD007078-01 (ARD), and the Burroughs-Wellcome Career Award at the Scientific Interface (ARD).
ABBREVIATIONS
- MMP-1
Matrix Metalloproteinase-1
Footnotes
ASSOCIATED CONTENT
Supporting Information. Detailed information on materials and methods used. This information is available free of charge via the Internet at http://pubs.acs.org/.
REFERENCES
- 1.Wozniak MA, Chen CS. Nat. Rev. Mol. Cell. Biol. 2009;10:34. doi: 10.1038/nrm2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krieg M, Arboleda-Estudillo Y, Puech PH, Kafer J, Graner F, Muller DJ, Heisenberg CP. Nat. Cell. Biol. 2008;10:429. doi: 10.1038/ncb1705. [DOI] [PubMed] [Google Scholar]
- 3.Sherwood DR. Trends. Cell. Biol. 2006;16:250. doi: 10.1016/j.tcb.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Latimer A, Jessen JR. Matrix. Biol. 2010;29:89. doi: 10.1016/j.matbio.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Ingber DE. Semin. Cancer. Biol. 2008;18:356. doi: 10.1016/j.semcancer.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiong W, Knispel R, MacTaggart J, Greiner TC, Weiss SJ, Baxter BT. J. Biol. Chem. 2009;284:1765. doi: 10.1074/jbc.M806239200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hahn C, Schwartz MA. Nat. Rev. Mol. Cell. Biol. 2009;10:53. doi: 10.1038/nrm2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Lullo GA, Sweeney SM, Korkko J, Ala-Kokko L, San Antonio JD. J. Biol. Chem. 2002;277:4223. doi: 10.1074/jbc.M110709200. [DOI] [PubMed] [Google Scholar]
- 9.Han S, Makareeva E, Kuznetsova NV, DeRidder AM, Sutter MB, Losert W, Phillips CL, Visse R, Nagase H, Leikin S. J. Biol. Chem. 2010;285:22276. doi: 10.1074/jbc.M110.102079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jimenez SA, Bashey RI, Benditt M, Yankowski R. Biochem. Biophys. Res. Commun. 1977;78:1354. doi: 10.1016/0006-291x(77)91441-3. [DOI] [PubMed] [Google Scholar]
- 11.Rojkind M, Giambrone MA, Biempica L. Gastroenterology. 1979;76:710. [PubMed] [Google Scholar]
- 12.Narayanan AS, Page RC, Meyers DF. Biochemistry. 1980;19:5037. doi: 10.1021/bi00563a016. [DOI] [PubMed] [Google Scholar]
- 13.Ehrlich HP, Brown H, White BS. Biochem. Med. 1982;28:273. doi: 10.1016/0006-2944(82)90080-1. [DOI] [PubMed] [Google Scholar]
- 14.Makareeva E, Han S, Vera JC, Sackett DL, Holmbeck K, Phillips CL, Visse R, Nagase H, Leikin S. Cancer Res. 2010;70:4366. doi: 10.1158/0008-5472.CAN-09-4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moro L, Smith BD. Arch. Biochem. Biophys. 1977;182:33. doi: 10.1016/0003-9861(77)90280-6. [DOI] [PubMed] [Google Scholar]
- 16.Shapiro FD, Eyre DR. J. Natl. Cancer Inst. 1982;69:1009. [PubMed] [Google Scholar]
- 17.Yamagata S, Yamagata T. J. Biochem. 1984;96:17. doi: 10.1093/oxfordjournals.jbchem.a134809. [DOI] [PubMed] [Google Scholar]
- 18.Asokan R, Puvanakrishnan R, Ravichandran LV, Kokila V, Reddy GK, Dhar SC. Mol. Cell. Biochem. 1993;121:99. doi: 10.1007/BF00925968. [DOI] [PubMed] [Google Scholar]
- 19.Page-McCaw A, Ewald AJ, Werb Z. Nat. Rev. Mol. Cell. Biol. 2007;8:221. doi: 10.1038/nrm2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung L, Dinakarpandian D, Yoshida N, Lauer-Fields JL, Fields GB, Visse R, Nagase H. EMBO J. 2004;23:3020. doi: 10.1038/sj.emboj.7600318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nerenberg PS, Stultz CM. J. Mol. Biol. 2008;382:246. doi: 10.1016/j.jmb.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 22.Minond D, Lauer-Fields JL, Nagase H, Fields GB. Biochemistry. 2004;43:11474. doi: 10.1021/bi048938i. [DOI] [PubMed] [Google Scholar]
- 23.Willett TL, Labow RS, Avery NC, Lee JM. Ann. Biomed. Eng. 2007;35:1961. doi: 10.1007/s10439-007-9375-x. [DOI] [PubMed] [Google Scholar]
- 24.Zareian R, Church KP, Saeidi N, Flynn BP, Beale JW, Ruberti JW. Langmuir. 2010;26:9917. doi: 10.1021/la100384e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robitaille MC, Zareian R, DiMarzio CA, Wan KT, Ruberti JW. Interface Focus. 2011;1:767. doi: 10.1098/rsfs.2011.0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nabeshima Y, Grood ES, Sakurai A, Herman JH. J. Orthop. Res. 1996;14:123. doi: 10.1002/jor.1100140120. [DOI] [PubMed] [Google Scholar]
- 27.Lotz JC, Hadi T, Bratton C, Reiser KM, Hsieh AH. Eur. Spine J. 2008;17:1149. doi: 10.1007/s00586-008-0721-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wyatt KE, Bourne JW, Torzilli PA. J. Biomech. Eng. 2009;131:051004. doi: 10.1115/1.3078177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruberti JW, Hallab NJ. Biochem. Biophys. Res. Commun. 2005;336:483. doi: 10.1016/j.bbrc.2005.08.128. [DOI] [PubMed] [Google Scholar]
- 30.Flynn BP, Bhole AP, Saeidi N, Liles M, Dimarzio CA, Ruberti JW. PLoS One. 2010;5:e12337. doi: 10.1371/journal.pone.0012337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhole AP, Flynn BP, Liles M, Saeidi N, Dimarzio CA, Ruberti JW. Philos. Transact. A. Math. Phys. Eng. Sci. 2009;367:3339. doi: 10.1098/rsta.2009.0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hadi MF, Sander EA, Ruberti JW, Barocas VH. Mech. Mater. 2012;44:72. doi: 10.1016/j.mechmat.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grytz R, Sigal IA, Ruberti JW, Meschke GJ, Downs C. Mech. Mater. 2012;44:99. doi: 10.1016/j.mechmat.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adhikari AS, Chai J, Dunn AR. J. Am. Chem. Soc. 2011;133:1686. doi: 10.1021/ja109972p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Camp RJ, Liles M, Beale J, Saeidi N, Flynn BP, Moore E, Murthy SK, Ruberti JW. J. Am. Chem. Soc. 2011;133:4073. doi: 10.1021/ja110098b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bertini I, Fragai M, Luchinat C, Melikian M, Toccafondi M, Lauer JL, Fields GB. J. Am. Chem. Soc. 2012;134:2100. doi: 10.1021/ja208338j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Minond D, Lauer-Fields JL, Cudic M, Overall CM, Pei D, Brew K, Moss ML, Fields GB. Biochemistry. 2007;46:3724. doi: 10.1021/bi062199j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mandl I, Zipper H, Ferguson LT. Arch. Biochem. Biophys. 1958;74:465. doi: 10.1016/0003-9861(58)90017-1. [DOI] [PubMed] [Google Scholar]
- 39.Van Wart HE, Steinbrink DR. Anal. Biochem. 1981;113:356. doi: 10.1016/0003-2697(81)90089-0. [DOI] [PubMed] [Google Scholar]
- 40.Weinberg M, Seguin P. C. R. Acad. Sci. Paris. 1916;163 [Google Scholar]
- 41.Stevens DL, Aldape MJ, Bryant AE. Anaerobe. 2011 [Google Scholar]
- 42.Werkmeister JA, Ramshaw JA, Ellender G. Eur. J. Biochem. 1990;187:439. doi: 10.1111/j.1432-1033.1990.tb15323.x. [DOI] [PubMed] [Google Scholar]
- 43.Sletten EM, Bertozzi CR. Angew. Chem. Int. Edit. 2009;48:6974. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gilmore JM, Scheck RA, Esser-Kahn AP, Joshi NS, Francis MB. Angew. Chem. Int. Edit. 2006;45:5307. doi: 10.1002/anie.200600368. [DOI] [PubMed] [Google Scholar]
- 45.Christman KL, Broyer RM, Tolstyka ZP, Maynard HD. J. Mater. Chem. 2007;17:2021. [Google Scholar]
- 46.Danilowicz C, Greenfield D, Prentiss M. Anal. Chem. 2005;77:3023. doi: 10.1021/ac050057+. [DOI] [PubMed] [Google Scholar]
- 47.Bell GI. Science. 1978;200:618. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 48.Dudko OK, Hummer G, Szabo A. Proc. Natl. Acad. Sci. U S A. 2008;105:15755. doi: 10.1073/pnas.0806085105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woodside MT, Behnke-Parks WM, Larizadeh K, Travers K, Herschlag D, Block SM. Proc. Natl. Acad. Sci. U S A. 2006;103:6190. doi: 10.1073/pnas.0511048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liphardt J, Onoa B, Smith SB, Tinoco I, Jr, Bustamante C. Science. 2001;292:733. doi: 10.1126/science.1058498. [DOI] [PubMed] [Google Scholar]
- 51.Evans E. Biophys. Chem. 1999;82:83. doi: 10.1016/s0301-4622(99)00108-8. [DOI] [PubMed] [Google Scholar]
- 52.Cecconi C, Shank EA, Bustamante C, Marqusee S. Science. 2005;309:2057. doi: 10.1126/science.1116702. [DOI] [PubMed] [Google Scholar]
- 53.Chen S, Springer TA. Proc. Natl. Acad. Sci. U S A. 2001;98:950. doi: 10.1073/pnas.98.3.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Forero M, Yakovenko O, Sokurenko EV, Thomas WE, Vogel V. PLoS Biol. 2006;4:e298. doi: 10.1371/journal.pbio.0040298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bjornham O, Axner O. Biophys. J. 2010;99:1331. doi: 10.1016/j.bpj.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Purcell TJ, Morris C, Spudich JA, Sweeney HL. Proc. Natl. Acad. Sci. U S A. 2002;99:14159. doi: 10.1073/pnas.182539599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Purcell TJ, Sweeney HL, Spudich JA. Proc. Natl. Acad. Sci. U S A. 2005;102:13873. doi: 10.1073/pnas.0506441102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chuan P, Spudich JA, Dunn AR. J. Mol. Biol. 2011;405:105. doi: 10.1016/j.jmb.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dunn AR, Chuan P, Bryant Z, Spudich JA. Proc. Natl. Acad. Sci. U S A. 2010;107:7746. doi: 10.1073/pnas.1002430107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dunn AR, Spudich JA. Nat. Struct. Mol. Biol. 2007;14:246. doi: 10.1038/nsmb1206. [DOI] [PubMed] [Google Scholar]
- 61.Frank S, Boudko S, Mizuno K, Schulthess T, Engel J, Bachinger HP. J. Biol. Chem. 2003;278:7747. doi: 10.1074/jbc.C200698200. [DOI] [PubMed] [Google Scholar]
- 62.Stultz CM. J. Mol. Biol. 2002;319:997. doi: 10.1016/S0022-2836(02)00421-7. [DOI] [PubMed] [Google Scholar]
- 63.Fiori S, Sacca B, Moroder L. J. Mol. Biol. 2002;319:1235. doi: 10.1016/S0022-2836(02)00365-0. [DOI] [PubMed] [Google Scholar]
- 64.Salsas-Escat R, Nerenberg PS, Stultz CM. Biochemistry. 2010;49:4147. doi: 10.1021/bi9021473. [DOI] [PubMed] [Google Scholar]
- 65.Chang SW, Flynn BP, Ruberti JW, Buehler MJ. Biomaterials. 2012;33:3852. doi: 10.1016/j.biomaterials.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eckhard U, Schonauer E, Nuss D, Brandstetter H. Nat. Struct. Mol. Biol. 2011;18:1109. doi: 10.1038/nsmb.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grossman M, Born B, Heyden M, Tworowski D, Fields GB, Sagi I, Havenith M. Nat. Struct. Mol. Biol. 2011;18:1102. doi: 10.1038/nsmb.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.O'Farrell TJ, Guo R, Hasegawa H, Pourmotabbed T. Biochemistry. 2006;45:15411. doi: 10.1021/bi060849d. [DOI] [PubMed] [Google Scholar]
- 69.Tzafriri AR, Bercovier M, Parnas H. Biophys. J. 2002;83:776. doi: 10.1016/S0006-3495(02)75208-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Varju I, Sotonyi P, Machovich R, Szabo L, Tenekedjiev K, Silva MM, Longstaff C, Kolev K. J. Thromb. Haemost. 2011;9:979. doi: 10.1111/j.1538-7836.2011.04203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Adhikari AS, Mekhdjian AH, Dunn AR. Biomacromolecules. 2012;13:499. doi: 10.1021/bm2015619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jackson A, Yao H, Brown MD, Yong Gu W. Spine (Phila Pa 1976) 2006;31:2783. doi: 10.1097/01.brs.0000245842.02717.1b. [DOI] [PubMed] [Google Scholar]
- 73.Chiu EJ, Newitt DC, Segal MR, Hu SS, Lotz JC, Majumdar S. Spine (Phila Pa 1976) 2001;26:E437. doi: 10.1097/00007632-200110010-00017. [DOI] [PubMed] [Google Scholar]
- 74.Travascio F, Gu WY. Ann. Biomed. Eng. 2011;39:53. doi: 10.1007/s10439-010-0138-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Flynn BP, Tilburey GE, Ruberti JW. Biomech. Model. Mechanobiol. 2012 doi: 10.1007/s10237-012-0399-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Paulsson M. Crit. Rev. Biochem. Mol. Biol. 1992;27:93. doi: 10.3109/10409239209082560. [DOI] [PubMed] [Google Scholar]
- 77.Asuwa N. Virchows Arch. B. Cell. Pathol. Incl. Mol. Pathol. 1988;55:345. doi: 10.1007/BF02896593. [DOI] [PubMed] [Google Scholar]
- 78.Woolley DE, Glanville RW, Crossley MJ, Evanson JM. Eur. J. Biochem. 1975;54:611. doi: 10.1111/j.1432-1033.1975.tb04173.x. [DOI] [PubMed] [Google Scholar]
- 79.Gonzalo P, Moreno V, Galvez BG, Arroyo AG. Biofactors. 2010;36:248. doi: 10.1002/biof.99. [DOI] [PubMed] [Google Scholar]
- 80.Wolf K, Wu YI, Liu Y, Geiger J, Tam E, Overall C, Stack MS, Friedl P. Nat. Cell Biol. 2007;9:893. doi: 10.1038/ncb1616. [DOI] [PubMed] [Google Scholar]
- 81.Wang Y, McNiven MA. J. Cell. Biol. 2012;196:375. doi: 10.1083/jcb.201105153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li F, Redick SD, Erickson HP, Moy VT. Biophys. J. 2003;84:1252. doi: 10.1016/S0006-3495(03)74940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Overall CM, Kleifeld O. Nat. Rev. Cancer. 2006;6:227. doi: 10.1038/nrc1821. [DOI] [PubMed] [Google Scholar]
- 84.Lia NG, Shib ZH, Tang YP, Duan JA. Curr. Med. Chem. 2009;16:3805. doi: 10.2174/092986709789178037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.