Abstract
Transcriptional signal cointegrators associate with transcription factors or nuclear receptors and coregulate tissue-specific gene transcription. We report on recessive loss-of-function mutations in two genes (TRIP4 and ASCC1) that encode subunits of the nuclear activating signal cointegrator 1 (ASC-1) complex. We used autozygosity mapping and whole-exome sequencing to search for pathogenic mutations in four families. Affected individuals presented with prenatal-onset spinal muscular atrophy (SMA), multiple congenital contractures (arthrogryposis multiplex congenita), respiratory distress, and congenital bone fractures. We identified homozygous and compound-heterozygous nonsense and frameshift TRIP4 and ASCC1 mutations that led to a truncation or the entire absence of the respective proteins and cosegregated with the disease phenotype. Trip4 and Ascc1 have identical expression patterns in 17.5-day-old mouse embryos with high expression levels in the spinal cord, brain, paraspinal ganglia, thyroid, and submandibular glands. Antisense morpholino-mediated knockdown of either trip4 or ascc1 in zebrafish disrupted the highly patterned and coordinated process of α-motoneuron outgrowth and formation of myotomes and neuromuscular junctions and led to a swimming defect in the larvae. Immunoprecipitation of the ASC-1 complex consistently copurified cysteine and glycine rich protein 1 (CSRP1), a transcriptional cofactor, which is known to be involved in spinal cord regeneration upon injury in adult zebrafish. ASCC1 mutant fibroblasts downregulated genes associated with neurogenesis, neuronal migration, and pathfinding (SERPINF1, DAB1, SEMA3D, SEMA3A), as well as with bone development (TNFRSF11B, RASSF2, STC1). Our findings indicate that the dysfunction of a transcriptional coactivator complex can result in a clinical syndrome affecting the neuromuscular system.
Keywords: TRIP4, ASCC1, spinal muscular atrophy, arthrogryposis multiplex congenita, respiratory distress, bone fractures, neuromuscular unit, exome sequencing, zebrafish model
Introduction
Arthrogryposis multiplex congenita (AMC) comprises a heterogeneous group of disorders that has a prevalence of 8.5/100,000 individuals and an incidence of 1/3,000–5,000 births and shares multiple congenital joint contractures as a defining feature.1, 2 AMC is caused by limited fetal movement (akinesia) resulting from conditions that impede intrauterine neuromuscular development.3 Much has been learned from AMC-causing gene defects about the development of the human neuromuscular system. Mutations can affect the genes of structural proteins of the contractile apparatus,4 trophic factors, proteins implicated in pathfinding5, 6 and myelination,7 receptor proteins,8, 9 or proteins involved in transmitter release.10
In six out of seven individuals from our study, AMC occurred together with congenital bone fractures. From the epidemiologic view, congenital bone fractures are an extremely rare condition and are mainly due to trauma or osteogenesis imperfecta.11 A retrospective birth register study of 343,941 newborns from Sweden uncovered 68 cases of multiple congenital contractures (incidence ∼1/5,100). Of these 68 children, 14 had amyoplasia (incidence ∼1/45,000), 18 had different neuromuscular disorders (incidence ∼1/19,100), and only 2 had perinatal fractures (incidence ∼1/172,000), although it was not mentioned to what group the latter two individuals belonged. Hall et al. analyzed the clinical symptoms of amyoplasia, the most frequent subform of AMC, and found a much higher frequency of congenital bone fractures among these children, in the range of ten percent.12 Other rare disorders in which AMC associates with congenital bone fractures occur with mutations in UBA1 (formerly UBE1, X-linked spinal muscular atrophy type 2 [SMAX2] [MIM: 301830]),13, 14 FKBP10 (Bruck syndrome type 1 [MIM: 259450]),15 and ERBB3 (lethal congenital contracture syndrome type 2 [LCCS2] [MIM: 607598])16 and in several forms of nemaline myopathy.17, 18, 19 Although the most severe form of infantile spinal muscular atrophy (SMA type 1, Werdnig Hoffmann disease [MIM: 253300]) is often associated with fetal akinesia, congenital bone fractures seem to be a rarity in this condition. In these children, fractures might occur later in life due to inactivity. We only found a single publication in which a case of SMA type 1 plus congenital bone fractures had been verified on the molecular level through detection of an SMN1 deletion.20 In most published cases of SMA plus congenital bone fractures, SMN1 deletions had been excluded.21, 22, 23, 24 Hence, we set out to search for mutations in other genes that might involve neuromuscular development as well as bone metabolism in three families with six affected children who had AMC plus congenital fractures. Gene mapping and whole-exome sequencing (WES) revealed mutations in two genes that encode subunits of a transcriptional signal cointegrator complex.
Transcriptional signal cointegrators associate with transcription factors or with nuclear receptors in multi-protein complexes and are able to bi-directionally affect the link between receptor and transcription machinery, either as corepressors or coactivators. They enable the functional integration of multiple transcription factors25 and thus fine-tune cell metabolism and transcription depending on environmental cues26 or provide tissue specificity.27 Over the past few years, a number of studies have shown that coactivator complexes are often bi-functional proteins that do not only coactivate transcription mediated by specific transcription factors, like nuclear hormone receptors, but also participate in pre-mRNA processing and regulation of splicing.28 The tetrameric ASC-1 transcriptional cointegrator complex is composed of the following four subunits.29 TRIP4 (thyroid receptor interacting protein 4 [MIM: 604501]) contains a conserved cysteine-rich Zn-chelating domain, which binds transcription factors,29 and a conserved C-terminal domain, which harbors a RNA-binding PUA domain30 thought to be an ancient structural motif for RNA-protein interactions. ASCC1 (ASC-1 complex, subunit 1 [MIM: 614215]) has an RNA-binding KH domain fused to a 2H RNA-phosphoesterase.29, 31 Not much is known about the ∼100 kDa subunit ASCC2 (ASC-1 complex subunit 2 [MIM: 614216]). The largest subunit of ∼200 kDa, ASCC3 (ASC-1 complex subunit 3 [MIM: 614217]), is an RNA helicase and shows paralogy to the small nuclear ribonucleoprotein 200, which is involved in RNA splicing. Hence, the ASC-1 complex is likely to be a ribonucleoprotein complex that participates in transcriptional coactivation, as well as in RNA processing events.
Methods
Ethics
Human samples were collected according to guidelines laid down in the Declaration of Helsinki in the amended version of 2013. All caretakers provided written informed consent for all investigations of the study (IRB approval of the Charité EA2/092/06). Zebrafish were raised and used in compliance with the guidelines approved by the animal care and use committee at the National Institute of Genetics (Japan).
Haplotype and Mutation Analysis
Autozygosity mapping32 was performed with members of families A, B, and D (Figures S3–S5, members indicated by an asterisk on the pedigrees in Figure 2). WES was done in three index case individuals (B.II_01, D.II_02, D.II_03), as previously described.33 DNA analyses were done with genomic DNA from blood leukocytes. For SNP analysis, we used the GeneChip Human Mapping 250K SNP Array (Affymetrix) and analyzed each separate family with the HomozygosityMapper software for autozygous regions that were only present in affected children and not in unaffected family members.32
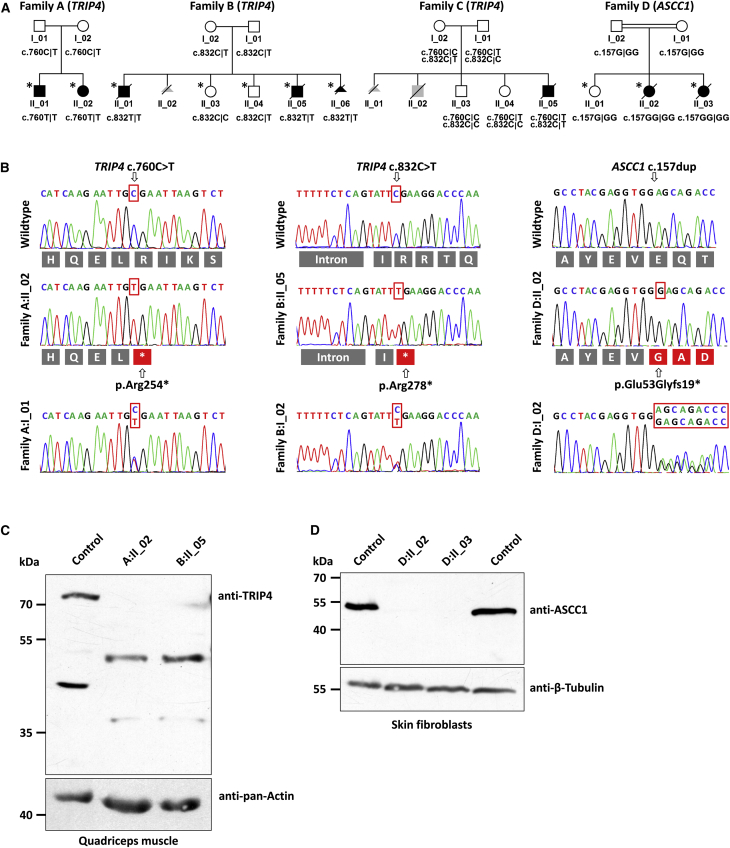
Figure 2.
Pedigrees of the Families and Molecular Genetic Findings
(A) The pedigrees of all investigated families with their respective genotypes are depicted below the symbols. Individuals marked with an asterisk were used for autozygosity mapping.
(B) Variants identified by WES were verified by Sanger sequencing for segregation in all family members. Below the electropherograms, the reading frame of the respective amino acids is provided in the three letter code. Both TRIP4 mutations resulted in a premature termination codon, whereas the ASCC1 mutation led to a frameshift with a termination codon after insertion of 19 non-original amino acids.
(C) Western blot of a muscle protein extract from two individuals with a TRIP4 mutation and from a control individual. The premature termination codons in both affected children lead to upregulation of an alternative splice isoform at ∼53 kDa that excludes both mutant positions. Anti-pan-Actin band density was used as a loading control.
(D) Western blot of a protein extract from cultured fibroblasts of both children from family D and of two control individuals. The blot demonstrates the complete absence of the ASCC1 band in the affected children. β-tubulin band density was used as a loading control.
For WES, we captured the exonic sequences by using the SeqCap EZ Human Exome Library v.3.0 (NimbleGen) and sequenced them on a HiSeq2000 (Illumina) machine. The paired-end reads were aligned to the 1000 Genomes GRCh37.p11 human reference sequence with the Burrows-Wheeler Aligner (BWA)-MEM v.0.7.134 and then fine-adjusted and called for deviations from the human reference with the Genome Analysis Toolkit (GATK) v.2.7 in all exonic ± 50 bp flanking regions.35 For coverage details, see Table S2. The resulting variants were filtered for homozygosity (e.g., the absence of more than four homozygotes in the 1000 Genomes Project or of more than 20 homozygotes in the Exome Aggregation Consortium [ExAC] database) and the presence of the variant in the autozygous interval. These variants were then assessed by the MutationTaster software for potential pathogenicity.36 Variants and their segregation were verified by bi-directional Sanger sequencing with the BigDye Terminator method (Applied Biosystems). Subsequently, we searched for TRIP4 and ASCC1 mutations in 11 unrelated children with AMC, respiratory distress, and congenital bone fractures by performing sequence analysis of the entire open reading frame and intron-exon borders of both genes; genomic primers are listed in Table S8.
Analysis of TRIP4 Splice Isoforms
In order to explain the identity of the additional bands on the TRIP4 western blot, we used RT-PCR. mRNA from muscle tissue of affected children and control indiviuduals was reversely transcribed into cDNA and amplified with various primer combinations with forward primers (located at exons 1–5) and reverse primers (located at exons 8–13). Resulting PCR products were subcloned and subjected to bi-directional automatic Sanger sequencing. Resulting sequence fragments were aligned to the TRIP4 reference sequence with the STAR v.2.4.0 software, which is able to identify novel splice junction sites.5 For the identification of different splicing patterns in RNA-seq datasets from the GEO repository (Figure 3), we used the STAR v.2.4.0 aligner followed by analysis with the Cufflinks v.2.2.1 software suite.37
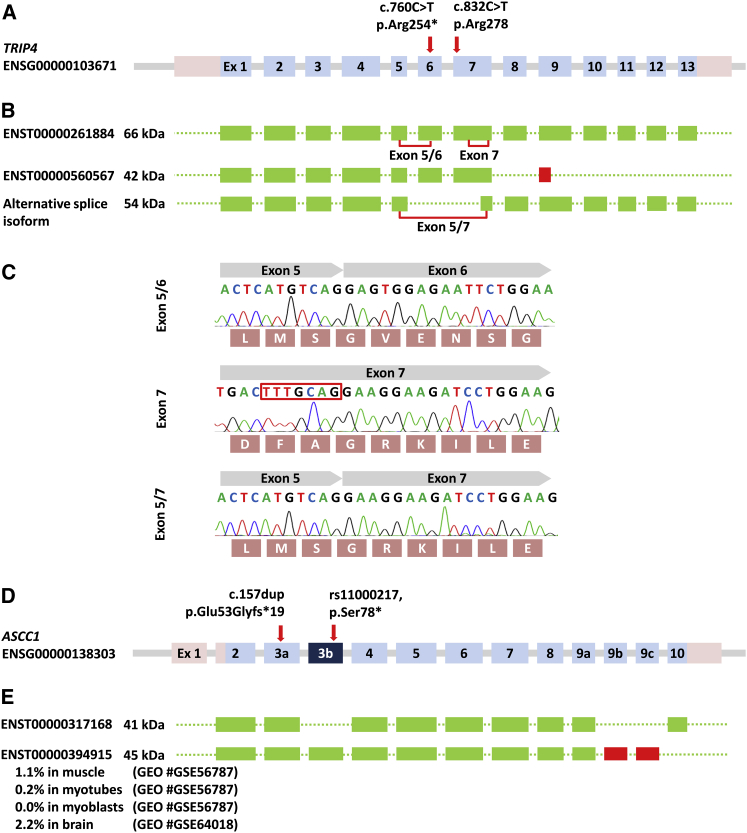
Figure 3.
Splice Isoforms of TRIP4 and ASCC1
(A) Genomic structure of TRIP4 (not drawn to scale). The introns are indicated by gray lines.
(B) The exons included into the various mRNA splice isoforms are depicted in green, and the red box indicates a frameshift.
(C) Sequence traces of the exon splice junctions. Their respective localization is marked on (B). The alternative splice acceptor site in exon 7 is highlighted by a red box.
(D) Genomic structure of ASCC1 (not drawn to scale).
(E) The transcript encoding the 41 kDa ASCC1 is the most abundant. Exon 3b, which contains a missense mutation in 5.4% of the ExAC alleles and leads to the truncation of the protein (p.Ser78∗), is only present in a rare ASCC1 45 kDa splice variant found in 0%–2% of the indicated GEO RNA-seq datasets.
Histology
Cryosections from muscle biopsy specimens were stained with Gömöri trichrome, H&E non-specific esterase, and ATPases pre-incubated at pH 4.3, 4.6, and 9.4, as well as with primary antibodies directed against myosin heavy chain: MHCslow, MHCfast, MHCneo, and MHCdev. Signals were visualized with appropriate secondary antibodies and the diaminobenzidine system. Zebrafish morphants were anesthetized at 48 hours post fertilization (hpf) in 0.02% tricaine (Sigma), fixed in cacodylate-buffered (pH 7.2) 2% glutaraldehyde, and prepared for electron microscopy as previously described.38 The skeletal muscle was likewise processed for electron microscopy. For antibodies, see Table S9.
Western Blot
Western blot was performed with protein extracts from muscle and fibroblasts as previously described.39 We used antibodies against TRIP4 and ASCC1. Anti-pan-Actin was used as loading control for muscle and β-tubulin for fibroblasts. Bands were visualized by chemiluminescence with peroxidase-labeled secondary antibodies. For antibodies, see Table S9.
Cell Sub-fractionation
Fibroblasts were yielded from nearly confluent plastic culture flasks by trypsination. Cross linking was done in DMEM with 1% formaldehyde for 10 min on ice. The reaction was stopped by adding glycine to a concentration of 130 mM, and the cells were spun down at 250 × g for 7 min. The pellet was resuspended in LF buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM EDTA, 0.5% NP40, 1.15% Triton X-100, one Complete tablet [Roche]) and incubated for 10 min on ice. The cell suspension was then transferred into a Dounce tissue grinder where cells were broken by 10 up and down strokes and centrifuged for 10 min at 4°C at 3,000 × g. The supernatant was kept at −80°C as the cytosolic fraction. The pellet containing the nuclei was resuspended in freshly prepared nuclear lysis buffer (10 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% Na-deoxycholate, 0.5% N-lauroylsarcosine, one Complete tablet) and sonicated 10 times for 10 s on ice. Finally, a 1/10 volume of Triton X-100 was added and a centrifugation at 16,000 × g for 1 min was performed to remove debris. The supernatant was kept as the nuclear fraction.
Immunoprecipitation
500 μg protein of the nuclear fraction were mixed with 2 μg of specific antibody and incubated in 500 μl nuclear lysis buffer on a rotator at 4°C overnight. Washed 50 μl Protein-G Sepharose beads were then added and incubated on a rotator at 4°C overnight to let the beads bind to the specific antibodies. Beads were then spun down at 2,000 × g at 4°C for 2 min. The supernatant contained the unbound fraction. The bead pellet was then washed six times with wash buffer (50 mM HEPES-KOH [pH 7.55], 1 mM EDTA, 1.0% NP40, 0.7% Na-deoxycholate, 500 mM LiCl). For western blot, the beads were finally washed once with TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 50 mM NaCl). Protein was eluted from the beads with elution buffer (50 mM Tris-HCl [pH 8.0], 10 mM EDTA, 1.0% SDS). Remaining beads were spun down at 2,000 × g at 4°C for 2 min, and the supernatant was used for SDS-PAGE. For mass spectrometry, the beads were finally washed once with 50 mM ammonium-bicarbonate (pH 8.0) and spun down at 2,000 × g for 2 min at 4°C. After removal of the supernatant, the pellets were frozen on dry ice.
Trip4 and Ascc1 In Situ Expression Analysis in Mice and Zebrafish
Studies of Trip4 and Ascc1 gene expression were done in axial and sagittal sections of whole C57BL6/J mouse embryos (embryonic day 17.5 [E17.5]) by in situ hybridization as previously described.40 Mouse embryos were embedded in TissueTek (Sakura), and longitudinal and axial sections were prepared on a cryostat. Riboprobes of ∼600 bp length were generated by RT-PCR from mouse muscle with tailed primers (Table S8) and cloned into the pCR-Script vector (Clontech). After linearization of the vector, antisense and sense DIG-labeled probes were generated with T3 and T7 RNA polymerase (DIG RNA Labeling Kit, Roche), respectively. Hybridization was performed overnight at 60°C followed by a colorimetric reaction at room temperature (24–48 hr). Slides were photographed with a 20× lens on a DMI4000 (Leica) microscope. Single visual fields were joined by the tiling-software (Image-Pro Premier v.9.1) of the microscope to generate the cross-sectional image of the spinal cord and adjacent structures (Figure 4, Figures S6 and S7). Whole-mount in situ hybridization of zebrafish was performed as previously published,6 with trip4 and ascc1 DIG-labeled antisense probes, and labeling with the respective sense probes did not detect any significant signal.
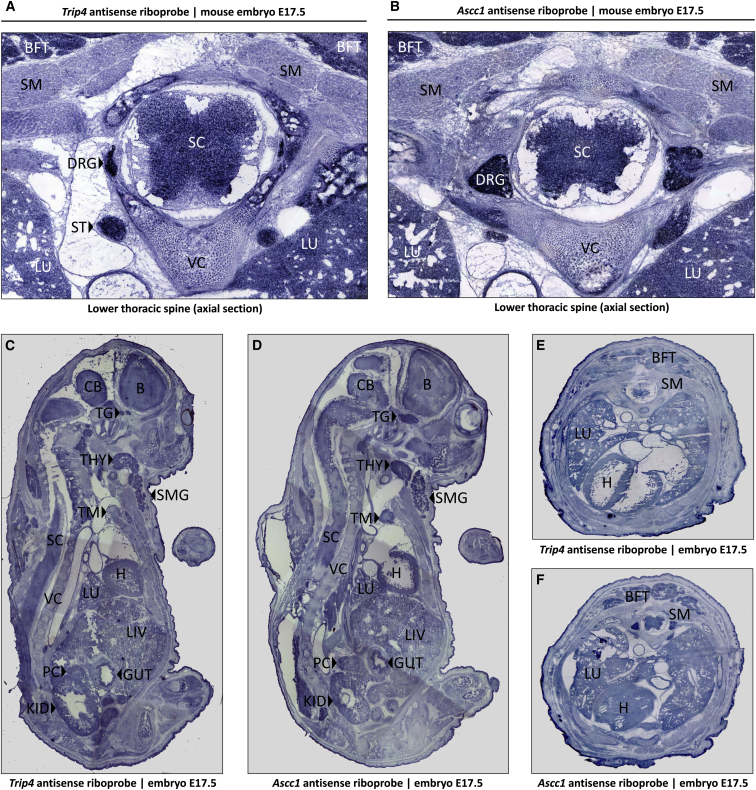
Figure 4.
Gene Expression Study in E17.5 Mouse Embryos
In situ hybridizations of cryosections from E17.5 mouse embryos demonstrate the nearly identical expression patterns of Trip4 (A, C, and E) and of Ascc1 (B, D, and F) mRNA. At the lower thoracic level (A and B), the highest expression levels were seen in the spinal cord, dorsal root ganglia, paraspinal sympathetic ganglia, muscle, lung, and brown fat tissue. In the parasagittal sections, the highest expression levels were seen on the thyroid and submandibular salivary gland and the trigeminal ganglion. Abbreviations are as follows: B, brain; BFT, brown fat tissue; CB, cerebellum; DRG, dorsal root ganglion; GUT, gut; H, heart; KID, kidney; LIV, liver; LU, lung; PC, pancreas; SC, spinal cord; SM, skeletal muscle; SMG, submandibular salivary gland; ST, sympathetic tract; TG, trigeminal ganglion; TG, thyroid gland; TM; thymus; VC, vertebral column.
Functional Analysis in Zebrafish
MO-Mediated Knockdown of trip4 and ascc1
Two different antisense morpholino oligonucleotides (MOs) were designed against the splice acceptor sites of zebrafish trip4 (boundary of intron 4 and exon 5) and ascc1 (boundary of intron 3 and exon 4) (Figures S8–S10). These antisense MOs and a standard negative control MO were purchased from Gene Tools. The sequences of these MOs and the control MO are depicted on Table S8. Zebrafish embryos were injected with 5 ng of MOs at the one- or two-cell stage in three independent trials and studied as previously published.6 At these dosages, the control MO produced no discernible phenotype change.
Quantification of MO-Mediated trip4 and assc1 Knockdown on the mRNA Level
Total RNA was extracted from a mixture of five MO-injected embryos and subjected to RT-PCR with the SuperScript III kit (Life Technologies) for reverse transcription. The number of reaction cycles for PCR was 23 for trip4 and ascc1 and 18 for bactin. All primer sequences are provided in Table S8. Quantitative analysis was done by measurement of band intensities of trip4, ascc1, and bactin with the ImageJ software, and p values were calculated with the t test.
Quantification of MO-Mediated trip4 and assc1 Knockdown on the Protein and Morphological Levels
For morphological analysis of α-motoneurons and neuromuscular junctions, zebrafish embryos (36 hpf) were anaesthetized in 0.02% tricaine (Sigma) and fixed in 4% paraformaldehyde (Sigma) at 4°C overnight and subjected to immunostaining as described previously (Figure 5).6 The following antibodies and labeling reagents were used: anti-synaptotagmin (znp-1, 1:100), AlexaFluor488-conjugated anti-mouse immunoglobulin G (IgG, 1:1,000), and AlexaFluor594-conjugated α-bungarotoxin (1:1,000). Fluorescent images were captured with a confocal microscope (SP5, Leica). For antibodies, see Table S9. To quantify the reduction of neuromuscular junctions in the morphants, we densitometrically measured the intensities of the α-bungarotoxin staining in control and morphant zebrafish larvae by using ImageJ. Statistic analysis was done by t test.
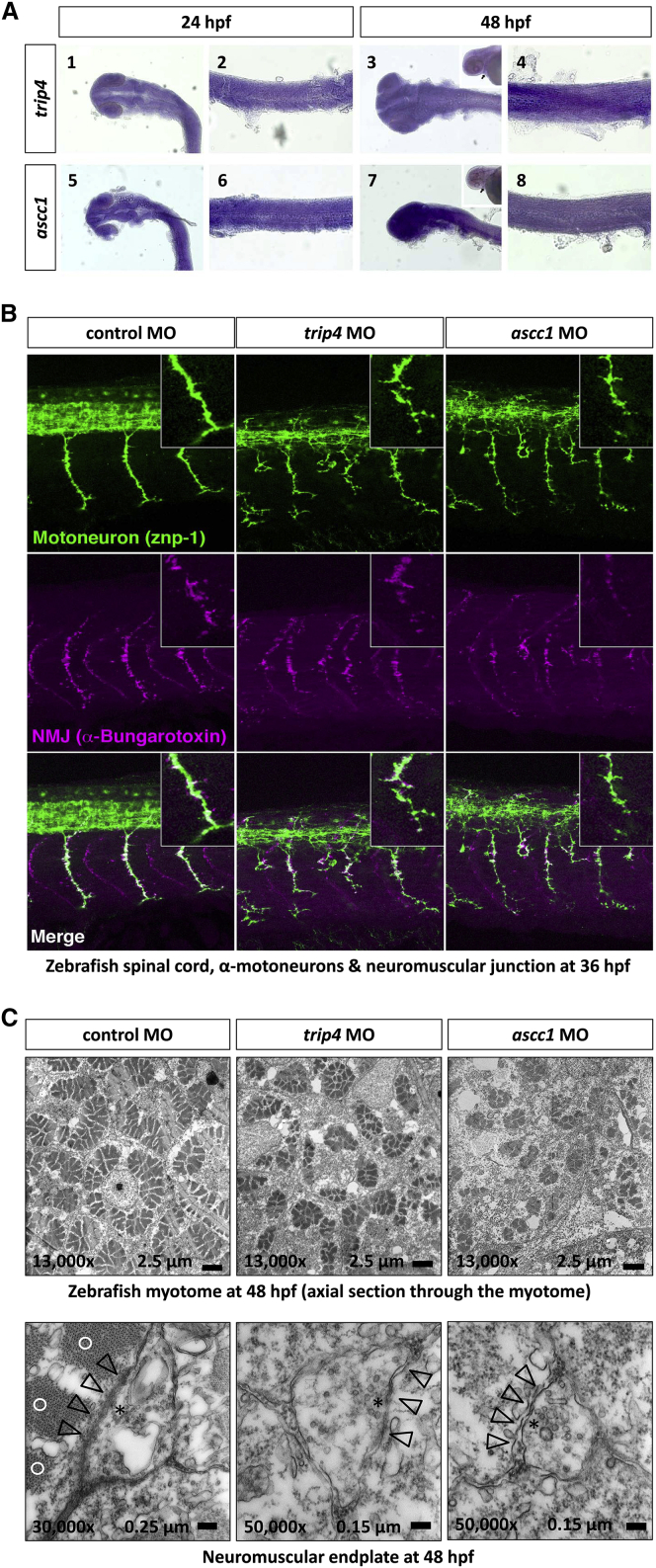
Figure 5.
Studies on Zebrafish Embryos
Expression of trip4 and ascc1 mRNA.
(A) trip4 is expressed ubiquitously in the head (A1) and trunk (A2) at 24 hpf and at 48 hpf (A3 and A4). ascc1 is also expressed ubiquitously in the head (A5) and trunk (A6) at 24 hpf and 48 hpf (A7 and A8). The arrowheads indicate the heart, verifying that both trip4 and ascc1 are expressed in cardiac muscles.
(B) MO-mediated trip4 and ascc1 knockdown in zebrafish larvae led to a severe derangement of α-motoneuron axons and the myotome. In the morphants, we found a perturbed outgrowth of α-motoneuron axons projecting to the trunk muscle in every somite segment at 36 hpf. The α-motoneurons were short, thin, and fragile with abnormal branches in trip4 morphants and ascc1 morphants. In these morphants, we additionally see ectopic outgrowth of motoneurons from the spinal cord. The α-motoneurons are labeled with the znp-1 antibody (green). Labeling with α-bungarotoxin (purple) displays the formation of neuromuscular junctions that form along with the α-motoneurons. The neuromuscular junctions were thin, reduced in number, and disorganized in the trip4 and ascc1 morphants.
(C) Electron-microscopic images of axial sections through the zebrafish myotome at 48 hpf. The rosette-like formation of myofibrils is greatly disturbed with reduced size and numbers. The lower panels show a neuromuscular endplate of the control morphants (left) with a normal thickened basal lamina, which is directly adjacent to the contractile elements of the myofibril. This is in contrast to the endplates from the trip4 and ascc1 morphants, which are smaller and have a disrupted basal lamina and no adjacent contractile elements. An asterisk denotes clusters of neurotransmitter vesicles (vesicle diameter 30–40 nm); open arrowheads denote the synaptic cleft and basal lamina of the neuromuscular endplate; open circles denote sarcomers (contractile elements) in the vicinity of the neuromuscular endplate. Note the higher magnification of the morphant endplate.
Movement Analysis of Zebrafish
At 1 dpf, the chorion of the embryos was removed to allow touch access to the embryos. Embryos showing retarded development or developmental malformations, which were even seen in un-injected wild-type clutch, were removed before assay. Tactile stimulation was applied to the tail with a forceps (Movies S1, S2, and S3). Touch responses of MO-injected embryos (morphants) were video recorded at 30 hpf with a high-speed camera at 200 frames per second (HAS-220, Ditect).41 The normal touch response of control morphants at 1 dpf is a vigorous coiling behavior, twisting their tail to the trunk (“full coil”), but ascc1 and trip4 morphants displayed only a faint twitch without coiling their tail to the trunk (“partial coil”). The ratio of abnormal embryos showing partial coil to total embryos in the three experimental trials is provided in Table S3. Statistic analysis was done by the χ2 test.
Rescue of MO-Knockdown with trip4 and ascc1 Constructs
Full-length zebrafish trip4 and ascc1 cDNAs were cloned into the pCS2+ zebrafish expression vector with the oligonucleotide primers detailed in Table S8. The trip4 nonsense mutation leading to a truncation of the protein (zebrafish, p.Lys265∗; corresponding alteration in humans, p.Arg278∗) and the ascc1 frameshift mutation, also leading to a truncation of the protein (zebrafish, p.Glu53Glyfs∗) were introduced by site-directed mutagenesis. 100 pg of trip4 or ascc1 wild-type and mutant RNA were then injected into zebrafish embryos along with trip4 or ascc1 MOs, and the morphants were assayed for touch response and motoneuron and neuromuscular junction labeling (Figure S11, Table S5).
Cell Culture
Low passage skin fibroblasts of two affected children and four age-matched control individuals were grown to semi-confluency in DMEM supplemented with 15% fetal bovine serum (FBS) at 37°C in a 5% CO2 atmosphere. After being washed with PBS, the cells were incubated with DMEM supplemented with 0.5% FBS over 24 hr for serum starvation. A fibroblast pellet was yielded after this period (time point = 00W). After being washed with PBS, the other culture dishes were incubated with DMEM supplemented with 15% FBS, and aliquots of cells were yielded exactly 30 min (time point = 30W) and 60 min (time point = 60W) later. All pellets were flash frozen in liquid nitrogen and stored at −80°C until further use.
Global mRNA Expression Profiling
Skin fibroblasts of affected children (D:II_2 and D:II_3) and of age-matched control indiviuduals (n = 4) were processed as described above. Total RNA was prepared from the frozen pellets with the standard TRIzol (Life Technologies) protocol. Quality of the RNA was controlled via the 2100 Bioanalyzer (Agilent). RNA was hybridized, according the manufacturer’s instructions, on the GeneChip Human Gene 2.0 ST Array (Affymetrix), which represents 40,716 human RefSeq transcripts, including 30,654 transcripts which are annotated as protein coding. The chips were scanned with the 428 Scanner (Affymetrix) and data were processed with the Affymetrix GeneChip Command Console Software (AGCC) to produce the CEL files that could be uploaded to the GenePattern platform for further downstream analysis and visualization.42 We first inspected those genes that are known downstream targets of serum response factor (SRF), which, however, failed to show any difference in serum-dependent regulation between affected children and control individuals (Figure S12). Next, we identified those genes that were significantly up- and downregulated in the samples from affected children in comparison to those from control individuals. In order to control for multiple testing, we only considered genes as significantly regulated if they had a false discovery rate (FDR) below 0.01. The raw data of this experiment can be accessed from the GEO database (GEO: GSE67627). Pathway and gene network analysis of the regulated genes was performed with AmiGO2.43
Statistics
For comparison of normally distributed values, we used the t test, and for not normally distributed values, either the χ2 or the nonparametric Mann-Whitney U test. To control for multiple testing in the gene expression studies, we calculated the FDR,44 and differences in gene expression were considered significant if the FDR was <0.01.
Results
Clinical Presentation in Eight Individuals from Four Families
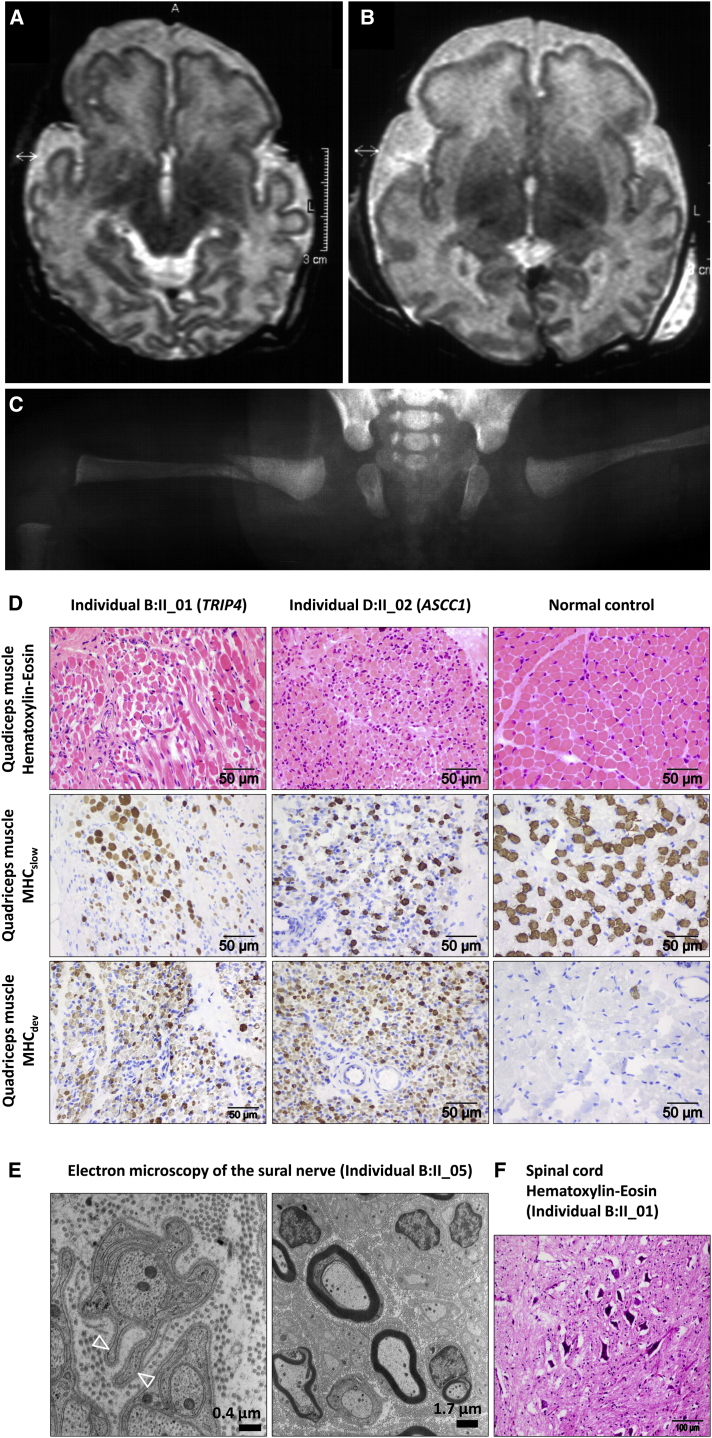
We describe here a cohort of seven individuals who were born with distal and proximal joint contractures, as well as generalized muscle atrophy and fractures of their long bones (humerus and femur), and one aborted fetus. Deep tendon reflexes were absent, muscles did not contract upon electrical nerve stimulation, and neurography revealed axonal neuropathy in these indiviudals. Respiratory distress, diaphragmatic eventration and pulmonary hypoplasia necessitated mechanical ventilation soon after birth. Dysphagia required gavage feeding. For a detailed comparison of clinical symptoms, see Table 1. Most children died from respiratory failure between 2 weeks and 16 months of life. One child died from acute heart failure. Additional variable features included premature birth, microretrognathia, hypertelorism, a high-arched palate, secundum atrial septal defect, patent ductus arteriosus, and cardiomyopathy. In the affected children from family D, cerebral MRI had been performed and revealed a simplified gyral pattern of the cerebral cortex (Figure 1).
Table 1.
Clinical Data of the Investigated Families
| Family A | Family B | Family C | Family D | Total | |
|---|---|---|---|---|---|
| Country of origin | Kosovo | Kosovo | Albania | Turkey | – |
| Gene involved | TRIP4 | TRIP4 | TRIP4 | ASCC1 | – |
| Mutation on the cDNA level | c.760C>T | c.832C>T | c.760C>T c.832C>T | c.157dupG | – |
| Predicted protein alteration | p.Arg254∗ | p.Arg278∗ | p.Arg254∗ p.Arg278∗ | p.Glu53Glyfs19∗ | – |
| Number of affected individuals |
n = 2 |
n = 2 + (1)a |
n = 1 |
n = 2 |
n = 7 + (1)a |
|
Prenatal Manifestation: HPO ID | |||||
| Decreased or absent fetal movement: 1558 | n = 2 | n = 2 | ND | n = 2 | n = 6/6, 100% |
| Polyhydramnios: 1561 | n = 0 | n = 0 | ND | n = 2 | n = 2/6, 33% |
| Oligohydramnios: 1562 | n = 2 | n = 1 | ND | n = 0 | n = 3/6, 50% |
| Premature birth (< 37 weeks): 1622 |
n = 1 |
n = 1 |
ND |
n = 2 |
n = 4/6, 67% |
|
Nervous System: HPO ID | |||||
| Severe muscular hypotonia: 6829 | n = 2 | n = 2 | n = 1 | n = 2 | n = 7/7, 100% |
| Areflexia: 1284 | n = 2 | ND | ND | n = 2 | n = 4/4, 100% |
| Muscle weakness: 1324 | n = 2 | n = 2 | ND | n = 2 | n = 6/6, 100% |
| Global developmental delay: 1263 | n = 2 | n = 2 | ND | n = 2 | n = 6/6, 100% |
| Dysphagia: 2015 | n = 2 | n = 2 | ND | n = 2 | n = 6/6, 100% |
| Abnormal cortical gyration: 2536 |
ND |
ND |
ND |
n = 2 |
n = 2/2, 100% |
|
Head and Neck: HPO ID | |||||
| Microretrognathia: 0308 | n = 2 | n = 2 | ND | n = 0 | n = 4/6, 67% |
| Hypertelorism: 0316 | n = 2 | n = 1 | ND | n = 0 | n = 3/6, 50% |
| High palate: 0218 | n = 2 | n = 1 | ND | n = 0 | n = 3/6, 50% |
| Narrow mouth: 0160 |
n = 2 |
n = 0 |
ND |
n = 0 |
n = 2/6, 33% |
|
Cardiovascular System: HPO ID | |||||
| Patent foramen ovale: 1655 | n = 0 | n = 1 | ND | n = 2 | n = 3/6, 50% |
| Secundum atrial septal defect: 1684 | n = 2 | n = 0 | ND | n = 0 | n = 2/6, 33% |
| Patent ductus arteriosus: 1643 | n = 2 | n = 1 | ND | n = 2 | n = 5/6, 83% |
| Cardiac failure: 1635 | n = 1 | n = 0 | ND | n = 0 | n = 1/6, 17% |
| Cardiomyopathy: 1638 |
n = 1 |
n = 1 |
ND |
n = 0 |
n = 2/6, 34% |
|
Respiratory System: HPO ID | |||||
| Neonatal respiratory distress: 2643 | n = 2 | n = 2 | n = 1 | n = 2 | n = 7/7, 100% |
| Pulmonary hypoplasia: 2089 |
n = 0 |
n = 2 |
ND |
n = 2 |
n = 4/6, 67% |
|
Musculo-skeletal System: HPO ID | |||||
| Multiple prenatal fractures: 5855 | n = 2 | n = 1 | ND | n = 2 | n = 5/6, 83% |
| Arthrogryposis multiplex congenita: 2804 | n = 2 | n = 3 | n = 1 | n = 2 | n = 8/8, 100% |
| Muscle fiber immaturity: NA | n = 1 | n = 2 | ND | n = 1 | n = 4/4, 100% |
| Muscle fiber size variation and atrophic fibers: 3557 | n = 1 | n = 2 | ND | n = 1 | n = 4/4, 100% |
Figure 1.
Clinical Presentation of the Affected Children
(A and B) Axial T2-weighted cranial MRI images of affected children D.II_02 (A) (at a corrected gestational age of 39 weeks) and D.II_03 (B) (at a corrected gestational age of 37 weeks) with a simplified gyral pattern of the frontal lobes and enlargement of the external CSF spaces. Myelination of the brain stem and the basal ganglia is normal.
(C) X-ray of the bilateral congenital femoral fractures of individual D.II_03.
(D) Muscle histology demonstrates a reduction in fiber size and an increase in fiber-size variation in two individuals with a TRIP4 and ASCC1 mutation, in contrast to fiber size and variation in an age-matched control individual. The grouping of the larger type I fibers (marked by MHCslow), in contrast to a normal checkerboard pattern in the control individual, is characteristic for prenatal SMA. The intense staining of the muscles of the affected children for MHCdev highlights their immaturity.
(E) Ultrastructure of a sural nerve biopsy specimen with normal myelinization but with loss of unmyelinated axons as documented by “empty” pouches (open triangles).
(F) Presence of multiple intensely stained apoptotic α-motoneurons in the anterior horn of the spinal cord in a postmortem sample.
Muscle biopsy samples from four children were available for re-analysis (Figure 1): the muscle fibers were reduced in size, showed increased fiber-size variation, and were much more immature than samples from age-matched control individuals. The affected childrens’ type I fibers were clustered in groups, in comparison to the checkerboard pattern in control individuals, which is a characteristic finding in prenatal SMA (Figure 1). In one individual, postmortem histology of the spinal cord was available and revealed numerous apoptotic α-motoneurons in the anterior horns at different levels (Figure 1). Sural nerve biopsy from two children showed normal density of myelinated fibers within the range of published reference values46, 47, 48 (Figures S1 and S2, Table S1), but also showed signs of unmyelinated axon loss (Figure 1). The average axon diameters were increased in one individual from family B, but in the range of a normal age-matched control in an individual from family D (Figure S2). On X-ray, the bones of individual D.II_3 had failed to develop callus after 14 days of splinting. Subperiostal new bone formation would be expected after 11 days at the latest, and even earlier in young infants.49 Bone mineralization and alkaline phosphatase activities were normal.
Autozygosity Mapping and WES
Even though families A and B denied consanguinity, we assumed a founder haplotype due to their common Kosovan origin and performed autozygosity mapping. Their shared autozygous region comprised 38 protein-coding genes (Figures S3–S5). Via WES of individual B.II_01, we found a homozygous nonsense mutation, c.832C>T (p.Arg278∗), in TRIP4 (chr15:64,701,816C>T [GRCh37], exon 7 [GenBank: NM_016213.4]). Genotype-phenotype segregation was verified by Sanger sequencing of the entire family, including DNA from a fetus, who had been aborted at 27 weeks and 3 days, after joint contractures were observed on ultrasound imaging (Figure 2). Sequencing of TRIP4 in family A also revealed a nonsense mutation (c.760C>T [p.Arg254∗]), albeit unexpectedly, at a different position (chr15:64,698,591C>T [GRCh37], exon 6), and homozygosity of this mutation segregated with the disease. Both mutations were absent in the 1000 Genomes Project as well as in 135 ancestry-matched in-house exomes. The p.Arg278∗ truncation was present in heterozygous form in 2/121,356 alleles of the ExAC database (accessed November 2015), in two individuals of European and African descent, whereas the p.Arg254∗ truncation was not listed.
Despite a similar clinical phenotype, autozygosity mapping located the disease locus of family D not on chr15, but on chr10. Using WES, we discovered a frameshift mutation, c.157dup (p.Glu53Glyfs∗19), in ASCC1 (chr10:73,970,544_5insC [GRCh37], exon 3 of ASCC1-003 [Ensembl: ENST00000317168] [GenBank: NM_001198800.2]) that was homozygous only in the affected children (Figure 2). This mutation is present in heterozygous form in 2/120,662 alleles of the ExAC database, in two individuals of European and Latino descent. Interestingly, the ExAC database lists a variant in ASCC1 (rs11000217) in 896/16,490 alleles (59 of them homozygotes), mainly in individuals from African or South Asian descent, that causes a truncation of ASCC1 (p.Ser78∗). Such a finding might put into question the essential importance of ASCC1 for the ASC-1 complex. However, a transcriptomic analysis of RNA-seq datasets from muscle and brain revealed that this variant (dbSNP: rs11000217) resides in an exon that is only included in 0%–2% of the transcripts from the ASCC1 locus (exon 3b, Figure 3), whereas the exon 3a that carries the mutation of the affected children in our study is present in >95% of the transcripts.
Cohort screening of TRIP4 and ASCC1 in 11 unrelated affected children with the appropriate phenotype revealed an additional individual (C:II_05), who also originated from the Balkans (Albania) and carried both TRIP4 mutations in compound heterozygous state (Figure 2).
In order to further exclude mutations in other (known) genes that might have an influence on the phenotype of our affected children, we specifically searched within the autozygous regions of families A, B, and D for variants in genes that were associated in the Human Phenotype Ontology45 with “arthrogryposis, HP:0002804,” “fractures, HP:0003084,” “spinal muscular atrophy, HP:0007269,” “muscle weakness, HP:0001324,” or with “abnormity of cortical gyration, HP:0002536,” but did not find any other potentially pathogenic variants, either in heterozygous or in homozygous states.
Given that cryo-muscle samples were available from individuals A:II_02 and B:II_05, we studied the effect of the mutation on the protein level by western blot. Whereas, in normal muscle, we found two bands at ∼40 and ∼70 kDa and an additional very weak band at ∼53 kDa, both individuals exhibited bands at ∼53 kDa and ∼36 kDa, with absence of the ∼40 kDa and ∼70 kDa bands (Figure 2). Interestingly, both mutant TRIP4 bands migrated at the same molecular size despite the fact that the premature termination codons were located at different positions. RT-PCR analysis of different TRIP4 splice isoforms (Figure 3) revealed the activation of a cryptic splice acceptor site leading to a novel isoform with skipping of exons 6 and part of exon 7 that preserved the reading frame but excluded both missense mutations. This splice isoform seems to be upregulated in mutant cells, whereas the full-length transcript was subjected to nonsense mediated messenger decay. Such corrective splicing events leading to the upregulation of a rare splice isoform have been described, albeit rarely, for other conditions, such as spinal muscular atrophy (MIM: 253300),50 Rothmund-Thomson syndrome (MIM: 268400),51 retinitis pigmentosa (MIM: 304020),52 Ullrich congenital muscular dystrophy (MIM: 254090),53 and epidermolysis bullosa (MIM: 226650).54 In these reported cases, the truncated proteins were expressed and seemed to retain some residual function that ameliorated the clinical phenotype. From family D, we only had cultured fibroblasts, which entirely lacked ASCC1 and in which we did not detect any corrective splice isoforms that excluded the mutant exon (Figure 2).
Subcellular Location of the ASC-1 Complex
TRIP4 had been shown previously by indirect immunofluorescence to be a nuclear protein that localized into the cytoplasm under conditions of serum deprivation.55 Because TRIP4 forms a tetrameric protein complex, we hypothesized that its correct subcellular localization might be compromised if one subunit (e.g., TRIP4 or ASCC1) of the ASC-1 complex was altered. Hence, we separated nuclear and cytosolic fractions of wild-type and ASCC1 mutant fibroblasts under normal growth conditions, after serum depletion, and after serum depletion with subsequent 30 and 60 min of serum re-challenge and probed them on western blot with antibodies directed against three subunits of the ASC-1 complex (anti-TRIP4, anti-ASCC1, anti-ASCC2). We were unable to confirm a translocation to the cytoplasm under serum starvation, neither of the entire ASC-1 complex nor of TRIP4 alone. The signals of three subunits of the ASC-1 complex remained stationary in the nuclear fraction independently of the presence or absence of serum (Figure S13).
Trip4 and Ascc1 Expression Analysis in Mouse Embryos by In Situ Hybridization
To determine the spatial expression pattern of Trip4 and Ascc1 in a later phase of embryonic development, we performed in situ hybridization on cryosections of E17.5 mouse embryos. Both genes showed nearly identical mRNA-expression patterns with ubiquitous expression, albeit at different levels (Figure 4). Highest expression was seen in dorsal root ganglia, the paraspinal sympathetic and trigeminal ganglia, and thyroid and submandibular glands, as well as the spinal cord. Expression in the cerebral cortex was comparable to the expression in the spinal cord.
MO Knockdown of trip4 and ascc1 in Zebrafish
The function of TRIP4 and ASCC1 in vertebrates has not been explored. To evaluate the physiological consequences of the absence of both mutant proteins, we performed a MO-mediated knockdown of trip4 and ascc1 in zebrafish, given that both proteins are conserved between humans and zebrafish (Figures S14 and S15). Based on the postmortem findings regarding the affected children, which revealed numerous apoptotic neurons in the anterior horn of the spinal cord, and the muscle histology of prenatal SMA, we focused our investigation on α-motoneuron and muscle development. Indeed, we found a severe impairment of axonal outgrowth, formation of the neuromuscular junction, and organization of the myotome (Figure 5, Table S3). Control MO-injected embryos responded to touch with typical coiling behavior, which consists of two to three vigorous contractions of trunk and tail at 30 hpf.41 Knockdown of trip4 or ascc1 resulted in a compromised response (Movies S1, S2, and S3, Table S3). The knockout coiling phenotype of zebrafish could be rescued by injection of the respective full-length wild-type trip4 and ascc1 mRNA, but not by injection of mRNA carrying the affected childrens’ mutations (Figure S11). To verify the specificity of the used MO, we used a second MO (MO2) aimed at a different splice site and were able to reproduce the original effect (Figure S10, Table S4). These loss-of-function analyses confirm that trip4 and ascc1 are indispensable for motor system development. In accordance with the observation of the motor behavior, the myotomes, as well as the neuromuscular endplates, appeared severely disorganized on electron microscopy (Figure 5).
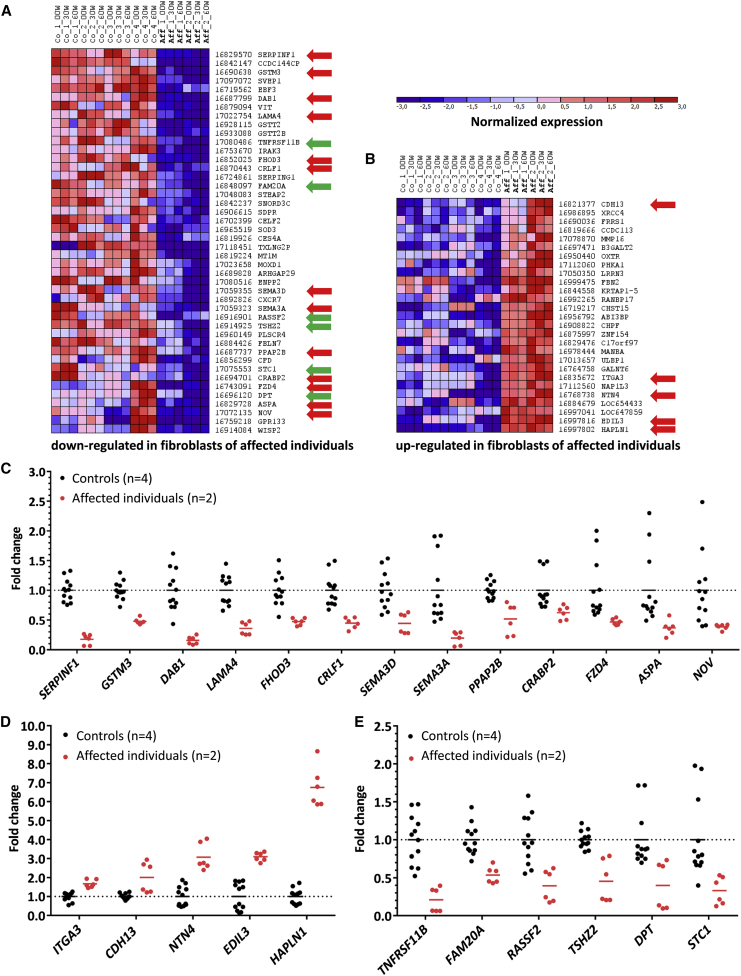
Identification of ASC-1 Target Genes
In order to identify those genes whose mRNA transcript levels are differently regulated in the absence of a functionally intact ASC-1 complex, we performed a global mRNA expression analysis of ASCC1 mutant and control fibroblasts after serum depletion and re-challenge (Figure 6, Table S6). Contrary to the originally published hypothesis that the ASC-1 complex might regulate transcription via the SRF,29 we discovered normal upregulation of the SRF-dependent downstream target genes FOS (MIM: 164810), IER2, FOSL1 (MIM: 1365150), FOSB (MIM: 164772), JUNB (MIM: 165161), and TRIB1 (MIM: 609461) after serum challenge that was indistinguishable between affected and control fibroblasts (Figure S12). Hypothesis-free analysis of expression patterns after stringent filtering against genes with a FDR >0.01 revealed downregulation of 44 genes (Figure 6, Table S6), 13 of which are involved in neurogenesis (SERPINF1 [MIM: 172860], NOV [MIM: 164958]), neuronal projection (SERPINF1, CRABP2 [MIM: 180231]), pathfinding (SEMA3A [MIM: 603961], SEMA3D [MIM: 609907]), migration (DAB1 [MIM: 603448]), and suppression of neuronal apoptosis (SERPINF1, CRLF1 [MIM: 604237]). On the other hand, genes that suppress neuronal plasticity (HAPLN1 [MIM: 115435]) and negatively regulate neuron projection (CDH13 [MIM: 601364]) were upregulated. Six of the downregulated genes (Figure 6, Table S6) are involved in the negative regulation of bone resorption (TNFRSF11B [MIM: 602643]), ossification and regulation of osteoblast differentiation (RASSF2 [MIM: 609492]), and collagen fibril organization (DPT [MIM: 125597]), as well as calcium ion homeostasis (FAM20A [MIM: 611062], STC1 [MIM: 601185]).
Figure 6.
Gene Expression Analysis of Wild-Type and ASCC1 Mutant Fibroblasts
(A) Clustergram of genes downregulated in fibroblasts of affected children with FDR < 0.01. Blue denotes normalized downregulation; red denotes upregulation (see attached scale above). The numbers on the right identify the Affymetrix Human GeneChip 2.0 probe set. The gene names are given on the far right column. The red arrows depict genes involved in neurogenesis; green arrows depict genes involved in bone metabolism. Abbreviations are as follows: Co, control; Aff, affected individual with ASCC1 mutation; 00W, serum starved for 12 hr; 30W, serum challenge for 30 min; 60W, serum challenge for 60 min; FDR, false discovery rate.
(B) Genes upregulated in fibroblasts of affected children with FDR < 0.01.
(C and D) Genes involved in neurogenesis that are downregulated (C) or upregulated (D) in ASCC1 mutant fibroblasts (red dots).
(E) Downregulated genes involved in bone metabolism and development (red dots). Each dot represents a separate hybridization experiment. Black dots denote control individuals; red dots denote affected individuals. The horizontal lines represent the arithmetic mean.
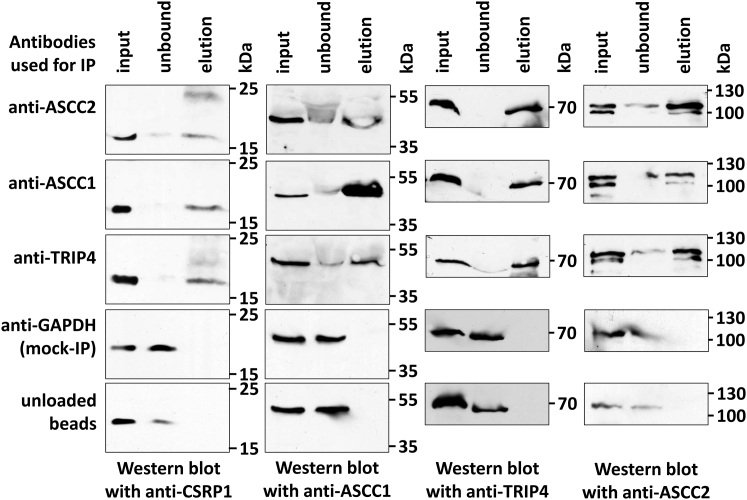
Identification of ASC-1 Binding Partners
Immunoprecipitation with three different antibodies (anti-TRIP4, anti-ASCC1, and anti-ASCC2) against subunits of the ASC-1 complex mutually copurified the other members of the holocomplex (Figures 7 and S16–S18, Table S7). Even in the absence of ASCC1, copurification between TRIP4 and ASCC2 was still possible (Figures S16 and S18), suggesting the stability of the complex even in the absence of one subunit. Mass spectrometric analysis of the immunoprecipitates consistently identified copurification of the cysteine and glycine rich protein 1 (CSRP1), a transcriptional cofactor that is known to be involved in spinal cord regeneration in adult zebrafish (Table S7, Figures S19–S23).56 Copurification of the CSRP1 protein was additionally verified by western blot of all three anti-ASC-1 immunoprecipitates with an anti-CSRP1 antibody (Figure 7).
Figure 7.
Coimmunoprecipitation of CSRP1 with Three Subunits of the ASC-1 Complex
Immunoprecipitation was done with anti-TRIP4, anti-ASCC1, and anti-ASCC2 antibodies. For mock immunoprecipitation, we used GAPDH antibodies, and for empty controls, we used unloaded Protein-G-Sepharose beads. Western blots from input, flow-through (unbound), and bound protein (elution) were serially incubated with anti-TRIP4, anti-ASCC1, anti-ASCC2, as well as with anti-CSRP1 antibodies.
Discussion
Our data show that mutations in two genes that encode different subunits (TRIP4 and ASCC1) of the activating signal cointegrator 1 (ASC-1) complex cause a profound disturbance of neuromotor unit development and result in prenatal onset spinal muscular atrophy with bone fractures. The notion that these two proteins form a functional complex and are involved in the same biological process in vivo is supported (1) by the nearly identical mRNA-expression pattern in the mouse embryo, (2) by the indistinguishable phenotypes of trip4 and ascc1 zebrafish morphants, (3) by mutual coimmunoprecipitation, and (4) ultimately by the similar phenotype of affected children and zebrafish. Such a phenomenon is known for other genetic disorders, such as leukoencephalopathy with vanishing white matter, which can be caused by alterations in five different subunits of the translation initiation factor eIF2B.57
The ASC-1 complex is composed of four subunits, which bind to nuclear receptors and coactivate the transcription of a wide range of transcripts. One subunit, TRIP4, was initially identified as a transcriptional coactivator of the thyroid hormone receptor.58 Later, it was shown that TRIP4 stably associates with three other polypeptides, the activating signal cointegrator subunits 1–3 (ASCC1–ASCC3), and interacts with a wider range of transcription factors such as activating protein 1 (AP-1), nuclear factor kappa-B (NF-κB), and SRF.29 In particular, the latter interaction between TRIP4 and SRF, which had been demonstrated by reporter gene assays,29 seemed to offer a functional link between genetic defect and disease phenotype because SRF is an essential transcription factor for muscle,59 nervous system,60, 61 and bone62 development. However, we detected neither physical interaction between SRF and ASC-1 (Figures S16–S18) nor abnormal SRF-dependent transcription of downstream target genes in affected childrens’ fibroblasts after serum depletion and re-challenge (Figure S12).
To explore alternative explanations for the pathomechanism seen in the affected children studied here and to search for proteins that bind to ASC-1, we performed an immunoprecipitation of the ASC-1 complex in fibroblasts of affected children and controls. Using mass spectrometry analysis, as well as western blot after immunoprecipitation, we consistently found coimmunopurification of CSRP1. CSRP1 localizes to the nucleus in the same manner as the ASC-1 complex (Figure S13).
CSRP1 is an evolutionarily highly conserved 23.4 kDa transcriptional coactivator that shares 83% identity between humans and zebrafish on the amino acid level. It contains a double zinc-finger motif, a nuclear localization signal, and a LIM motif, suggesting its involvement in developmental processes.63, 64 In the E14.5 mouse embryo, strong expression can be found in the CNS, especially in the spinal cord.65 MO-mediated knockdown of csrp1 in zebrafish during early development resulted in abnormal axis formation and severe deformities of midline structures. Miyasaka et al. also demonstrated that Csrp1 interacts with dishevelled 2 and diversin, thereby controlling cell morphology and the generation of pseudopodial processes via the non-canonical Wnt and JNK pathways.66 Both pathways largely influence the generation of filopodia at axonal growth cones and are deemed essential for neuronal polarity and axon outgrowth and branching, as well as the navigation to their final targets. Interestingly, in adult zebrafish, which have the capability to regrow severed descending spinal axons after spinal injury and regain locomotor activity, Ma et al.56 demonstrate a direct influence of Csrp1 on the re-growth behavior of axons. In contrast to mammals, zebrafish are able to reactivate specific developmental programs for regenerative purposes, and Ma et al. found a significant upregulation of csrp1 in those neurons that regrew their axons, whereas MO-mediated csrp1 knockdown impaired axonal regeneration and subsequent locomotor recovery.
We are aware that the conjectures about ASC-1 and CSRP1 interaction have to be proven in a mammalian system. It would thus be an interesting line of research to investigate how the simultaneous presence of ASC-1 and CSRP1 would shape neuronal patterning, outgrowth, pathfinding, and endplate formation. Because a complete knockout of Csrp1 would most likely be embryonically lethal,66 such experiments would have to be done in conditional knockout mice and with an α-motoneuron specific Cre-driver line.
Comparison of gene expression between mutant and wild-type cells shows that the ASC-1 complex exerts an influence on entire modules of genes (Figure 6, Table S6), many of which either enhance neurodevelopment and pathfinding or whose products suppress other genes that negatively regulate neuron projection, cell proliferation, or neuronal plasticity. Despite the fact that the mRNA expression analyses could only be done in the available fibroblasts of the affected children, it has been shown for another disease (MCT8-deficiency) that analysis of genome-wide expression data from fibroblasts derived from affected individuals was able to identify the dysregulation of coexpressed gene modules67 that are disease specific and mirror those seen in the human brain transcriptome.68
As an example from our cohort, SERPINF1 (serum peptidase inhibitor F), the most prominently downregulated gene (−5.7-fold) in mutant fibroblasts, has been shown to be involved in the positive regulation of spinal axon sprouting,69 neuroprotection,70 and survival of spinal motoneurons,71 whereas the extracellular matrix protein HAPLN1 (hyaluronan and proteoglycan link protein), the most prominently upregulated gene (+6.7-fold) in mutant fibroblasts suppresses neuronal plasticity.72 The same modular phenomenon can be seen for genes involved in bone development. Here, the TNFRSF11B (tumor necrosis factor receptor superfamily, member 11b; osteoprotegerin) gene was most prominently downregulated (−4.8-fold). Osteoprotegerin functions as an inhibitor of osteoclastogenesis73 and bone resorption74 and positively regulates bone mass.75
In conclusion, the ASC-1 complex seems to be involved in the highly regulated development of the neuromuscular unit and possibly of the neighboring bony structures as well. However, further research is needed to elucidate the exact signaling pathways that are involved in the patterning and development of the neuromuscular unit via the ASC-1 complex. Knowledge about the factors involved might have implications for regenerative medicine.
Acknowledgments
We thank the parents and their children for participation at this study, Hannah Plückhan for excellent technical assistance in electron microscopy, and Beata Lukaszewska-McGreal for LC-MS/MS sample preparation. The project was funded by grants from the Deutsche Forschungsgemeinschaft (DFG; SFB 665 TP C4) and the Einstein Foundation Berlin (A-2011-63) to M.S., the Grant-in-Aid for Scientific Research (25920008) of the Ministry of Education, Culture, Sports, Science, and Technology of Japan and funding from the Takeda Science Foundation, the Mochida Memorial Foundation for Medical and Pharmaceutical Research, the Naito Foundation, and the Suzuken Memorial Foundation to H.H., by DFG (Ru746/1-2) and Interdisziplinäres Zentrum für Klinische Forschung Aachen (NP 5-4) grants to S.R.S., and by funding from the Planck Institute for Molecular Genetics to D.M. Additionally, M.S. is member of the NeuroCure Center of Excellence (grant no. Exc 257).
Published: February 25, 2016
Footnotes
Supplemental Data include 23 figures, 9 tables, and 3 movies and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.01.006.
Contributor Information
Hiromi Hirata, Email: hihirata@chem.aoyama.ac.jp.
Markus Schuelke, Email: markus.schuelke@charite.de.
Accession Numbers
Raw data from the gene expression analysis of ASCC1 mutant versus control fibroblasts can be accessed under the accession number GEO: GSE67627.
Web Resources
The URLs for data presented herein are as follows:
1000 Genomes, http://browser.1000genomes.org
1000 Genomes reference sequence, ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/technical/reference/human_g1k_v37.fasta.gz
CRAPome database, http://www.crapome.org
Cufflinks v.2.2.0 software, http://cole-trapnell-lab.github.io/cufflinks/
Ensembl Genome Browser, human genome, http://useast.ensembl.org/Homo_sapiens/Info/Index
ExAC Browser, http://exac.broadinstitute.org/
GenePattern, http://genepattern.broadinstitute.org/gp/pages/login.jsf
HomozygosityMapper software, http://www.homozygositymapper.org/
Mouse Genome Informatics, http://www.informatics.jax.org/image/MGI:5329998
MutationTaster, http://www.mutationtaster.org/StartQueryEngine.html
OMIM, http://www.omim.org/
STAR v.2.4.0 software, https://code.google.com/archive/p/rna-star/
T-Coffee, http://tcoffee.crg.cat/
Supplemental Data
Normal response to touch of a 30 hpf zebrafish control morphants with coiling and several vigorous contractions.
Reduced motor response in a 30 hpf zebrafish trip4 morphant with just a single contraction executed with a weak twitch.
Reduced motor response in a 30 hpf zebrafish ascc1 morphant with just a single contraction executed with a weak twitch.
References
- 1.Hoff J.M., Loane M., Gilhus N.E., Rasmussen S., Daltveit A.K. Arthrogryposis multiplexa congenita: an epidemiologic study of nearly 9 million births in 24 EUROCAT registers. Eur. J. Obstet. Gynecol. Reprod. Biol. 2011;159:347–350. doi: 10.1016/j.ejogrb.2011.09.027. [DOI] [PubMed] [Google Scholar]
- 2.Hall J.G. Arthrogryposis (multiple congenital contractures): diagnostic approach to etiology, classification, genetics, and general principles. Eur. J. Med. Genet. 2014;57:464–472. doi: 10.1016/j.ejmg.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Bamshad M., Van Heest A.E., Pleasure D. Arthrogryposis: a review and update. J. Bone Joint Surg. Am. 2009;91(Suppl 4):40–46. doi: 10.2106/JBJS.I.00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beck A.E., McMillin M.J., Gildersleeve H.I.S., Kezele P.R., Shively K.M., Carey J.C., Regnier M., Bamshad M.J. Spectrum of mutations that cause distal arthrogryposis types 1 and 2B. Am. J. Med. Genet. A. 2013;161A:550–555. doi: 10.1002/ajmg.a.35809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McMillin M.J., Below J.E., Shively K.M., Beck A.E., Gildersleeve H.I., Pinner J., Gogola G.R., Hecht J.T., Grange D.K., Harris D.J., University of Washington Center for Mendelian Genomics Mutations in ECEL1 cause distal arthrogryposis type 5D. Am. J. Hum. Genet. 2013;92:150–156. doi: 10.1016/j.ajhg.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirata H., Nanda I., van Riesen A., McMichael G., Hu H., Hambrock M., Papon M.-A., Fischer U., Marouillat S., Ding C. ZC4H2 mutations are associated with arthrogryposis multiplex congenita and intellectual disability through impairment of central and peripheral synaptic plasticity. Am. J. Hum. Genet. 2013;92:681–695. doi: 10.1016/j.ajhg.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laquérriere A., Maluenda J., Camus A., Fontenas L., Dieterich K., Nolent F., Zhou J., Monnier N., Latour P., Gentil D. Mutations in CNTNAP1 and ADCY6 are responsible for severe arthrogryposis multiplex congenita with axoglial defects. Hum. Mol. Genet. 2014;23:2279–2289. doi: 10.1093/hmg/ddt618. [DOI] [PubMed] [Google Scholar]
- 8.Abicht A., Müller J., Lochmüller H. Congenital Myasthenic Syndromes. In: Pagon R.A., Adam M.P., Ardinger H.H., Wallace S.E., Amemiya A., Bean L.J., Bird T.D., Dolan C.R., Fong C.-T., Smith R.J., editors. Congenital Myasthenic Syndromes. University of Washington, Seattle; Seattle, WA: 2012. [Google Scholar]
- 9.Vogt J., Harrison B.J., Spearman H., Cossins J., Vermeer S., ten Cate L.N., Morgan N.V., Beeson D., Maher E.R. Mutation analysis of CHRNA1, CHRNB1, CHRND, and RAPSN genes in multiple pterygium syndrome/fetal akinesia patients. Am. J. Hum. Genet. 2008;82:222–227. doi: 10.1016/j.ajhg.2007.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cullinane A.R., Straatman-Iwanowska A., Zaucker A., Wakabayashi Y., Bruce C.K., Luo G., Rahman F., Gürakan F., Utine E., Özkan T.B. Mutations in VIPAR cause an arthrogryposis, renal dysfunction and cholestasis syndrome phenotype with defects in epithelial polarization. Nat. Genet. 2010;42:303–312. doi: 10.1038/ng.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morgan J.A., Marcus P.S. Prenatal diagnosis and management of intrauterine fracture. Obstet. Gynecol. Surv. 2010;65:249–259. doi: 10.1097/OGX.0b013e3181dbc50b. [DOI] [PubMed] [Google Scholar]
- 12.Hall J.G., Aldinger K.A., Tanaka K.I. Amyoplasia revisited. Am. J. Med. Genet. A. 2014;164A:700–730. doi: 10.1002/ajmg.a.36395. [DOI] [PubMed] [Google Scholar]
- 13.Greenberg F., Fenolio K.R., Hejtmancik J.F., Armstrong D., Willis J.K., Shapira E., Huntington H.W., Haun R.L. X-linked infantile spinal muscular atrophy. Am. J. Dis. Child. 1988;142:217–219. doi: 10.1001/archpedi.1988.02150020119045. [DOI] [PubMed] [Google Scholar]
- 14.Ramser J., Ahearn M.E., Lenski C., Yariz K.O., Hellebrand H., von Rhein M., Clark R.D., Schmutzler R.K., Lichtner P., Hoffman E.P. Rare missense and synonymous variants in UBE1 are associated with X-linked infantile spinal muscular atrophy. Am. J. Hum. Genet. 2008;82:188–193. doi: 10.1016/j.ajhg.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shaheen R., Al-Owain M., Sakati N., Alzayed Z.S., Alkuraya F.S. FKBP10 and Bruck syndrome: phenotypic heterogeneity or call for reclassification? Am. J. Hum. Genet. 2010;87:306–307. doi: 10.1016/j.ajhg.2010.05.020. author reply 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Narkis G., Ofir R., Landau D., Manor E., Volokita M., Hershkowitz R., Elbedour K., Birk O.S. Lethal contractural syndrome type 3 (LCCS3) is caused by a mutation in PIP5K1C, which encodes PIPKI γ of the phophatidylinsitol pathway. Am. J. Hum. Genet. 2007;81:530–539. doi: 10.1086/520771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ryan M.M., Schnell C., Strickland C.D., Shield L.K., Morgan G., Iannaccone S.T., Laing N.G., Beggs A.H., North K.N. Nemaline myopathy: a clinical study of 143 cases. Ann. Neurol. 2001;50:312–320. doi: 10.1002/ana.1080. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Angarita N., Kirschner J., Heiliger M., Thirion C., Walter M.C., Schnittfeld-Acarlioglu S., Albrecht M., Müller K., Wieczorek D., Lochmüller H., Krause S. Severe nemaline myopathy associated with consecutive mutations E74D and H75Y on a single ACTA1 allele. Neuromuscul. Disord. 2009;19:481–484. doi: 10.1016/j.nmd.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 19.Ravenscroft G., Miyatake S., Lehtokari V.-L., Todd E.J., Vornanen P., Yau K.S., Hayashi Y.K., Miyake N., Tsurusaki Y., Doi H. Mutations in KLHL40 are a frequent cause of severe autosomal-recessive nemaline myopathy. Am. J. Hum. Genet. 2013;93:6–18. doi: 10.1016/j.ajhg.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.García-Cabezas M.A., García-Alix A., Martín Y., Gutiérrez M., Hernández C., Rodríguez J.I., Morales C. Neonatal spinal muscular atrophy with multiple contractures, bone fractures, respiratory insufficiency and 5q13 deletion. Acta Neuropathol. 2004;107:475–478. doi: 10.1007/s00401-004-0825-3. [DOI] [PubMed] [Google Scholar]
- 21.Kelly T.E., Amoroso K., Ferre M., Blanco J., Allinson P., Prior T.W. Spinal muscular atrophy variant with congenital fractures. Am. J. Med. Genet. 1999;87:65–68. doi: 10.1002/(sici)1096-8628(19991105)87:1<65::aid-ajmg13>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 22.Courtens W., Johansson A.-B., Dachy B., Avni F., Telerman-Toppet N., Scheffer H. Infantile spinal muscular atrophy variant with congenital fractures in a female neonate: evidence for autosomal recessive inheritance. J. Med. Genet. 2002;39:74–77. doi: 10.1136/jmg.39.1.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Toorn R., Davies J., Wilmshurst J.M. Spinal muscular atrophy with congenital fractures: postmortem analysis. J. Child Neurol. 2002;17:721–723. doi: 10.1177/088307380201700916. [DOI] [PubMed] [Google Scholar]
- 24.Felderhoff-Mueser U., Grohmann K., Harder A., Stadelmann C., Zerres K., Bührer C., Obladen M. Severe spinal muscular atrophy variant associated with congenital bone fractures. J. Child Neurol. 2002;17:718–721. doi: 10.1177/088307380201700915. [DOI] [PubMed] [Google Scholar]
- 25.Spiegelman B.M., Heinrich R. Biological control through regulated transcriptional coactivators. Cell. 2004;119:157–167. doi: 10.1016/j.cell.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 26.Mouchiroud L., Eichner L.J., Shaw R.J., Auwerx J. Transcriptional coregulators: fine-tuning metabolism. Cell Metab. 2014;20:26–40. doi: 10.1016/j.cmet.2014.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hahm J.B., Privalsky M.L. Research resource: identification of novel coregulators specific for thyroid hormone receptor-β2. Mol. Endocrinol. 2013;27:840–859. doi: 10.1210/me.2012-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Auboeuf D., Dowhan D.H., Kang Y.K., Larkin K., Lee J.W., Berget S.M., O’Malley B.W. Differential recruitment of nuclear receptor coactivators may determine alternative RNA splice site choice in target genes. Proc. Natl. Acad. Sci. USA. 2004;101:2270–2274. doi: 10.1073/pnas.0308133100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jung D.-J., Sung H.-S., Goo Y.-W., Lee H.M., Park O.K., Jung S.-Y., Lim J., Kim H.-J., Lee S.-K., Kim T.S. Novel transcription coactivator complex containing activating signal cointegrator 1. Mol. Cell. Biol. 2002;22:5203–5211. doi: 10.1128/MCB.22.14.5203-5211.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iyer L.M., Burroughs A.M., Aravind L. The ASCH superfamily: novel domains with a fold related to the PUA domain and a potential role in RNA metabolism. Bioinformatics. 2006;22:257–263. doi: 10.1093/bioinformatics/bti767. [DOI] [PubMed] [Google Scholar]
- 31.Mazumder R., Iyer L.M., Vasudevan S., Aravind L. Detection of novel members, structure-function analysis and evolutionary classification of the 2H phosphoesterase superfamily. Nucleic Acids Res. 2002;30:5229–5243. doi: 10.1093/nar/gkf645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seelow D., Schuelke M. HomozygosityMapper2012--bridging the gap between homozygosity mapping and deep sequencing. Nucleic Acids Res. 2012;40:W516–W520. doi: 10.1093/nar/gks487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.von Renesse A., Petkova M.V., Lützkendorf S., Heinemeyer J., Gill E., Hübner C., von Moers A., Stenzel W., Schuelke M. POMK mutation in a family with congenital muscular dystrophy with merosin deficiency, hypomyelination, mild hearing deficit and intellectual disability. J. Med. Genet. 2014;51:275–282. doi: 10.1136/jmedgenet-2013-102236. [DOI] [PubMed] [Google Scholar]
- 34.Li, H. (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv, arXiv:1303.3997, http://arxiv.org/pdf/1303.3997.pdf.
- 35.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwarz J.M., Rödelsperger C., Schuelke M., Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 37.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schuelke M., Cervós-Navarro J. Degenerative changes in unmyelinated nerve fibers in late-infantile neuronal ceroidlipofuscinosis. A morphometric study of conjunctival biopsy specimens. Acta Neuropathol. 1998;95:175–183. doi: 10.1007/s004010050784. [DOI] [PubMed] [Google Scholar]
- 39.Rajab A., Straub V., McCann L.J., Seelow D., Varon R., Barresi R., Schulze A., Lucke B., Lützkendorf S., Karbasiyan M. Fatal cardiac arrhythmia and long-QT syndrome in a new form of congenital generalized lipodystrophy with muscle rippling (CGL4) due to PTRF-CAVIN mutations. PLoS Genet. 2010;6:e1000874. doi: 10.1371/journal.pgen.1000874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wirtz S., Schuelke M. Region-specific expression of mitochondrial complex I genes during murine brain development. PLoS ONE. 2011;6:e18897. doi: 10.1371/journal.pone.0018897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Naganawa Y., Hirata H. Developmental transition of touch response from slow muscle-mediated coilings to fast muscle-mediated burst swimming in zebrafish. Dev. Biol. 2011;355:194–204. doi: 10.1016/j.ydbio.2011.04.027. [DOI] [PubMed] [Google Scholar]
- 42.Kuehn H., Liberzon A., Reich M., Mesirov J.P. Current Protocols in Bioinformatics. John Wiley & Sons; 2008. Using GenePattern for Gene Expression Analysis. bi0712s22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., The Gene Ontology Consortium Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reiner A., Yekutieli D., Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- 45.Robinson P.N., Köhler S., Bauer S., Seelow D., Horn D., Mundlos S. The Human Phenotype Ontology: a tool for annotating and analyzing human hereditary disease. Am. J. Hum. Genet. 2008;83:610–615. doi: 10.1016/j.ajhg.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Origuchi Y. Quantitative histological study in the sural nerves of children. Brain Dev. 1981;3:395–402. doi: 10.1016/s0387-7604(81)80068-x. [DOI] [PubMed] [Google Scholar]
- 47.Schröder J.M., Bohl J., Brodda K. Changes of the ratio between myelin thickness and axon diameter in the human developing sural nerve. Acta Neuropathol. 1978;43:169–178. doi: 10.1007/BF00685012. [DOI] [PubMed] [Google Scholar]
- 48.Shield L.K., King R.H.M., Thomas P.K. A morphometric study of human fetal sural nerve. Acta Neuropathol. 1986;70:60–70. doi: 10.1007/BF00689515. [DOI] [PubMed] [Google Scholar]
- 49.Halliday K.E., Broderick N.J., Somers J.M., Hawkes R. Dating fractures in infants. Clin. Radiol. 2011;66:1049–1054. doi: 10.1016/j.crad.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 50.Sossi V., Giuli A., Vitali T., Tiziano F., Mirabella M., Antonelli A., Neri G., Brahe C. Premature termination mutations in exon 3 of the SMN1 gene are associated with exon skipping and a relatively mild SMA phenotype. Eur. J. Hum. Genet. 2001;9:113–120. doi: 10.1038/sj.ejhg.5200599. [DOI] [PubMed] [Google Scholar]
- 51.Colombo E.A., Fontana L., Roversi G., Negri G., Castiglia D., Paradisi M., Zambruno G., Larizza L. Novel physiological RECQL4 alternative transcript disclosed by molecular characterisation of Rothmund-Thomson Syndrome sibs with mild phenotype. Eur. J. Hum. Genet. 2014;22:1298–1304. doi: 10.1038/ejhg.2014.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmid F., Glaus E., Cremers F.P.M., Kloeckener-Gruissem B., Berger W., Neidhardt J. Mutation- and tissue-specific alterations of RPGR transcripts. Invest. Ophthalmol. Vis. Sci. 2010;51:1628–1635. doi: 10.1167/iovs.09-4031. [DOI] [PubMed] [Google Scholar]
- 53.Demir E., Sabatelli P., Allamand V., Ferreiro A., Moghadaszadeh B., Makrelouf M., Topaloglu H., Echenne B., Merlini L., Guicheney P. Mutations in COL6A3 cause severe and mild phenotypes of Ullrich congenital muscular dystrophy. Am. J. Hum. Genet. 2002;70:1446–1458. doi: 10.1086/340608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ruzzi L., Pas H., Posteraro P., Mazzanti C., Didona B., Owaribe K., Meneguzzi G., Zambruno G., Castiglia D., D’Alessio M. A homozygous nonsense mutation in type XVII collagen gene (COL17A1) uncovers an alternatively spliced mRNA accounting for an unusually mild form of non-Herlitz junctional epidermolysis bullosa. J. Invest. Dermatol. 2001;116:182–187. doi: 10.1046/j.1523-1747.2001.00229.x. [DOI] [PubMed] [Google Scholar]
- 55.Kim H.-J., Yi J.-Y., Sung H.-S., Moore D.D., Jhun B.H., Lee Y.C., Lee J.W. Activating signal cointegrator 1, a novel transcription coactivator of nuclear receptors, and its cytosolic localization under conditions of serum deprivation. Mol. Cell. Biol. 1999;19:6323–6332. doi: 10.1128/mcb.19.9.6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ma L., Yu Y.-M., Guo Y., Hart R.P., Schachner M. Cysteine- and glycine-rich protein 1a is involved in spinal cord regeneration in adult zebrafish. Eur. J. Neurosci. 2012;35:353–365. doi: 10.1111/j.1460-9568.2011.07958.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van der Knaap M.S., Leegwater P.A.J., Könst A.A.M., Visser A., Naidu S., Oudejans C.B.M., Schutgens R.B.H., Pronk J.C. Mutations in each of the five subunits of translation initiation factor eIF2B can cause leukoencephalopathy with vanishing white matter. Ann. Neurol. 2002;51:264–270. doi: 10.1002/ana.10112. [DOI] [PubMed] [Google Scholar]
- 58.Lee J.W., Choi H.S., Gyuris J., Brent R., Moore D.D. Two classes of proteins dependent on either the presence or absence of thyroid hormone for interaction with the thyroid hormone receptor. Mol. Endocrinol. 1995;9:243–254. doi: 10.1210/mend.9.2.7776974. [DOI] [PubMed] [Google Scholar]
- 59.Li S., Czubryt M.P., McAnally J., Bassel-Duby R., Richardson J.A., Wiebel F.F., Nordheim A., Olson E.N. Requirement for serum response factor for skeletal muscle growth and maturation revealed by tissue-specific gene deletion in mice. Proc. Natl. Acad. Sci. USA. 2005;102:1082–1087. doi: 10.1073/pnas.0409103102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wickramasinghe S.R., Alvania R.S., Ramanan N., Wood J.N., Mandai K., Ginty D.D. Serum response factor mediates NGF-dependent target innervation by embryonic DRG sensory neurons. Neuron. 2008;58:532–545. doi: 10.1016/j.neuron.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stern S., Haverkamp S., Sinske D., Tedeschi A., Naumann U., Di Giovanni S., Kochanek S., Nordheim A., Knöll B. The transcription factor serum response factor stimulates axon regeneration through cytoplasmic localization and cofilin interaction. J. Neurosci. 2013;33:18836–18848. doi: 10.1523/JNEUROSCI.3029-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen J., Yuan K., Mao X., Miano J.M., Wu H., Chen Y. Serum response factor regulates bone formation via IGF-1 and Runx2 signals. J. Bone Miner. Res. 2012;27:1659–1668. doi: 10.1002/jbmr.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang X., Lee G., Liebhaber S.A., Cooke N.E. Human cysteine-rich protein. A member of the LIM/double-finger family displaying coordinate serum induction with c-myc. J. Biol. Chem. 1992;267:9176–9184. [PubMed] [Google Scholar]
- 64.Chang D.F., Belaguli N.S., Iyer D., Roberts W.B., Wu S.-P., Dong X.-R., Marx J.G., Moore M.S., Beckerle M.C., Majesky M.W., Schwartz R.J. Cysteine-rich LIM-only proteins CRP1 and CRP2 are potent smooth muscle differentiation cofactors. Dev. Cell. 2003;4:107–118. doi: 10.1016/s1534-5807(02)00396-9. [DOI] [PubMed] [Google Scholar]
- 65.Visel A., Thaller C., Eichele G. GenePaint.org: an atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 2004;32:D552–D556. doi: 10.1093/nar/gkh029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miyasaka K.Y., Kida Y.S., Sato T., Minami M., Ogura T. Csrp1 regulates dynamic cell movements of the mesendoderm and cardiac mesoderm through interactions with Dishevelled and Diversin. Proc. Natl. Acad. Sci. USA. 2007;104:11274–11279. doi: 10.1073/pnas.0702000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oldham M.C., Konopka G., Iwamoto K., Langfelder P., Kato T., Horvath S., Geschwind D.H. Functional organization of the transcriptome in human brain. Nat. Neurosci. 2008;11:1271–1282. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Visser W.E., Swagemakers S.M.A., Őzgűr Z., Schot R., Verheijen F.W., van Ijcken W.F.J., van der Spek P.J., Visser T.J. Transcriptional profiling of fibroblasts from patients with mutations in MCT8 and comparative analysis with the human brain transcriptome. Hum. Mol. Genet. 2010;19:4189–4200. doi: 10.1093/hmg/ddq337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Roet K.C.D., Franssen E.H.P., de Bree F.M., Essing A.H.W., Zijlstra S.-J.J., Fagoe N.D., Eggink H.M., Eggers R., Smit A.B., van Kesteren R.E., Verhaagen J. A multilevel screening strategy defines a molecular fingerprint of proregenerative olfactory ensheathing cells and identifies SCARB2, a protein that improves regenerative sprouting of injured sensory spinal axons. J. Neurosci. 2013;33:11116–11135. doi: 10.1523/JNEUROSCI.1002-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Unterlauft J.D., Eichler W., Kuhne K., Yang X.M., Yafai Y., Wiedemann P., Reichenbach A., Claudepierre T. Pigment epithelium-derived factor released by Müller glial cells exerts neuroprotective effects on retinal ganglion cells. Neurochem. Res. 2012;37:1524–1533. doi: 10.1007/s11064-012-0747-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bilak M.M.P., Corse A.M., Bilak S.R.P., Lehar M., Tombran-Tink J., Kuncl R.W.M. Pigment epithelium-derived factor (PEDF) protects motor neurons from chronic glutamate-mediated neurodegeneration. J. Neuropathol. Exp. Neurol. 1999;58:719–728. doi: 10.1097/00005072-199907000-00006. [DOI] [PubMed] [Google Scholar]
- 72.Carulli D., Pizzorusso T., Kwok J.C.F., Putignano E., Poli A., Forostyak S., Andrews M.R., Deepa S.S., Glant T.T., Fawcett J.W. Animals lacking link protein have attenuated perineuronal nets and persistent plasticity. Brain. 2010;133:2331–2347. doi: 10.1093/brain/awq145. [DOI] [PubMed] [Google Scholar]
- 73.Wise G.E., Yao S., Zhang Q., Ren Y. Inhibition of osteoclastogenesis by the secretion of osteoprotegerin in vitro by rat dental follicle cells and its implications for tooth eruption. Arch. Oral Biol. 2002;47:247–254. doi: 10.1016/s0003-9969(01)00109-1. [DOI] [PubMed] [Google Scholar]
- 74.Roudier M.P., Bain S.D., Dougall W.C. Effects of the RANKL inhibitor, osteoprotegerin, on the pain and histopathology of bone cancer in rats. Clin. Exp. Metastasis. 2006;23:167–175. doi: 10.1007/s10585-006-9026-x. [DOI] [PubMed] [Google Scholar]
- 75.Simonet W.S., Lacey D.L., Dunstan C.R., Kelley M., Chang M.-S., Lüthy R., Nguyen H.Q., Wooden S., Bennett L., Boone T. Osteoprotegerin: a novel secreted protein involved in the regulation of bone density. Cell. 1997;89:309–319. doi: 10.1016/s0092-8674(00)80209-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Normal response to touch of a 30 hpf zebrafish control morphants with coiling and several vigorous contractions.
Reduced motor response in a 30 hpf zebrafish trip4 morphant with just a single contraction executed with a weak twitch.
Reduced motor response in a 30 hpf zebrafish ascc1 morphant with just a single contraction executed with a weak twitch.