Abstract
Genomic mosaicism arising from post-zygotic mutation has recently been demonstrated to occur in normal tissue of individuals ascertained with varied phenotypes, indicating that detectable mosaicism may be less an exception than a rule in the general population. A challenge to comprehensive cataloging of mosaic mutations and their consequences is the presence of heterogeneous mixtures of cells, rendering low-frequency clones difficult to discern. Here we applied a computational method using estimated haplotypes to characterize mosaic megabase-scale structural mutations in 31,100 GWA study subjects. We provide in silico validation of 293 previously identified somatic mutations and identify an additional 794 novel mutations, most of which exist at lower aberrant cell fractions than have been demonstrated in previous surveys. These mutations occurred across the genome but in a nonrandom manner, and several chromosomes and loci showed unusual levels of mutation. Our analysis supports recent findings about the relationship between clonal mosaicism and old age. Finally, our results, in which we demonstrate a nearly 3-fold higher rate of clonal mosaicism, suggest that SNP-based population surveys of mosaic structural mutations should be conducted with haplotypes for optimal discovery.
Main Text
Although post-zygotic mosaic mutations have been traditionally associated with cancer, they have recently been invoked in explanations of pathways of other diseases as well. For example, “selfish selection” in spermatogonial cells for clones carrying certain activating mutations of genes in the MAPK/RAS pathway provides a parsimonious explanation for the paternal age effect for several RASopathies and neurodegenerative disease.1 Another example is the observation that individuals with type 2 diabetes (T2D) have a 5-fold higher risk of blood mosaicism than individuals without T2D and that the risk is even higher in the subset of T2D individuals with vascular complications, suggesting that the “accelerated aging” phenotype associated with T2D may be the secondary consequence of genetic instability mediated by inflammation.2 On the other hand, multiple recent large-scale studies have revealed that apparently healthy individuals harbor detectable mosaic mutations; the frequencies are low in young individuals but increase to frequencies of 2%–3% in elderly (> 70+ years) individuals.3, 4, 5, 6 These rates represent the detectable mutations only.
These examples and others7, 8, 9, 10 highlight that mosaic mutations create a spectrum of phenotypes, in addition to being a prognostic indicator for hematological cancer risk (in blood samples),3 and that the effect of any particular mutation depends on multiple factors, such as the cell type in which it arises and the number of cells carrying the mutation. A detailed picture of the landscape of somatic mosaic mutations, i.e., their prevalence among individuals as well as their frequencies among cells of specific tissues, is therefore of significant value. The low end of the intra-tissue frequency spectrum might be the most dense and dynamic, given that all mutations will start out at very low frequency and some mutations might be suppressed as a result of intra-tissue selection pressures. It is difficult to detect mutations at low frequency by agnostic whole-genome methods, and it is widely acknowledged that mosaic mutations in the low end of the frequency spectrum have been under-characterized.
The goal of our study was to investigate the prevalence of low-frequency somatic structural mosaicism in healthy tissue by applying a haplotype-based method to SNP array data from 31,100 individuals. Several reports have cited the potential increase in sensitivity from using haplotype information.11, 12 Below, we summarize the genomic locations of our discovered aberrations and describe characteristics of these aberrations in comparison to those discovered in a previous analysis of these data, and we report on the association between risk of mosaicism and age.
We obtained SNP microarray data from ten large genome-wide association studies (Table S1) that were all previously analyzed for somatic structural mosaicism by the GENEVA consortium.3 These were case-control studies investigating the role of genetic variation and gene-environment interaction in a wide range of disease phenotypes, including cancer and non-cancer phenotypes. To these data we applied hapLOH13 for an orthogonal assessment of mosaicism due to acquired chromosomal mutations that create allelic imbalance, or a departure from the inherited 1:1 ratio of maternal and paternal alleles. The method targets segmental (megabase-scale to whole-chromosome) alterations by using a powerful and robust haplotype-based approach to sensitively detect somatic hemizygous deletions, copy-neutral loss of heterozygosity (CNLOH), and duplications (collectively, somatic chromosomal and copy-number alterations, SCNAs).
The DNA samples were collected from blood or buccal cells, or from blood-derived cell lines, and were genotyped with Illumina arrays. Genotypes, B allele frequencies (BAFs), and log R Ratios (LRRs) were downloaded from dbGaP (study accession numbers: Table S1). We considered data from bi-allelic SNP markers from both case and control samples after applying basic quality-control procedures. Specifically, we excluded duplicate samples, samples derived from whole-genome-amplified DNA or cell-line DNA, or samples with a LRR waviness score (calculated with PennCNV14) such that . Within each study, we excluded markers with a missing rate greater than 10% or that departed from Hardy-Weinberg proportions (Chi-square or exact test p value < 10−5).
Genotypes were phased with fastPHASE15 or Beagle.16 The hapLOH hidden Markov model (HMM) was set to 2 states. Transition parameters for each sample were set to correspond to an expected imbalance event size of 20 Mb and a genome-wide imbalance rate of 0.1%. We performed two runs of the EM algorithm with starting values for the emission probabilities defined as (pn, pn + 0.05) and (pn, 0.95), where pn is the sample-specific average phase concordance rate calculated from all informative (germline heterozygous) markers. Each EM run continued until the log-likelihood increase was smaller than 0.0001 (usually between 4 and 20 iterations), and the parameter set with the highest likelihood was used for calculating posterior probabilities. To create a list of discrete event calls, we applied a threshold of 0.95 to the probability of being in the aberrant state and defined an event as a run of intervals with probabilities exceeding this value. We used a three-state HMM to reanalyze samples with an event call to improve discovery in samples with multiple events at possibly varying levels of imbalance. The start and end base positions for each SCNA were defined by the left-side marker of the first interval and the right-side marker of the last interval of the run.
We applied additional quality filters after obtaining output from hapLOH. First, we excluded samples with values > 0.52 for , the HMM emission parameter corresponding to the “normal” state. Elevated values of this parameter might indicate a sample-level quality issue, such as a low level of inter-sample contamination, that could create a false positive signal of mosaicism. We also excluded any events overlapping the HLA region (genomic coordinates chr6: 29,677,984–33,485,677, taken from17) because the BAF and LRR data from markers in this region show atypically high variation and might not be reliable. For one sample, more than 75% of the genome was called as imbalance. This sample is most likely a case of inter-sample contamination but did not fail the threshold. We excluded this sample from analysis. We also excluded four calls that had fewer than 15 informative markers and were artifacts of the calling procedure.
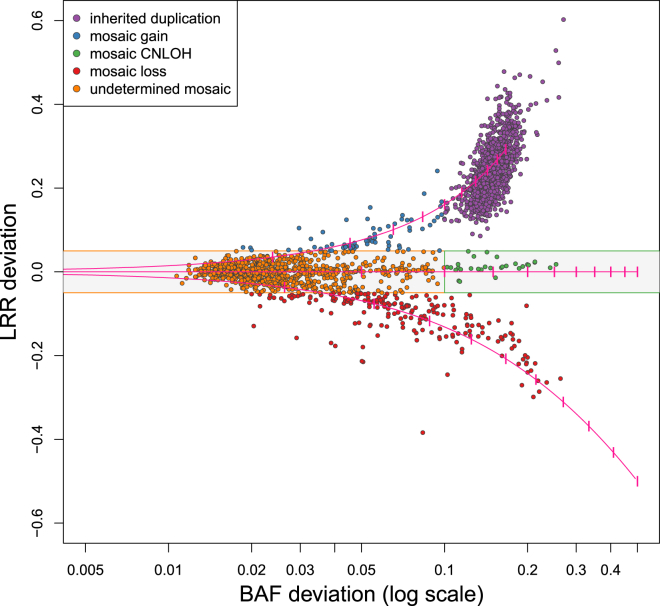
We calculated a BAF and LRR deviation for each discrete event call that passed the above quality-control steps. These data types can be considered a function of the specific SCNA type and the proportion of cells harboring the alteration in the sample. The BAF deviation was defined as the average of the absolute value of the differences between the median heterozygote BAF for the sample and the heterozygote BAFs within the event call. The LRR deviation was defined as the average difference between the median LRR for the sample and the LRRs within the event call. We used the observed deviations to identify 1,507 calls from the preliminary set as likely inherited duplications and removed these from subsequent analyses. Specifically, we applied a simple thresholding procedure to classify calls with LRR deviation > 0.08 and BAF deviation < 0.10 as likely inherited duplications. We expect that with this procedure we might misidentify some true high-frequency somatic events as inherited duplications; we accept this loss of sensitivity to maintain specificity. The remaining calls are putative SCNAs.
We identified 1,141 unique SCNAs in 901 of 31,100 samples (2.9% of samples). Those with LRR deviation > 0.05 or LRR deviation < 0.05 were classified as gains or losses, respectively. The remaining calls included the CNLOH events and events involving very low cell fractions, for which we expect the LRR deviation will be small even if there is a copy-number change. Events with BAF deviation > 0.1 were classified as CNLOH, and the remaining events (with small LRR deviation and small BAF deviation) were left as “undetermined.” Of the 1,141 SCNAs, we classified 70 as single-copy gain, 202 as hemizygous loss, and 30 as CNLOH and left 839 unclassified (Figure 1). Ninety-four (94) samples (0.3%) exhibited two SCNAs, and 44 samples (0.14%) exhibited three or more SCNAs; one exhibited 18 SCNAs that ranged in size from less than 0.3 Mb to 92 Mb. These 138 subjects carrying multiple SCNAs represent a 5.3-fold enrichment over what would be expected by chance, consistent with the existence of individual-level factors that affect the likelihood of observing a mutation. SCNA locations are presented in Table S2.
Figure 1.
BAF and LRR Deviations per SCNA
The pink lines indicate the expected values for mosaic hemizygous deletions (lower line), mosaic CNLOH (middle horizontal line), and mosaic single-copy duplications (upper line) for aberrations present in 10% to 100% of the sampled cells (dashes are at 10% increments). The gray shaded area indicates the area within the thresholds used to define an SCNA as a copy-number gain or copy-number loss. Each point is colored according to the copy-number classification on the basis of these deviations.
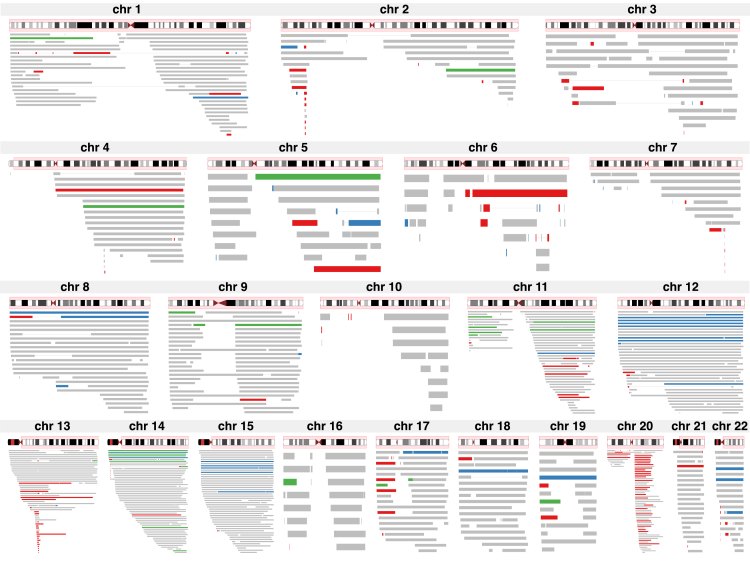
The rate of mutation and inferred copy numbers of SCNAs varied substantially by genomic region (Figure 2). As a measure of the local mutation rate, we compared the SCNA overlap count for each gene for 24,383 genes (we used the largest transcript from RefSeq18 to represent gene location). For this assessment, we used the SCNAs observed in the 26,927 blood samples only (we excluded buccal samples and samples without annotation on DNA source) because aberration patterns might differ by tissue. Only 1,318 genes were not covered by an SCNA in any of the samples. The most frequently overlapped gene was PTPRT (MIM: 608712) on chromosome 20, which was overlapped in 60 samples; nearby genes in the surrounding region had the next highest overlap counts. Multiple chromosomes exhibited similar sharp peaks in SCNA overlap counts (Figure S1 and Table S3), the most notable being chromosome 13, which had a peak overlap count of 49 SCNAs covering the contiguous genes DLEU1 (MIM: 605765) and DLEU7. Other chromosomes showed broader peaks in SCNA overlap counts. For example, 17 contiguous genes on chromosome 14 were overlapped by SCNAs in 57 samples. Chromosomes 5, 6, 10, and 16 had the lowest SCNA overlap counts, and indeed the fewest counts in general; fewer than ten SCNAs covered any gene.
Figure 2.
SCNAs by Chromosome
The red shading on each ideogram indicates the range of the plotted mutations. SCNAs are plotted as horizontal bars, colored by inferred copy-number: red, loss; green, CNLOH; blue, gain; and gray, undetermined. A thin line connecting SCNAs within a chromosome indicates the SCNAs occur in the same sample.
In a recent meta-analysis of SNP array data from more than 127,000 subjects, Machiela et al.5 reported that SCNAs aggregated on chromosomes by copy number. They cited chromosomes 8, 12, and 15 as carrying the majority of somatic gains, chromosomes 13 and 20 as carrying the majority of somatic losses, and chromosomes 9 and 14 as carrying the majority of somatic CNLOH. They also pointed out that focal deletions on 13q and 20q are frequent. As we describe below, many of the SCNAs we observed are low frequency (carried in a small proportion of cells) and do not create strong enough deviations in the BAF and LRR data to allow determination of copy number. However, most recurrent loci (those at which SCNAs were observed at relatively high frequency) that harbored SCNAs with determinable copy number demonstrated a particular mutation type. For example, we observed deletions on chromosomes 13 and 20 in regions that are commonly deleted in hematological cancer, and we observed multiple instances wherein the entire chromosome 12 was duplicated, in accord with previous studies.3, 4, 5 We also observed large chromosome 15 duplications that span at least the entire q arm, or possibly the entire chromosome (these two possibilities are indistinguishable in our data because none of the SNP arrays included markers on the p arm). Some loci do harbor classifiable SCNAs of multiple copy-number classes; for example, at 14q (or possibly the entirety of chromosome 14) we observe both duplications and CNLOH.
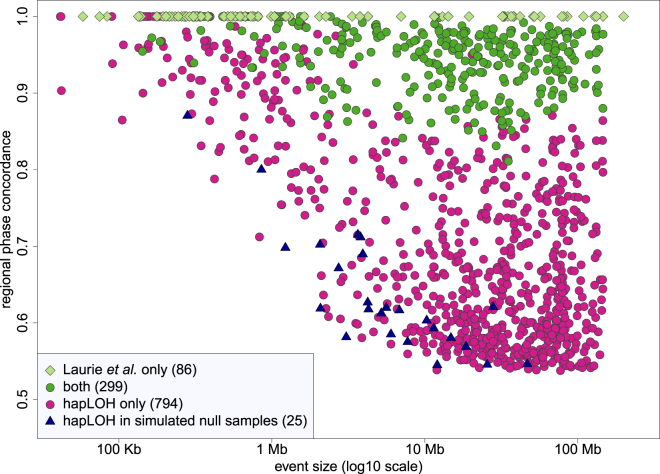
A large subset of our dataset (30,208 samples) was analyzed previously for SCNAs by a different method.3 Laurie et al. applied a method designed for discovering SCNAs on the basis of the magnitude of BAF and LRR deviations (without using haplotype information). Within samples common to both analyses, our analysis identified far more SCNAs (1,093 versus 379). We used the genomic positions to define the extent of overlap between hapLOH and Laurie et al. calls in these samples. More than 90% of overlapping events had more than 80% overlap with events in the other analysis, although there were instances in which one analysis called one event but the other split the same region into multiple events, so that the overlap with an individual event could be low but the total overlap when all overlapping events were considered was high. To make a comparison of the sets of calls in the two analyses, we deemed calls to be concordant if they had any overlap with sample-specific calls in the other analysis and ignored copy-number classifications, although our conclusions do not change qualitatively for other overlap criteria (Table S4). Using these criteria, we classified 299 hapLOH SCNAs and 293 Laurie et al. SCNAs as concordant (the counts are not equal because some calls overlapped multiple calls in the other analysis). A total of 794 SCNAs were unique to our analysis, and 86 SCNAs were unique to Laurie et al. Ten of the SCNAs unique to Laurie et al. were part of the initial hapLOH call set but were excluded as possible inherited duplications or because they overlapped the HLA region. Another 33 of the SCNAs were short (spanning fewer than 200 markers, mean size 415 Kb), and for the remaining 43 SCNAs the mutant cell fraction was high enough that that there were almost no called heterozygous genotypes, upon which our method is based; thus, these mutations were outside the range of events targeted in our analysis. hapLOH uses phase concordance (a measure of the switch accuracy between the statistical haplotypes and the BAFs; see Vattathil and Scheet13) to detect SCNAs. The observed phase concordance is a function of several factors, including the copy number of the mutant cells, mutant cell fraction, and the accuracy of the statistical phasing, yet can roughly be interpreted as a level of allelic imbalance created by the mutation, particularly at lower cell fractions. All of the SCNAs present in both call sets had phase concordance exceeding 0.8, whereas three-fourths of the SCNAs uniquely identified in our analysis had phase concordance values less than 0.8 (Figure 3). This is in line with expectations because the haplotype-based method we employed is especially sensitive for low-cell-fraction SCNAs.
Figure 3.
Phase Concordance versus Genomic Size
The circles and diamonds represent SCNAs called by hapLOH only, Laurie et al. only, or both in samples that were included in both analyses. Many of the SCNAs called by Laurie et al. only had few or no heterozygous calls, so the phase-concordance values were incalculable or imprecise; all of these were plotted with phase concordance = 1. The triangles represent the calls made in the simulated null samples.
An important characteristic of our method is that the sensitivity increases with both the magnitude of the phase concordance and the size of the event (in terms of number of heterozygous genotypes). SCNAs inducing subtle allelic imbalance are therefore detectable, but only if their size is large enough. The lack of SCNAs in the lower left corner of Figure 3 demonstrates this point. By the same token, short regions are detectable, but only if the phase concordance is high enough (upper left corner of Figure 3). The sensitivity for short events is also restricted in this analysis by the specific parameter settings we employed; we did not enforce a minimum size threshold for SCNA identification but chose parameters that would provide sensitivity for subtle events yet keep the false-positive rate low. Using this setting, one can identify kilobase-range SCNAs, but probably only when the phase concordance is high. We expect that many SCNAs with low phase concordance exist at small genomic size, but our analysis was not designed for their discovery.
One question regarding low-cell-fraction events is whether they occur randomly across the genome or show spatial and copy-number patterns similar to those of higher-cell-fraction events. To address this, we looked at the location and copy-number assignments of hapLOH-exclusive calls (Figure S2). We considered only calls in blood samples, as we did for the spatial-distribution analysis of the total call set. Out of the 698 hapLOH-exclusive calls in blood samples, only 74 were assigned a copy number (56 gains, 18 deletions, and 0 CNLOH). These included five deletions on 13q and four deletions on 20q that overlapped the commonly deleted regions reported by Machiela et al. One gene on chromosome 7, MTRNR2L6, also was overlapped by a deletion in four samples. These were the most common recurrent deletions in this set. No region was overlapped by more than two gain events. To get a rough sense of how well our 624 “undetermined” calls match the Machiela et al. set in terms of chromosomal aggregation by copy number, we calculated the average LRR deviation per chromosome for these calls. The averages are consistent with the copy-number distribution by Machiela et al.—chromosomes 8, 12, and 15 showed the highest average LRR deviation for undetermined calls, whereas chromosomes 10, 13, and 20 showed the lowest average LRR deviation. Of note, chromosome 10 had the fewest calls (16), so sampling variation might explain its unexpected ranking.
We used the observed BAF and haplotype data to perform a permutation-based simulation to estimate the false-positive rate of the method. Specifically, for each of the 31,328 samples that passed our initial quality-control steps, we permuted the observed BAFs at the informative markers (that is, the subset of markers at which the sample had heterozygous genotype calls; this subset was unique for each sample), and then applied our analysis protocol to these data. Permuting the BAFs at informative markers disrupts the dependence in the BAF deviations that would arise from somatic imbalance while preserving the level of random variation originally present in the data. So, any calls made in these “simulated null” samples represent false positives arising from chance stretches of increased phase concordance. Because there could be other sources of false positives (although we have attempted to rule these out by quality-control procedures), the call rate estimated here is effectively a lower bound on the false-positive rate. Application of our analysis protocol to data generated from a single permutation of each of the 31,328 samples yielded 25 SCNA calls in 25 samples, or about 0.08% of samples. Thus, the rate of 2.9% we observed in the original data represents an approximately 37-fold enrichment over the estimated null rate and a false discovery rate of <3%. The 25 calls in the permuted data display a very different distribution in terms of phase concordance and genomic size than the calls from the real data (Figure 3); they reside along a gradient of lower values for these features. Therefore, in practice this false-discovery rate will vary as a function of attributes of the event call. Of note, none of the simulated null samples failed the -based quality-control filter, which is the expected result if elevated values reflect biological contamination and are not simply due to poor parameter estimation.
Because the BAF and LRR deviations depend on the mutant cell fraction, we could theoretically attempt to infer this quantity for each SCNA. However, just as with the inference of copy number, the low magnitude of the deviations for most of the SCNAs interfered with precise characterization. We conjecture that the vast majority of the SCNAs we observed were present in less than 10% of the cell population in each sample. It is worth emphasizing that even when SCNAs displayed small BAF and LRR deviations, the statistical evidence for AI, based on the phase concordance, was still exceptionally high for all of the called events. We also note that a majority of large SCNAs we discovered coincided with chromosomes even though the HMM is applied to ordered marker data for all 22 autosomes concatenated into a single input vector without regard to specific marker locations or chromosomal annotation; this observation favors a molecular rather than a stochastic source.
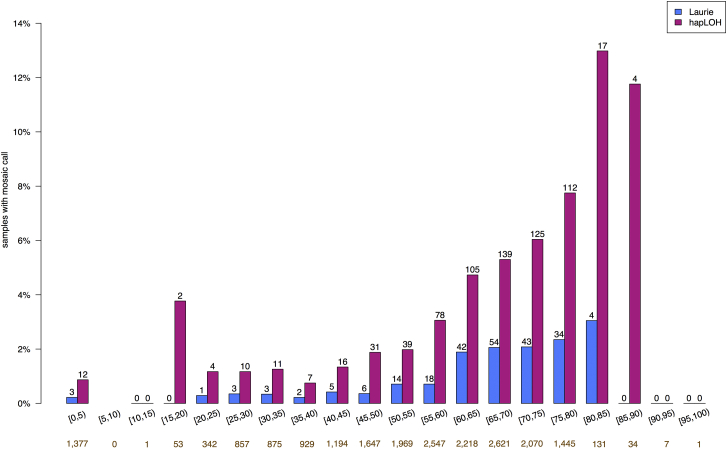
In previous analyses, SCNA prevalence (that is, the frequency of individuals with one or more SCNAs) was strongly positively associated with age. In our results, the prevalence of SCNAs among individuals older than 80 years of age was approximately 12% (Figure 4). Although the sample size at this age range is modest, the increase in SCNA rate compared to that in middle age is quite large. To formally examine the relationship between age and our observed SCNAs while accounting for the possible confounding effect of samples being genotyped in different studies, we applied the Mantel extension test for trend by using only the 20,727 samples derived from blood DNA from individuals for whom we had age information. We found that age was a significant predictor of the presence of one or more observed SCNAs (p value = 10−26). We generally detected two to four times as many SCNAs per age category as Laurie et al. did. It is interesting to note that low-cell-fraction clones seemingly went undetected in every age category.
Figure 4.
Mosaic Rate by Age
The gold numbers below each age bin indicate the sample size for that bin. The numbers above each bar are the number of samples that fall in that age bin and have at least one SCNA.
These results corroborate and augment the current observational evidence of somatic mosaicism in apparently healthy tissue and suggest that the rate of mosaicism in phenotypically normal individuals is higher than was reported in recent large-scale studies. Our analysis was specifically motivated to detect mosaicism from low-cell-fraction mutations. This part of the landscape of somatic mutations is important because it is likely that the majority of somatic mutations exist at low cell fractions. Indeed, our analysis supports this notion even though lower-frequency mutations are more difficult to detect. By using a haplotype-based method that leverages the dependence among BAFs in imbalanced regions, we detected a larger number of low-cell-fraction aberrations than in previous analyses of these data. Even so, low-cell-fraction SCNAs create a weak signal that is difficult to discern from background noise, and when they cover short genomic regions there is insufficient statistical evidence for their detection. An analogy is detecting a subtly unfair coin, which is possible only with a sufficiently large number of coin flips. In the case of detecting SCNAs with a subtle signal, we need a large number of informative loci. Thus, to maintain high specificity in our study, we targeted large aberrations. Small events with a high cell fraction do create a strong enough signal that they are also picked up with this setting. This bias for aberrations of certain sizes and phase-concordance ranges must be kept in mind when one interprets the observed distribution of SCNAs—the lack of observations that are small in size and exhibit low phase concordance is clearly due to the lack of power to detect this category of aberrations. We can easily rationalize that large aberrations will be expected to exist mostly at low cell fractions because they are more likely than smaller aberrations to have a negative impact on cell fitness. Interestingly, we do observe a number of large SCNAs with cell fractions that are likely to exceed 15%; these might comprise mutations that increase cell fitness in the balance, at least for the sampled tissue at the post-developmental stage of the organism.
Our results support previous reports3, 5 of a sharp increase in the rate of detected mosaicism in elderly individuals compared to younger individuals. This observation may indicate a higher rate of somatic mutation in the elderly, which is consistent with the hypothesis that mutation rate increases with age as a result of a reduction in DNA-repair activity or an increase in the incidence of errors (for example, an increase in the incidence of structural rearrangements and aneuploidy resulting from telomere attrition19). An alternative explanation is that the mutation rate is largely constant over time but that detectable mosaicism is associated with age because in older individuals there has been more time for viable mutant clones to initiate and expand by drift or selection. Further investigation of mosaicism in youth and middle age, by methods tuned for low-frequency mosaic mutations, might shed light on the relative impact of factors influencing somatic mutation rates.
The nonrandom distribution of SCNAs and mutation types across the genome suggests highly preferential mutation initiation or selection for or against mutations in certain regions. Several of the recurrently imbalanced regions include genes that have been associated with cancer. Because all of the blood samples analyzed were collected from individuals without diagnosed hematological cancer, we can conclude that observed aberrations are generally insufficient to initiate transformation, but how important are their potential impacts on proliferation? One exciting possibility is that low-frequency clones can be used as valuable early-disease cancer biomarkers. Indeed, Laurie et al.3 established such a relationship in these data, and this has been observed elsewhere as well.4, 7 Although somatic mutation is a driving force in cancer, the extreme level of genomic aberration observed in many cancers highlights the high level of robustness of the human genome and supports the notion that sporadic random somatic mutations can be of little consequence and should be expected at a low frequency in normal tissues. In fact, mathematical modeling demonstrates that large fractions of the single-nucleotide mutations observed in tumors of self-renewing tissues are passenger mutations acquired during normal tissue maintenance that happened to be carried by the initiating tumor cell,20 and a recent study found that the large variation in lifetime risk among cancers of different tissues is explained in large part by variation in the number of normal cell divisions among tissues.21 These observations underscore the need for further characterization of the landscape of somatic mutation in normal tissue to improve our understanding of the significance of mutations observed in cancer. Because the landscape of tolerated and functional somatic mutations is likely to vary by tissue, studies using samples from other tissues would complement the largely blood-based studies that have been recently conducted.
Acknowledgments
We thank C. Laurie for sharing sample identifiers for mutations called in their analysis. X. Xiao and J. Fowler provided assistance with array processing and workflows. L. Huang performed haplotype phasing and ran hapLOH on multiple data sets. C. Huff and Y. M. Chen provided helpful comments on analyses. This work was supported by NIH grants R01HG005859 and R01HG005855.
Published: March 3, 2016
Footnotes
Supplemental data include three figures and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.02.003.
Web Resources
The URLs for data presented herein are as follows:
hapLOH software, http://scheet.org/software.html
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org
Supplemental Data
References
- 1.Goriely A., McGrath J.J., Hultman C.M., Wilkie A.O., Malaspina D. “Selfish spermatogonial selection”: A novel mechanism for the association between advanced paternal age and neurodevelopmental disorders. Am. J. Psychiatry. 2013;170:599–608. doi: 10.1176/appi.ajp.2013.12101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonnefond A., Skrobek B., Lobbens S., Eury E., Thuillier D., Cauchi S., Lantieri O., Balkau B., Riboli E., Marre M. Association between large detectable clonal mosaicism and type 2 diabetes with vascular complications. Nat. Genet. 2013;45:1040–1043. doi: 10.1038/ng.2700. [DOI] [PubMed] [Google Scholar]
- 3.Laurie C.C., Laurie C.A., Rice K., Doheny K.F., Zelnick L.R., McHugh C.P., Ling H., Hetrick K.N., Pugh E.W., Amos C. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat. Genet. 2012;44:642–650. doi: 10.1038/ng.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jacobs K.B., Yeager M., Zhou W., Wacholder S., Wang Z., Rodriguez-Santiago B., Hutchinson A., Deng X., Liu C., Horner M.-J. Detectable clonal mosaicism and its relationship to aging and cancer. Nat. Genet. 2012;44:651–658. doi: 10.1038/ng.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Machiela M.J., Zhou W., Sampson J.N., Dean M.C., Jacobs K.B., Black A., Brinton L.A., Chang I.S., Chen C., Chen C. Characterization of large structural genetic mosaicism in human autosomes. Am. J. Hum. Genet. 2015;96:487–497. doi: 10.1016/j.ajhg.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forsberg L.A., Rasi C., Razzaghian H.R., Pakalapati G., Waite L., Thilbeault K.S., Ronowicz A., Wineinger N.E., Tiwari H.K., Boomsma D. Age-related somatic structural changes in the nuclear genome of human blood cells. Am. J. Hum. Genet. 2012;90:217–228. doi: 10.1016/j.ajhg.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forsberg L.A., Rasi C., Malmqvist N., Davies H., Pasupulati S., Pakalapati G., Sandgren J., Diaz de Ståhl T., Zaghlool A., Giedraitis V. Mosaic loss of chromosome Y in peripheral blood is associated with shorter survival and higher risk of cancer. Nat. Genet. 2014;46:624–628. doi: 10.1038/ng.2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodríguez-Santiago B., Malats N., Rothman N., Armengol L., Garcia-Closas M., Kogevinas M., Villa O., Hutchinson A., Earl J., Marenne G. Mosaic uniparental disomies and aneuploidies as large structural variants of the human genome. Am. J. Hum. Genet. 2010;87:129–138. doi: 10.1016/j.ajhg.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biesecker L.G., Spinner N.B. A genomic view of mosaicism and human disease. Nat. Rev. Genet. 2013;14:307–320. doi: 10.1038/nrg3424. [DOI] [PubMed] [Google Scholar]
- 10.Bruder C.E.G., Piotrowski A., Gijsbers A.A.C.J., Andersson R., Erickson S., Diaz de Ståhl T., Menzel U., Sandgren J., von Tell D., Poplawski A. Phenotypically concordant and discordant monozygotic twins display different DNA copy-number-variation profiles. Am. J. Hum. Genet. 2008;82:763–771. doi: 10.1016/j.ajhg.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nik-Zainal S., Van Loo P., Wedge D.C., Alexandrov L.B., Greenman C.D., Lau K.W., Raine K., Jones D., Marshall J., Ramakrishna M., Breast Cancer Working Group of the International Cancer Genome Consortium The life history of 21 breast cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baugher J.D., Baugher B.D., Shirley M.D., Pevsner J. Sensitive and specific detection of mosaic chromosomal abnormalities using the parent-of-origin-based detection (POD) method. BMC Genomics. 2013;14:367. doi: 10.1186/1471-2164-14-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vattathil S., Scheet P. Haplotype-based profiling of subtle allelic imbalance with SNP arrays. Genome Res. 2013;23:152–158. doi: 10.1101/gr.141374.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang K., Li M., Hadley D., Liu R., Glessner J., Grant S.F.A., Hakonarson H., Bucan M. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scheet P., Stephens M. A fast and flexible statistical model for large-scale population genotype data: Applications to inferring missing genotypes and haplotypic phase. Am. J. Hum. Genet. 2006;78:629–644. doi: 10.1086/502802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Browning S.R., Browning B.L. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shiina T., Hosomichi K., Inoko H., Kulski J.K. The HLA genomic loci map: Expression, interaction, diversity and disease. J. Hum. Genet. 2009;54:15–39. doi: 10.1038/jhg.2008.5. [DOI] [PubMed] [Google Scholar]
- 18.Pruitt K.D., Brown G.R., Hiatt S.M., Thibaud-Nissen F., Astashyn A., Ermolaeva O., Farrell C.M., Hart J., Landrum M.J., McGarvey K.M. RefSeq: An update on mammalian reference sequences. Nucleic Acids Res. 2014;42:D756–D763. doi: 10.1093/nar/gkt1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aviv A., Aviv H. Telomeres, hidden mosaicism, loss of heterozygosity, and complex genetic traits. Hum. Genet. 1998;103:2–4. doi: 10.1007/s004390050774. [DOI] [PubMed] [Google Scholar]
- 20.Tomasetti C., Vogelstein B., Parmigiani G. Half or more of the somatic mutations in cancers of self-renewing tissues originate prior to tumor initiation. Proc. Natl. Acad. Sci. USA. 2013;110:1999–2004. doi: 10.1073/pnas.1221068110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomasetti C., Vogelstein B. Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science. 2015;347:78–81. doi: 10.1126/science.1260825. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.